Open Access Article
Open Access ArticleCarotenoid biosynthesis of pak choi (Brassica rapa ssp. chinensis) sprouts grown under different light-emitting diodes during the diurnal course†
K.
Frede
 *ab,
M.
Schreiner
*ab,
M.
Schreiner
 a,
R.
Zrenner
a,
J.
Graefe
a,
R.
Zrenner
a,
J.
Graefe
 a and
S.
Baldermann
a and
S.
Baldermann
 *ab
*ab
aLeibniz Institute of Vegetable and Ornamental Crops, Plant Quality and Food Security, Theodor-Echtermeyer-Weg 1, 14979 Großbeeren, Germany. E-mail: frede@igzev.de; baldermann@igzev.de
bUniversity of Potsdam, Institute of Nutritional Science, Department of Food Chemistry, Arthur-Scheunert-Allee 114-116, 14558 Nuthetal, Germany
First published on 1st August 2018
Abstract
Light-emitting diodes (LEDs) are considered the future of greenhouse lighting. This study investigates the carotenoid concentrations of pak choi sprouts after growth under blue, red and white LEDs at six different time points. Furthermore, the diurnal changes of RNA transcripts of key genes of the carotenoid biosynthesis pathway as well as of the carotenoid cleavage dioxygenase 4 (CCD4) gene and of the transcription factor genes elongated hypocotyl 5 (HY5) and circadian clock associated 1 (CCA1) were investigated. The carotenoid concentrations were steady throughout the day, but showed a small maximum in the afternoon. An average total carotenoid concentration of 536 ± 29 ng mg−1 DM produced under white LEDs was measured, which is comparable to previously described field-grown levels. The carotenoid concentrations were slightly lower under blue or red LEDs. Moreover, the diurnal RNA transcript rhythms of most of the carotenoid biosynthesis genes showed an increase during the light period, which can be correlated to the carotenoid maxima in the afternoon. Blue LEDs caused the highest transcriptional induction of biosynthetic genes as well as of CCD4, thereby indicating an increased flux through the pathway. In addition, the highest levels of HY5 transcripts and CCA1 transcripts were determined under blue LEDs.
Introduction
Leading a healthy lifestyle is becoming increasingly more important for consumers, thereby increasing demand for vegetables throughout the whole year. This consumer demand can only be met through greenhouse farming for which lighting systems are needed. In the past, lighting systems with high pressure sodium lamps or metal–halide lamps were commonly used, whereas nowadays, light-emitting diodes (LEDs) are a very attractive alternative for greenhouse lighting.1 Since 10–30% of the production costs of greenhouse farming are caused by energy consumption, energy-saving, long-lived LEDs could contribute to significant cost reductions. Moreover, narrow-band LEDs characterized by specific wavelengths relevant for plant growth could be used, thereby avoiding wavelengths less efficient for vegetable production.1 In addition to optimizing quantum yield of photosynthesis,2 LEDs producing specific light wavelengths can be used to trigger desirable plant responses aimed at enhancing vegetable quality. For example, light quality can influence the dry mass3 or ascorbic acid content4 in lettuce (Lactuca sativa ‘Grand Rapids’3 or ‘Redfire’4) and specific light wavelengths can also be used to induce changes in the secondary metabolite profile of vegetables, including carotenoids.5Carotenoids are yellow, orange, and red pigments colouring leaves, flowers, fruits, and vegetables.6 They are known to confer health-promoting properties to humans, thereby making them of key interest to consumers.7 In detail, in lettuce (Lactuca sativa ‘Banchu Red Fire’8 or ‘Red Cross’9), blue light was shown to increase the carotenoid concentrations.8,9 However, the influence of different light qualities on carotenoid concentrations seems to be species-dependent. For example, Ohashi-Kaneko et al. investigated spinach (Spinacia oleracea ‘Okame’), lettuce (Lactuca sativa ‘Redfire’) and komatsuna (Brassica campestris ‘Komatsuna’) in regards to their carotenoid concentrations after growth under different light qualities.4 Total carotenoid concentrations were highest under blue fluorescent lamps in spinach, under white and red fluorescent lamps in lettuce, and under white fluorescent lamps in komatsuna. This light effect highlights the fact that different light qualities influence carotenoid concentrations. However, there are currently only a few studies on the effects of specific wavelengths on regulating mechanisms involved in the carotenoid biosynthesis pathway. In detail, an increase in carotenoids was measured in white mustard (Sinapis alba),10Arabidopsis thaliana10 and tobacco (Nicotiana tabacum ‘Samsun’)11 during the de-etiolation process initiated by light. Furthermore, in tomato fruits (Solanum lycopersicum ‘Moneymaker’), the overexpression of the blue light photoreceptor cryptochrome-2 (CRY2) gene led to an increase of lycopene and a decrease of β-cyclase (βLCY) transcripts, an enzyme of the carotenoid biosynthesis pathway (Fig. 1).12 Further, also in tomato fruits (Solanum lycopersicum ‘Ailsa Craig’ and ‘Rapsodie’), Schofield and Paliyath measured an increase in phytoene synthase-1 (PSY1) activity, the enzyme catalysing the first step in carotenoid biosynthesis, under red light conditions.13 Thus, there is an accumulating body of evidence to suggest that carotenoid metabolism is influenced by different light wavelengths, although the exact mechanisms by which light regulates carotenoid concentrations are still currently poorly characterized.
Here, we used pak choi (Brassica rapa ssp. chinensis ‘Black Behi’) to investigate the effect of three different light wavelengths on carotenoid concentrations. Pak choi is an Asian leafy vegetable especially rich in both carotenoids and chlorophylls.14 Since the consumption of sprouts containing high contents of such health-promoting compounds has recently increased,15 we focussed on this development stage in pak choi and used LEDs with different light qualities during their growth (emission spectra with maxima at blue or red wavelengths, and broad-band emission spectra for white light).
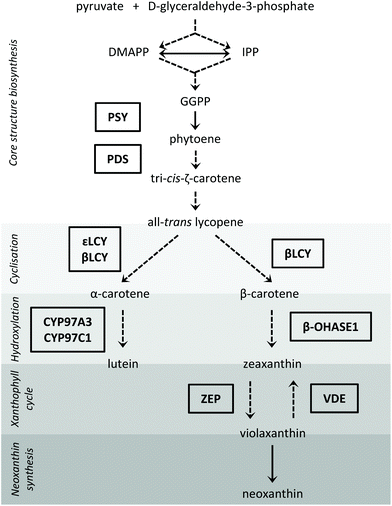
Since carotenoid steady-state concentrations are determined by their biosynthesis, storage and degradation, we studied the transcript levels of selected enzymes of the carotenoid biosynthesis pathway as well as of a carotenoid degrading enzyme. The steps of the carotenoid biosynthesis pathway investigated in our study are depicted in Fig. 1 (for a review on carotenoid biosynthesis see Cazzonelli, 20116). With regard to the hydroxylases of the β-branch, only the β-carotene hydroxylase 1 gene (β-OHASE1) was analysed because PCR products of β-OHASE2 could not be detected in the tested pak choi cultivar ‘Black Behi’. The degradation of the carotenoids can take place chemically or enzymatically. The enzymatic degradation in plants is performed by two large enzyme families: the nine-cis-epoxy-carotenoid dioxygenases (NCEDs), which are involved in abscisic acid production from carotenoids, and those carotenoid cleavage dioxygenases (CCDs) that yield hormones, pigments and short-chained apocarotenoids.16 To date, five CCD groups have been identified (CCD1, CCD2, CCD4, CCD7, and CCD8). CCD1 is only found in the cytosol,17 which probably limits its access to carotenoids in the chloroplasts. In other words, it is unlikely that CCD1 degrades carotenoids that are present in the thylakoids. Furthermore, a CCD1 loss-of-function mutant did not show a changed carotenoid content in Arabidopsis thaliana leaves, thus confirming this assumption.17 To date, CCD2 has only been identified in Crocus species.18 Moreover, CCD7 and CCD8 are both known to be involved in the biosynthesis of strigolactones, which are plant hormones derived from carotenoids.17 In Arabidopsis thaliana, there is evidence that CCD4 is involved in the carotenoid degradation in leaves.19,20 Thus, we hypothesize that CCD4 enzymes are the ones most likely responsible for the carotenoid turnover in chloroplasts. Therefore, we measured CCD4 transcript levels in this study. In addition, we also studied two transcription factors, namely elongated hypocotyl 5 (HY5), a transcription factor activated by different light qualities and that has many other transcription factor genes amongst its binding targets,21 and circadian clock associated 1 (CCA1), which is part of the circadian clock.22 HY5 levels are positively regulated by light through transcriptionally and posttranslationally mechanisms. It is a key regulator of photomorphogenesis and mediates up- and down-regulation of gene expression by light.22 The circadian clock consists of several autoregulatory negative-feedback loops, is synchronized by external stimuli, such as light and temperature, and regulates the circadian rhythm of many genes. CCA1 participates in the feedback regulation of the circadian clock and also regulates the expression of many genes which show a diurnal oscillation by binding to their promotors. The transcription of CCA1 is induced by light.22 Therefore, both transcription factors are key regulators for light-mediated gene expression.
Thus, since the exact mechanisms by which light regulates carotenoid concentrations are not yet well understood, this study aimed at providing novel and new insights into light quality-regulated processes of the carotenoid metabolism that determine the carotenoid concentrations in leafy vegetables.
Materials and methods
Plant material and growth conditions
The pak choi (Brassica rapa ssp. chinensis) cultivar ‘Black Behi’ was used in all experiments (Allied Botanical Corporation, Quezon City, Philippines). Aluminium foil trays (33 × 10 cm) were filled with perlite that was wet with water. One gram of seeds (∼370 seeds) was sown per replicate on a wet strip of fleece which was placed onto the perlite and sprayed with water. The sprouts were grown on the moist fleece in the dark for five days (25 °C, 75% air humidity). Afterwards, some of the dark-grown sprouts (one strip with sprouts was pooled per replicate) were harvested just above the perlite surface (cotyledons and hypocotyl). These dark-grown samples were frozen immediately in liquid nitrogen and stored at −80 °C until further analysis. The rest of the sprouts were transferred to a growth chamber (25 °C, 75% air humidity) equipped with blue, red and white LEDs. The light of the white LEDs was adjusted with a filter to reduce the amount of blue light. The properties of the filter were determined with a Jasco spectrometer V670 (Jasco, Tokyo, Japan) at 3 points of the filter. The spectra of the LEDs were measured with an EPP2000 StellarNet spectrometer (StellarNet, Tampa, FL, USA). The presented spectra are the means of 12 measuring spots per LED type. The photoperiod in the growth chamber was 12 h from 06:00 h to 18:00 h. A photosynthetic active radiation of 80 μmol m−2 s−1 was confirmed for each light quality (LI-250 Light Meter, LI-COR, Lincoln, NE, USA). Sprouts were grown for six days under the three different light qualities and were watered as needed. The trays were spatially rotated every day, in order to optimize the uniformity of light treatment. Throughout the final day of the light treatment, the sprouts were harvested as aforementioned at six different time points (from 18:00 h of day 5 until 18:00 h of day 6) with three biological replicates for each time point per treatment (two independent series of experiments: two replicates from series #1 and one replicate from series #2). For further analysis, the frozen samples were ground to a fine powder under liquid nitrogen and kept at −80 °C. Carotenoid analysis was performed using freeze-dried material.Analysis of carotenoids and chlorophylls by LC-ToF-MS
About 20 mg freeze-dried plant material per sample were used for the analysis. Carotenoids and chlorophylls were extracted by adding 750 μL methanol/tetrahydrofuran (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) (Th. Geyer, Berlin, Germany; VWR International, Radnor, PA, USA), shaking for 5 min at 1400 rpm followed by centrifugation for 5 min at 4000g. The supernatant was transferred into a new 4 mL glass vial. The extraction step was repeated three times using 500 μL of methanol/tetrahydrofuran (1
1, v/v) (Th. Geyer, Berlin, Germany; VWR International, Radnor, PA, USA), shaking for 5 min at 1400 rpm followed by centrifugation for 5 min at 4000g. The supernatant was transferred into a new 4 mL glass vial. The extraction step was repeated three times using 500 μL of methanol/tetrahydrofuran (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The combined supernatant was concentrated under N2 to dryness and re-dissolved in 100 μL of dichloromethane (Merck, Darmstadt, Germany) and 400 μL of 2-propanol (Merck, Darmstadt, Germany). The extract was filtered (0.2 μm, PTFE) and kept at 10 °C in the autosampler. Carotenoids and chlorophylls were analysed as previously described.23 Separation was performed on a C30 column (YMC Co. Ltd, Kyoto, Japan, YMC C30, 100 × 2.1 mm, 3 μm) on an Agilent Technologies 1290 Infinity UHPLC. The column temperature was maintained at 20 °C. The mobile phases were (A) methanol/water (96
1, v/v). The combined supernatant was concentrated under N2 to dryness and re-dissolved in 100 μL of dichloromethane (Merck, Darmstadt, Germany) and 400 μL of 2-propanol (Merck, Darmstadt, Germany). The extract was filtered (0.2 μm, PTFE) and kept at 10 °C in the autosampler. Carotenoids and chlorophylls were analysed as previously described.23 Separation was performed on a C30 column (YMC Co. Ltd, Kyoto, Japan, YMC C30, 100 × 2.1 mm, 3 μm) on an Agilent Technologies 1290 Infinity UHPLC. The column temperature was maintained at 20 °C. The mobile phases were (A) methanol/water (96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v) and (B) methanol/tert-butyl methyl ether (Roth, Karlsruhe, Germany)/water (6
4, v/v) and (B) methanol/tert-butyl methyl ether (Roth, Karlsruhe, Germany)/water (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90
90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v/v). To increase the ionization, 20 mM ammonium acetate (Roth, Karlsruhe, Germany) was added to the mobile phases. The flow rate was 0.2 mL min−1. Elution was carried out with the following gradient: 100% A for 10 min, 100% A to 80% A in 7 min, 80% A for 28 min, 80% A to 0% A in 10 min and 0% A to 100% A in 2 min. The compounds were detected with an Agilent Technologies 6230 TOF LC/MS equipped with an APCI ion source in positive ionization mode. The gas temperature was set to 325 °C at a flow rate of 8 L min−1, the vaporizer was set to 350 °C and the nebulizer pressure was set to 35 psig. The voltage was set to 3500 V and a fragmentor voltage of 175 V was applied at a corona current of 6.5 μA. Carotenoid (CaroteNature GmbH, Münsingen Switzerland) and chlorophyll (Sigma-Aldrich, St Louis, MO, USA) standards were prepared. Their concentrations were determined spectrophotometrically using the specific absorption coefficients.24 The standards were used for external calibration. Quantification was performed at a detection wavelength of 450 nm.
4, v/v/v). To increase the ionization, 20 mM ammonium acetate (Roth, Karlsruhe, Germany) was added to the mobile phases. The flow rate was 0.2 mL min−1. Elution was carried out with the following gradient: 100% A for 10 min, 100% A to 80% A in 7 min, 80% A for 28 min, 80% A to 0% A in 10 min and 0% A to 100% A in 2 min. The compounds were detected with an Agilent Technologies 6230 TOF LC/MS equipped with an APCI ion source in positive ionization mode. The gas temperature was set to 325 °C at a flow rate of 8 L min−1, the vaporizer was set to 350 °C and the nebulizer pressure was set to 35 psig. The voltage was set to 3500 V and a fragmentor voltage of 175 V was applied at a corona current of 6.5 μA. Carotenoid (CaroteNature GmbH, Münsingen Switzerland) and chlorophyll (Sigma-Aldrich, St Louis, MO, USA) standards were prepared. Their concentrations were determined spectrophotometrically using the specific absorption coefficients.24 The standards were used for external calibration. Quantification was performed at a detection wavelength of 450 nm.
RT-qPCR analysis
Total RNA extraction was performed from about 50 mg ground, frozen plant material using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) as described by the manufacturer, including on-column DNase I digestion. RNA was quantified spectrophotometrically at 260 nm (Nanodrop ND1000, Thermo Fisher Scientific, Waltham, MA, USA). The purity was tested using the ratios of absorbance at 260/280 nm (desired value: ∼2.0) and 260/230 nm (desired value: ∼2.0–2.2). cDNA was synthesized from 1 μg total RNA using SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) with oligo (dT)12−18 primers according to the manufacturer's instructions. The reaction mix per well consisted of 3 μL cDNA (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), 5 μL 2× SensiMix SYBR Low-ROX (Bioline, Luckenwalde, Germany) and 2 μL 2 μM primer (forward and reverse primer). The primers were designed based on the sequence information available in the Brassica database BRAD (http://brassicadb.org/brad/) (for primer pair sequences, see Table S1 in the ESI†). In cases when more than a single gene encoded a protein, the primers were designed to cover all of them. Transcript quantification was carried out in triplicate. A CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for RT-qPCR. The thermal cycling conditions were 50 °C for 2 min, 95 °C for 10 min, 39 cycles of 95 °C for 15 s and 60 °C for 1 min, followed by a melting curve analysis. The amplification efficiency for each primer pair was tested using a cDNA dilution series. Gene expression was calculated as n-fold induction of the gene of interest in light-grown sprouts in comparison to dark-grown sprouts by the ΔΔCq method using the geometric mean of three reference genes.25,26 Reference genes were actin 2, ubiquitin-conjugating enzyme E2 30 and elongation factor 1-alpha. Their expression stability in the samples was tested (M value <0.5; coefficient variance <0.25). No-template controls were also used.
10), 5 μL 2× SensiMix SYBR Low-ROX (Bioline, Luckenwalde, Germany) and 2 μL 2 μM primer (forward and reverse primer). The primers were designed based on the sequence information available in the Brassica database BRAD (http://brassicadb.org/brad/) (for primer pair sequences, see Table S1 in the ESI†). In cases when more than a single gene encoded a protein, the primers were designed to cover all of them. Transcript quantification was carried out in triplicate. A CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for RT-qPCR. The thermal cycling conditions were 50 °C for 2 min, 95 °C for 10 min, 39 cycles of 95 °C for 15 s and 60 °C for 1 min, followed by a melting curve analysis. The amplification efficiency for each primer pair was tested using a cDNA dilution series. Gene expression was calculated as n-fold induction of the gene of interest in light-grown sprouts in comparison to dark-grown sprouts by the ΔΔCq method using the geometric mean of three reference genes.25,26 Reference genes were actin 2, ubiquitin-conjugating enzyme E2 30 and elongation factor 1-alpha. Their expression stability in the samples was tested (M value <0.5; coefficient variance <0.25). No-template controls were also used.
Statistical analyses
For statistical analyses, IBM SPSS Statistics 22 (IBM Deutschland, Ehningen, Germany) was used. Treatment differences were analysed using one-way ANOVA followed by Tukey HSD post hoc test. If the assumption of homogeneity of variances was violated, Welch's ANOVA followed by Games-Howell post hoc test was used. If the normality assumption was violated, the Kruskal–Wallis-test was applied. A p-value ≤0.05 was considered as statistically significant. Data are presented as means ± standard deviation.Results
Measurement of LED spectra
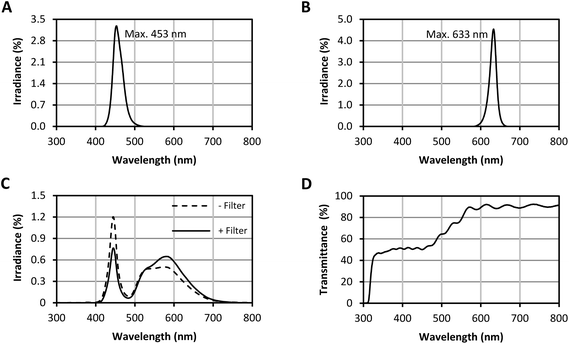
Fig. 2 depicts the measured spectra of the used LEDs. The blue LEDs had a wavelength range of 416–528 nm with a wavelength peak at 453 nm (90% of the irradiance emitted between 438 nm and 480 nm; Fig. 2A). The red LEDs showed a wavelength range of 581–669 nm with a wavelength peak at 633 nm (90% of the irradiance emitted between 613 nm and 647 nm; Fig. 2B). The white LEDs had a prominent blue portion (Fig. 2C). To gain a more evenly distributed spectral irradiance, a filter was applied (Fig. 2D). The transmittance of the filter was ∼50% in the range 400–500 nm, which successfully reduced the blue portion of the white LED spectrum. The resulting wavelength range with the installed filter was 404–789 nm.Diurnal changes in carotenoid concentrations
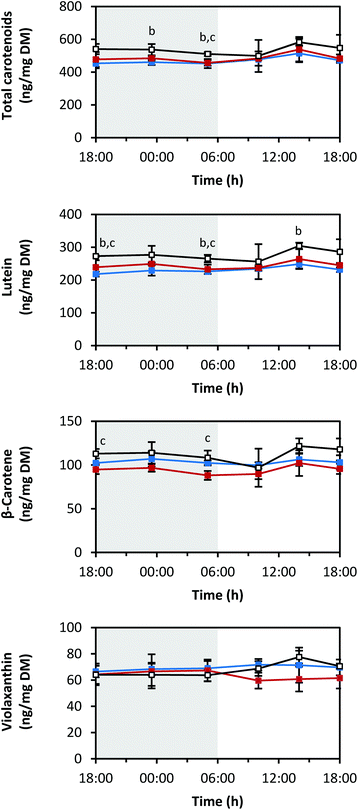
The measured carotenoid concentrations of the light-grown pak choi sprouts were quite steady throughout the whole day (Fig. 3). The average total carotenoid concentration of the six analysed time points under blue LEDs was 471 ± 23 ng mg−1 DM, under red LEDs 487 ± 27 ng mg−1 DM and under white LEDs 536 ± 29 ng mg−1 DM. Thus, the total carotenoid concentration was highest under white LEDs compared to blue and red LEDs. There was a small peak under all light qualities at 14:00 h in the afternoon. Lutein and β-carotene were the two main carotenoids found (Fig. S1 in ESI†). Lutein accounted for about half of the total carotenoids, whereas β-carotene accounted for about one-fifth. Violaxanthin and neoxanthin were present in lower amounts. Neither zeaxanthin nor antheraxanthin were detected (LOD: 0.04 ng mg−1 DM and 0.01 ng mg−1 DM, respectively).Diurnal changes in RNA transcript levels of carotenoid biosynthesis genes
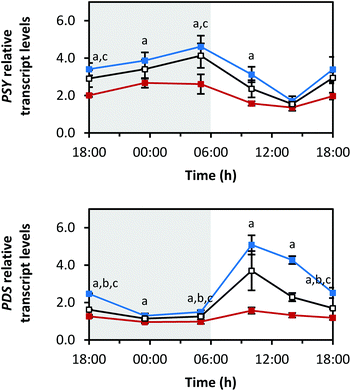
Fig. 4–7 show the transcript levels of key carotenoid biosynthesis genes under different light qualities throughout the day (from 18:00 h of day 5 until 18:00 h of day 6 of light treatment) as n-fold expression of the dark-grown sprouts (harvested after growth in darkness for five days). All three light qualities caused the same diurnal rhythms in steady-state transcript abundance of the different carotenoid biosynthesis genes, but with differences in the amplitudes. For most genes, growth under blue LEDs led to the highest transcript levels, whereas during growth under red LEDs, the transcript levels were the lowest. The transcript levels under white LEDs were in-between. The biosynthetic genes showed differences in the phases and amplitudes of their diurnal rhythms.The first two reactions of the carotenoid biosynthesis pathway are catalysed by PSY and phytoene desaturase (PDS) and contribute to the core structure biosynthesis of carotenoids (Fig. 1). PSY forms phytoene from two geranylgeranyl diphosphates and is considered a rate-limiting enzyme of carotenoid biosynthesis,27 whereas PDS is involved in the production of lycopene together with three other enzymes.28 The RNA transcript levels of PSY increased in the dark period with the highest levels at the end of the dark period (Fig. 4) (max: blue 4.6 ± 0.6-fold at 05:00 h, red 2.7 ± 0.3-fold at 23:30 h, white 4.1 ± 0.7-fold at 05:00 h). With the beginning of the light period, PSY transcript levels decreased within 8 h (min: at 14:00 h with blue 1.7 ± 0.3-fold, red 1.4 ± 0.2-fold, white 1.5 ± 0.2-fold) and started to increase again after 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00 h. In contrast, PDS transcripts decreased during the dark period (min: at 23:30 h with blue 1.3 ± 0.1-fold, red 1.0 ± 0.1-fold, white 1.1 ± 0.1-fold), strongly increased after the beginning of the light period with a maximum after 4 h of illumination (max: at 10:00 h with blue 5.1 ± 0.5-fold, red 1.6 ± 0.2-fold, white 3.7 ± 1.0-fold), and finally, started to decrease again after 10:00 h.
00 h. In contrast, PDS transcripts decreased during the dark period (min: at 23:30 h with blue 1.3 ± 0.1-fold, red 1.0 ± 0.1-fold, white 1.1 ± 0.1-fold), strongly increased after the beginning of the light period with a maximum after 4 h of illumination (max: at 10:00 h with blue 5.1 ± 0.5-fold, red 1.6 ± 0.2-fold, white 3.7 ± 1.0-fold), and finally, started to decrease again after 10:00 h.
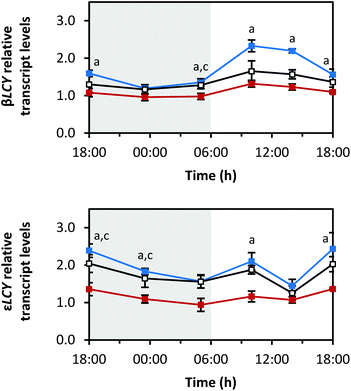
The diurnal rhythms of both cyclases, which are both responsible for the cyclisation of lycopene and the formation of α-carotene and β-carotene (Fig. 1),29 also differed (Fig. 5). βLCY showed a comparable expression pattern to PDS, but with smaller amplitudes (min: at 23:30 h with blue 1.2 ± 0.0-fold, red 1.0 ± 0.1-fold, white 1.2 ± 0.1-fold; max: at 10:00 h with blue 2.3 ± 0.2-fold, red 1.3 ± 0.1-fold, white 1.7 ± 0.3-fold). εLCY transcript levels decreased until the end of the dark period, then increased with a small peak at 10:00 h, and finally, increased again after 14:00 h until the end of the light period (min: blue 1.4 ± 0.2-fold at 14:00 h, red 0.9 ± 0.2-fold at 05:00 h, white 1.3 ± 0.2-fold at 14:00 h; max: blue 2.4 ± 0.4-fold at 18:00 h (day 6), red 1.4 ± 0.0-fold at 18:00 h (day 6), white 2.0 ± 0.2-fold at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00 h (day 5)).
00 h (day 5)).
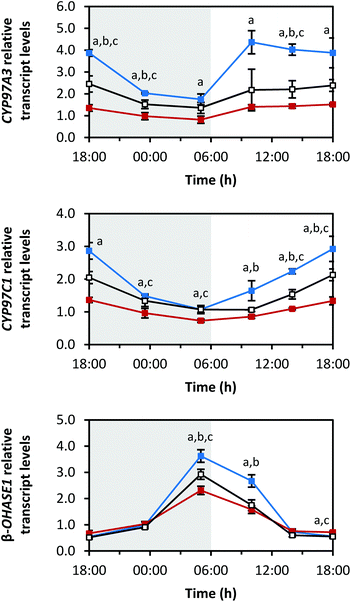
The oxygen-containing xanthophylls are formed from the carotenes by non-haem di-iron hydroxylases (β-OHASEs) and by two haem-containing cytochrome P450 enzymes (CYP97A3 and CYP97C1) (Fig. 1).30 CYP97A3 and CYP97C1 preferentially hydroxylate the β- and ε-ionone rings of α-carotene yielding lutein, while the β-OHASEs form zeaxanthin from β-carotene. The transcripts of all 3 investigated hydroxylases, namely CYP97A3, CYP97C1 and β-OHASE1, differed in their diurnal rhythms (Fig. 6). CYP97A3 decreased during the dark period (min: at 05:00 h with blue 1.7 ± 0.2-fold, red 0.8 ± 0.2-fold, white 1.4 ± 0.3-fold), increased after the beginning of the light period, and then stayed quite steady until the beginning of the dark period (max: blue 4.4 ± 0.5-fold at 10:00 h, red 1.5 ± 0.1-fold at 18:00 h (day 6), white 2.5 ± 0.4-fold at 18:00 h (day 5)). CYP97C1 declined during the dark period (min: at 05:00 h with blue 1.1 ± 0.1-fold, red 0.7 ± 0.0-fold, white 1.1 ± 0.1-fold) and accumulated during the light period (max: blue 2.9 ± 0.4-fold at 18:00 h (day 6), red 1.4 ± 0.1-fold at 18:00 h (day 5), white 2.1 ± 0.2-fold at 18:00 h (day 6)). In stark contrast, β-OHASE1 transcript levels were opposite to those of CYP97C1. In detail, the transcript levels accumulated during the dark period (max: at 05:00 h with blue 3.6 ± 0.2-fold, red 2.3 ± 0.2-fold, white 2.9 ± 0.2-fold) and declined during the light period (min: at 18:00 h of day 5 with blue 0.5 ± 0.0-fold, red 0.7 ± 0.1-fold, white 0.5 ± 0.1-fold). Remarkably, the decrease during the light period was slower under red LEDs than under blue or white LEDs, thereby leading to higher transcript levels under red LEDs than under blue or white LEDs at the end of the light period. Thus, β-OHASE1 was the only enzyme, except for the zeaxanthin epoxidase (ZEP) (Fig. 7), which had the highest transcript levels under red light at one time point, thus indicating that this enzyme may differ in its transcriptional regulation from the others investigated in this study.
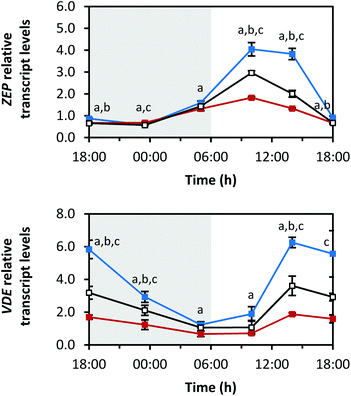
ZEP and violaxanthin de-epoxidase (VDE) are the enzymes of the so-called xanthophyll cycle (Fig. 1).31 Zeaxanthin is formed by VDE and mediates the thermal dissipation of excess light energy, thereby protecting the photosystems.32ZEP showed a similar diurnal rhythm to PDS and βLCY (Fig. 7; min: at 23:30 h with blue 0.6 ± 0.0-fold, red 0.7 ± 0.0-fold, white 0.6 ± 0.0-fold; max: at 10:00 h with blue 4.0 ± 0.3-fold, red 1.8 ± 0.0-fold, white 3.0 ± 0.1-fold). Interestingly, ZEP was higher expressed under red LEDs than under blue or white LEDs at 23:30 h. This is of note because usually exposure to blue LEDs led to the highest transcript levels with other transcripts reported here. For the transcript levels of VDE, the opposite diurnal rhythm of PSY was observed. The transcript levels decreased during the dark period (min: at 05:00 h with blue 1.2 ± 0.2-fold, red 0.7 ± 0.2-fold, white 1.1 ± 0.2-fold), increased after the beginning of the light period (max: at 14:00 h with blue 6.3 ± 0.3-fold, red 1.9 ± 0.1-fold, white 3.6 ± 0.6-fold) and then started to decrease again after 14:00 h.
Diurnal changes in RNA transcript levels of the carotenoid cleavage dioxygenase 4
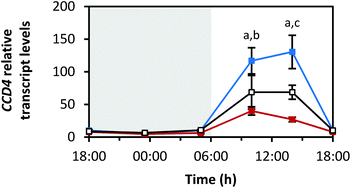
Fig. 8 depicts the diurnal rhythm of the transcript levels of CCD4, a carotenoid degrading enzyme, as n-fold expression of the dark-grown sprouts. As in the case of the carotenoid biosynthesis genes, blue LEDs caused the highest and red LEDs the smallest changes in the transcript levels. Furthermore, the transcript levels stayed low during the dark period (min: at 23:30 h with blue 5.4 ± 0.4-fold, red 4.7 ± 1.1-fold, white 6.6 ± 0.6-fold), but very strongly increased during the light period (max: blue 130.4 ± 25.5-fold at 14:00 h, red 40.0 ± 6.3-fold at 10:00 h, white 68.7 ± 10.7-fold at 14:00 h). During the end of the light period, the transcript levels returned to low values.Diurnal changes in RNA transcript levels of the transcription factors elongated hypocotyl 5 and circadian clock associated 1
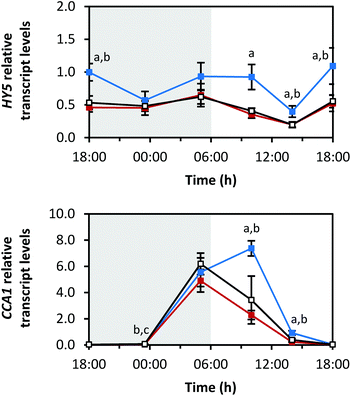
For transcription factor HY5, all light qualities led to comparable diurnal rhythms. Illumination by blue LEDs caused the highest amplitude. Interestingly, there was no difference in transcript levels between sprouts grown under red and white LEDs (Fig. 9). The maxima of the transcript levels were at the transitions from dark to light periods (min: at 14:00 h with blue 0.4 ± 0.1-fold, red 0.2 ± 0.0-fold, white 0.2 ± 0.0-fold; max: blue 1.1 ± 0.3-fold at 18:00 h (day 6), red 0.6 ± 0.2-fold at 05:00 h, white 0.6 ± 0.1-fold at 18:00 h (day 6)).The transcription factor CCA1 is part of the circadian clock. The transcript levels of CCA1 increased at the end of the dark period (Fig. 9; max: blue 7.4 ± 0.6-fold at 10:00 h, red 4.9 ± 0.9-fold at 05:00 h, white 6.2 ± 0.8-fold at 05:00 h) and decreased during the light period (min: blue 0.03 ± 0.01-fold at 23:30 h, red 0.04 ± 0.01-fold at 18:00 h (day 5), white 0.04 ± 0.01-fold at 18:00 h (day 5)).
Interestingly, growth under blue LEDs delayed the transcript level decrease of both transcription factors at the beginning of the light period.
Discussion
LEDs emitting different light spectra are an attractive light source for cultivating vegetables under greenhouse conditions. The measurement of the LED spectra confirmed that all three LED colours studied here had wavelength ranges suitable for photosynthesis. The action spectrum of photosynthesis, which depicts the ability of light of different wavelengths to support photosynthesis, shows that plants are able to use light over a wavelength range from 400 nm to 700 nm with peaks at wavelengths representing blue and red light.33 The narrow-band spectra of the blue and red LEDs had a very small wavelength range of ∼40 nm. Using such narrow-band spectra LEDs allows to selectively activate blue light photoreceptors (cryptochromes, phototropins and Zeitlupes) or red light photoreceptors (phytochromes),34 whereas white LEDs activate both. Of note is that there is already some evidence for the involvement of cryptochromes and phytochromes in carotenoid biosynthesis regulation.10,12 However, the exact influence of cryptochromes and phytochromes on the light signalling transduction to carotenoid biosynthesis pathway and subsequent metabolism has to be further investigated.The measured total carotenoid concentrations were highest under white LEDs and slightly lower under blue and red LEDs. This was also shown for lettuce (Lactuca sativa ‘Redfire’) and komatsuna (Brassica campestris ‘Komatsuna’), although for lettuce, red light caused the same content as white light.4 In contrast, further studies showed an increase of carotenoid levels under blue light in other lettuce cultivars (Lactuca sativa ‘Banchu Red Fire’ or ‘Red Cross’)8,9 and spinach (Spinacia oleracea ‘Okame’),4 thereby confirming the species-dependent aspects of carotenoid biosynthesis and subsequent metabolism.
In our study, total carotenoid concentrations of ∼500 ng mg−1 DM were reached under all three different LEDs. These values are in line with the carotenoid concentrations of field-grown pak choi during the dry and wet season in Taiwan.35 Indeed, our growth conditions matched the wet season in which total carotenoid levels ranging from 440 to 1110 ng mg−1 DM were reached. This result was unexpected since the photosynthetic active radiation in our experiments was quite low compared to field conditions with sunlight. The measured chlorophyll concentrations correlated with the carotenoid levels (Fig. S2 in ESI†). Carotenoids are central for photosynthesis because they are required for the correct assembly of photosystems, light-harvesting and photoprotection.6 It is known that carotenoid biosynthesis genes and other photosynthesis-related genes are co-expressed.36 In tobacco seedlings (Nicotiana tabacum ‘Samsun’), the chlorophyll biosynthesis gene protoporphyrinogen IX oxidase (PPOX) was induced together with carotenoid biosynthesis genes during the de-etiolation process. In the same study, the expression pattern of a light-harvesting protein of photosystem II resembled the transcript patterns of ZEP and β-OHASE.11 Furthermore, in tomato leaves (Solanum lycopersicum ‘Ailsa Craig’), ZEP transcripts oscillated with a phase very similar to light-harvesting complex II (LHCII) transcripts during the course of the day.37 Since ZEP belongs to the xanthophyll cycle which protects photosystem II, this result is in line with the assumption that diurnal RNA transcript patterns are coordinated for individual complexes such as photosystem II. Therefore, we hypothesize that chlorophyll concentrations and photosynthesis should be targeted to change carotenoid levels in green tissues.
Since both biosynthesis and degradation determine overall total carotenoid concentrations, the transcripts of biosynthetic genes and one degrading gene were investigated. In this context, it is important to consider that photosynthetically active and non-active tissues differ in their regulation of carotenoid concentrations. In detail, PSY is often modulated to increase carotenoids since it catalyses the first step of carotenoid biosynthesis. However, while PSY is the limiting factor in non-active tissues as shown by overexpression of PSY in cassava (Manihot esculenta) roots,38 in tomato (Solanum lycopersicum ‘Aisla Craig’) during fruit ripening39 as well as in rice (Oryza sativa) endosperm,40 it does not seem to be equally important in photosynthetically active tissues. In Arabidopsis thaliana seedlings, PSY overexpression did not affect the carotenoid concentrations in leaves, whereas in the calli and roots, higher carotenoid levels were detected.41 Thus, the regulation of the steady-state carotenoid levels differs in green tissues compared to non-green tissues. One reason might be that the stoichiometry between carotenoids and chlorophylls has to be tightly regulated in chloroplasts to guarantee optimal photosynthesis. Therefore, our RNA transcript results were better compared to studies performed with photosynthetically active tissues.
The same diurnal changes of RNA transcripts were measured under all three light qualities. In Arabidopsis thaliana seedlings grown with continuous illumination, gene expression patterns under different light qualities were quite similar.42 The highest transcripts of carotenoid biosynthesis genes were measured under blue LEDs, thereby also implying that the highest carotenoid concentrations are obtained under blue rather than white LEDs. Thus, other mechanisms might counterbalance an increase in carotenoids under blue LEDs. All biosynthesis genes were transcriptionally induced during the light period, except for PSY and β-OHASE1. However, PSY and β-OHASE1 were already highly expressed at the beginning of the light period. The light-induced transcription correlates to the small carotenoid maxima measured at 14:00 h. In contrast, all RNA transcripts were reduced during the dark period, except for PSY and β-OHASE1. Although the RNA transcripts of the carotenoid biosynthesis genes conformed to these general trends found for during the light and dark periods, they differed in their phases and amplitudes. For example, PDS, βLCY and ZEP transcriptional rhythms were quite comparable, indicating a shared regulation of these three genes. ZEP and VDE are part of the xanthophyll cycle31 that protects the photosystems against excessive energy and for which zeaxanthin is also necessary. Thus, it is not unexpected that ZEP transcripts started to decline during the middle of the light period, whereas the maximum of VDE transcripts was reached later during the light period. However, ZEP and VDE transcripts were incongruent with the concentrations of the xanthophyll cycle carotenoids since violaxanthin remained quite constant and no zeaxanthin was detected. Hence, investigations on posttranscriptional regulation and the flux through the xanthophyll cycle should be performed in the future. In this context, it is already known that the VDE activity is posttranslationally regulated by the transthylakoid pH gradient and the luminal ascorbate content of which the pH gradient is modulated through the light intensity.32 Furthermore, β-OHASE1 and ZEP were the only genes whose transcripts were found in a higher abundance under red LEDs at one of the time points analysed. Thus, more information is needed about transcriptional regulation and also transcript stability of carotenoid biosynthesis genes.
To date, it is known that HY5 and phytochrome-interacting factors (PIFs) act as antagonistic regulators of PSY transcription.43 Furthermore, changes in PSY transcripts have also been investigated during the de-etiolation process. In detail, during de-etiolation, PSY transcripts were reported to increase in Arabidopsis thaliana and Sinapis alba.10 In tobacco seedlings (Nicotiana tabacum ‘Samsun’), PSY transcripts accumulated most under red and white light conditions and less under blue light during de-etiolation.11 Nevertheless, although the de-etiolation process is initiated by light, the induced changes are probably differently regulated compared to our study, since our sprouts were grown already for several days under light conditions.
Degradation is also known to influence the steady-state carotenoid concentrations. In this study, we focussed on CCD4 in this regard. In Arabidopsis thaliana, β-carotene and total carotenoids of a CCD4 loss-of-function mutant were investigated in leaves. The mutant possessed slightly higher levels of β-carotene and total carotenoids compared to wild-type plants. Moreover, during dark-induced senescence, the carotenoid levels were significantly higher compared to wild-type. These effects were not observed in a CCD1 mutant. Thus, CCD4 seems to be important for the β-carotene turnover in leaves.19 Lätari et al. overexpressed PSY in Arabidopsis thaliana leading to an increase of β-carotene in roots and non-green calli, but interestingly, not in leaves. Instead, they found higher levels of C13 apocarotenoid glycosides (AGs). Of note is that in the CCD4 mutant, the AGs were reduced in the leaves. The authors hypothesized that CCD4 is responsible for AG formation and that the glycosylation serves as a detoxification pathway for apocarotenoids.20 Hence, both studies provide evidence that CCD4 is involved in the degradation of carotenoids in leaves. In our study, CCD4 transcripts were accumulated during the light period like the RNA transcripts of the carotenoid biosynthesis genes. This finding confirms the role of CCD4 as an antagonist to carotenoid biosynthesis in leaves. The highest CCD4 transcript levels were measured under illumination with blue LEDs, thereby indicating increased degradation, and thus, a higher carotenoid turnover under blue LEDs. This result could explain why the highest carotenoid concentrations were not measured under blue LEDs despite the fact that the carotenoid biosynthesis genes were the most highly expressed under blue light. However, it is necessary to further investigate the flux through the carotenoid biosynthesis pathway as well as to measure relevant protein levels and to assess activities to confirm this hypothesis.
The transcript levels of HY5 were also highest under blue LEDs. HY5 is a transcription factor that is downstream of light-activated signalling pathways. Thus, this transcription factor could be directly (as in the case of PSY) or indirectly be involved in the transcriptional regulation of carotenoid biosynthesis and degrading genes. This would explain why the RNA transcript levels of the carotenoid biosynthesis and degrading genes studied here were highest under blue LEDs like those of HY5.
The investigated RNA transcripts also showed distinct diurnal rhythms. In plants, many genes with a specific diurnal rhythm are under the control of the circadian clock. For example, about one-third of the expressed genes are circadian regulated in Arabidopsis thaliana.44 In addition, other genes involved in the photosynthesis process are circadian regulated such as photosystem II chlorophyll a–b binding protein (LHCB).45 Until now, the circadian regulation of carotenoid biosynthesis genes has not been comprehensively investigated. However, there is evidence that some of these genes are under circadian control in Arabidopsis thaliana.44 Furthermore, circadian regulation was observed for PSY transcripts in poplar (Populus x canescens) leaves46 and for ZEP transcripts in tomato leaves (Solanum lycopersicum ‘Ailsa Craig’).37 In our study, the transcripts of the transcription factor CCA1 showed a diurnal rhythm, which is in line with the literature.22,47 CCA1 controls the diurnal transcription of target genes. Of interest is that the highest CCA1 RNA transcript levels were measured once again under blue LEDs. In case that the carotenoid biosynthesis genes are also regulated by the circadian clock in pak choi, this could be a further explanation for why they too are most highly induced under blue light conditions. However, investigations regarding the circadian regulation of carotenoid biosynthesis genes were not performed in our study and should be part of future experiments, such as testing whether the RNA transcript levels of carotenoid biosynthesis genes still oscillate in continuous darkness and/or light as well as testing for CCA1-binding to their promotors.
Conclusions
This study confirmed that LEDs are suitable to produce pak choi that are rich in carotenoids. Although LEDs were used as sole light sources, the carotenoid concentrations were similar to values described for field-grown pak choi. The transcripts of most carotenoid biosynthesis genes increased during the light period which was in line with the small carotenoid maximum in the afternoon. However, the transcript levels of individual genes were incongruent to the measured carotenoid profiles. Thus, further investigations regarding the posttranscriptional regulation are necessary. Interestingly, the RNA transcripts involved in the carotenoid biosynthesis pathway were highest under blue lighting, whereas the carotenoid content was highest under white light, thereby suggesting a higher turnover of carotenoids by CCD4 under blue light conditions. Furthermore, this study investigated the metabolism of carotenoids in vegetables for a whole day which has been rarely studied for secondary metabolites to date. Future research using LEDs with specific wavelengths during the growth of various vegetables would contribute to fully benefit from LED technology in vegetable production.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank I. Hauschild and M. Skoruppa for the LED module installations, and A. Maikath, A. Platalla and A. Jankowsky for their excellent technical assistance.References
- C. A. Mitchell, A.-J. Both, C. M. Bourget, J. F. Burr, C. Kubota, R. G. Lopez, R. C. Morrow and E. S. Runkle, LEDs: The future of greenhouse lighting!, Chron. Horticult., 2012, 52, 5–12 Search PubMed.
- K. J. McCree, Action spectrum, absorptance and quantum yield of photosynthesis in crop plants, Agric. Meteorol., 1972, 9, 191–216 CrossRef.
- T. A. O. Dougher and B. Bugbee, Differences in the response of wheat, soybean and lettuce to reduced blue radiation, Photochem. Photobiol., 2001, 73, 199–207 CrossRef PubMed.
- K. Ohashi-Kaneko, M. Takase, N. Kon, K. Fujiwara and K. Kurata, Effect of light quality on growth and vegetable quality in leaf lettuce, spinach and komatsuna, Environ. Control Biol., 2007, 45, 189–198 CrossRef.
- M. M. Hasan, T. Bashir, R. Ghosh, S. K. Lee and H. Bae, An overview of LEDs’ effects on the production of bioactive compounds and crop quality, Molecules, 2017, 22, E1420 CrossRef PubMed.
- C. I. Cazzonelli, Goldacre Review: Carotenoids in nature: insights from plants and beyond, Funct. Plant Biol., 2011, 38, 833–847 CrossRef.
- J. Fiedor and K. Burda, Potential role of carotenoids as antioxidants in human health and disease, Nutrients, 2014, 6, 466–488 CrossRef PubMed.
- M. Johkan, K. Shoji, F. Goto, S.-n. Hashida and T. Yoshihara, Blue light-emitting diode light irradiation of seedlings improves seedling quality and growth after transplanting in red leaf lettuce, HortScience, 2010, 45, 1809–1814 Search PubMed.
- Q. Li and C. Kubota, Effects of supplemental light quality on growth and phytochemicals of baby leaf lettuce, Environ. Exp. Bot., 2009, 67, 59–64 CrossRef.
- J. von Lintig, R. Welsch, M. Bonk, G. Giuliano, A. Batschauer and H. Kleinig, Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings, Plant J., 1997, 12, 625–634 CrossRef PubMed.
- S. Woitsch and S. Römer, Expression of xanthophyll biosynthetic genes during light-dependent chloroplast differentiation, Plant Physiol., 2003, 132, 1508–1517 CrossRef PubMed.
- L. Giliberto, G. Perrotta, P. Pallara, J. L. Weller, P. D. Fraser, P. M. Bramley, A. Fiore, M. Tavazza and G. Giuliano, Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content, Plant Physiol., 2005, 137, 199–208 CrossRef PubMed.
- A. Schofield and G. Paliyath, Modulation of carotenoid biosynthesis during tomato fruit ripening through phytochrome regulation of phytoene synthase activity, Plant Physiol. Biochem., 2005, 43, 1052–1060 CrossRef PubMed.
- C. Reif, E. Arrigoni, F. Berger, D. Baumgartner and L. Nyström, Lutein and β-carotene content of green leafy Brassica, species grown under different conditions, LWT–Food Sci. Technol., 2013, 53, 378–381 CrossRef.
- F. Di Gioia and P. Santamaria, The nutritional properties of microgreens, in Microgreens, Eco-logica, Bari, 2015, pp. 41–50 Search PubMed.
- O. Ahrazem, L. Gómez-Gómez, M. J. Rodrigo, J. Avalos and M. C. Limón, Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions, Int. J. Mol. Sci., 2016, 17, E1781 CrossRef PubMed.
- M. E. Auldridge, A. Block, J. T. Vogel, C. Dabney-Smith, I. Mila, M. Bouzayen, M. Magallanes-Lundback, D. DellaPenna, D. R. McCarty and H. J. Klee, Characterization of three members of the Arabidopsis, carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family, Plant J., 2006, 45, 982–993 CrossRef PubMed.
- S. Frusciante, G. Diretto, M. Bruno, P. Ferrante, M. Pietrella, A. Prado-Cabrero, A. Rubio-Moraga, P. Beyer, L. Gomez-Gomez, S. Al-Babili and G. Giuliano, Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12246–12251 CrossRef PubMed.
- S. Gonzalez-Jorge, S. H. Ha, M. Magallanes-Lundback, L. U. Gilliland, A. Zhou, A. E. Lipka, Y. N. Nguyen, R. Angelovici, H. Lin, J. Cepela, H. Little, C. R. Buell, M. A. Gore and D. Dellapenna, Carotenoid cleavage dioxygenase4 is a negative regulator of beta-carotene content in Arabidopsis seeds, Plant Cell, 2013, 25, 4812–4826 CrossRef PubMed.
- K. Lätari, F. Wüst, M. Hübner, P. Schaub, K. G. Beisel, S. Matsubara, P. Beyer and R. Welsch, Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves, Plant Physiol., 2015, 168, 1550–1562 CrossRef PubMed.
- J. Lee, K. He, V. Stolc, H. Lee, P. Figueroa, Y. Gao, W. Tongprasit, H. Zhao, I. Lee and X. W. Deng, Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development, Plant Cell, 2007, 19, 731–749 CrossRef PubMed.
- Y. Jiao, O. S. Lau and X. W. Deng, Light-regulated transcriptional networks in higher plants, Nat. Rev. Genet., 2007, 8, 217–230 CrossRef PubMed.
- V. Mageney, S. Baldermann and D. C. Albach, Intraspecific variation in carotenoids of Brassica oleracea, var. sabellica, J. Agric. Food Chem., 2016, 64, 3251–3257 CrossRef PubMed.
- G. Britton, S. Liaaen-Jensen and H. Pfander, Carotenoids - Handbook, Birkhäuser Verlag, Basel, Switzerland, 2004 Search PubMed.
- J. Vandesompele, K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe and F. Speleman, Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes, Genome Biol., 2002, 3, research0034 CrossRef PubMed.
- M. W. Pfaffl, Quantification strategies in real-time PCR, in A-Z of quantitative PCR, ed. S. A. Bustin, IUL Biotechnol. Ser., La Jolla, CA, USA, 2004, ch. 3, pp. 87–112 Search PubMed.
- P. D. Fraser, W. Schuch and P. M. Bramley, Phytoene synthase from tomato (Lycopersicon esculentum) chloroplasts - partial purification and biochemical properties., Planta, 2000, 211, 361–369 CrossRef PubMed.
- P. A. Scolnik and G. E. Bartley, Phytoene desaturase from Arabidopsis, Plant Physiol., 1993, 103, 1475 CrossRef PubMed.
- F. X. Cunningham, B. Pogson, Z. Sun, K. A. McDonald, D. DellaPenna and E. Gantt, Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation, Plant Cell, 1996, 8, 1613–1626 Search PubMed.
- J. Kim, J. J. Smith, L. Tian and D. Dellapenna, The evolution and function of carotenoid hydroxylases in Arabidopsis, Plant Cell Physiol., 2009, 50, 463–479 CrossRef PubMed.
- R. C. Bugos, A. D. Hieber and H. Y. Yamamoto, Xanthophyll cycle enzymes are members of the lipocalin family, the first identified from plants, J. Biol. Chem., 1998, 273, 15321–15324 CrossRef PubMed.
- D. C. Rockholm and H. Y. Yamamoto, Violaxanthin de-epoxidase, Plant Physiol., 1996, 110, 697–703 CrossRef PubMed.
- H. Lodish, A. Berk, S. L. Zipursky, P. Matsudaira, D. Baltimore and J. Darnell, Photosynthetic stages and light-absorbing pigments, in Molecular Cell Biology, 2000, https://www.ncbi.nlm.nih.gov/books/NBK21598/, (accessed January, 2018) Search PubMed.
- C. Kami, S. Lorrain, P. Hornitschek and C. Fankhauser, Light-regulated plant growth and development, Curr. Top. Dev. Biol., 2010, 91, 29–66 Search PubMed.
- P. Hanson, R.-y. Yang, L.-c. Chang, L. Ledesma and D. Ledesma, Contents of carotenoids, ascorbic acid, minerals and total glucosinolates in leafy brassica pak choi (Brassica rapa, L. chinensis) as affected by season and variety, J. Sci. Food Agric., 2009, 89, 906–914 CrossRef.
- C. Stange and C. Flores, Carotenoids and photosynthesis - regulation of carotenoid biosyntesis by photoreceptors, in Advances in photosynthesis – Fundamental aspects, ed. M. Najafpour, InTech, Rijeka, Croatia, 2012, ch. 4, pp. 77–96 Search PubMed.
- A. J. Thompson, A. C. Jackson, R. A. Parker, D. R. Morpeth, A. Burbidge and I. B. Taylor, Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid, Plant Mol. Biol., 2000, 42, 833–845 CrossRef PubMed.
- R. Welsch, J. Arango, C. Bar, B. Salazar, S. Al-Babili, J. Beltran, P. Chavarriaga, H. Ceballos, J. Tohme and P. Beyer, Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene, Plant Cell, 2010, 22, 3348–3356 CrossRef PubMed.
- P. D. Fraser, E. M. Enfissi, J. M. Halket, M. R. Truesdale, D. Yu, C. Gerrish and P. M. Bramley, Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism, Plant Cell, 2007, 19, 3194–3211 CrossRef PubMed.
- X. Ye, S. Al-Babili, A. Klöti, J. Zhang, P. Lucca, P. Beyer and I. Potrykus, Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm, Science, 2000, 287, 303–305 CrossRef PubMed.
- D. Maass, J. Arango, F. Wust, P. Beyer and R. Welsch, Carotenoid crystal formation in Arabidopsis, and carrot roots caused by increased phytoene synthase protein levels, PLoS One, 2009, 4, e6373 CrossRef PubMed.
- L. Ma, J. Li, L. Qu, J. Hager, Z. Chen, H. Zhao and X. W. Deng, Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways, Plant Cell, 2001, 13, 2589–2607 CrossRef.
- G. Toledo-Ortiz, H. Johansson, K. P. Lee, J. Bou-Torrent, K. Stewart, G. Steel, M. Rodriguez-Concepcion and K. J. Halliday, The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription, PLoS Genet., 2014, 10, e1004416 CrossRef PubMed.
- M. F. Covington, J. N. Maloof, M. Straume, S. A. Kay and S. L. Harmer, Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development, Genome Biol., 2008, 9, R130 CrossRef PubMed.
- C. Andronis, S. Barak, S. M. Knowles, S. Sugano and E. M. Tobin, The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis, Mol. Plant, 2008, 1, 58–67 CrossRef PubMed.
- M. Loivamäki, S. Louis, G. Cinege, I. Zimmer, R. J. Fischbach and J. P. Schnitzler, Circadian rhythms of isoprene biosynthesis in grey poplar leaves, Plant Physiol., 2007, 143, 540–551 CrossRef PubMed.
- A. Pokhilko, J. Bou-Torrent, P. Pulido, M. Rodríguez-Concepción and O. Ebenhöh, Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis, New Phytol., 2015, 206, 1075–1085 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Primer pairs used for gene expression analysis by RT-qPCR, accession numbers of the nucleotide sequences used for primer design, characteristic chromatogram of carotenoids and chlorophylls in pak choi, diurnal chlorophyll concentrations in pak choi sprouts grown under three different light qualities. See DOI: 10.1039/c8pp00136g |
| This journal is © The Royal Society of Chemistry and Owner Societies 2018 |