Open Access Article
Open Access ArticleTracking down protein–protein interactions via a FRET-system using site-specific thiol-labeling†
B.
Söveges
a,
T.
Imre
b,
Á. L.
Póti
c,
P.
Sok
c,
Zs.
Kele
a,
A.
Alexa
c,
P.
Kele
 a and
K.
Németh
a and
K.
Németh
 *a
*a
aResearch Centre for Natural Sciences of the Hungarian Academy of Sciences, Institute of Organic Chemistry, Chemical Biology Research Group, Magyar tudósok krt. 2, H-1117 Budapest, Hungary. E-mail: nemeth.krisztina@ttk.mta.hu; Tel: +36 1 382 6659
bResearch Centre for Natural Sciences of Hungarian Academy of Sciences, Instrumentation Center, MS Metabolomics Research Group, Magyar tudósok krt. 2., H-1117 Budapest, Hungary
cResearch Centre for Natural Sciences of the Hungarian Academy of Sciences, Institute of Enzymology, Protein Research Group, Magyar tudósok krt. 2, H-1117 Budapest, Hungary
First published on 19th June 2018
Abstract
Förster resonance energy transfer is among the most popular tools to follow protein–protein interactions. Although limited to certain cases, site-specific fluorescent labeling of proteins via natural functions by means of chemical manipulations can redeem laborious protein engineering techniques. Herein we report on the synthesis of a heterobifunctional tag and its use in site-specific protein labeling studies aiming at exploring protein–protein interactions. The oxadiazole-methylsulfonyl functionality serves as a thiol specific warhead that enables easy and selective installation of fluorescent labels through a bioorthogonal motif. Mitogen activated protein kinase (MAPK14) and its substrate mitogen activated protein kinase activated kinase (MAPKAP2) or its docking motif, a 22 amino acid-long peptide fragment, were labeled with a donor and an acceptor, respectively. Evolution of strong FRET signals upon protein–protein interactions supported the specific communication between the partners. Using an efficient FRET pair allowed the estimation of dissociation constants for protein–protein and peptide–protein interactions (145 nM and 240 nM, respectively).
Introduction
The human genome consists of nearly 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 genes, coding over half a million proteins. According to current estimations ca. 80% of proteins operate in complexes.1 Understanding the interactions in such protein complexes is important in the exploration of biochemical processes e.g. antibody–antigen interactions, signal transduction and regulation or hormone-receptor binding.2 Nowadays many robust methods are available for the detection of protein–protein interactions (PPI) (e.g., affinity blotting, immunoprecipitation, quantitative proteomics, chemical crosslinking).3,4 To investigate protein communication in live cells, however, fluorescence methods e.g. bimolecular fluorescence complementation (BiFC)5,6 or Förster resonance energy transfer (FRET) are the methods of choice.
000 genes, coding over half a million proteins. According to current estimations ca. 80% of proteins operate in complexes.1 Understanding the interactions in such protein complexes is important in the exploration of biochemical processes e.g. antibody–antigen interactions, signal transduction and regulation or hormone-receptor binding.2 Nowadays many robust methods are available for the detection of protein–protein interactions (PPI) (e.g., affinity blotting, immunoprecipitation, quantitative proteomics, chemical crosslinking).3,4 To investigate protein communication in live cells, however, fluorescence methods e.g. bimolecular fluorescence complementation (BiFC)5,6 or Förster resonance energy transfer (FRET) are the methods of choice.
Due to the time requirement for the reporter protein to mature BiFC cannot provide real-time information on PPIs, not to mention that the reconstituted fluorescent protein (FP) remains stable; thus the interaction is irreversible making BiFC less suitable to determine the dynamic features or spatio-temporal changes.7
The phenomenon of FRET is a highly distance-dependent process where an excited-state donor molecule transfers its energy to the acceptor molecule via non-radiative, coulombic interactions. Because the efficiency of FRET is inversely proportional to the sixth power of the distance between the donor and the acceptor, it proves to be a very sensitive and dynamic tool for the investigation and characterization of the dynamics of protein–protein interactions.8,9
Selective fluorescence modulation of the proteins of interest can be considered one of the major challenges in the FRET technology. Fused FPs overcome this limitation in terms of selectivity; however, the comparable size of the fusion tags often hamper the dynamics of the interaction studied. Selective fluorescent tagging with small organic reporters can be achieved by the use of smaller fusion tags (SNAP, CyS4etc.) or by encoding fluorescent or bioorthogonalized non-canonical amino acids.10 While these techniques are powerful they are often laborious and require special skills and advanced infrastructure. In certain cases, purely chemical modification can be effected e.g. with reagents harboring a warhead that selectively targets rare functions.11 Among the 20 protein-building amino acids, cysteine is one of the most suitable targets for such selective reactions because of the low abundance and good nucleophilicity of the free sulfhydryl group.12 Hence, various methods were developed for cysteine bioconjugation,13 such as reactions with electrophiles (α-halo carbonyl derivatives),14 use of Michael acceptors (maleimide derivatives,15 vinyl sulfone16), thiol–yne or –ene reactions,17 disulfide exchange,18 nucleophilic aromatic substitution of perfluorobenzene derivatives,19 or use of allenamide derivatives.20 The majority of these approaches suffer from some limitations, such as some of the reagents being able to also modify other nucleophilic amino acid side chains. In the case of Michael-acceptors the resulting products are often unstable, due to retro-Michael reactions or in the case of maleimides due to the fact that the maleimide linkage is prone to hydrolysis.13 Moreover, the thioether group suffers from reversible exchange reactions with reactive thiols. Disulfide conjugates also tend to participate in exchange reactions,21 while traditional thiol–ene reactions require nucleophilic or UV-light activation.22 The vinyl sulfone derivatives successfully addressed these problems; however, their reaction kinetics is still too slow for certain in vivo biological experiments.23 Recently, Toda and coworkers investigated the Julia–Kocienski-like methylsulfonyl-functionalized heteroaromatic derivatives and presented a new thiol-selective reagent, which was stable in biological environments and offered superior reaction kinetics compared to other thiol-specific reactions.24
In this study, we aimed at investigating protein–protein interactions using FRET as an indicator of the communication of model protein p38α (MAPK14) and its direct substrate MK2 (MAPKAP2). Both proteins are the key components of the mitogen-activated protein kinase signaling pathway.25 p38α is the main transponder of the inflammatory response, and MK2 plays an important role in the production of inflammatory cytokines.2
Based on our previous studies26 with p38 and calculations of solvent accessible surface areas of the proteins, both of them contain accessible cysteines on their surface regions. Along the above considerations we designed a bifunctional chemical reporter tag carrying a thiol-reactive oxadiazole-methylsulfone and a bioorthogonally reactive cyclooctyne motif that creates a platform for fluorescent modification. The fluorescently labeled protein constructs were then used in the exploration of protein–protein interactions using FRET.
Results and discussion
Synthetic procedures
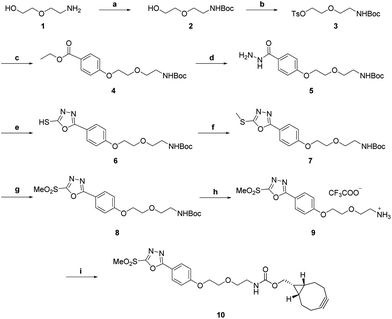
First, we wanted to screen for FRET pairs from our azide and tetrazine derivatized dye-pool.27–30 Since photophysical characteristics may substantially change upon conjugation to proteins, we needed a heterobifunctional tag that would enable Cys-specific targeting on the one hand and allow sequential one-pot high yielding modification both with azide- and tetrazine-modified fluorescent probes. To this end we designed cyclooctyne bearing methylsulfonyl-oxadiazole 10 (Scheme 1). The synthesis started with Boc-protection of aminoethoxyethanol, 1, followed by the activation of the hydroxyl group with tosyl-chloride to provide 3. The product was reacted with ethyl-hydroxybenzoate under basic conditions to obtain 4 in excellent yields. Hydrazide 5 was accessed upon treatment of 4 with hydrazine monohydrate.Oxadiazole 6 was then formed in good yields by a reaction of hydrazide 5 with carbon disulfide. Methylation of the sulfhydryl moiety was effected by treatment of 6 with methyl iodide to furnish 7. Oxidation of 7 with m-chloroperoxybenzoic acid gave 8. Acidic removal of the Boc-protecting group quantitatively afforded amine 9. Compound 9 was functionalized with commercially available BCN–NHS to result in the Cys-specific heterobifunctional linker 10.
Protein labeling and FRET pair screening
For protein–protein interactions we chose a mutant variant of p38α where the cysteine at position 162 was replaced by serine (p38C162S). This site should remain minimally perturbed, as it is located near the docking site of the protein. For an interacting partner we selected its downstream substrate, the wild type MAPKAP2 (MK2). Besides mutant p38C162S and MK2 we extended our study to a 22 amino acid oligopeptide representing the docking motif of MK2 (CQIKIKKIEDASNPLLLKRRKK, pepMK2).25In order to screen for potential FRET pairs, we aimed at comparing the excitation and emission spectra of our dyes in their conjugate forms. To this end, we created a bioorthogonally targetable protein scaffold using heterobifunctional tag 10, and p38C162S (Scheme 2).
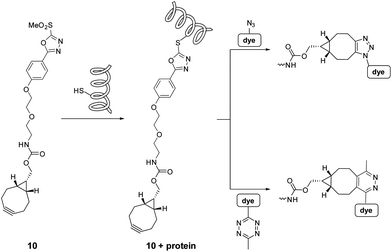 | ||
| Scheme 2 Sequential chemical labeling strategy using the heterobifunctional linker and the bioorthogonalized fluorescent dyes. | ||
Before labeling, we confirmed that 10 reacts solely with sulfhydryls. To do so we allowed 10 to react with glutathione (GSH). MS measurements (Fig. S1†) affirmed that only the SH-group participated in the reaction and no N-terminal modification was observed. These results are in agreement with the conclusions of Toda et al.24
Intact whole protein MS measurements verified that complete conversion of p38C162S could be achieved within 10 minutes by adding 2 equivalents of 10 to the protein, resulting in a mono-conjugated form of p38C162S with a negligible amount of any multiply labeled species (Fig. S2 and 3†).
In order to identify the labeling site, tandem mass-spectrometry measurements were performed following trypsin digestion of p38C162S treated with 9 (Fig. S11†). A comparison of untreated and derivatized p38C162S samples revealed that the linker reacted mainly at Cys119, which is in accordance with our former results on calculated accessible surface area values (Table S1†).26
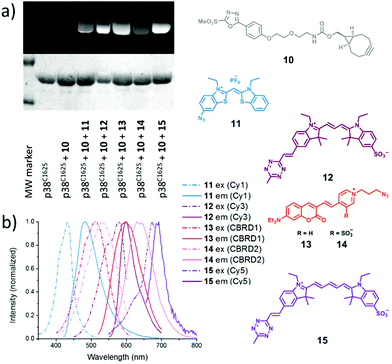
Next, we wanted to screen for the most promising FRET pairs in our bioorthogonal dye-pool. For this, we applied the bifunctional, cyclooctyne bearing chemical reporter 10 on mutant p38 in sequential labeling experiments with a series of azide or tetrazine bearing fluorescent or fluorogenic dyes (11–15). The labeling reactions were conducted in one-pot with sequential administration of 10 and the respective dyes to the solution of p38C162S, without any intermediate purification steps. Such bioorthogonal conjugation of the dyes was very efficient and in each case the conversion rates reached ca. 100% within 10 minutes, according to intact protein MS analyses (Fig. S2–8†). Due to the fluorogenic nature of the dyes, no non-specific signal was observed even if non-specific adhesion of the dyes onto the surface of the proteins occurred (Table S3†). The labeled proteins were separated using SDS-PAGE and imaged under different excitation sources. In each case the dyes labeled the protein successfully (Fig. 1).
Next, we analyzed the excitation and emission spectra of the dye–protein conjugates to identify donor–acceptor pairs with overlapping emission and excitation bands. After evaluating the spectra, we identified a potential FRET pair of 11–12 (Fig. S13a†).
The efficiency and selectivity of the conjugation of 10 to MK2 were also assessed. In this case, addition of 2 equivalents of 10 to MK2 resulted in mono- and double-conjugated species as seen from intact whole protein MS measurements (Fig. S10†). Comparative MS-analysis of labeled and unlabeled, trypsin digested MK2 samples revealed that the two most accessible cysteines i.e., Cys224 and Cys98 were involved (Table S2†).
FRET measurements
To explore the FRET efficiency between 11 (Cy1) and 12 (Cy3), the interacting partners were modified with the dyes applying the same sequential scheme using 10 as a heterobifunctional linker (Scheme 2). This way, mutant p38 was labeled with 11, while MK2 and pepMK2 were tagged with 12 (Fig. S12†).Following the purification of the fluorescently labeled proteins and peptide, we studied the interaction between Cy1-conjugated p38C162S and Cy3-conjugated pepMK2 or MK2. The FRET interaction between labeled p38C162S (0.5 μM) and increasing amounts of pepMK2 (0 to 8 μM) was monitored upon excitation of the donor fluorophore (λexc = 420 nm). The evolution of the FRET signal resulted in a gradual decrease of the donor fluorescence with a simultaneous increase of the acceptor's fluorescence (Fig. 2). FRET efficiency was calculated using the below equation:
| EFRET = 100(1 − D/D0), |
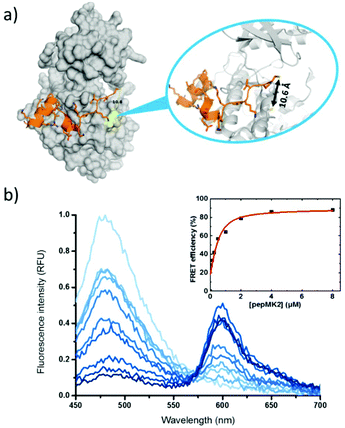 | ||
| Fig. 2 (a) The structure of the complex of protein p38C162S (based on PDB code 2OZA) and peptide MK2. The accessible cysteines are highlighted in yellow. (b) Changes in fluorescence upon interaction between the Cy1-labeled p38C162S and Cy3-labeled peptide MK2 (inset shows the calculated FRET-efficiency). | ||
In the case of the protein–protein interactions similar conditions were applied (Fig. 3). Signal changes were monitored at the same wavelengths upon interaction of fluorescently labeled Cy1-p38C162S with increasing amounts of Cy3-labeled MK2 (0 to 4 μM). For the calculation of the FRET efficiency the same equation was used; however, effects of protein interaction on donor fluorescence were taken into account as the presence of MK2 creates a less-polar environment for the donor resulting in an increase of the fluorescence intensity. Thus, the fluorescence intensity of the donor fluorophore in the presence of acceptor labeled MK2 was always normalized with the fluorescence intensity of the donor, in the presence of unlabeled MK2 at the respective concentrations (Fig. S15†). The FRET efficiency went to saturation at 74% (Fig. 3b). Both cysteines were located within the critical distance i.e., 100 Å necessary for efficient FRET (dCys224 = 29.8 Å and dCys98 = 23.7 Å according to the structure of the protein complex PDB code 2OZA).31 The decreased FRET efficiency compared to the p38C162S–pepMK2 interaction could be explained by the longer distance between the labeling sites. The binding affinity between p38 and MK2 protein was found to be Kd = 145 nM (see the ESI†). Estimated Kd values in the literature vary between 1 and 100 nM depending on the amino acid sequence, the size of the given fragment, the activity (phosphorylation) state of the interacting molecules and the ionic strength of the media.32
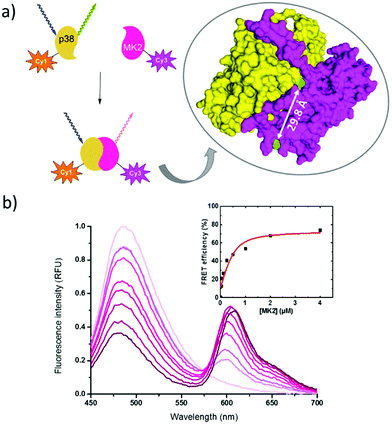 | ||
| Fig. 3 (a) The structure of the complex of protein p38C162S and protein MK2 (based on PDB code 2OZA). The accessible cysteines are highlighted in green. (b) The FRET signal between Cy1-labeled p38C162S and Cy3-labeled protein MK2 (inset shows the calculated FRET-efficiency). | ||
The high salt concentration – applied in order to prevent the aggregation of MK2 – was concomitant of lower affinity in our setup.
Conclusions
In conclusion, we demonstrated the synthesis and use of a heterobifunctional linker that enables highly specific and efficient modification of proteins via free-sulfhydryl functionalities of cysteine residues through its methylsulfonyl-oxadiazole warhead. The cyclooctyne functionalized bifunctional methylsulfonyl linker readily creates a bioorthogonal platform that offers easy and high-yielding modification with azide or tetrazine modified fluorescent probes. First, we used this heterobifunctional linker to screen for possible FRET pairs from our existing bioorthogonalized fluorescent pool. Following the identification of a promising FRET pair, we used this Cys-specific linker to modify p38C162S and its interacting partner MK2 or its docking motif pepMK2 with a FRET donor and acceptor, respectively. Tandem-MS measurements verified the selective and specific labeling of both protein components. FRET studies involving donor and acceptor labeled p38C162S–MK2 or p38C162S–pepMK2 partners confirmed the specific interactions of the proteins. Such heterobifunctional linkers are suitable for the selective modification of proteins possessing accessible free sulfhydryl functionalities. On the other hand, the high FRET efficiency between the identified dye pairs (ca. 80%) is suitable for the detection of protein–protein interactions and estimation of the strength of affinity.Experimental
Organic syntheses
Peptide synthesis and protein expression and purification
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 Å, LC Column 250 × 10 mm; Phenomenex, Torrance, CA, USA).
300 Å, LC Column 250 × 10 mm; Phenomenex, Torrance, CA, USA).
Conjugation processes
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The size of the polyacrylamide gels was 8.5 cm × 7.5 cm × 0.1 cm. They were the combination of 4% concentration and 10% separation PAGE gels (acrylamide/bisacrylamide ratio was 29/1; and concentration gel buffer was 125 mM Tris-HCl + 0.1% SDS (pH 6.8) and the separation gel buffer was 375 mM Tris-HCl + 0.1% SDS (pH 8.8)). 7.5 μg of protein was loaded per lane. A Prestained PAGERuler Plus (Thermo Fisher Scientific) was applied as the molecular weight standard. Separations were carried out in a Mini-Protean Tetra Cell (Bio-Rad, Hercules, CA, USA) using 25 mM Tris/192 mM glycine + 0.1% SDS (pH 8.3) as the running buffer and 180 V voltage for 40 min at 25 °C. The gels were documented by using a Typhoon Trio Imaging System (GE Healthcare, Uppsala, Sweden). Furthermore, the gels were stained for proteins with Coomassie Brilliant-Blue.
1 ratio. The size of the polyacrylamide gels was 8.5 cm × 7.5 cm × 0.1 cm. They were the combination of 4% concentration and 10% separation PAGE gels (acrylamide/bisacrylamide ratio was 29/1; and concentration gel buffer was 125 mM Tris-HCl + 0.1% SDS (pH 6.8) and the separation gel buffer was 375 mM Tris-HCl + 0.1% SDS (pH 8.8)). 7.5 μg of protein was loaded per lane. A Prestained PAGERuler Plus (Thermo Fisher Scientific) was applied as the molecular weight standard. Separations were carried out in a Mini-Protean Tetra Cell (Bio-Rad, Hercules, CA, USA) using 25 mM Tris/192 mM glycine + 0.1% SDS (pH 8.3) as the running buffer and 180 V voltage for 40 min at 25 °C. The gels were documented by using a Typhoon Trio Imaging System (GE Healthcare, Uppsala, Sweden). Furthermore, the gels were stained for proteins with Coomassie Brilliant-Blue.
FRET measurements
The labeled p38C162S samples were prepared in 1.0 μM concentration in PBS (pH 7.4) buffer. The labeled MK2 peptide and protein samples were prepared in 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625, and 0.03125 μM concentrations for the titration. From the appropriate solutions 10 + 10 μL were transferred into the wells. The FRET interactions between the two labeled partners were monitored based on the generation of the fluorescence signal (λex: 420 nm; λem: 450–700 nm) by using a PerkinElmer Enspire Multimode Reader (PerkinElmer Inc.; Waltham, MA, USA) in a 384-well round bottom black plate (Corning, Corning, NY, USA) at room temperature with 100 flashes per point. Data were acquired and handled by Corning Enspire Manager 4.10 software.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The help of Gábor Glatz with SDS-PAGE gel imaging is greatly acknowledged. The present work was supported by the Hungarian Scientific Research Fund (OTKA-NN-116265), the “Lendület” Programme of the Hungarian Academy of Sciences (LP2013-55/2013) and the National Research, Development and Innovation Office, Hungary (VEKOP-2.3.3.-15-20016-00011).References
- T. Berggård, S. Linse and P. James, Proteomics, 2007, 7, 2833–2842 CrossRef PubMed.
- Y. Yang, H. Liu and X. Yao, Mol. BioSyst., 2012, 8, 2106–2118 RSC.
- Y. Song, V. G. J. Rodgers, J. S. Schultz and J. Liao, Biotechnol. Bioeng., 2012, 109, 2875–2883 CrossRef PubMed.
- E. M. Phizicky and S. Fields, Microbiol. Rev., 1995, 59, 94–123 Search PubMed.
- T. K. Kerppola, Annu. Rev. Biophys., 2008, 37, 465–487 CrossRef PubMed.
- K. E. Miller, Y. Kim, W. Huh and H. Park, J. Mol. Biol., 2015, 427, 2039–2055 CrossRef PubMed.
- S. Xing, N. Wallmeroth, K. W. Berendzen and C. Grefen, Plant Physiol., 2016, 171, 727–758 Search PubMed.
- D. W. Piston and G. J. Kremers, Trends Biochem. Sci., 2007, 32, 407–414 CrossRef PubMed.
- H. Sahoo, J. Photochem. Photobiol., C, 2011, 12, 20–30 CrossRef.
- G. Zhang, S. Zheng, H. Liuc and P. R. Chen, Chem. Soc. Rev., 2015, 44, 3405–3417 RSC.
- N. Krall, F. P. da Cruz, O. Boutureira and G. J. Bernardes, Nat. Chem., 2016, 8, 103–113 CrossRef PubMed.
- X. Chen and Y.-W. Wu, Org. Biomol. Chem., 2016, 14, 5417–5439 RSC.
- S. B. Gunnoo and A. Madder, ChemBioChem, 2016, 17, 529–553 CrossRef PubMed.
- J. M. Chalker, G. J. L. Bernardes, Y. A. Lin and B. G. Davis, Chem. – Asian J., 2009, 4, 630–640 CrossRef PubMed.
- A. D. Baldwin and K. L. Kiick, Bioconjugate Chem., 2011, 22, 1946–1953 CrossRef PubMed.
- G. B. Cserép, Zs. Baranyai, D. Komáromy, K. Horváti, Sz. Bősze and P. Kele, Tetrahedron, 2014, 70, 5961–5965 CrossRef.
- C. E. Hoyle and C. N. Bowman, Angew. Chem., Int. Ed., 2010, 49, 1540–1573 CrossRef PubMed.
- S. van Kasteren, Biochem. Soc. Trans., 2012, 40, 929–944 CrossRef PubMed.
- A. M. Spokoyny, Y. Zou, J. J. Ling, H. Yu, Y.-S. Lin and B. L. Pentelute, J. Am. Chem. Soc., 2013, 135, 5946–5949 CrossRef PubMed.
- A. Abbas, B. Xing and T. P. Loh, Angew. Chem., Int. Ed., 2014, 53, 74914–77494 CrossRef PubMed.
- J. M. Chalker, G. J. L. Bernardes and B. G. Davis, Acc. Chem. Res., 2011, 44, 730–741 CrossRef PubMed.
- S. P. S. Koo, M. M. Stamenovic, R. A. Prasath, A. J. Inglis, F. E. Du Prez, C. Barner-Kowollik, W. van Camp and T. Junkers, J. Polym. Sci., Part A: Polym. Chem., 2010, 48, 1699–1713 CrossRef.
- J. Morales-Sanfrutos, J. Lopez-Jaramillo, M. Ortega-Muñoz, A. Megia-Fernandez, F. Perez-Balderas, F. Hernandez-Mateo and F. Santoyo-Gonzalez, Org. Biomol. Chem., 2010, 8, 667–675 RSC.
- N. Toda, S. Asano and C. F. Barbas III, Angew. Chem., Int. Ed., 2013, 52, 12592–12596 CrossRef PubMed.
- Á. Garai, A. Zeke, G. Gógl, I. Törő, F. Fördős, H. Blankenburg, T. Bárkai, J. Varga, A. Alexa, D. Emig, M. Albrecht and A. Reményi, Sci. Signaling, 2012, 5, ra74 CrossRef PubMed.
- B. Söveges, T. Imre, T. Szende, Á. L. Póti, G. B. Cserép, T. Hegedűs, P. Kele and K. Németh, Org. Biomol. Chem., 2016, 14, 6071–6078 RSC.
- R. Petrovics, B. Söveges, A. Egyed, G. Knorr, A. Kormos, T. Imre, Gy. Török, A. Zeke, É. Kocsmár, G. Lotz, P. Kele and K. Németh, Org. Biomol. Chem., 2018, 16, 2997–3005 RSC.
- G. Knorr, E. Kozma, J. M. Schaart, K. Németh, G. Török and P. Kele, Bioconjugate Chem., 2018, 29, 1312–1318 CrossRef PubMed.
- P. Kele, X. Li, M. Link, K. Nagy, A. Herner, K. Lőrincz, Sz. Béni and O. S. Wolfbeis, Org. Biomol. Chem., 2009, 7, 3486–3490 RSC.
- K. Nagy, E. Orbán, Sz. Bősze and P. Kele, Chem. – Asian J., 2010, 5, 773–777 CrossRef PubMed.
- A. White, C. A. Pargellis, J. M. Studts, B. G. Werneburg and B. T. Farmer II, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6353–6358 CrossRef PubMed.
- S. M. Lukas, R. R. Kroe, J. Wildeson, G. W. Peet, L. Frego, W. Davidson, R. H. Ingraham, C. A. Pargellis, M. E. Labadia and B. G. Werneburg, Biochemistry, 2004, 43, 9950–9960 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Accessible surface area calculations, Kd calculation, mass spectrometric analysis of labeled proteins, spectral overlap of the fluorescent proteins, NMR spectra of the synthetic products. See DOI: 10.1039/c8ob00742j |
| This journal is © The Royal Society of Chemistry 2018 |