Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein denaturation caused by heat inactivation detrimentally affects biomolecular corona formation and cellular uptake†
Johanna
Simon
 ab,
Julius
Müller
ab,
Artur
Ghazaryan
b,
Svenja
Morsbach
ab,
Julius
Müller
ab,
Artur
Ghazaryan
b,
Svenja
Morsbach
 b,
Volker
Mailänder‡
b,
Volker
Mailänder‡
 *ab and
Katharina
Landfester‡
*ab and
Katharina
Landfester‡
 b
b
aDermatology Clinic, University Medical Center of the Johannes Gutenberg-University Mainz, Langenbeckstr. 1, 55131 Mainz, Germany. E-mail: volker.mailaender@unimedizin-mainz.de
bMax Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany
First published on 1st November 2018
Abstract
Adsorption of blood proteins to the surface of nanocarriers is known to be the critical factor influencing cellular interactions and eventually determining the successful application of nanocarriers as drug carriers in vivo. There is an increasing number of reports summarizing large data sets of all identified corona proteins. However, to date our knowledge about the multiple mechanisms mediating interactions between proteins and nanocarriers is still limited. In this study, we investigate the influence of protein structure on the adsorption process and focus on the effect of heat inactivation of serum and plasma, which is a common cell culture procedure used to inactivate the complement system. As in general routine lab procedure, heat inactivation was performed at 56 °C for 30 min in order to denature heat labile proteins. When nanocarriers were exposed to native versus heat inactivated serum, we saw that the cellular uptake by macrophages was significantly affected. These results were then correlated with an altered corona composition that depended on the treatment of the protein source. In summary, we were able to prove that the protein structure is one of the key parameters determining protein corona formation.
Introduction
Considerable progress has been made in the field of nanotechnology towards the development of novel nanocarrier formulation.1,2 Innovative nanomaterials with varied surface modifications,3 high encapsulation efficiencies,4 and controlled release properties5 are promising candidates for use in targeted drug delivery.6 However, to date there are very few studies that actually prove their successful performance in vivo.7 This implies that there is still a significant gap between the synthetic design of nanocarriers and their in vivo behavior.8–10Over the last decade, scientists have recognized that the properties of nanocarriers are significantly altered after coming in contact with biological fluids (e.g. blood plasma).11–13 It has already been shown that various biomolecules rapidly interact with the nanocarrier and immediately cover its surface (this is termed the ‘biomolecular corona’).14 Due to this process, the physico-chemical properties of the nanocarriers such as size, charge, or aggregation behavior are altered15 and this further determines cellular uptake,16 toxicity,17 and body distribution.18 Several studies have shown that due to corona formation the intended targeting properties can even be completely lost, emphasizing the crucial influence of the corona formation.19,20 Understanding and controlling this process has become increasingly important.21,22
In cell culture fetal bovine or human serum are commonly used for cell maintenance and in vitro experiments.23 Mirshafiee et al.24 and Schöttler et al.25 have already reported that protein sources such as fetal calf serum, human serum, human heparin plasma or human citrate plasma can significantly influence cellular uptake of nanocarriers and their protein adsorption patterns. Besides using the different possible cell culture supplements, it is a common procedure to inactivate the respective protein source prior to use by applying heat.26,27 During this procedure, serum is typically heated up to 56 °C for 30 min. This procedure, is aimed at inactivating heat labile proteins (e.g. complement proteins), which can interfere in immunological assays.28 Complement proteins have been identified as components of the biomolecule corona effective in a great variety of different nanocarrier formulations and they are also known to mediate interactions with immune cells.29,30 First studies regarding the influence of temperature on the formation of the protein corona were conducted by Lesniak et al.31 and Mahmoudi et al.32 In general, they found that the temperature is a crucial parameter, which affects both the protein corona composition and coverage density.
However, up to now there is still a limited knowledge about the principal mechanism, which are involved in nanoparticle–protein interaction. Next to this, the majority of literature reports do not specify whether native or heat inactivated serum was used in the experimental setup.33 In order to bridge this gap, our study specifically focuses on the effect of heat inactivation on cellular uptake and corona formation of different nanocarriers. In this study, we have been able to shed light on the role of protein structure, which critically influences the protein adsorption process. This basic knowledge is needed to unravel the process of corona formation, to streamline in vitro studies of nanocarriers, and to transfer the knowledge gained from the in vitro studies to further in vivo experiments.
Results and discussion
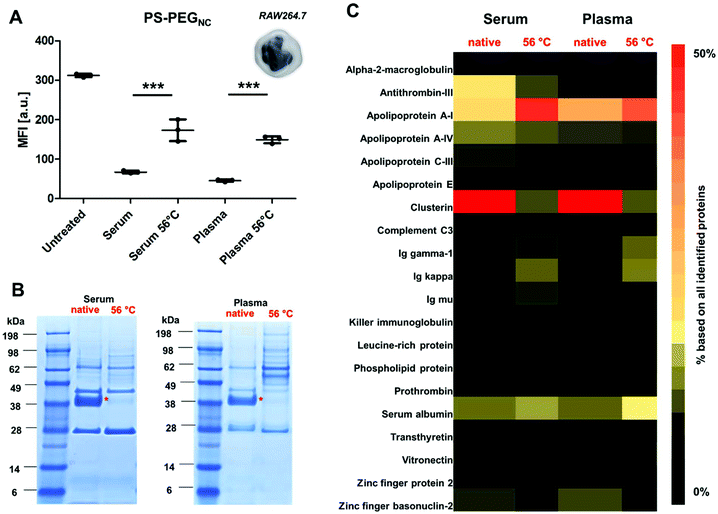
Phagocytic cells such as macrophages (Fig. 1) play an important role in the immune system as they engulf foreign material (e.g. nanocarriers).34 In this study, we chose non-covalently PEGylated polystyrene nanocarriers (named as PS-PEGNC, Table S1†) as a model system to investigate the cellular interactions with a macrophage cell line (RAW264.7), as well as to study the protein adsorption behavior with regard to the heat treatment of the protein source.Nanocarriers were incubated with native or heat inactivated serum and plasma (56 °C, 30 min) prior to cellular uptake studies. As a reference, cellular interaction of nanocarriers with macrophages in a protein-free medium were investigated (‘untreated’). The results corresponded with previous findings: in the presence of native serum or plasma the cellular uptake decreased strongly compared to untreated nanocarriers (Fig. 1A). This points to stealth properties induced by corona proteins.35 However, when nanocarriers were pre-coated with heat inactivated serum or plasma; we observed a significant increase in cellular uptake (***p < 0.001).
Coating the surface of nanocarriers with PEG is up to now the gold standard to circumvent their rapid clearance from the blood stream. In general, the adsorption of blood proteins towards PEGylated surfaces is strongly reduced.36 This effect was thought to be the decisive factor to avoid a cellular recognition of nanocarriers by phagocytic cells. However, recently several research groups reported that protein adsorption is not completely prevented for PEGylated nanocarriers.37–39 In fact, in our previous studies, we were able to show that distinct proteins preferably adsorb to stealth nanocarriers.35 These so called ‘stealth’ proteins (e.g. apolipoprotein J known as clusterin)40 need to be present in order to reduce non-specific cellular uptake.41
Based on this, we investigated the protein corona pattern of the PS-PEGNC after incubation with native or heat inactivated human serum or plasma (Fig. 1B and C). As visualized by SDS PAGE, there was a pronounced change in the corona pattern when nanocarriers were incubated with heat inactivated serum compared to native serum (Fig. 1B). We carried out a detailed proteomic investigation and highlighted the distinct variations in the hard corona pattern. Clusterin was the major protein identified after incubation with native serum or plasma (>50%). Second to this, apolipoprotein AI (Apo AI) contributed to 15% (Fig. 1C and Fig. S3†). A list of all proteins is provided as a supplement in a separate Excel File (Table S5†). These results are in line with the relevant literature reporting the specific interaction of PEGylated nanocarriers and clusterin42 and a strong enrichment of lipoproteins in the corona.43
In strong contrast to this, we detected only minor amounts of clusterin in the corona (∼3%) after incubation with heat inactivated serum or plasma. Interestingly, the amounts of ApoAI remained high (Fig. 1C). In addition, the corona after incubation with heat inactivated serum and plasma was enriched with immunoglobulins (∼15%) and acute phase proteins (∼12%). Minor amounts of these proteins were identified after incubation with the native protein source (Fig. S3†).
However, there was no significant difference in the absolute amount of proteins that adsorbed to the nanocarriers after incubation with native or heat inactivated serum (Table S2†). This indicates that it is actually the protein structure (altered due to heat inactivation) that controls the distinct protein corona pattern and leads to the mediation of cellular interaction. Therefore, the increased cellular uptake after incubation with heat inactivated serum or plasma is a direct result of the altered protein corona composition (Fig. 1A).
In addition to the corona composition, the structure of proteins surrounding the nanocarrier is also known to influence cellular uptake.44 The interaction of proteins with nanocarriers can cause conformational changes in the protein structure, leading to unfolding and subsequently causing surface exposure of unknown epitopes.45 Additionally, temperature changes induce unfolding and denaturation of certain proteins.46
By using label-free differential scanning fluorimetry (nanoDSF) we were able to study the unfolding process of proteins upon heating.47 The fluorescence intensity of tryptophan or tyrosine residues in a protein strongly depends on the surroundings of these amino acids and therefore changes through temperature-induced denaturation.48 In these experiments, the characteristic melting point of a protein TM is defined at a specific temperature, which is needed to unfold 50% of the protein. To explore the difference in protein corona composition when exposed to heat, the unfolding and refolding behavior of ApoAI and clusterin was investigated with nanoDSF (Fig. 2). Here, we found that clusterin had a significantly lower TM ∼ 46 °C (Fig. 2B) compared to ApoAI TM ∼ 58 °C (Fig. 2C). In addition, ApoAI was able to refold upon cooling in contrast to clusterin.
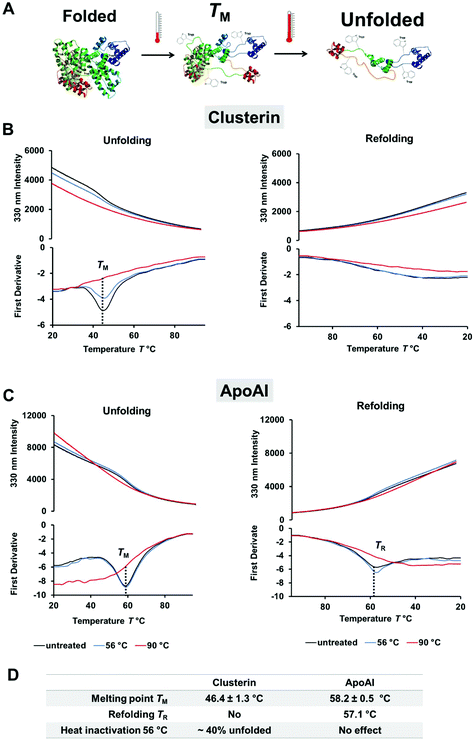 | ||
| Fig. 2 Clusterin is a heat sensitive protein and readily unfolds. Differential scanning fluorimetry (nanoDSF) was used to monitor protein folding upon heating. (A) Schematic illustration of the unfolding process and exposure of tryptophan residues (here shown for human serum albumin PDB structure 1AO6). The melting temperature TM is defined as the point where 50% of the protein is unfolded. (B) Unfolding and refolding curves of clusterin in PBS (0.5 mg mL−1). The fluorescence intensity at 330 nm and the corresponding first derivative are shown. The melting temperature TM is determined from the minimum of the first derivative. One representative measurement is shown. The experiment was repeated three independent times yielding similar results. (C) Monitoring the unfolding and refolding of ApoAI in PBS (0.5 mg mL−1). One representative measurement is shown. The experiment was repeated three independent times yielding similar results. (D) Protein structure properties of clusterin and ApoAI as determined by nanoDSF. The melting point TM is given mean ± SD from three independent measurements (n = 3). The amount of unfolded protein was calculated based on the distance between the native and the denatured protein sample (90 °C) at the TM point. The denatured protein sample (90 °C) was defined as 100% unfolded. | ||
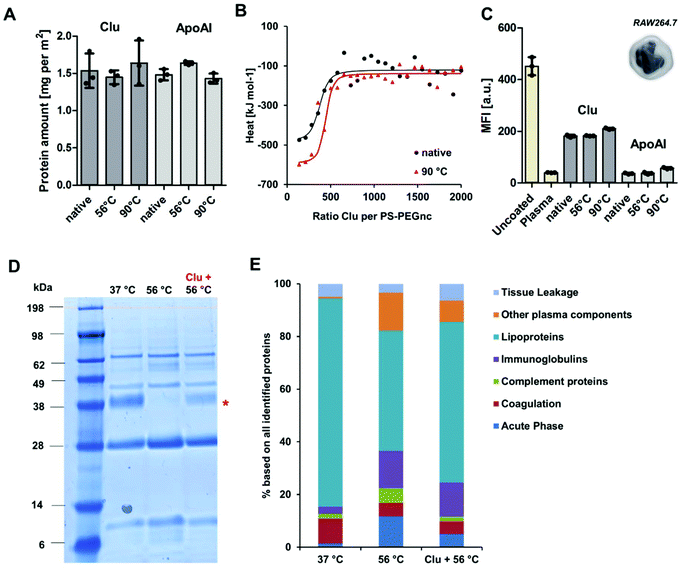
For comparison, both proteins were incubated at 56 °C as well as 90 °C prior to taking measurements. This was done in order to determine the effect of the heat inactivation procedure and complete denaturation for reference purposes. During heat inactivation at a temperature of 56 °C for 30 min, we found that about 40% of clusterin had already unfolded, whereas the structure of ApoAI had not yet been affected. Heat inactivation at 90 °C denatured both proteins completely and irreversibly. The influence of the temperature on the protein structure was additionally analyzed by circular dichroism measurements and confirmed the results obtained from the nanoDSF measurements (Fig. S1 and S2†).
To investigate the structural involvement in protein–nanocarrier interactions still further, PS-PEGNC were incubated with the native single proteins – clusterin and ApoAI. Additionally, proteins were heated up to 56 °C or 90 °C prior to incubation in order to investigate whether temperature induced denaturation affects the binding efficiency to the nanocarriers and whether it influences cellular interaction (Fig. 3). The absolute amount of clusterin and ApoAI adsorbed to PS-PEGNC was quantified via a Pierce Assay. Here, we found that heat inactivation of the single proteins did not affect the total amount of adsorbed proteins (Fig. 3A). In addition to this, we carried out isothermal calorimetry measurements. Native or heat inactivated (90 °C) and therefore completely denatured clusterin was titrated to PS-PEGNC. Interestingly, we determined no significant differences in the binding affinity and all other binding parameters of heat inactivated clusterin compared to native clusterin with PS-PEGNC (Fig. 3B and ESI Table 4†).
In order to investigate whether the stealth properties of clusterin and ApoAI (meaning the ability to prevent interaction with phagocytic cells) are affected by structural alterations, we subsequently analyzed the cellular uptake with macrophages of PS-PEGNC pre-incubated with native and heat inactivated ApoAI and clusterin. We found that pre-incubation with both proteins effectively reduced cellular uptake compared to uncoated nanocarriers. Significantly, there was no difference as to whether proteins were heat inactivated or native (Fig. 3C).
Nanocarrier behavior after incubation with single proteins contrasts strongly to incubation with the complex protein mixture (Fig. 1A). We were able to show that the interactions with native or heat inactivated serum and PEGylated nanocarriers differed strongly. We found that the abundance of clusterin within the heat inactivated serum mixture decreased dramatically in comparison to native serum.
To understand the structural involvement responsible for mediating the corona composition, serum was heat inactivated and native clusterin was re-added to the protein mixture. The protein pattern was visualized by SDS PAGE (Fig. 3D) and quantitatively analyzed by LC-MS (Fig. 3E). We found that native clusterin, which was re-added to heat inactivated serum, adsorbed to PS-PEGNC (clusterin band marked with a red star). This proves that in a complex protein mixture it is the protein structure that critically determines the adsorption behavior, which eventually affects the cellular outcome.
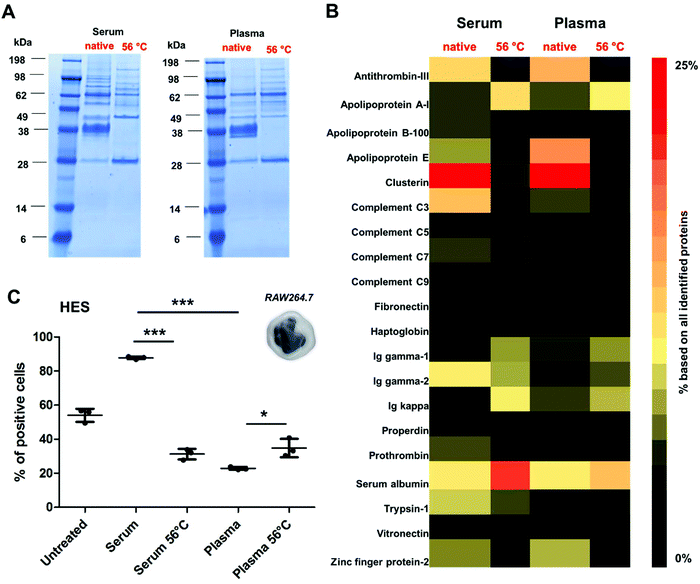
To further reveal the influence of the protein structure on corona formation, additional experiments were performed using a broad range of different nanocarrier systems. Here, we chose covalently PEGylated PS-NPs (PS-PEGC; Fig. S5†), carboxy- or amino-functionalized PS-NPs (PS-COOH and PS-NH2; Fig. S6†) as well as biodegradable hydroxyethyl starch nanocapsules (HES; Fig. 4 and Table S3†). All nanocarriers were incubated with native or heat inactivated serum and plasma. As in the case of PS-PEGNC there was a significant difference when particles were incubated with native serum/plasma in comparison to the heat inactivated protein source (Fig. 4A).
Proteomic analysis highlighted the major differences in the corona composition for HES nanocapsules incubated with the respective protein source (Fig. 4B). As shown above, the adsorption of clusterin was prevented when HES nanocapsules were incubated with heat inactivated serum or plasma. Additionally, we found that the corona of HES nanocapsules incubated with native serum nanocapsules was enriched with complement C3 (∼6.8%). After heat inactivation of the serum, the amount of complement C3 was strongly decreased (∼0.8%).
As widely reported, complement proteins are heat labile and undergo structural changes at 56 °C.28,49 This indicates that interactions with the nanocarriers are prevented due to the structural alterations of complement proteins (Fig. 4B).
In addition, we found major differences in the corona pattern for serum compared to plasma (Fig. 4B). As already reported in the literature, the protein source can influence corona formation.24,25 In the case of the HES nanocapsules investigated here, we detected a strong enrichment of complement proteins in the corona after serum incubation (∼15%). In contrast, significantly lower amounts of complement proteins were identified in the corona after plasma incubation (∼7%). This can be explained by the fact that complement proteins require calcium to maintain their native structure and function.50 Citrate, which was used as an anticoagulant for plasma generation in this study, specifically binds calcium.51 This suggests that the structure of the complement proteins in human plasma is different compared to that of human serum, which eventually affects the interactions of protein–nanocarriers.
Additionally, cellular uptake studies of HES nanocapsules coated with serum or plasma (native vs. heat inactivated) were able to link the distinct role of the corona composition and cellular interactions. Serum incubation strongly enhanced cellular uptake, which is mainly attributed to the involvement of complement proteins. Due to heat inactivation of serum or plasma the overall amount of complement proteins was decreased strongly, which resulted in decreased cellular interaction. Heat inactivation of plasma slightly increased cellular uptake compared to native plasma (*p < 0.05). This can probably be attributed to the lower amount of clusterin in the corona after heat inactivation, which is comparable to the results presented for PS-PEGNC.
Conclusion
In this study we were able to show that heat inactivation of serum or plasma critically affects cellular uptake and protein corona formation. We determined that the protein structure is, in fact, a key factor, which mediates the adsorption process. Clusterin and complement proteins were identified as heat labile proteins, which readily undergo temperature induced structural changes. Therefore, in a complex protein environment, the binding affinity of the denatured proteins to the nanocarriers was significantly decreased. In summary, the present study reveals the major role played by the protein structure and this process needs to be considered in order to unravel protein corona formation and to understand cellular interactions.Experimental
Polystyrene nanoparticles
Polystyrene nanoparticles stabilized with the surfactant Lutensol-AT50 (BASF) were synthesized in accordance to previous reports using free-radical miniemulsion polymerization.52,53 BOPIPY was in cooperated in to the nanoparticles for cell uptake studies.54Hydroxyethyl starch (HES) nanocapsules
HES nanocapsules were obtained by the inverse mini emulsion process.55 The fluorescent dye sulforhodamine 101 (SR101) was in cooperated into the nanocapsules.Cell culture
RAW264.7 were bought from DSMZ (Deutsche Sammlung für Mikroorganismen und Zellen, Braunschweig, Germany). The supplier provides an authentication testing. To ensure no cross-contamination, the original cells were expanded and frozen down in roughly 30 vials. Old cells were discarded every 2–3 month or after passage 25 and a new starting vial was used. Cells were maintained in Dulbecco's modified eagle medium (DMEM) supplemented with 10% FBS, 100 U ml−1 penicillin, 100 mg ml−1 streptomycin and 2 mM glutamine (Thermo Fisher) and passaged 2–3 times per week.Cell uptake experiments by flow cytometry
Cells (150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per well) were seeded out in 24-well plates overnight. The medium was changed to serum-free cell culture conditions for 2 h. Further nanoparticles (75 μg mL−1) were added to cells for 2 h, 37 °C. Cells were washed with PBS, detached with 0.25% Trypsin-EDTA (Gibco) and resuspended with PBS. Flow cytometry measurements were performed on a CyFlow ML cytometer (Partec). Data was analyzed by FCS Express V4 software (DeNovo Software).
000 per well) were seeded out in 24-well plates overnight. The medium was changed to serum-free cell culture conditions for 2 h. Further nanoparticles (75 μg mL−1) were added to cells for 2 h, 37 °C. Cells were washed with PBS, detached with 0.25% Trypsin-EDTA (Gibco) and resuspended with PBS. Flow cytometry measurements were performed on a CyFlow ML cytometer (Partec). Data was analyzed by FCS Express V4 software (DeNovo Software).
Pre-coating experiments for cellular uptake
Nanoparticles (0.05 m2) were incubated with 100 μg clusterin (Biozol) or apolipoprotein AI (EMD Millipore) for 1 h at 37 °C, centrifuged and further used in cell uptake experiments. Proteins were either directly used (‘native’) or heat inactivated (56 °C or 90 °C for 30 min) as indicated in the figure legend.Human plasma and serum
Serum and plasma (each bag contains a volume of ∼300 mL) was obtained from the Department of Transfusion Medicine Mainz from anonymous, healthy donors. To minimize the effect of donor specific protein alterations, serum and plasma from ten donors was pooled. The serum and plasma pools were stored in 15 mL aliquots at −80 °C.Heat inactivation
Serum or plasma was heated up to 56 °C for 30 min and centrifuged afterwards for 30 min, 4 °C (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g).
000g).
Protein corona preparation
Nanoparticles were incubated with the respective protein source and hard corona analysis was carried out as previously described.53 To detach proteins adsorbed to the nanoparticles, the pellet was re-suspended in 2% SDS (62.5 mM Tris·HCl). The sample was heated up to 95 °C for 5 min, centrifuged (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 4 °C, 1 h) and the supernatant was further used for protein analysis.
000g, 4 °C, 1 h) and the supernatant was further used for protein analysis.
Pierce Assay
Nanoparticles (0.05 m2) were incubated with 100 μg Clusterin or ApoAI (native or heat inactivated as described above) for 1 h at 37 °C. Protein coated nanoparticles were purified via repetitive centrifugation and redispersion (1 mL PBS, 3 times) to remove unbound proteins. The adsorbed proteins were detached with 2% SDS (62.5 mM Tris·HCl) and the protein amount was quantified via Pierce Assay (Pierce 660 nm Protein Assay) according to the manufactures’ instruction (Thermo Fisher).SDS PAGE
Hard corona proteins (7 μg in 26 μL) were mixed with 4 μL of NuPage Reducing Agent and 10 μL of NuPage LDS Sample Buffer NuPage. The mixture was heated up (70 °C, 10 min) and applied to a 10% Bis–Tris–Protein Gel. NuPAGE MES SDS Running Buffer was used as buffer, See Blue Plus2 Pre-Stained as molecular marker and the gel was run for 1 h at 120 V. Bands were stained with SimplyBlue SafeStain overnight and destained with distilled water for 2 h (all products Thermo Fisher Scientific).LC-MS analysis
Protein digestion was carried out as described in former reports.56,57 Isolated peptide were diluted with 0.1% formic acid and spiked with 50 fmol μL−1 Hi3 Ecoli (Waters) for absolute protein quantification.58 Samples were analyzed using a nanoACQUITY UPLC system couple to a Synapt G2-Si mass spectrometer. Data was processed with MassLynx 4.1. Protein identification was carried out with Progenesis QI (2.0) using a reviewed human data base (Uniprot). For peptide and protein identification the parameter were set as described elsewhere.53,59Nano differential scanning fluorimetry (nanoDSF)
Proteins (Clusterin and ApoAI, 0.5 mg mL−1 in PBS) were loaded into nanoDSF High Sensitivity capillaries (NanoTemper Technologies) and applied into a Prometheus NT·48 instrument. A linear thermal ramp program starting 20 °C to 95 °C (1 °C min−1) was set and the tryptophan fluorescence was measured at 330 and 350 nm. Thermal unfolding curves of the single wavelength at 330 nm and the first derivative of the fluorescence ratio (330 nm) are plotted against the temperature. The melting temperature TM is determined from the minimum of the first derivative.Isothermal titration calorimetry (ITC)
Measurements were performed with a NanoITC Low Volume from TA Instruments (Eschborn) and experiments were conducted as previously described.42,60 Native or heat-treated Custerin or ApoAI was titrated towards the nanoparticle dispersion. Data was analyzed using the software NanoAnalyze.Circular dichroism spectroscopy (CD)
CD measurements were carried out in a Jasco J-815 (Circular Dichroism Spectropolarimeter). Clusterin (150 μg mL−1) was dissolved in PBS. All CD spectra were recorded at wavelengths ranging from 190 to 260 nm and analyzed with DichroWeb.61,62Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors wish to thank Katja Klein for the synthesis and characterization of the nanoparticles. The authors gratefully acknowledge the financial support from the Deutsche Forschungsgemeinschaft (DFG Sonderforschungsbereich SFB 1066). Open Access funding is provided by the Max Planck Society.References
- X. Xu, W. Ho, X. Zhang, N. Bertrand and O. Farokhzad, Cancer nanomedicine: from targeted delivery to combination therapy, Trends Mol. Med., 2015, 21(4), 223–232 CrossRef CAS PubMed.
- H. Krug, Nanosafety research—are we on the right track?, Angew. Chem., Int. Ed., 2014, 53(46), 12304–12319 CAS.
- D. A. Richards, A. Maruani and V. Chudasama, Antibody fragments as nanoparticle targeting ligands: a step in the right direction, Chem. Sci., 2017, 8(1), 63–77 RSC.
- S.-B. Sun, P. Liu, F.-M. Shao and Q.-L. Miao, Formulation and evaluation of PLGA nanoparticles loaded capecitabine for prostate cancer, Int. J. Clin. Exp. Med., 2015, 8(10), 19670 CAS.
- A. Rodzinski, R. Guduru, P. Liang, A. Hadjikhani, T. Stewart, E. Stimphil, C. Runowicz, R. Cote, N. Altman and R. Datar, Targeted and controlled anticancer drug delivery and release with magnetoelectric nanoparticles, Sci. Rep., 2016, 6, 20867 CrossRef CAS PubMed.
- X. Yu, I. Trase, M. Ren, K. Duval, X. Guo and Z. Chen, Design of nanoparticle-based carriers for targeted drug delivery, J. Nanomater., 2016, 3, 1–15 Search PubMed.
- D. Bobo, K. J. Robinson, J. Islam, K. J. Thurecht and S. R. Corrie, Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date, Pharm. Res., 2016, 33(10), 2373–2387 CrossRef CAS PubMed.
- E. Polo, M. Collado, B. Pelaz and P. del Pino, Advances toward more efficient targeted delivery of nanoparticles in vivo: understanding interactions between nanoparticles and cells, ACS Nano, 2017, 11(3), 2397–2402 CrossRef CAS PubMed.
- G. Caracciolo, O. C. Farokhzad and M. Mahmoudi, Biological identity of nanoparticles in vivo: clinical implications of the protein corona, Trends Biotechnol., 2017, 35(3), 257–264 CrossRef CAS PubMed.
- M. Mahmoudi, Debugging Nano–Bio Interfaces: Systematic Strategies to Accelerate Clinical Translation of Nanotechnologies, Trends Biotechnol., 2018, 36(8), 755–769 CrossRef CAS PubMed.
- P. C. Ke, S. Lin, W. J. Parak, T. P. Davis and F. Caruso, A Decade of the Protein Corona, ACS Nano, 2017, 11(12), 11773–11776 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2050–2055 CrossRef CAS PubMed.
- I. Lynch, A. Salvati and K. A. Dawson, Protein-nanoparticle interactions: What does the cell see?, Nat. Nanotechnol., 2009, 4(9), 546–547 CrossRef CAS PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7(12), 779–786 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, S. Rosfa, A. Wlodarski, J. Kuharev, A. Rekik, S. K. Knauer, C. Bantz, T. Nawroth, C. Bier, J. Sirirattanapan, W. Mann, L. Treuel, R. Zellner, M. Maskos, H. Schild and R. H. Stauber, Nanoparticle Size Is a Critical Physicochemical Determinant of the Human Blood Plasma Corona: A Comprehensive Quantitative Proteomic Analysis, ACS Nano, 2011, 5(9), 7155–7167 CrossRef CAS PubMed.
- Y. Li and N. A. Monteiro-Riviere, Mechanisms of cell uptake, inflammatory potential and protein corona effects with gold nanoparticles, Nanomedicine, 2016, 11(24), 3185–3203 CrossRef CAS PubMed.
- L. Landgraf, C. Christner, W. Storck, I. Schick, I. Krumbein, H. Dähring, K. Haedicke, K. Heinz-Herrmann, U. Teichgräber and J. R. Reichenbach, A plasma protein corona enhances the biocompatibility of Au@ Fe3O4 Janus particles, Biomaterials, 2015, 68, 77–88 CrossRef CAS PubMed.
- A. B. Chinen, C. M. Guan, C. H. Ko and C. A. Mirkin, The Impact of Protein Corona Formation on the Macrophage Cellular Uptake and Biodistribution of Spherical Nucleic Acids, Small, 2017, 13(16), 1–8 CrossRef PubMed.
- V. Mirshafiee, M. Mahmoudi, K. Lou, J. Cheng and M. L. Kraft, Protein corona significantly reduces active targeting yield, Chem. Commun., 2013, 49(25), 2557–2559 RSC.
- A. Salvati, A. S. Pitek, M. P. Monopoli, K. Prapainop, F. B. Bombelli, D. R. Hristov, P. M. Kelly, C. Åberg, E. Mahon and K. A. Dawson, Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface, Nat. Nanotechnol., 2013, 8(2), 137–143 CrossRef CAS PubMed.
- P. Jain, R. Pawar, R. Pandey, J. Madan, S. Pawar, P. Lakshmi and M. Sudheesh, In vitro in vivo correlation (IVIVC) in nanomedicine: Is protein corona the missing link?, Biotechnol. Adv., 2017, 35(7), 889–904 CrossRef CAS PubMed.
- G. Caracciolo, S. Palchetti, L. Digiacomo, R. Zenezini Chiozzi, A. L. Capriotti, H. Amenitsch, P. M. Tentori, V. Palmieri, M. Papi, F. Cardarelli, D. Pozzi and A. Laganà, The human biomolecular corona of liposomal doxorubicin: The overlooked factor in anticancer drug delivery, ACS Appl. Mater. Interfaces, 2018, 10(27), 22951–22962 CrossRef CAS PubMed.
- M. Arora, Cell Culture Media: A Review, Mater. Methods, 2013, 175, 1–29 Search PubMed.
- V. Mirshafiee, R. Kim, M. Mahmoudi and M. L. Kraft, The importance of selecting a proper biological milieu for protein corona analysis in vitro: Human plasma versus human serum, Int. J. Biochem. Cell Biol., 2016, 75, 188–195 CrossRef CAS PubMed.
- S. Schöttler, K. Klein, K. Landfester and V. Mailänder, Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake, Nanoscale, 2016, 8(10), 5526–5536 RSC.
- E. Cancado, L. S. Vilas-Boas, C. P. Abrantes-Lemos, N. F. Novo, G. Porta, L. C. Da Silva and A. A. Laudanna, Heat serum inactivation as a mandatory procedure for antiactin antibody detection in cell culture, Hepatology, 1996, 23(5), 1098–1104 CrossRef CAS PubMed.
- R. Soltis, D. Hasz, M. Morris and I. Wilson, The effect of heat inactivation of serum on aggregation of immunoglobulins, Immunology, 1979, 36(1), 37 CAS.
- R. Soltis, D. Hasz, M. Morris and I. Wilson, Studies on the nature of heat-labile anti-complementary activity in normal human serum, Clin. Exp. Immunol., 1979, 37(2), 310 CAS.
- L. Shen, S. Tenzer, W. Storck, D. Hobernik, V. K. Raker, K. Fischer, S. Decker, A. Dzionek, S. Krauthauser, M. Diken, A. Nikolaev, J. Maxeiner, P. Schuster, C. Kappel, A. Verschoor, H. Schild, S. Grabbe and M. Bros, Protein corona-mediated targeting of nano-carriers to B cells allows redirection of allergic immune responses, J. Allergy Clin. Immunol., 2018, 142(5), 1558–1570 CrossRef CAS PubMed.
- F. Chen, G. Wang, J. I. Griffin, B. Brenneman, N. K. Banda, V. M. Holers, D. S. Backos, L. Wu, S. M. Moghimi and D. Simberg, Complement proteins bind to nanoparticle protein corona and undergo dynamic exchange in vivo, Nat. Nanotechnol., 2017, 12(4), 387 CrossRef CAS PubMed.
- A. Lesniak, A. Campbell, M. P. Monopoli, I. Lynch, A. Salvati and K. A. Dawson, Serum heat inactivation affects protein corona composition and nanoparticle uptake, Biomaterials, 2010, 31(36), 9511–9518 CrossRef CAS PubMed.
- M. Mahmoudi, A. M. Abdelmonem, S. Behzadi, J. H. Clement, S. Dutz, M. R. Ejtehadi, R. Hartmann, K. Kantner, U. Linne and P. Maffre, Temperature: the “ignored” factor at the nanobio interface, ACS Nano, 2013, 7(8), 6555–6562 CrossRef CAS PubMed.
- H. F. Krug, Nanosafety research—are we on the right track?, Angew. Chem., Int. Ed. Engl., 2014, 53(46), 12304–12319 CAS.
- H. H. Gustafson, D. Holt-Casper, D. W. Grainger and H. Ghandehari, Nanoparticle uptake: the phagocyte problem, Nano Today, 2015, 10(4), 487–510 CrossRef CAS PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Protein adsorption is required for stealth effect of poly (ethylene glycol)-and poly (phosphoester)-coated nanocarriers, Nat. Nanotechnol., 2016, 11(4), 372 CrossRef PubMed.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, PEGylation as a strategy for improving nanoparticle-based drug and gene delivery, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- D. Pozzi, V. Colapicchioni, G. Caracciolo, S. Piovesana, A. L. Capriotti, S. Palchetti, S. De Grossi, A. Riccioli, H. Amenitsch and A. J. N. Laganà, Effect of polyethyleneglycol (PEG) chain length on the bio–nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells, Nanoscale, 2014, 6(5), 2782–2792 RSC.
- S. Palchetti, D. Pozzi, A. L. Capriotti, G. La Barbera, R. Z. Chiozzi, L. Digiacomo, G. Peruzzi, G. Caracciolo and A. J. C. Laganà, Influence of dynamic flow environment on nanoparticle-protein corona: From protein patterns to uptake in cancer cells, Colloids Surf., B, 2017, 153, 263–271 CrossRef CAS PubMed.
- M. Papi, D. Caputo, V. Palmieri, R. Coppola, S. Palchetti, F. Bugli, C. Martini, L. Digiacomo, D. Pozzi and G. J. N. Caracciolo, Clinically approved PEGylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cells, Nanoscale, 2017, 9(29), 10327–10334 RSC.
- M. Aoyama, K. Hata, K. Higashisaka, K. Nagano, Y. Yoshioka and Y. Tsutsumi, Clusterin in the protein corona plays a key role in the stealth effect of nanoparticles against phagocytes, Biochem. Biophys. Res. Commun., 2016, 480(4), 690–695 CrossRef CAS PubMed.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailänder and I. Lieberwirth, Visualization of the protein corona: towards a biomolecular understanding of nanoparticle-cell-interactions, Nanoscale, 2017, 9(25), 8858–8870 RSC.
- J. Müller, K. N. Bauer, D. Prozeller, J. Simon, V. Mailänder, F. R. Wurm, S. Winzen and K. Landfester, Coating nanoparticles with tunable surfactants facilitates control over the protein corona, Biomaterials, 2017, 115, 1–8 CrossRef PubMed.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. M. Lima, E. A. Appel, F. Dormont, J.-M. Lim, R. Karnik, R. Langer and O. C. Farokhzad, Mechanistic understanding of in vivo protein corona formation on polymeric nanoparticles and impact on pharmacokinetics, Nat. Commun., 2017, 8(1), 777 CrossRef PubMed.
- C. C. Fleischer and C. K. Payne, Secondary structure of corona proteins determines the cell surface receptors used by nanoparticles, J. Phys. Chem. B, 2014, 118(49), 14017–14026 CrossRef CAS PubMed.
- Y. Kim, S. M. Ko and J. M. Nam, Protein–Nanoparticle Interaction-Induced Changes in Protein Structure and Aggregation, Chem. – Asian J., 2016, 11(13), 1869–1877 CrossRef CAS PubMed.
- A. M. Tsai, J. H. van Zanten and M. J. Betenbaugh, I. Study of protein aggregation due to heat denaturation: a structural approach using circular dichroism spectroscopy, nuclear magnetic resonance, and static light scattering, Biotechnol. Bioeng., 1998, 59(3), 273–280 CrossRef CAS PubMed.
- W. Strutz, Exploring Protein Stability by NanoDSF, Biophys. J., 2016, 110(3), 393a CrossRef.
- R. Chaudhuri, Y. Cheng, C. R. Middaugh and D. B. Volkin, High-throughput biophysical analysis of protein therapeutics to examine interrelationships between aggregate formation and conformational stability, AAPS J., 2014, 16(1), 48–64 CrossRef CAS PubMed.
- C. Hamsten, L. Skattum, L. Truedsson, U. von Döbeln, M. Uhlén, J. M. Schwenk, L. Hammarström, P. Nilsson and M. Neiman, Heat differentiated complement factor profiling, J. Proteomics, 2015, 126, 155–162 CrossRef CAS PubMed.
- R. Snyderman and M. C. Pike, Interaction of complex polysaccharides with the complement system: effect of calcium depletion on terminal component consumption, Infect. Immun., 1975, 11(2), 273–279 CAS.
- K. Mann, M. Whelihan, S. Butenas and T. Orfeo, Citrate anticoagulation and the dynamics of thrombin generation, J. Thromb. Haemostasis, 2007, 5(10), 2055–2061 CrossRef CAS PubMed.
- K. Landfester, Synthesis of colloidal particles in miniemulsions, Annu. Rev. Mater. Res., 2006, 36(1), 231–279 CrossRef CAS.
- M. Kokkinopoulou, J. Simon, K. Landfester, V. Mailänder and I. Lieberwirth, Visualization of the Protein Corona: towards a biomolecular understanding of nanoparticle-cell-interactions, Nanoscale, 2017, 9(25), 8858–8870 RSC.
- I. García-Moreno, A. Costela and L. Campo, 8-Phenyl-Substituted Dipyrromethene·BF2 Complexes as Highly Efficient and Photostable Laser Dyes, J. Phys. Chem. A, 2004, 108(16), 3315–3323 CrossRef.
- G. Baier, D. Baumann, J. r. M. Siebert, A. Musyanovych, V. Mailänder and K. Landfester, Suppressing unspecific cell uptake for targeted delivery using hydroxyethyl starch nanocapsules, Biomacromolecules, 2012, 13(9), 2704–2715 CrossRef CAS PubMed.
- D. Hofmann, S. Tenzer, M. B. Bannwarth, C. Messerschmidt, S.-F. Glaser, H. Schild, K. Landfester and V. Mailänder, Mass Spectrometry and Imaging Analysis of Nanoparticle-Containing Vesicles Provide a Mechanistic Insight into Cellular Trafficking, ACS Nano, 2014, 8(10), 10077–10088 CrossRef CAS PubMed.
- S. Tenzer, Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis, ACS Nano, 2011, 5, 7155–7167 CrossRef CAS PubMed.
- R. A. Bradshaw, A. L. Burlingame, S. Carr and R. Aebersold, Reporting protein identification data: the next generation of guidelines, Mol. Cell. Proteomics, 2006, 5(5), 787–788 CrossRef CAS PubMed.
- L. K. Müller, J. Simon, S. Schöttler, K. Landfester, V. Mailänder and K. Mohr, Pre-coating with protein fractions inhibits nano-carrier aggregation in human blood plasma, RSC Adv., 2016, 6(99), 96495–96509 RSC.
- S. Winzen, S. Schoettler, G. Baier, C. Rosenauer, V. Mailaender, K. Landfester and K. Mohr, Complementary analysis of the hard and soft protein corona: sample preparation critically effects corona composition, Nanoscale, 2015, 7(7), 2992–3001 RSC.
- L. Whitmore and B. A. Wallace, Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases, Biopolymers, 2008, 89(5), 392–400 CrossRef CAS PubMed.
- L. Whitmore and B. Wallace, DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data, Nucleic Acids Res., 2004, 32(suppl_2), W668–W673 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: All identified proteins are summarized in a separate Excel File. See DOI: 10.1039/c8nr07424k |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2018 |