Open Access Article
Open Access ArticleStudies on a landscape of perfluoroaromatic-reactive peptides†
Ethan D.
Evans
 and
Bradley L.
Pentelute
and
Bradley L.
Pentelute
 *
*
Department of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA. E-mail: blp@mit.edu
First published on 26th July 2018
Abstract
We investigated 26 midsized peptides (∼30 amino acids in length) selected using mRNA display to perform a nucleophilic aromatic substitution reaction (SNAr). Analysis suggested a diverse set of reactive sequences with significant differences in primary sequence, secondary structure and even predicted tertiary structural features. Several of the sequences displayed rapid kinetics allowing for near complete labeling in under one hour. Rosetta ab initio structure prediction of these sequences suggested a landscape of structural features, ranging from beta-sheet-based sequences to those possessing more alpha-helical-like character. Circular dichroism spectroscopy confirmed elements of the structure predictions for the majority of peptides. This analysis additionally uncovered that several peptides underwent secondary structure alterations upon reaction. These results suggest a broad sequence and structural landscape of SNAr active peptides along with a potentially important feature of these biopolymers.
Introduction
Nature has evolved a host of protein-based reaction mechanisms to perform the chemical transformations that sustain life. In cases of convergent evolution, multiple sequence and structural protein families evolved to perform a common chemistry.1,2 A well-known example of this convergence is the glycoside hydrolase group of enzymes.3 Within the broad category of glycoside hydrolases are the cellulases which exist in over 10 different sequence-based families and possess distinct folds and may hydrolyse a range of substrates.4,5 As another example, in the case of the various carbonic anhydrases, separate folds perform identical overall chemical reactions using the same substrates.6In general, for a given chemical transformation performed by proteins, similar or drastically different mechanistic themes may exist. For instance, while the metal center in carbonic anhydrases ranges from iron and zinc to even cadmium, the use of a metal ion is a common theme and appears indispensable for this reaction.7,8 In contrast, for the cleavage of an amide bond, a host of mechanisms have evolved. Some rely on direct amino acid-based nucleophiles (serine or cysteine for example) while still others leverage metal ions or residues to activate water for bond cleavage.9–11 Therefore, when considering an abiotic reaction of interest, it is unclear whether similar or different themes will emerge in a population of functional biopolymers.
Significant efforts have been devoted to identifying peptides capable of reacting with small molecules.12,13 Much of the focus on discovering reactive sequences has been on leveraging selection and screening based approaches.14–19 Focusing on perfluoroaromatic-based nucleophilic aromatic substitution (SNAr) reactions, two separate functional peptides have been reported. The first, termed the ‘π-clamp’ is a four-residue peptide (FCPF),14 while the other ‘MP01’ is 29-residues in length, originated from an mRNA display selection and displayed no obvious similarities to the π-clamp aside from the use of an active site cysteine.15
The selection used to discover MP01 assayed a large, predominantly randomized library of ∼30-amino-acid long peptides for SNAr activity. Five rounds of selection were performed and followed by high-throughput sequencing, revealing a predominantly unconverged landscape with millions of unique sequences. The primary isolate, MP01, significantly stood out from the majority of the sequences and was thus studied. This selection, however, presented a large, diverse and uncharacterized landscape of other potentially reactive sequences that were only partially enriched.
Here, we probed what features emerged from the aforementioned selection by studying a host of the discovered midsized peptides. In particular, we investigated whether multiple routes to reactivity were possible—from primary amino acid sequence to structural features—and if common mechanistic themes occurred. Using chemical synthesis, structure prediction and circular dichroism (CD) spectroscopy, we uncovered several routes to SNAr reactivity with disparate reaction rates. This work found that there are many mixed alpha-helix/random coil sequences in addition to reactive beta-sheet peptides. Furthermore, many peptides underwent secondary structure alterations upon reaction with the perfluoroaromatic probe. These results may suggest that while there are several different solutions to the problem of reactivity in the space of midsized peptides, common themes may still emerge.
Results and discussion
The sequence landscape is diverse
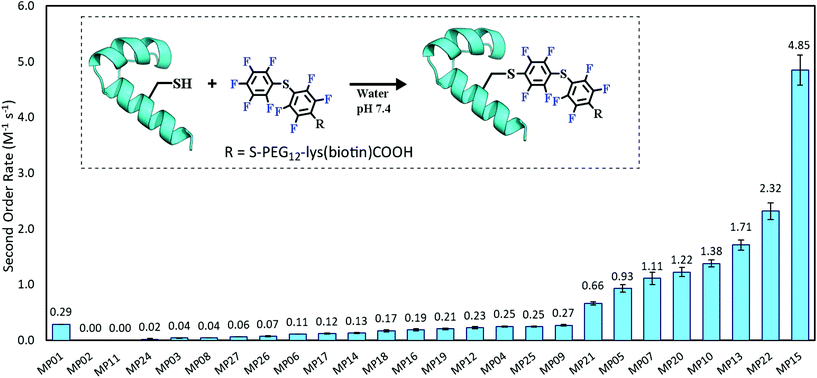
The sequence space of peptides (referred to as MPs) outside of the previously characterized isolate (MP01) showed minimal similarity and low sequence counts (Table 1).15 In this diverse set of MPs, the proposed reactive cysteine was found throughout the length of the sequence—from more N-terminal placements to C-terminal locations—with the majority found in a central location due to the initial library biasing. Peptides appeared with as few as two counts in the high-throughput sequencing to 30 for MP03 (one count corresponds to a single observation of the complete sequence). The majority appeared at or below six counts. As with MP01, many of the sequences did not possess the full 30 amino acid library region (only 11 out of 26 sequences did).‡ No major similarities between sequences were observed in terms of long sequence motifs and only two sequences contained the FCPF motif that was initially doped into the library (MP04 and 27). However, there were several related sequences like FCPI (MP03), WCPF (MP05) and FCPS (MP06). Notably, this set of peptides contained two sequences each with two cysteines (MP08) and another with three (MP05). Similarly, we included a sequence possessing a high sequence count, albeit lacking a cysteine (MP02).| Name | Sequence | Count |
|---|---|---|
| a Peptide synthesized with the full C-terminal constant region from the selection (–SGSLGHHHHHHRL). b Required refolding procedure. | ||
| MP02a | MPNYG PLSPS QPSHG YTFWM VPIWD NSHNA AG | 25 |
| MP03 | MTSVT ASLLM HF PI RAHIT NKPSF NPSG PI RAHIT NKPSF NPSG |
30 |
| MP04 | MRTPI KFAPR LSQPF  PFRK QHQLH LHPLI EG PFRK QHQLH LHPLI EG |
21 |
| MP05 | MRP A RRDRT LW A RRDRT LW PF DSPAW FLLSG FS PF DSPAW FLLSG FS G G |
5 |
| MP06 | MGIVH NATRF PKR F YSFIA TRQSK NSIRV SG F YSFIA TRQSK NSIRV SG |
6 |
| MP07 | MKTFS SDQRF SKK Y RIYFH KLRQR NHNTS VG Y RIYFH KLRQR NHNTS VG |
6 |
| MP08 | MQHED L TWY GF TWY GF PS GNFTP RNLRG DSDG PS GNFTP RNLRG DSDG |
9 |
| MP09 | MRYIY VLRLK SW GG ASARS SPRS GG ASARS SPRS ATKLL G ATKLL G |
10 |
| MP10 | MHNAY LRKSM RQL Y FRRTL HNIHV MSHRG Y FRRTL HNIHV MSHRG |
6 |
| MP11 | MSTGD  HIQH LRPFH GNIAW MRSGN MDG HIQH LRPFH GNIAW MRSGN MDG |
4 |
| MP12 | MVKLS GKERT TRN F FSFLA SRRTK KFNNL SG F FSFLA SRRTK KFNNL SG |
4 |
| MP13 | MGHLH I MVW RVNTS GHILS VGHKS YSSHK TG MVW RVNTS GHILS VGHKS YSSHK TG |
4 |
| MP14b | MSSGT HYGIL NMVIR  HLVK NQTSQ MVVLT TG HLVK NQTSQ MVVLT TG |
2 |
| MP15 | MHHY SKMKR RILMH YLFAN TMAHR DLGTN G SKMKR RILMH YLFAN TMAHR DLGTN G |
5 |
| MP16b | MHLRM IRYLN RRRHL  HVVE IRHGL FASRE IG HVVE IRHGL FASRE IG |
6 |
| MP17b | MNGHY P YLI TSVLV GATTS GVPVV VHLRV G YLI TSVLV GATTS GVPVV VHLRV G |
5 |
| MP18b | MRHYH LT FQ GFRIF RRTVD SLEME ISLG FQ GFRIF RRTVD SLEME ISLG |
5 |
| MP19b | MHMHK TTSYR IRVLV GVDVY RMSHT  LTSS SG LTSS SG |
5 |
| MP20 | MHTSL RSRAK SHSRS FGK A SIYTR YLKMG A SIYTR YLKMG |
5 |
| MP21 | MQNSK HRPRR  LRLL PLLRG HLHRM FRERG LRLL PLLRG HLHRM FRERG |
6 |
| MP22 | MRSTH QRVRR PRNL SFKHK WLIKF LKTLT G SFKHK WLIKF LKTLT G |
5 |
| MP23 | MRFFA H LSI DSSYM WANFS VDRQT RG LSI DSSYM WANFS VDRQT RG |
6 |
| MP24 | MRRTP STRAR GRVFL LPTLR FFITL  NLNG NLNG |
5 |
| MP25b | MNRIF HKRST YQMVF GR SD FTSTY HVLIS YG SD FTSTY HVLIS YG |
5 |
| MP26 | MTATS SSTSR G RPS TAQVV QRLRG LLLVV G RPS TAQVV QRLRG LLLVV G |
6 |
| MP27 | MLFMR LTKKT MATKF  PFRR KRKHR ERRAL YG PFRR KRKHR ERRAL YG |
5 |
Following chemical synthesis using an Automated Flow Peptide Synthesizer20 and purification, several of the sequences required a guanidinium chloride-based refolding procedure (MP14, 16–19 and 25, ESI 2.3†). After this protocol most were readily soluble with the exception of MP14 which was marginally soluble. MP23 was dropped as it proved challenging to both cleanly synthesize and purify.
The functional landscape is broad with reaction rate constants spanning two orders of magnitude
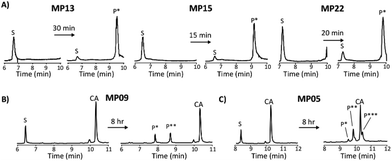
Each peptide was assayed for reactivity, revealing a broad functional landscape. Briefly, individual peptides (100 μM) were incubated with an excess of the perfluoroaromatic probe (500 μM) from the initial selection, referred to as the capture agent (CA) under reducing conditions (5 mM tris(2-carboxyethyl)phosphine, TCEP) at pH 7.4 and room temperature (Fig. 1, inset). Kinetic analysis of the reactions revealed that many peptides were capable of reactivity (Fig. 1). 15 sequences displayed low to moderate reactivity (0.02–0.27 M−1 s−1) while eight possessed second order rate constants significantly higher than that of the previously characterized primary isolate from the selection (MP01, 185 sequencing counts, 0.29 M−1 s−1).15 With the exception of MP02 and 11, all of the sequences displayed rates greater than that of a previously characterized, random cysteine-containing peptide (0.007 M−1 s−1).15 While MP11 did not show reactivity on the time scale of the kinetics measurements, with longer incubations MP11 could minimally react, akin to the reactivity of an unselected cysteine containing peptide (Fig. S34B†).For the most reactive peptides, no sequence similarities were observed and high conversion to a singly labeled product was possible in roughly 30 minutes (Fig. 2). These data support the notion that the absolute copy number from sequencing is not the best indicator of function as previously suggested.21 We observed that sequences that showed up many times (MP01–04 for example) were not as reactive as some that only appeared a handful of times (MP13, 15 and 22 appeared 5 or fewer times).
The multi-cysteine containing peptides displayed varying levels of reactivity along with heterogeneous labeling states. Possessing two cysteines, MP08 and MP09 both showed single and double labeled products while MP05 was labeled between one and three times within eight hours (Fig. 2B and C). These sequences may have required longer reaction times and higher CA concentrations to drive the reaction to a fully labeled state. Additionally, MP05 and 09 were not soluble following reaction.
The reactive peptides exist in a diverse structural landscape
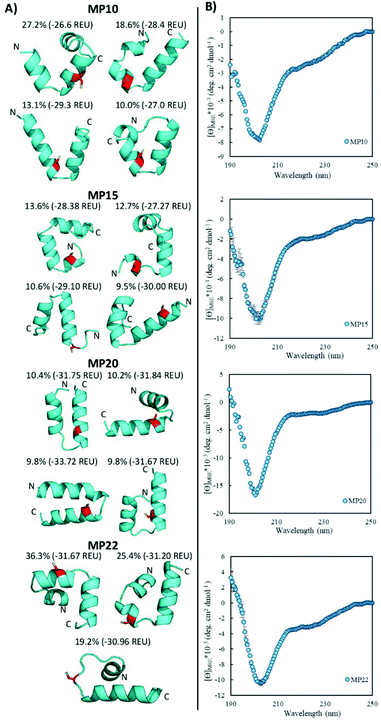
Rosetta ab initio structure prediction22,23 and CD spectroscopy suggested a diverse structural landscape in which both alpha-helical and beta-sheet peptides existed. For the Rosetta analysis, the presented structures were the best representative structures of the major structural families (by percent size of the total population) determined by having both a low energy but also low in-family root-mean-square deviation. This suggested the structure to be a reasonable representation of the family.Structure prediction was used to suggest general topologies capable of performing the SNAr chemistry. While short structured peptides exist,24 we chose this modeling paradigm because peptides in this size range often need to be specifically designed (typically with cyclization or disulfide bonds) for structural stability.25,26 Additionally, cluster analysis suggested that several structural families may be important. This typically occurred for the predicted helical peptides, where for a given sequence, the structural landscape contained families of similar population percentages and comparable energies. For such sequences, the actual structure in solution is likely an ensemble of these states and not a single conformation as one structure would suggest. In light of these observations, and without additional experimental structural data, we analysed the structural landscape itself and do not present a single structure.
Helical peptides exist and display partial random coil elements
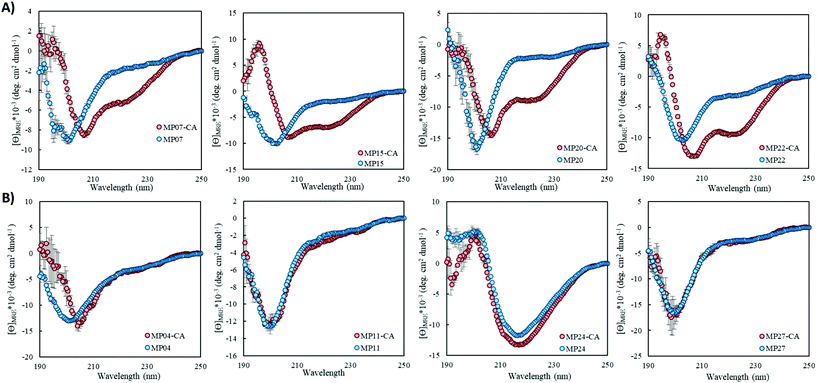
Of the sequences studied, fifteen were predicted to primarily possess helical components (MP03–5, 7–10, 12, 15, 20–22, 24, 26 and 27, Fig. 3A and ESI 3.5†). Four out of the top five most reactive peptides came from this set of sequences, all of which contained a single cysteine. For each of these top sequences, several key structural families were observed that appeared unique for each sequence. For a given peptide, some families displayed similar elements. As an example, in the case of MP10, three of the representative structures suggested a common cysteine placement (Fig. 3A). Across all of the sequences and structures, several structural themes regarding the placement of the active site cysteine emerged; the two most notable being that the cysteine was often observed near the N-terminus of an alpha-helix (MP07–9, 12, 15, 20 and 21) and that it is commonly seen in loop regions (MP03–5, 8, 26 and 27 for example). The placement of the cysteine near the N-terminus of an alpha-helix may contribute to elevated reactivity due to the macromolecular dipole induced by the helix as has been suggested.27 However, these are not the only predicted locations, as placement near the C-terminus of a helix was sometimes observed.CD spectroscopy revealed that the majority of the predicted helical peptides displayed both coil and helix-like secondary structures. This suggested that they either adopted a defined structure with significant random coil elements or did not exist with a long lived, defined helical structure. In line with our structural prediction analysis, we believe the lack of a defined structure to be a more plausible explanation. We note that while many of the representative (predicted) structures display significant structural elements, structure in general may have been over stabilized, perhaps due to the scoring function or the large 9-mer fragments used in the prediction protocol. Finally, we find it pertinent to mention that MP24 displayed a predominantly beta-sheet like secondary structure, in contrast to the predicted helical structure. This may suggest that the structure prediction over-stabilized helical peptides or that MP24 was in an aggregated state (which might be the cause of its low reactivity).
Beta-sheet structures exist in the landscape of reactive peptides
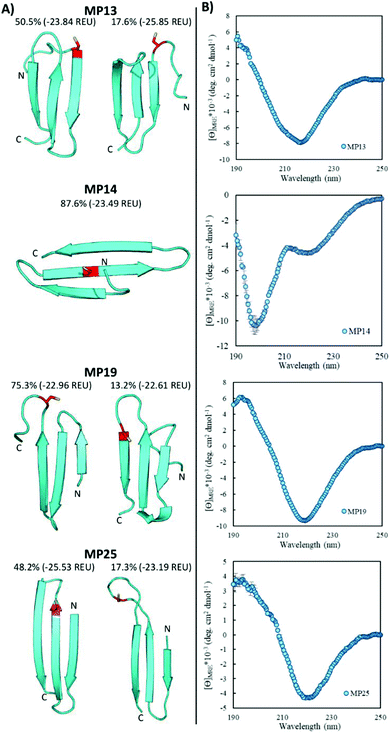
Pointing to the diversity of structures able to undergo SNAr reactions, several beta-sheet-based peptides were discovered. A total of seven sequences were predicted to fall into this category (MP11, 13, 14, 17–19, 25, Fig. 4 and ESI 3.5†). One of these peptides displayed significant reactivity (MP13), arriving at number three in the list of most reactive sequences with a rate constant of 1.71 M−1 s−1. The majority of other beta-sheet predicted peptides displayed lower levels of reactivity, with MP25 being the next fastest at 0.25 M−1 s−1. Perhaps the most common element of this class of peptide was the observation of three-stranded beta-sheets in which the active site cysteine was seen closer to or in one of the loop regions. The prime exception to this was MP14 in which the cysteine was located in the center of the middle beta-strand; however, it was one of the least reactive beta-sheet peptides. The beta-sheet peptides in general also possessed primary structural families that were represented at much higher percentages than families seen in the helical peptides (50.5% and 75.3% for the main families of MP13 and MP19 respectively).In addition to their predicted structural elements, the secondary structures seen for many of these peptides were predominantly beta-sheet-like with the characteristic minimum near 218 nm. Relative to the helical peptides, the secondary structural features seen in many of these beta-sheet sequences were more pronounced (Fig. 4, ESI 3.3 and 3.4†). However, not all of the peptides displayed only beta-sheet-like structure. Two of the peptides (MP11 and MP14) displayed significant random coil components in their secondary structure. As a point of interest, we note that the majority of the beta-sheet peptides required a chemical refolding protocol, while none of the alpha-helical peptides did.
Structural alteration upon reaction is a common feature in SNAr active midsized peptides
Structural alteration upon reaction emerged as a common feature. CD analysis of select MPs following an approximately 24 hour incubation with the CA revealed that many sequences underwent significant alterations to their secondary structure (Fig. 5A, ESI 3.3†). This feature was quite common in the predicted helical peptides, like MP07, 15 and 20, that exhibited varying degrees of structural alteration. In general, helical features became more pronounced following reaction with the CA. We note that many CA-labeled peptides were not soluble enough to perform CD analysis – particularly the beta-sheet peptides like MP13, 19 and 25. This may be the root cause as to why the observed trend is an increase of alpha-helicity.There is a general trend (aside from MP05, 10 and 13 for which we do not have data due to solubility issues) that the most reactive peptides undergo structural alterations (MP07, 15 and 20–22). This observation may point to structural alterations being important for high levels of reactivity. However, it may simply be a common feature of reactive peptides in this size range, as sequences like MP08 and 26 also display alteration but are much less reactive.
We do not believe these alterations are due to the CA acting as simply a structure promoting additive. Several peptides displayed negligible change upon reaction with (MP04 and 27) or in the presence of CA (MP11 and 24, Fig. 5B). Additionally, the concentrations of CA used (250 μM) are lower than those used with agents like trifluoroethanol or trimethylamine N-oxide to impart structural alterations (commonly used in the mM–M range).28
Experimental
All materials and methods are outlined in detail in the ESI.† Key methods include peptide synthesis and handling, kinetics measurements, CD spectroscopy and structure prediction.Conclusions
Three main findings have come out of this work. First, by studying a portion of the selected sequences, we suggest that there are potentially hundreds of reactive peptides in a library of 1013 members. Second, the peptides in this subspace spanned diverse primary sequences, secondary structural elements and perhaps tertiary features that were capable of performing the SNAr reaction with rates enhanced relative to unselected peptides. The final observation is that structural alterations mediated by or in response to reaction with a small molecule is a common feature of these peptides.Our experiments demonstrate that the landscape of SNAr active peptides is broad. Not only is reactivity readily achievable, but high levels of reactivity are possible, evidenced by sequences like MP13, 15 and 22. Even among these more reactive sequences, differences still emerge in terms of structural features, akin to the diversity of structures seen in enzymes. This work demonstrated that peptides from both major secondary structural families (alpha-helices and beta-sheets) are capable of rapid reactivity. Expanding from here and with additional engineering, the multi-cysteine peptides (MP05 and 09 for instance) may enable fusion proteins to be labeled a desired number of times in a kinetically-controlled biorthogonal manner. Looking forward, we believe there may yet be other, highly reactive sequences in this space. Additional experiments will more fully characterize the breadth of sequences, structures and functions.
The frequent observation of CA-mediated structural alterations is a notable property emergent from the selection. It may even begin to suggest a common feature used by (reactive) peptides in this space. In this regard, these peptides are reminiscent of natural proteins and peptides that undergo disorder-to-order transitions upon performing their function like the cyclin-dependent kinase inhibitor p21 or ACTR and CBP that significantly alter their structures upon binding.29,30 However, it is still an open question whether or not this property is important for reactivity.
Taken together these results portray a rich and dynamic landscape of functional biomaterials. Additional investigations are necessary and may reveal common routes to high reactivity as well as general design rules to guide future reactive peptide development.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank A. Mijalis, J. Wolfe and A. Loftis for critical editing. We thank the Massachusetts Green High Performance Computing Center and the Commonwealth Computational Cloud for Data Driven Biology. The Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-0070319) is gratefully acknowledged. E. D. E. was supported by an NSF graduate research fellowship (#112237) and then the Martin Family Society of Fellows for Sustainability. DARPA (Award #023504-001) supported B. L. P.Notes and references
- H. Hegyi and M. Gerstein, J. Mol. Biol., 1999, 288, 147–164 CrossRef CAS.
- A. E. Todd, C. A. Orengo and J. M. Thornton, J. Mol. Biol., 2001, 307, 1113–1143 CrossRef CAS.
- G. Davies and B. Henrissat, Structure, 1995, 3, 853–859 CrossRef CAS.
- V. Lombard, H. Golaconda Ramulu, E. Drula, P. M. Coutinho and B. Henrissat, Nucleic Acids Res., 2014, 42, D490–D495 CrossRef CAS.
- J. L. A. Brás, A. Cartmell, A. L. M. Carvalho, G. Verzé, E. A. Bayer, Y. Vazana, M. A. S. Correia, J. A. M. Prates, S. Ratnaparkhe, A. B. Boraston, M. J. Romão, C. M. G. A. Fontes and H. J. Gilbert, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5237–5242 CrossRef.
- C. T. Supuran, Biochem. J., 2016, 473, 2023–2032 CrossRef CAS.
- J. G. Ferry, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1804, 374–381 CrossRef CAS.
- Y. Xu, L. Feng, P. D. Jeffrey, Y. Shi and F. M. M. Morel, Nature, 2008, 452, 56–61 CrossRef CAS.
- L. Hedstrom, Chem. Rev., 2002, 102, 4501–4524 CrossRef CAS.
- N. D. Rawlings and A. J. Barrett, in Methods in Enzymology, Academic Press, 1995, vol. 248, pp. 183–228 Search PubMed.
- K. Suguna, E. A. Padlan, C. W. Smith, W. D. Carlson and D. R. Davies, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 7009–7013 CrossRef CAS.
- B. A. Griffin, S. R. Adams and R. Y. Tsien, Science, 1998, 281, 269–272 CrossRef CAS.
- S. R. Adams, R. E. Campbell, L. A. Gross, B. R. Martin, G. K. Walkup, Y. Yao, J. Llopis and R. Y. Tsien, J. Am. Chem. Soc., 2002, 124, 6063–6076 CrossRef CAS.
- C. Zhang, M. Welborn, T. Zhu, N. J. Yang, M. S. Santos, T. Van Voorhis and B. L. Pentelute, Nat. Chem., 2016, 8, 120–128 CrossRef CAS PubMed.
- E. D. Evans and B. L. Pentelute, ACS Chem. Biol., 2018, 13, 527–532 CrossRef CAS.
- R. K. V. Lim, N. Li, C. P. Ramil and Q. Lin, ACS Chem. Biol., 2014, 9, 2139–2148 CrossRef CAS.
- T. Kawakami, K. Ogawa, N. Goshima and T. Natsume, Chem. Biol., 2015, 22, 1671–1679 CrossRef CAS.
- C. P. Ramil, P. An, Z. Yu and Q. Lin, J. Am. Chem. Soc., 2016, 138, 5499–5502 CrossRef CAS.
- F. Tanaka, R. Fuller, L. Asawapornmongkol, A. Warsinke, S. Gobuty and C. F. Barbas III, Bioconjugate Chem., 2007, 18, 1318–1324 CrossRef CAS.
- A. J. Mijalis, D. A. Thomas III, M. D. Simon, A. Adamo, R. Beaumont, K. F. Jensen and B. L. Pentelute, Nat. Chem. Biol., 2017, 13, 464–466 CrossRef CAS.
- M. Cho, Y. Xiao, J. Nie, R. Stewart, A. T. Csordas, S. S. Oh, J. A. Thomson and H. T. Soh, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15373–15378 CrossRef CAS.
- P. Bradley, K. M. S. Misura and D. Baker, Science, 2005, 309, 1868–1871 CrossRef CAS.
- K. T. Simons, C. Kooperberg, E. Huang and D. Baker, J. Mol. Biol., 1997, 268, 209–225 CrossRef CAS PubMed.
- A. G. Cochran, N. J. Skelton and M. A. Starovasnik, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5578–5583 CrossRef CAS.
- G. Bhardwaj, V. K. Mulligan, C. D. Bahl, J. M. Gilmore, P. J. Harvey, O. Cheneval, G. W. Buchko, S. V. S. R. K. Pulavarti, Q. Kaas, A. Eletsky, P.-S. Huang, W. A. Johnsen, P. J. Greisen, G. J. Rocklin, Y. Song, T. W. Linsky, A. Watkins, S. A. Rettie, X. Xu, L. P. Carter, R. Bonneau, J. M. Olson, E. Coutsias, C. E. Correnti, T. Szyperski, D. J. Craik and D. Baker, Nature, 2016, 538, 329–335 CrossRef CAS.
- M. D. Struthers, R. P. Cheng and B. Imperiali, Science, 1996, 271, 342–345 CrossRef CAS.
- W. G. J. Hol, P. T. van Duijnen and H. J. C. Berendsen, Nature, 1978, 273, 443–446 CrossRef CAS.
- S. A. Celinski and J. M. Scholtz, Protein Sci., 2002, 11, 2048–2051 CrossRef CAS PubMed.
- R. W. Kriwacki, L. Hengst, L. Tennant, S. I. Reed and P. E. Wright, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 11504–11509 CrossRef CAS.
- S. J. Demarest, M. Martinez-Yamout, J. Chung, H. Chen, W. Xu, H. J. Dyson, R. M. Evans and P. E. Wright, Nature, 2002, 415, 549–553 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/C8OB01678J |
| ‡ The starting Met and trailing Gly were part of the fixed region and are thus not included in the length analysis. Thus a ‘full length’ sequence would possess 32 residues in Table 1. |
| This journal is © The Royal Society of Chemistry 2019 |