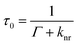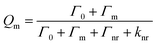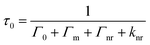DOI:
10.1039/C8NR06156D
(Review Article)
Nanoscale, 2018,
10, 20914-20929
Metal enhanced fluorescence biosensing: from ultra-violet towards second near-infrared window
Received
31st July 2018
, Accepted 12th September 2018
First published on 16th October 2018
Abstract
To increase disease survival rates, there is a vital need for diagnosis at very preliminary stages. Then, low concentrations of biomarkers are present which must be effectively detected and quantified for reliable diagnosis. Fluorescent biosensing is commonly enabled through the labelling of these biomarkers with nanostructures and fluorophores. Metal Enhanced Fluorescence (MEF) is a phenomenon whereby the intensity of a fluorescent biosensor signal can be considerably enhanced by placing a metallic nanostructure and fluorophore in close proximity. Importantly, this allows for an even lower detection limit and thus earlier diagnosis. In recent years, extraordinary efforts have been made in the understanding of how the chemical and physical properties of nanomaterials may be exploited advantageously. Via precise nanoscale engineering, it is possible to optimize the optical properties of plasmonic nanomaterials, which now need to be refined and applied in diagnostics. Through MEF, the intensity of this signal can be related in direct proportion to analyte concentration, allowing for diagnosis of disease at an earlier stage than previously. This review paper outlines the potential and recent progress of applied MEF biosensors, highlighting their substantial clinical potential. MEF biosensors are presented both upon assay-based platforms and in solution, with comments on the various metallic nanoparticle morphologies available. This is explored across various emission wavelengths from ultra-violet to the second near infrared window (NIR-II), emphasising their wide applicability. Further to this, the importance of near infrared (NIR-I and NIR-II) biosensing is made clear as it allows for higher penetration in biological media. Finally, by developing multiplexing techniques, multiple and simultaneous analyses of analytes can be achieved. Through the incorporation of metal enhanced fluorescence into biosensing, it will be possible to diagnose disease more rapidly and more reliably than before, with the potential to save countless lives.
 Sarah Madeline Fothergill | Sarah Madeline Fothergill is currently a PhD student in the Materials Department of Imperial College London. In 2017 she received a distinction in MSc Material Science and Engineering also from Imperial. Prior to this in 2016, she received a first class with honours in BSc Physics with Inorganic and Material Chemistry from University College London (UCL). Sarah's research focuses on utilising the fluorescence enhancing effects of plasmonic nanostructures for early diagnostic biosensing. She currently works under the supervision of Dr Fang Xie. |
 Caoimhe Joyce | Caoimhe Joyce received a first class honours in her BSc in Chemistry with Biophysical Chemistry from University College Dublin in 2016. In 2017 she received a Master of Research degree in Nanomaterials with distinction. She is currently enrolled in the Centre for Doctoral Training in the Advanced Characterisation of Materials (CDT-ACM), where she works in the Materials Department of Imperial College London as a PhD student, under the supervision of Dr Fang Xie. Caoimhe's research is focused on the fluorescence enhancing effects of plasmonic nanostructures in solution, working towards the design of an efficient fluorescent probe for image-guided surgery. |
 Fang Xie | Dr Fang Xie is a Senior Lecturer at Department of Materials, Imperial College London. She has expertise in functional nanomaterials including metal, semiconducting, and oxide nanomaterials synthesis, as well as the applications of the functional materials in energy and life sciences. Her current research interests include plasmonic nanostructures for efficient light harvesting for solar cells and solar fuels, as well as in ultrasensitive biosensing and bioimaging applications. She has over 60 publications including five patents. She has also delivered a number of keynote and invited talks in the international conferences. |
Introduction
Diagnosis of disease is important, so the earlier a diagnosis can be made, the greater the likelihood of patient survival, due to earlier possible treatment.1 Unfortunately, expressed symptoms can often be linked to more than one disease and can frequently show at later stages of disease onset, whereby treatment is less effective. It is therefore important to have a diagnosis in early stages, when lower concentrations of biomarkers are present in the body. Biosensing is one possible route to detect low concentrations of biomarkers.2 In its simplest form, a biosensor consists of a bioreceptor and a transducer. Here, the bioreceptor is a molecule which recognizes the target analyte, whereas the transducer converts the recognition event into a useful, measurable signal. To be commercially successful, a biosensor must meet many requirements, including: accuracy, repeatability, fast response, safety and cost effectiveness.3 Biosensing is not a new concept and it can rely on different recognition elements and transducers.4 It has already proven successful in diagnosis of various diseases, such as HIV5 and diabetes.6 Biosensing can employ many strategies, including enzymes,7 DNA8 and piezoelectric sensors.9 Fluorescent biosensing is one of the most popular biosensing techniques,2 which relies of the emission of radiation following excitation by incident radiation. This is a growing research field, combining the high sensitivity of fluorescence detection with the high selectivity of binding biomarkers. In order to lower detection limits further, metal enhanced fluorescence may be incorporated. The potential for fluorescent biosensing based on metal enhanced fluorescence (MEF) has previously been extensively reviewed10,11 with much focus on the synthesis of suitable nanostructures to produce high fluorescence enhancement factors. However, beyond simply the fabrication of these nanostructures, this review focuses primarily on instances in which MEF has already been incorporated successfully for biosensing applications. Fig. 1 demonstrates the fundamental development stages for the production of a MEF based biosensor. Comments on key variables, such as nanostructure morphology, choice of fluorophore and quantification method, are made throughout this review, highlighting their clinical potential and applicability. More importantly no review had comprehensively covered emissions from ultraviolet to second near infrared window. Therefore this perspective article will provide a complete review from a spectrum point of view, emphasizing the vast potential of MEF when applied to fluorescence biosensing across many wavelengths.
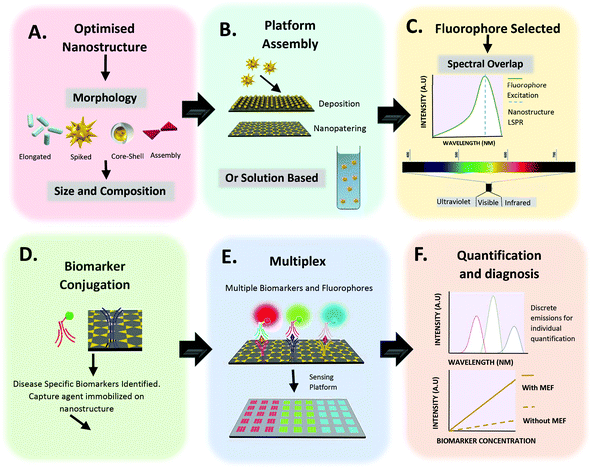 |
| | Fig. 1 Fundamental steps for the design of a MEF based biosensor. (A) Optimization of nanostructure. Suitable electromagnetic field enhancements are required for specific disease biomarker detection. The morphology, materials and size must be selected accordingly. (B) Platform assembly or solution preparation of optimised nanostructure. (C) Choice of fluorophore and spectral region. For increased fluorescence intensity, an overlap between fluorophore emission and metallic nanostructure must be present. (D) Capture agent conjugation to metallic nanostructure. (E) A move towards multiplexing, allowing for multiple biomarkers to be simultaneously analysed. (F) Quantification and early diagnosis of disease with enhanced fluorescence intensity. | |
Brief theory of metal enhanced fluorescence
MEF can improve the quantum efficiency and photostability of fluorophores.11 When a fluorophore is placed in the proximity of a metallic nanostructure, the free electrons within the metal surface (surface plasmon) couple with the fluorophore electrons.12 The metal here can be considered as an optical antenna at which localized hot spots of propagating electromagnetic radiation can be confined. As this confinement is close to the metallic nanoparticle surface, it is dependent on the particle's morphology, composition and surrounding environment. Fluorescent materials are widely used in biosensing.13 MEF makes it possible to detect much lower concentrations of the biomarkers used in biosensing or bioimaging. Metallic nanoparticles (MNPs) posess free surface electrons which resonate when illuminated with light of appropriate wavelength. This light causes the metal's electrons to be displaced relative to the particle's core, setting up a restoring force which leads to oscillations of the charge density,12 which is termed localised surface plasmon resonance (LSPR). This resonance condition is dependent on the size, morphology, distance and dielectric of the metallic nanoparticles and surrounding medium, as each will cause a shift in the electromagnetic field density at the metal nanoparticle surface. Upon placing a fluorophore in the near field of a metallic surface, both the emission and excitation characteristics of the fluorophore can be modified. MEF allows for an increase in the excitation rate due to an increase in the local electric field experienced by the fluorophore and the resultant coupling between this fluorophore and nearby MNPs. This can be more easily understood when considering the simplified Jablonski diagram (Fig. 2C). In the absence of a metal, a fluorophore is excited from the ground state (S0) to the excited states (S1, S2etc.). From here internal conversion happens as the fluorophores are able to relax to lower vibrational states, whereby they can subsequently emit energy as fluorescence and return to the ground state. In addition to this, another process called spin conversion can occur, which causes transitions from the singlet to the triplet state (T1 – not displayed in Fig. 2C). Both processes result in a radiative decay rate (Γ). In addition to this, non-radiative decay (knr) is also present and a quenching process (rate kq – not displayed in Fig. 2C) may occur. The fluorescence lifetime is measured as the time the fluorophore spends in an excited state before returning to its original ground state (τ0). The quantum yield defines the number of times fluorescence occurs per photon absorbed by the system (Q0). During this process the overall fluorescence rate Ψ may be expressed as the product of both the excitation rate and the quantum yield. In the presence of a metal the fluorophore is excited at a wavelength λex when exposed to incident radiation. It is possible that the near field around a metallic nanoparticle can be used to further excite a nearby fluorophore, which will emit at an emission wavelength, λem. In a similar manner to standard fluorescence, photon emission may then decay either radiatively (Γ) or non-radiatively (knr). As stated, it is possible to consider metal enhanced fluorescence as a coupled system between a fluorophore and a metallic nanoparticle. In this system, when a metallic nanoparticle is present beyond the quenching distance, the decay rate is modified to (Γ + Γm), where Γm represents the modified decay rate in the presence of the metal. Importantly, the metal is modifying the radiative decay rate and creating new channels of non-radiative decay through energy and charge transfer between the fluorophore and the metal. As a result, there is also an increase in quantum yield and a decrease in lifetime. This can be thought of as the metallic nanoparticle acting as an optical antenna such that the propagating radiation can be converted to near field energy. It is possible to express this phenomenon mathematically using well-documented formulae:14,15| | 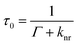 | (2) |
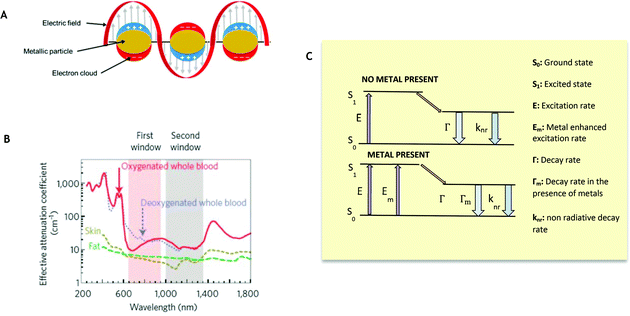 |
| | Fig. 2 Biological windows and Metal Enhanced Fluorescence. (A) Local Surface Plasmon Resonance (LSPR). (B) First and second biological window demonstrating increased transparency for NIR I (first window) and NIR II (second window). (Reprinted by permission from Springer Nature: Nature Nanotechnology, copyright (2009), from ref. 43.) (C) Classical Jablonski diagram for the free-space condition (top) and the modified form in the presence of metallic nanoparticles (bottom) accompanying eqn (1)–(4). | |
Eqn (1) and (2) demonstrate the quantum yield and lifetime for an isolated fluorophore. When considering the coupled system the modified decay rate is included. The modified quantum yield (Qm) and the lifetime may therefore be expressed as:
| | 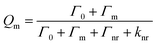 | (3) |
| | 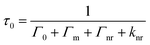 | (4) |
Fluorophores are sensitive to environmental changes, such as changes in pH, polarity, oxidation and temperature as well as distance. At large separation, the effectiveness of Γnr decreases so that the original quantum yield (unmodified) is again obtained. It has also been shown experimentally that as the separation distance between the metal nanoparticle and the fluorophore decreases to approximately 5 nm, quenching of the fluorophore occurs.16
General criteria for a MEF based biosensor
To date, many structures fabricated for MEF have consisted primarily of silver or gold, due to their LSPRs already being within the visible to near infrared wavelengths.17,18 In addition, their ease of fabrication and functionalization, as well as their stability make them a preferable, practical choice.19 Nevertheless, MEF is not limited to these two metals, with ZnO,20 copper21 and aluminum22 also being reported, for example. Similarly, the correct choice of fluorophore is essential in providing an overlap in the optical properties of both the metal and fluorophore.23 In the absence of any overlap between their emission and absorption spectra, there is no common excitation wavelength to allow for simultaneous excitation of both parts of the coupled system. Interestingly, instead of complete overlap, it has been shown that enhanced fluorescence occurs when the fluorophore emission peaks at a slightly longer wavelength than the LSPR peak.23,24 In the past decade, there have been plentiful novel nanoparticle structures showing an ability to improve both the fluorescence intensity and the photostability of fluorophores via this mechanism, including nanorods,25 nanocubes,26 nanodisks,27 nanotriangles28 and nanostars29 (Fig. 3A30). By tuning the morphology and size of the metallic nanoparticles, it is possible to alter the LSPR and the position and intensity of the localized ‘hot spots’, thus tuning the effect on the fluorophore's emission. As the separation distance between the metal and fluorophore must also be considered, it is necessary to consider a spacer material as well as an assembly method.31Fig. 3B demonstrates some planar MEF platforms with different spacer materials, including dielectrics, Langmuir–Blodgett films, polymers and biomolecules, as taken from an extensive review paper.32 Similarly, it is possible to translate these spacing techniques to the solution phase through coating of the MNPs.32 This review will explore the successful use of metal enhanced fluorescence biosensing applications, demonstrating the progress from UV and visible through to NIR wavelengths. One frequent example is the use of immunoassays for biosensing, which forms the majority of the basic principles of this review. Here, a capture agent (such as an aptamer or antibody) is able to trap target analytes (e.g. an antigen) on a substrate surface with antibodies bound to fluorophores. It is possible to analyze the concentration of analyte present in accordance with the resulting fluorescence intensity.
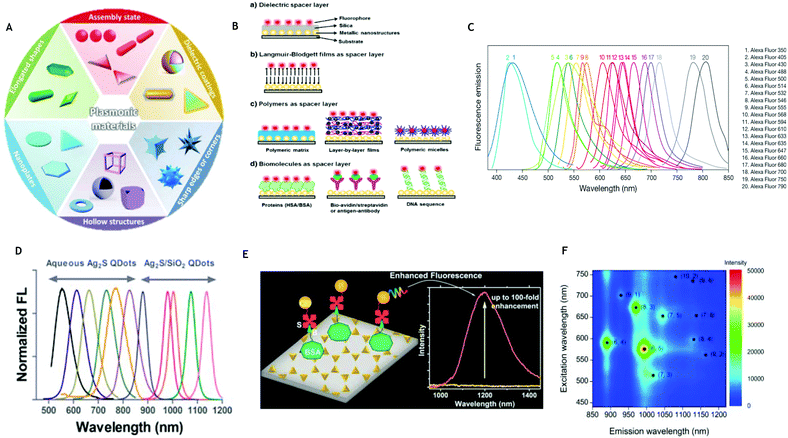 |
| | Fig. 3 (A) Morphological options for plasmonic materials. (Reprinted from Material Horizons, with permission from Royal Society of Chemistry, copyright (2014), from ref. 30.) (B) Schematic representation of the common kinds of MEF platforms on planar surfaces based on different spacer layers. (Reprinted from Advances in Colloid and Interface Science, with permission from Elsevier, from ref. 32.) (C) Wide ranging emission spectra for Alexa Fluor™ dyes (from www.thermofisher.com – Alexa Fluor Dyes Across the Spectrum). (D) Normalized emission spectra of Ag2S QDs and Ag2S/SiO2 QDs. (Reprinted with permission from ACS, DOI: 10.1021/nn5071183, further permissions directed to ACS, from ref. 66.) (E) Ag2S quantum dots conjugated to a nanotriangles array demonstrating up to 100-fold enhancement. (Reprinted from Nanoscale, with permission from Royal Society of Chemistry, copyright (2009), from ref. 67.) (F) Extinction and emission wavelengths and relative intensity of various (n, m) of single walled carbon nanotubes. (Reprinted from Sensors, https://doi.org/10.3390/s17112569, from ref. 58.) | |
Fluorescent detection labels
UV fluorophores
There are few reports of MEF biosensing with UV wavelengths available in the literature, most likely due to metallic nanostructures typically showing extinction in the visible to NIR range. Nevertheless, nanostructures which can support plasmons in the UV regime are still of interest for biosensing, as many organic molecules fluoresce naturally within the UV region. Many biomolecules, for example, absorb light in the range of 220–280 nm, which allows for label-free detection, with these molecules acting as natural fluorophores. This is hindered, however, due to the low efficiency of native biomolecular fluorescence. As previously highlighted, the choice of metal is limited, but aluminum is able to support plasmons within UV range33 and has therefore been reported most for UV MEF. As early as 2005, a six-fold enhancement in Rhodamine-6G molecules was observed using an aluminum film by Rigneault et al.34 Lakowicz et al. also explored how aluminum nanostructured films significantly enhance DNA base analogues and hence act as label-free detectors of amino acids.22 Similarly, it has been computationally verified that aluminum nanoparticles can enhance tryptophan protein fluorescence and that this degree of enhancement can be further increased by placing the fluorophore in a dimer of nanoparticles.35 Moving away from aluminum, in 2009, Henryk Szmacinski et al. demonstrated that silver nanostructures could be used to enhance the natural fluorescence of tryptophan protein in the UV range to allow label-free detection and 3-fold increased intensity compared to proteins on bare quartz.36 In addition to these examples, MEF has also been shown to increase the fluorescence intensity of carbon dots in the visible range (emission 440 nm) when coupled to gold nanoparticles.37
Visible fluorophores
The use of visible fluorophores is advantageous in allowing for ease of detection and quantification. Fluorescein isothiocyanate (FITC) is an organic dye with excitation and emission spectrum peak wavelength emissions at approximately 495 nm and 519 nm respectively. Derivatives of FITC are the most common fluorescence reagents for biological research because of their high absorptivity, excellent fluorescence quantum yield and good water solubility. FITCs have been used in Alexa 488 and DyLight 488, offering high photostability and fluorescence intensity. Perhaps the most noteworthy of the fluorophores spanning the UV to visible wavelengths are the commonly used Alexa Fluor family of fluorescent dyes. These are a series of dyes which are regularly used in fluorescence imaging and sensing due to their increased photostability and wide range emissions from 442 nm through to 814 nm. The full range of Alexa Flour dye emissions can be seen in Fig. 3C, showing distinct peaks allowing for multiplexed use. Cyanines (Cy) yield a brighter and more stable fluorescence than FITC that can be detected by the naked eye.38,39 Cy3 and Cy5 are the most popular, emitting at 570 nm and 670 nm respectively which can be easily distinguished and quantified by scanners.
NIR I and NIR II fluorophores
When considering applicability for biosensing through biological matter or fluids, the presence of hemoglobin and water limit the possible fluorescent molecules (and thus wavelengths) that may be used for in vivo biosensing due to their absorption of shorter wavelengths and thus low transparency (Fig. 2B). Therefore, we propose a shift towards MEF in NIR wavelengths for fluorescent biosensing. Here, two emission bands that have been identified as areas in which there is higher transparency of soft tissue and biological fluid40–42 are NIR-I (650–900 nm) and NIR-II (1000 nm–1700 nm).43 Within these two regions auto-fluorescence is lower in organic molecules compared to that in the visible region and hence results in a lower background signal and thus enhanced contrast. Reports of NIR fluorophores are limited, with far fewer NIR-II fluorophores being reported than their NIR-I counterparts. Ideally, fluorophores within NIR-II are desired such that minimal-to-no background signal is present to allow penetration through blood without any pre-treatment. As already stated, it would appear desirable to focus on these two regions for future fluorescence biosensing. Unfortunately, current NIR fluorophores show limitations, such as low quantum yield and photostability, particularly compared to visible or UV dyes,44 limiting their applicability. Although it has been proposed that new photostable NIR dyes be synthesized, this has proven to be challenging in practice and thus not a promising route to improving detection sensitivity.45 The incorporation of MEF into NIR biosensing platforms is crucial in improving their applicability. So far organic and inorganic materials, such as small molecules, conjugated polymers, carbon nanotubes, rare earth nanoparticles and quantum dots have been used in NIR imaging and sensing.46–50 A selection is outlined in more detail below.
Organic NIR fluorophores
Organic fluorophores that span the electromagnetic spectrum are now available, and when covalently conjugated to targeting molecules, sensitive contrast agents for NIR imaging and sensing are created.51 A few examples of NIR fluorescent dyes include classical cyanine dyes, rhodamine dyes and squaraine dyes.52 As it currently stands, commercially available NIR dyes (ICG,53 dyelite 800, AF750, AF790) suffer from low quantum yields and poor photo stability and thus the available range is limited. Synthesis of new NIR dyes is challenging; nevertheless there have been slow advances. For example, in 2017, a novel small molecule NIR-II dye was synthesized with an improved synthetic protocol and an improved quantum efficiency (quantum yield 2%) and high-resolution imaging of blood vessels of tumors was used for the first time in NIR-II image-guided surgery.49 Similarly, the functional design of a clickable NIR-II small molecule dye (800 nm–1700 nm) was used in histological brain tissue.54 Finally, in 2018, an efficient 1064 nm NIR-II excitation fluorescent molecular dye for deep-tissue high-resolution dynamic bioimaging was synthesized, allowing for higher penetration depth and superior resolution compared to previously reported NIR excitation from 650 nm to 980 nm.55 By coupling with metallic nanoparticle systems, there is the potential that this organic NIR fluorophore could be improved even further for biosensing applications.
Carbon nanotubes
Carbon nanotubes are already proving successful in in vivo imaging applications, offering deep imaging and high resolution.56 Likewise, they are also an option as a fluorophore for incorporation into MEF biosensing systems. Single walled carbon nanotubes (SWNTs) were first coupled to MEF substrates by Hong et al., demonstrating 10-fold enhancement57 in NIR-II using an Au film. SWNTs have been shown to be characterized by inherent photoluminescence between 650 nm and 1400 nm.43,58 Unfunctionalized SWNTs possess low fluorescence stability, intensity and biocompatibility, yet, in contrast, surface functionalization and environmental change affect their fluorescence emission significantly (both wavelength and intensity)59 widening their applicability. Upon surface functionalization or interaction with target molecules, the wavelength and intensity of emission of nanotubes have been shown to vary significantly, making them well suited to fluorescent-based sensing applications.60 In addition to this, the quantum yield of these SWNTs is comparable to that of quantum dots in NIR and they remain stable to photobleaching.61 Although there is limited literature, it would theoretically be possible to incorporate carbon nanotubes even further into MEF systems.
Specific example 1: quantum dots
Quantum dots (QDs) are promising probes for incorporation into MEF based biosensors both in the visible and NIR range.62 As a result of quantum confinement, QDs possess several advantages over organic dyes, such as a long excited-state lifetime, high photostability and high quantum yields. In addition to this, QD emissions can be tuned across a wide range of wavelengths, and multiple dots of different emission wavelengths can be stimulated by a single light source, reducing the requirement for more than one excitation per different fluorophore. Beneficially, the bandwidths of QDs are narrow and, as such, can reduce the spectral overlap between different quantum dot emissions, which allows for potential multiplexing and detection of multiple discrete signals.63 Typically, the choice of QDs suitable for in vivo biomedical applications is limited, due to concerns about high toxicity.64 However, this concern can be largely dismissed when considering biosensing on a chip substrate (in vitro). Ag2S QDs are an attractive option for use in biosensing due to their optical properties in the near infrared regime. Their size-dependent excited-state optical properties were systematically investigated by Yejun Zhang et al. in 2014,65 demonstrating that they may be tuned across NIR. Similarly, in 2015, Ag2S and Ag2S/SiO2 QDs were prepared covering a wide spectral window from 500 to 1200 nm by Tang et al., which were successfully conjugated to a tumor-avid peptide (Fig. 3D).66 Since then, Ag2S quantum dots have been incorporated into numerous MEF systems. For example, in 2016 I.G Theodorou et al. demonstrated significant fluorescence intensity enhancement of these probes in the NIR-II region when coupled with Au nanotriangular structures67 (Fig. 3E). As the quantum yield of QDs is lower in the NIR range than in the UV or visible regions, MEF is beneficial in allowing for their wider application. Beyond just Ag2S, additional quantum dots have been investigated. For example, Au nanoparticles have been shown to enhance NIR II fluorescence of PbS quantum dots by a factor of 2.68
Specific example 2: upconversion nanoparticles
Upconversion nanoparticles (UCNPs) are a class of nanoparticles of around 20–50 nm which have the ability to emit light with a shorter wavelength than the excitation light. This phenomenon is based on an anti-Stokes process known as two photon conversion. More explicitly, two or more low-energy photons may be absorbed, followed by the emission of a high-energy photon.69 With its capacity to convert NIR light into visible light, upconversion can be applied to biosensing, allowing for deeper biological medium penetration. Inorganic UCNPs are the most common, where lanthanide ions are dispersed as guests in an appropriate dielectric host.70 These feature a wealth of electronic transitions within the 4f electron shell.71 To achieve high upconversion efficiency, sensitizers must be co-doped alongside activator ions which have a closely matched intermediate excited state. Compared to QDs, UCNPs contain variable intermediate levels and by increasing the concentration of co-dopants, it is possible to directly improve their brightness. Although efforts have been made to improve the upconversion efficiency of UCNPs by tuning their phase, composition and size, back energy transfer still severely limits the achieveable quantum yield.72 In 2015, it was shown that UCNPs could be enhanced 22.6-fold when coupled to nanorods at a controlled distance.73 Also in 2015, a DNA biosensor based on UCNPs and graphene oxide was generated. Here, when DNA functionalised UCNPs were in the presence of complementary DNA, hybridization prevented interaction with graphene oxide, reducing quenching and allowing a detectable fluorescence signal.74 Further work in 2018 allowed for multiplexing with upconversion nanoparticles for the first time by manipulating the luminescence emission and decay lifetimes, allowing for detection of human papillioma virus (HPV).75 Hence the possibility for inclusion of UCNPs as part of MEF biosensing systems has been highlighted.
Progress in MEF biosensors
To date, the incorporation of metal enhanced fluorescence into biosensing systems has been limited, with most publications focusing on the fabrication of metallic nanostructures or substrates to increase fluorescence enhancement. Nevertheless, these findings can easily be applied to biosensing applications via the incorporation of biomolecule receptors, increasing their clinical applicability. The following work demonstrates some of the most recent advances in biosensing using metal enhanced fluorescence from UV to NIR wavelengths.
UV to visible substrate biosensors
There is a narrow literature available on the applicability of MEF within UV. As discussed, much of this relies on the inherent fluorescence of biomolecules. Nevertheless, in 2010 it was demonstrated computationally that nanoapertures in aluminum could significantly enhance the emission of dyes with 266 nm excitation.76 Experimentally, further work by Ono et al. used an aluminum thin film with QDs to enhance fluorescence with 266 nm excitation, reporting a high field enhancement of approximately 40 times.77 More recently, in 2017, Pourreza et al. presented a novel metal enhanced fluorescence bioprobe for the sensing of insulin based on a poly vinyl hydrogel functionalized by Ag dots. In this work fluorophores of insulin protein coupled to the Ag nanoparticles with an emission peak of around 300 nm were observed. The fluorescence intensity was enhanced in the presence of functionalized Ag with increasing insulin and a detection limit of 0.1 ng mL−−1 was achieved in human serum samples from both healthy and diabetic patients.78 Visible MEF biosensing is more commonly compared to its UV counterpart. Although possessing low penetration through biological matter, visible biosensing does have the advantage of relatively inexpensive equipment for analysis. As early as 2011, Goldman et al. showed the viability of silver nanoparticles to detect bio-macromolecular complexes such as ribosomes. Here, MEF increased the total photon emission of Cy3 and Cy5 (emission 570 nm and 670 nm respectively) labelled ribosomal complexes near 50 nm silver nanoparticles, improving the signal to noise ratio and demonstrating a 4.7-fold fluorescence enhancement.79 Also in 2011, the detection of troponin I (TnI), which is used as a biomarker for myocardial damage, was performed using a sensor chip incorporating Ag nanoparticles.80 Later, in 2016, Xiaofan Ji et al. investigated the metal enhanced fluorescence of Ag Zig Zag nanorod arrays made by oblique angle deposition for biomolecule-protein integration and DNA hybridization (Fig. 4B). By varying the folding number and the substrate deposition temperature, a 14-fold enhancement factor was obtained and a significant signal enhancement was observed with a detection limit as low as 0.01 pM (ref. 81) using Alexa 448 (emission at 519 nm). Although the use of nanorods in MEF has previously been documented in the literature,81–85 this work demonstrated their potential as biosensors using MEF. Here, the plasmonic response of the nanorod can be tuned through its size to overlap with the fluorophore excitation, making it advantageous to allow for a lower detection limit. Also in the visible region, a gold leaf based assay has been used for virus detection by using QDs (515 nm emission) as fluorophores86 (Fig. 4C). This film was made by dealloying a bimetallic film of gold with antibodies bound to amine-terminated QDs with a resulting detection limit twice as sensitive as a commercially available influenza diagnostic test. Further, in 2017, nanohole array substrates were used to provide up to 1600-fold higher field intensity enhancement for the generation of a sandwich assay for prostate specific antigen detection, reducing the limit of detection to 140 fM.87 In further work, a metal enhanced fluorescence biosensor was fabricated using a novel hydrogel microarray using Ag@SiO2 nanoparticles decorated with QDs, increasing the intensity by 5-fold compared to the system without MEF (Fig. 4D). Paraoxon and ACheE were detected based on amplified fluorescence quenching upon exposure.88 This work presented a fast and readily applicable strategy for microarray production which could also be used in multiplexing (see sixth section). Finally, a novel flower-like silver-enhanced fluorescence platform for the ultrasensitive detection of multiple miRNAs was successfully constructed based on the principle of multi-channel microfluidic paper-based analytical devices. This biosensor allowed for detection as low as 0.03 fM and demonstrated good capability in recycling and enhanced fluorescence in the visible range,89 again moving towards multiplexed applications.
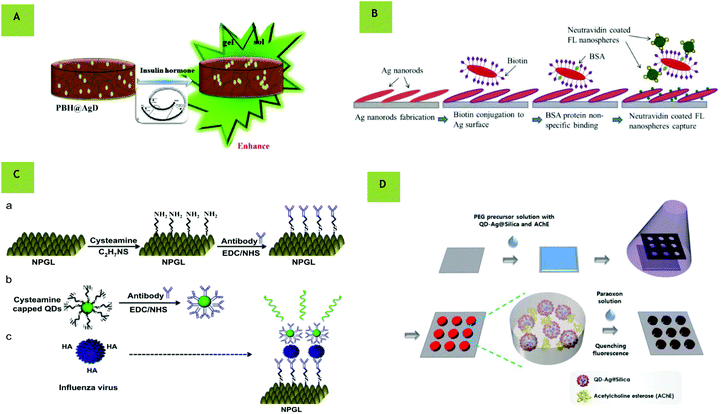 |
| | Fig. 4 (A) Polyvinyl alcohol and borax hydrogel (PBH) @AgD bioprobe for insulin sensing with enhanced fluorescence upon insulin hormone binding. (Reprinted from Sensors and Actuators B: Chemical, with permission from Elsevier, copyright (2017), from ref. 78.) (B) Process of neutravidin coated fluorescent nanospheres binding onto Ag nanorods. (Reprinted from Biosensors and Bioelectronics, with permission from Elsevier, copyright (2016), from ref. 81.) (C) Schematic of virus detection using nanoporous gold leaf (NPGL) film. The NPGL (a) and quantum dots (QDs) (b) were firstly conjugated with anti-hemagglutinin (HA) antibodies (anti-HA Ab, Y shape) by the reaction of ethylcarbodiimide (EDC)/N-hydroxysuccinimide (NHS). Then anti-HA Ab-conjugated with NPGL and QDs forms complex (c) in the presence of HA on the surface of influenza virus, finally enhancing fluorescence intensity. (Reprinted from Biosensors and Bioelectronics, with permission from Elsevier, copyright (2014), from ref. ref. 86.) (D) Schematic illustration of fabrication of hydrogel microarrays entrapping QD-Ag@SiO2 and AChE for paraoxon detection. (Reprinted with author permission from, Biofabrication, IOP Publishing, DOI: 10.1088/1758-5090/aab004, from ref. 88.) | |
Solution-based MEF biosensing
To date, nearly all of the work with metal enhanced fluorescence for biosensing has focused on fluorophores in close proximity to planar surfaces and the literature on solution-based biosensing is limited. This may be because random orientations in solution make it difficult to account for the interactions between nanoparticles at any given time. Nevertheless, there is still some literature available. The earliest work on solution-based biosensing dates back to 2004, when silver-based colloids provided a solution-based enhanced fluorescence sensing platform with up to 5-fold enhancement.90 Here, silver colloids were labelled with Cy3 (emission 570 nm), demonstrating that MEF biosensing is also applicable in solution. Around the same time, oligonucleotide-modified silver particles and a fluorophore-labeled complementary oligonucleotide were used to allow for increased emission after hybridization in the presence of metallic nanoparticles.91 Other reported solution biosensing techniques involve core–shell nanoparticles. These structures have attracted much attention in MEF over the past decade,26,92–94 as coating with a dielectric material provides a natural spacer for the optimal positioning of a fluorophore. Their biosensing potential has already been demonstrated in the literature. In 2014, Yuanfeng Pang et al. presented a MEF-based fluorescent aptasensor95 using core–shell Ag@SiO2 nanoparticles for the sensitive detection of the recombinant hemagglutinin (rHA) protein of the H5N1 influenza virus with a detection limit of 2 ng mL−1 (Fig. 5A). Here the anti-rHA aptamers were immobilized on the surface of the MNPs, which performed as a metal-enhanced fluorescent platform using a thiazole orange fluorescent tag (TO)96 with emission in the visible range (532 nm). Importantly, this detection process required only 30 minutes, making a rapid self-contained diagnostic kit. Similarly, in 2016, a sensitive ‘on/off’ fluorescent protocol for thrombin detection was demonstrated using aptamers.97 Unusually, rather than enhancing the fluorescence, this method worked by quenching the signal upon target binding. Here, thrombin aptamers hybridized to Cy5-labelled DNA were immobilized on the surface of Ag@SiO2 nanoparticles. Upon addition of the thrombin and graphene oxide, the thrombin displaced the Cy5-labelled DNA (670 nm emission) causing it to bind to the surface of the graphene oxide resulting in fluorescence quenching (Fig. 5B). These findings were used to design an assay that had a 0.05 nM detection limit and excellent selectivity, which could be easily adapted to additional analytes for which appropriate aptamers are available. This work is similar to previous methods whereby Cy5-labeled aptamers confined to the Ag@SiO2 nanoparticle surface dissociate from complementary DNA in solution, again leading to a reduction in fluorescence.98 Here, Lu et al. presented MEF of core–shell nanoflares for affinity biosensing via target-induced structure switching of the aptamer with a detection limit of 8 μM.98 One novel approach that has been taken by Dang-Dang Xu et al. is the combination of MEF of dye-doped silica nanoparticles with magnetic separation as a sensitive platform for the one-step fluorescence detection of prostate specific antigen99 (PSA). Here, a core–shell composite fluorescent nanoparticle Ag@SiO2@SiO2-RuBpy provided a 3-fold enhancement. This highly sensitive, specific and rapid strategy allowed for magnetic separation of PSA both in buffer and in serum using immunomagnetic nanospheres and immunofluorescent nanoparticles, allowing for a detection limit of 27 pg mL−1 (Fig. 5C). In 2014, a biosensing platform using Ag@SiO2-DNA-fluorophore nanostructures was used for metal ion and small organic molecule detection. In this work, DNA hybridization occurred in the presence of target molecules and the Cy3 fluorophore was brought to the surface of the core–shell nanostructures.100 Although most solution-based MEF biosensors have focused on silica-coated nanosphere probes, it is important to note that they are not limited to these morphologies. For example, core–shell nanorods have similarly been shown to be useful for highly sensitive detection of pyrophosphate, moving towards NIR. Beyond silica coating of nanoparticles, a DNA sensor for the detection of E. coli (O157:H7 eaeA gene) was constructed using Au@Ag nanorods with Cy3-labeled single stranded DNS (ssDNA) with a low detection limit of 3 × 10−18 M and high specificity (Fig. 5E).101 As stated, compared to substrate-based sensing, solution-based MEF biosensing presents lower electromagnetic field enhancements due to the reduced proximity to adjacent metallic nanoparticles. One possibility is the creation of dimers and clusters of metal nanoparticles in solution. Although there appears to be no literature regarding this, fluorescent enhancements of metal nanoparticle dimer systems have been reported in a few papers.102–105
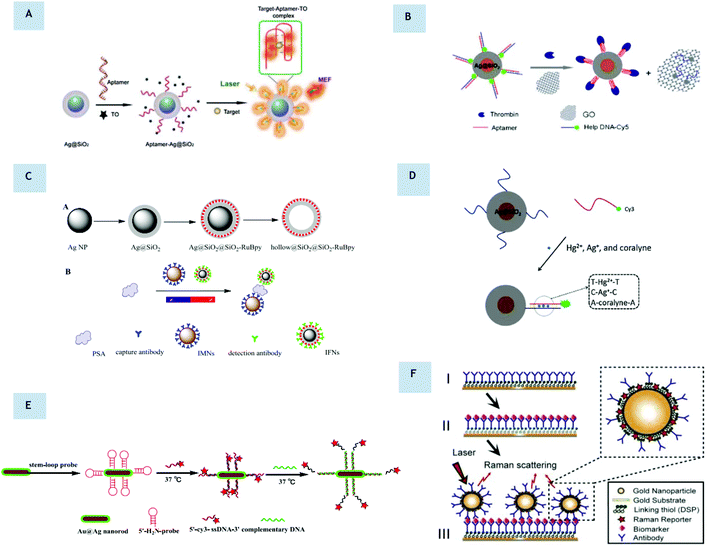 |
| | Fig. 5 (A) Schematic illustration of the preparation of the aptamer-Ag@SiO2 sensor and the determination of the rHA protein H5N1. (Reprinted from Biosensors and Electronics, with permission from Elsevier, copyright (2015), from ref. 95.) (B) Ultrasensitive aptamer-based thrombin assay based on on/off fluorescence. Thrombin displaces Cy5 and binds to graphene oxide causing fluorescence quenching. (Reprinted by permission from Microchimica Acta: Springer Nature, copyright (2016), from ref. 97.) (C) (A) Schematic illustration for the synthesis of Ag@SiO2@SiO2-RuBpy and hollow@SiO2@SiO2-RuBpy. (B) Schematic illustration for the construction of a detector for PSA with immunomagnetic nanospheres (IMNs) and immunofluorescent nanoparticles (IFNs). (Reprinted from Biosensors and Bioelectronics, with permission from Elsevier, copyright (2017), from ref. 99.) (D) Operating principle of a Ag@SiO2-DNA-Cy3 sensor. (Reprinted from Sensors and Actuators B: Chemical, with permission from Elsevier, copyright (2014), from ref. ref. 100.) (E) Schematic illustration of a DNA sensor using Au@Ag nanorods for the detection of E. coli genes. (Reprinted from Biosensors and Bioelectronics, with permission from Elsevier, copyright (2015), from ref. 101.) (F) Example of SERS-based immunoassay for biomarker quantification. (Reprinted with author permission, from Nanotechnology, IOP Science, https://doi.org/10.1088/1361-6528/aa8e8c, from ref. 107.) | |
Adaptation and incorporation of SERS technology
It is important to note that the technology currently available for surface enhanced Raman scattering (SERS) biosensing has the ability to be adapted to MEF systems. SERS spectroscopy utilizes enhanced optical excitation and scattering at metal surfaces associated with surface plasmon polarizations – the coherent coupling of light with the free electron plasma of the metal. For example, a multiplex SERS frequency shift immunoassay for liver and pancreatic cancer biomarkers has already been developed with high sensitivity.106 Nanoparticle films with an excellent SERS response have been fabricated capable of multiplexed detection of liver cancer biomarkers by recording two frequency shifts upon an assay.106 In a similar manner, a separate study demonstrated that immobilization of functionalized gold nanoshells on a gold-coated silicon substrate led to a significant improvement in the SERS signal for pancreatic cancer biomarker detection.107 This work was used to detect three cancer biomarkers at concentrations as low as 2 ng ml−1. What is more novel in this work is the combination of both a solid substrate and solution nanoparticles. As both SERS and MEF rely on the same fundamental properties of metallic nanostructure, a combination of both techniques could be possible (multimodal). By incorporating two methods into biomarker analysis, the reliability of a resultant biosensor could be improved.
Towards NIR I and NIR II biosensing
As stated, it is desirable to move towards near infra-red wavelengths to allow for higher penetration through biological media, and a lower background signal. A move towards NIR-II (1000–1700 nm) biosensing would result in a negligible background signal. NIR wavelengths have already attracted much attention in biological imaging from tumor imaging to image-guided surgery and could be similarly advantageous for biosensing applications.42,49,108 For example in 2018, NIR-II nanoprobes were shown to improve image-guided surgery for metastatic ovarian cancer.109 Also in 2018, NIR-II lanthanide nanoparticles were engineered to create distinct lifetime channels for multiplexed in vivo imaging.110 As highlighted, MEF can be performed both in solution and on an array.11 In comparison to solution-phased particle sensing, substrate-based assays offer an advantage in that the electromagnetic field is further enhanced between adjacent metallic nanostructures.111 It is important to note that for successful biosensing application, the level of the enhancement factor must be tuned according to a suitable level of sensitivity for a diagnosis to be made. Typically, many of the substrate-based platforms have focused on ‘bottom up’ fabrication methods in order to tightly control the resultant morphology with high reproducibility. It is preferable to have homogeneous substrate fabrication methods to increase the clinical applicability and reliability of diagnoses. Further to this, it allows for computational modelling and prediction of electromagnetic field enhancement.104,112,113 However, cases for successful inhomogenous substrates have also been presented. For example in 2010, the first case of metal enhanced fluorescence of surfactant-coated carbon nanotubes was demonstrated on solution-grown gold films with extinction towards the NIR window.57 Colloidal lithography allows for the deposition of periodic arrays of metal nanostructures and is a promising low-cost, easy fabrication method.114 The variability of structures that can be produced is still expanding and ranges from rod arrays to rings, discs and crescents.27,115–117 In 2016, the amplification of light in NIR-II from Ag2S quantum dots was reported for the first time, achieving an enhancement of over 100 times when conjugated to nanotriangular arrays as produced via colloidal lithography.67 Most notably, in 2017, protein microarrays based on NIR plasmonic gold nanostructures were reported for the highly sensitive detection of the pancreatic biomarker CA19-9.118 Here optically tunable substrates were fabricated with up to two orders of magnitude enhancement in fluorescence using DyLight800 (emission 794 nm). In this case, gold nanotriangular arrays were fabricated with a nanoscale controlled structure using colloidal lithography, as previously demonstrated in the literature111 (Fig. 6A). A ‘sandwich’ protein microarray procedure was performed on the nanotriangular arrays, allowing for a limit of detection of CA19-9 as low as 7.7 × 10−7 U mL−1, an improvement compared to commercially available CA19-9 immunoassays. Lithography is easy to implement, and the morphologies formed can easily be modified to be optically tuned as well as being repeatable and allowing for large areal coverage. As the level of fluorescence enhancement is proportional to the distance between adjacent triangles and the overlap with the fluorophore, nanotriangular substrates offer a simple yet effective route to controlling the fluorescence intensity, as is required in multiplexing. This is not dissimilar to an approach using gold nanorods as a substrate119 for DNA sensing (Fig. 6C). For MEF biosensing within NIR-II, nanostructures with their LSPR within this region are required. In 2017, gold nanostar substrates were shown to also enhance fluorescence through the first and second near-infrared windows with a fluorescence enhancement of up to 320 times being achieved using nanostar substrates.29 Star-shaped Au nanoparticles are a relatively new morphology of nanoparticle that are receiving attention due to their potential applications in bionanotechnology120–122 with their sharp features allowing strong localized surface plasmon resonances.29 It has further been numerically and experimentally shown that Au nanostar clusters can concentrate up to 460-fold higher energy than Au nanosphere clusters, with the results of this work validated in model assays for protein biomarker detection,123 highlighting their potential for biosensing applications. The incorporation of a dielectric core could increase enhancements even further in NIR-II (Fig. 6B). As previously mentioned, it would easily be possible to translate many successful existing MEF based substrates into NIR biosensing systems.
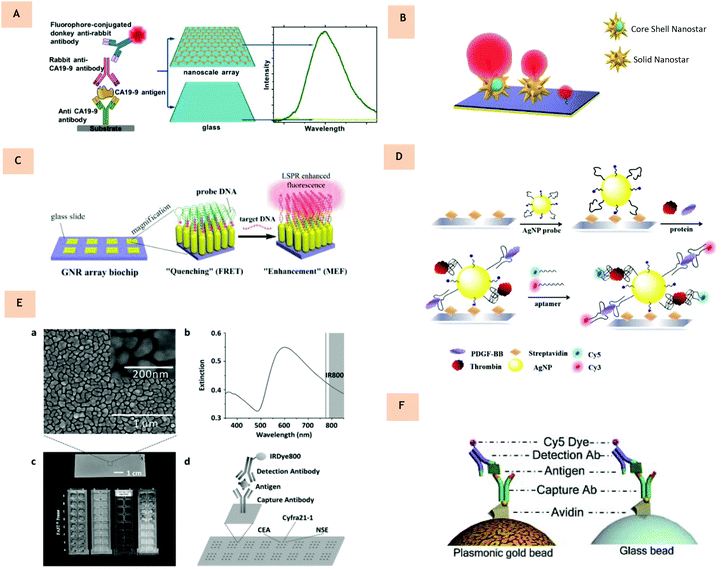 |
| | Fig. 6 (A) Near-infrared fluorescence enhancement immunoassay for the quantification of the pancreatic biomarker CA19-9, demonstrating superior sensitivity compared to glass controls. Schematic diagram of the CA19-9 ‘sandwich’ protein microarray performed on Au-nanotriangular substrates, resulting in up to two orders of magnitude enhancement in the fluorescence intensity compared to glass controls. (Reprinted from Nature Scientific Reports, https://doi.org/10.1038/s41598-017-14688-z, from ref. 118.) (B) Graphic illustration of core-shell nanostars and solid nanostars. (C) Gold nanorod DNA probe array. In the absence of the target, fluorescence is quenched. Upon target binding, fluorescence enhancement occurs. (Reprinted with permission from ref. 119, copyright (2017), American Chemical Society.) (D) Illustration of the aptamer-based sandwich assay for multiplex detection of thrombin and PDGF-BB. (Reprinted from Analytica Chimica Acta, with permission from Elsevier, copyright (2016), from ref. 130.) (E) Biomarker detection on a pGOLD chip. (a) Scanning electron image and high-resolution image of a pGOLD chip showing the nanoscopic gold island morphology. (b) Extinction spectrum of pGOLD chip overlaid with the excitation (grey line) and emission (shaded area) regions of near IR800 dye. (c) Digital photographs of pGOLD chip (top) and integrated device for experiments. (d) Schematic illustration of CEA, CYfra21-1, and NSE detection on pGOLD chip using a sandwich assay. (Reprinted from Advanced Functional Materials, with permission from John Wiley and Sons, copyright (2016), from ref. 132.) (F) Protein quantification on plasmonic gold beads. Sandwich assay schemes for protein detection on a plasmonic Au bead and glass bead. (Reprinted from Chemical Science, with permission from Royal Society of Chemistry, from ref. 136.) | |
Multiplexed biosensing
A multiplexed assay is a type of assay which can be used to measure multiple analytes in a single run of the assay.124 Example multiplex assay techniques include protein-based125 and nucleic acid multiplexing.126 As stated, for many diseases there may not be one definitive biomarker which can reliably diagnose its early stages with high specificity and sensitivity. Measuring many different biomarkers simultaneously is favorable since one biomarker may be indicative of more than one disease, and as related diseases can manifest with similar symptoms, so monitoring requires detection of subtle differences in biomarkers over time.127 In addition to this, as driven by demands for cost efficiency, there is an increasing need to quantify large numbers of species from minute sample volumes, quantify biomarkers and acquire greater information from a single experiment.128 Therefore, instead of a single analyte, a combination of biomarkers may be chosen and a diagnosis can be made based on their presence in patients. In order to do this effectively, different fluorescent signals must be received, identified individually, calibrated and quantified. An additional benefit of multiplexing is the requirement for much smaller sample volumes as low as hundreds of μL. Obtaining large volumes of sample from a patient (particularly the elderly, children or the critically ill) may not always be feasable or comfortable.129 Through multiplexing, small sample volumes could enable the use of capilary blood taken from a finger for example, rather than venous blood. Although limited in the literature, some successful MEF multiplexing methods are outlined.
In 2015, Wang et al. presented novel aptamer-modified silver nanoparticles designed to detect proteins on a microarray based on a sandwich assay130 (Fig. 6D). So far, the majority of aptamer-based methods of detection are designed for single target analysis and, hence, the development of new probes that are based on multiple aptamer-modified nanomaterials would be an effective way to enhance detection sensitivity. Here, aptamer-modified silver nanoparticles were first used as capture probes in a sandwich assay. From here secondary aptamers, labeled with fluorophores Cy3 and Cy5 were used as report probes. Interestingly, it was found that, compared to using aptamers alone as capture probes, the detection limit decreased by 80 and 8 times when detecting thrombin and growth factor PDGF-BB, respectively. This work demonstrated multiplex detection with high sensitivity and high throughput analysis. It would be possible to transfer this setup to different metal nanoparticle morphologies and fluorophores for further optimization. Additional multiplexed work was presented by S. M. Tabakman et al. as they demonstrated plasmonic substrates for multiplexed protein microarrays with femtomolar sensitivity and broad dynamic range.131 Here, protein microarrays were presented on a novel nanostructured plasmonic gold film with NIR fluorescence enhancement up to 100-fold, extending the dynamic range of protein detection by three orders of magnitude towards the fM range. In this work, a solution-phase, bottom-up growth procedure of Au films was formed in a scalable, simple process. It was demonstrated that a multiplexed autoantigen array for human autoantibodies in a range of autoimmune diseases showed superior signal to noise ratios and a broader dynamic range than controls and commercial nitrocellulose substrates. The high sensitivity, broad dynamic range and ease of adaptability of this plasmonic protein chip may present benefits in diagnostic applications. Similarly, work in 2016 presented multiplexed antibody microarrays on a similar plasmonic gold chip for detecting three circulating biomarkers associated with lung cancer.132 Again, nano gold islands were fabricated on the substrate with gaps of around 10 nm (Fig. 6E). These were synthesized via a solution-phase synthesis approach of seeding and growth of densely packed gold islands similar to that of S. M. Tanakman. In this work, it was reported that the nanostructured plasmonic gold islands allow for up to two orders of magnitude enhancement in fluorescence (by 50–100 compared to controls) over glass chips and gold controls. The platform was divided into 16 separate blocks, allowing for detection of 16 samples on each of four gold slides assembled in a microwell plate-like format in a single run, both reducing cost and sample volume required. Here, for the assay, a sandwich assay scheme was used and three different capture antibodies were directly printed onto the surface of the platform and labelled with IRDye 800. When compared to commercially available Luminex technology (based on bead and flow cytometry), the calibration curves here demonstrated an improved limit of detection and limit of quantification of between 5 and 20-fold. In order to perform multiplexed biomarker detection, capture antibodies were then printed onto 3 × 3 spot matrices and following incubation with a mixture of the three protein biomarkers, a single detection antibody or a mix of detection antibodies were applied as the final layer for fluorescence labelling and detection. These results demonstrated the high specificity of multiplexed assay capability on a plasmonic gold chip. In addition to this, the reproducibility at low concentrations also increased. This method allowed for a high throughput, and easily performed diagnosis for lung cancer, the first of its kind utilizing NIR fluorescent enhancement. Similar work within the same group uses these nano gold islands for the diagnosis of both diabetes and the zika virus.133,134 Multiplexed assays for the mass screening of toxoplasma using IgG, IgM and IgA antibodies has also been presented using a plasmonic gold (pGOLD) substrate using only 1 μL of serum or whole blood sample.135 Even further, this group has demonstrated that this principle can be extended to microbeads via coating Au nanostructures on glass microbeads, enhancing the ability to measure low-abundance protein biomarkers such as cytokines (Fig. 6F).136 Currently, work with pGOLD lacks optical tuning of the plasmonic gold chip, limiting control over morphology and hence overlap with the optical properties of the fluorophore chosen. Nevertheless, this research group have extensively demonstrated the potential for MEF in biosensing using multiple analytes, a principle that could easily be extended to additional morphologies.
Conclusions
The potential for MEF in biosensing has been firmly established. However, beyond just scientific challenges, there are also added additional engineering challenges that will need to be addressed before any assay can be taken from lab to clinic. For the generation of a substrate-based sensing chip, the size presented should be small so as to lessen the amount of patient sample required, while also permitting a sufficient surface for multiplexed sensing. To ease commercialization, it is important to consider whether multiple arrays can be produced rapidly and simultaneously on a single platform, which would assist manufacture. One possible route could be to incorporate photolithography to selectively deposit structures onto a substrate. As MEF based sensors are sensitive to variations in nanoparticle or substrate uniformity, size, composition and shape, the reliability of any resultant biosensor will potentially be hindered. Therefore, this highlights the requirement for uniform synthesis or separation techniques to be integrated in manufacture. An additional consideration is the long-term stability of any MEF biosensing system, e.g. from the effect of any solvents, annealing or oxidizing agents. To overcome these constraints, coating or encapsulation could be implemented and thus improve shelf life. Presently, most work regarding metal enhanced fluorescence has focused on attaining high enhancement factors, with extraordinary advancements in the generation of nanomaterials with distinctive and controlled properties. Nevertheless, this review has presented numerous examples of metal enhanced fluorescence for biosensing applications, ranging from UV to NIR wavelengths. From a medical viewpoint, different biomarkers will be present from patient to patient at diverse concentrations and this will result in altered levels of the enhancement factor required for diagnosis. Thus there will need to be calibration between the quantity of biomarker present and a ‘positive result’ value, requiring collaboration with physicians. Through a combination of different techniques, such as SERS and MEF, it would be possible to increase the reliability of biomarker detection even further. Evidentially, it is necessary for a concentrated effort to focus on the ideal attributes of a clinically viable assay. As indicated, a future move towards NIR wavelengths is desirable, benefitting from low to no background signal through biological matter: for example, fluorescence diagnostics through a patient's blood. By synthesizing varying metallic nanostructures, both in solution and on a substrate, it is possible to optimize plasmonic properties to couple to different fluorophores and thus enhance signal and sensitivity. It is hoped that through selecting examples that have to date applied MEF for varying biomolecule detection, the potential for clinical transfer has been highlighted as the next step forward. Although there are still many challenges that must be overcome before their incorporation into healthcare, it is anticipated that these metal enhanced fluorescence based diagnostic systems will provide a paradigm shift in a clinical setting in coming years.
Conflicts of interest
There are no conflicts of interest to declare.
Acknowledgements
S. M. F. acknowledges the EPSRC funding to support her PhD study. C. J. acknowledges the support from EPSRC CDT Advanced Characterisation. F. X. acknowledges funding support from British Council Newton Fund Institutional Links (216239013). We acknowledge the support from the Henry Royce Institute made through EPSRC grant EP/R00661X/1.
Notes and references
- K. Hanada, A. Okazaki, N. Hirano, Y. Izumi, T. Minami, J. Ikemoto, K. Kanemitsu and F. Hino, Best Pract. Res. Clin. Gastroenterol., 2015, 29, 929–939 CrossRef PubMed.
- J. N. Anker, W. P. Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. Van Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- A. Agah, A. Hassibi, J. D. Plummer, P. B. Griffin and I. Systems, Proc. SPIE, 2005, 5699, 403–413 CrossRef.
- B. Bohunicky and S. A. Mousa, Nanotechnol. Sci. Appl., 2011, 4, 1–10 CAS.
- M. A. Lifson, M. O. Ozen, F. Inci, S. Q. Wang, H. Inan, M. Baday, T. J. Henrich and U. Demirci, Adv. Drug Delivery Rev., 2016, 103, 90–104 CrossRef CAS PubMed.
- E. H. Yoo and S. Y. Lee, Sensors, 2010, 10, 4558–4576 CrossRef PubMed.
- O. Syshchyk, V. A. Skryshevsky, O. O. Soldatkin and A. P. Soldatkin, Biosens. Bioelectron., 2014, 66, 89–94 CrossRef PubMed.
- J. Wang, Nucleic Acids Res., 2000, 28, 3011–3016 CrossRef CAS PubMed.
- S. Babacan, P. Pivarnik, S. Letcher and A. G. Rand, Biosens. Bioelectron., 2000, 15, 615–621 CrossRef CAS PubMed.
- S. Abalde-Cela, S. Carregal-Romero, J. P. Coelho and A. Guerrero-Martínez, Adv. Colloid Interface Sci., 2016, 233, 255–270 CrossRef CAS PubMed.
- K. Aslan, I. Gryczynski, J. Malicka, E. Matveeva, J. R. Lakowicz and C. D. Geddes, Curr. Opin. Biotechnol., 2005, 16, 55–62 CrossRef CAS PubMed.
- C. D. Geddes and J. R. Lakowicz, J. Fluoresc., 2002, 12, 121–129 CrossRef.
-
S. Undenfriend, Fluorescence assay in biology and medicine, 1969, vol. 2 Search PubMed.
- K. Aslan, I. Gryczynski, J. Malicka, E. Matveeva, J. R. Lakowicz and C. D. Geddes, Curr. Opin. Biotechnol., 2005, 16, 55–62 CrossRef CAS PubMed.
- T. L. Jennings, M. P. Singh and G. F. Strouse, J. Am. Chem. Soc., 2006, 128, 5462–5467 CrossRef CAS PubMed.
- W. L. Barnes, J. Mod. Opt., 1998, 45, 661–699 CrossRef CAS.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef.
- A. Centeno, F. Xie and N. Alford, J. Opt. Soc. Am. B, 2011, 28, 325–330 CrossRef CAS.
- S. K. Balasubramanian, L. Yang, L. Y. L. Yung, C. N. Ong, W. Y. Ong and L. E. Yu, Biomaterials, 2010, 31, 9023–9030 CrossRef CAS PubMed.
- T. Wang, J. Costan, A. Centeno, J. S. Pang, D. Darvill, M. P. Ryan and F. Xie, J. Mater. Chem. C, 2015, 3, 2656–2663 RSC.
- K. Sugawa, T. Tamura, H. Tahara, D. Yamaguchi, T. Akiyama, J. Otsuki, Y. Kusaka, N. Fukuda and H. Ushijima, ACS Nano, 2013, 7, 9997–10010 CrossRef CAS PubMed.
- K. Ray, M. H. Chowdhury and J. R. Lakowicz, Anal. Chem., 2007, 79, 6480–6487 CrossRef CAS PubMed.
- Y. Chen, K. Munechika and D. S. Ginger, Nano Lett., 2007, 7, 690–696 CrossRef CAS PubMed.
- P. Bharadwaj and L. Novotny, Phys. Rev. Lett., 2000, 308, 9–21 Search PubMed.
- J. Gao, C. M. Bender and C. J. Murphy, Langmuir, 2003, 19, 9065–9070 CrossRef CAS.
- B. Sun, C. Wang, S. Han, Y. Hu and L. Zhang, RSC Adv., 2016, 6, 61109–61118 RSC.
- J. Pang, I. G. Theodorou, A. Centeno, P. K. Petrov, N. M. Alford, M. P. Ryan and F. Xie, J. Mater. Chem. C, 2017, 5, 917–925 RSC.
- F. Xie, M. S. Baker and E. M. Goldys, Chem. Mater., 2008, 20, 1788–1797 CrossRef CAS.
- I. G. Theodorou, Z. A. R. Jawad, Q. Jiang, E. O. Aboagye, A. E. Porter, M. P. Ryan and F. Xie, Chem. Mater., 2017, 29, 6916–6926 CrossRef CAS.
- L. Zhu, M. Gao, C. K. N. Peh and G. W. Ho, Mater. Horiz., 2018, 5, 323–343 RSC.
- K. Ray, R. Badugu and J. R. Lakowicz, Langmuir, 2006, 22, 8374–8378 CrossRef CAS PubMed.
- Q. Cui, F. He, L. Li and H. Möhwald, Adv. Colloid Interface Sci., 2014, 207, 164–177 CrossRef CAS PubMed.
- D. Gérard and S. K. Gray, J. Phys. D: Appl. Phys., 48, 184001 CrossRef.
- H. Rigneault, J. Capoulade, J. Dintinger, J. Wenger, N. Bonod, E. Popov, T. W. Ebbesen and P. F. Lenne, Phys. Rev. Lett., 2005, 95, 117401 CrossRef PubMed.
- M. H. Chowdhury, K. Ray, S. K. Gray, J. Pond and J. R. Lakowicz, Anal. Chem., 2009, 81, 1397–1403 CrossRef CAS PubMed.
- H. Szmacinski, K. Ray and J. R. Lakowicz, Anal. Biochem., 2009, 385, 358–364 CrossRef CAS PubMed.
- J. Jana, T. Aditya, M. Ganguly, S. K. Mehetor and T. Pal, Spectrochim. Acta, Part A, 2018, 188, 551–560 CrossRef CAS PubMed.
- P. L. Southwick, L. A. Ernst, E. W. Tauriello, S. R. Parker, R. B. Mujumdar, S. R. Mujumdar, H. A. Clever and A. S. Waggoner, Cytometry, 1990, 11, 418–430 CrossRef CAS PubMed.
- R. B. Mujumdar, L. A. Ernst, S. R. Mujumdar, C. J. Lewis and A. S. Waggoner, Bioconjugate Chem., 1993, 4, 105–111 CrossRef CAS PubMed.
- J. Bartelmess, S. J. Quinn and S. Giordani, Chem. Soc. Rev., 2015, 44, 4672–4698 RSC.
- M. Holzinger, A. Le Goff and S. Cosnier, Front. Chem., 2014, 2, 1–10 CAS.
- G. Hong, A. L. Antaris and H. Dai, Nat. Biomed. Eng., 2017, 1, 10 CrossRef.
- A. M. Smith, M. C. Mancini and S. Nie, Nat. Nanotechnol., 2009, 4, 710–711 CrossRef CAS PubMed.
- L. Guo, J. A. Jackman, H. H. Yang, P. Chen, N. J. Cho and D. H. Kim, Nano Today, 2015, 10, 213–239 CrossRef CAS.
- D. D. Nolting, J. C. Gore and W. Pham, Curr. Org. Synth., 2011, 8, 521–534 CrossRef CAS PubMed.
- Y. Zhang, G. Hong, Y. Zhang, G. Chen, F. Li, H. Dai and Q. Wang, ACS Nano, 2012, 6, 3695–3702 CrossRef CAS PubMed.
- D. J. Naczynski, M. C. Tan, M. Zevon, B. Wall, J. Kohl, A. Kulesa, S. Chen, C. M. Roth, R. E. Riman and P. V. Moghe, Nat. Commun., 2013, 4, 2199 CrossRef CAS PubMed.
- Y. Sun, C. Qu, H. Chen, M. He, C. Tang, K. Shou, S. Hong, M. Yang, Y. Jiang, B. Ding, Y. Xiao, L. Xing, X. Hong and Z. Cheng, Chem. Sci., 2016, 7, 6203–6207 RSC.
- Y. Sun, M. Ding, X. Zeng, Y. Xiao, H. Wu, H. Zhou, B. Ding, C. Qu, W. Hou, A. Er-Bu, Y. Zhang, Z. Cheng and X. Hong, Chem. Sci., 2017, 8, 3489–3493 RSC.
- G. Hong, Y. Zou, A. L. Antaris, S. Diao, D. Wu, K. Cheng, X. Zhang, C. Chen, B. Liu, Y. He, J. Z. Wu, J. Yuan, B. Zhang, Z. Tao, C. Fukunaga and H. Dai, Nat. Commun., 2014, 5, 4206 CrossRef CAS PubMed.
- J. V. Frangioni, Curr. Opin. Chem. Biol., 2003, 7, 626–634 CrossRef CAS PubMed.
- L. Yuan, W. Lin, K. Zheng, L. He and W. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- Z. Starosolski, R. Bhavane, K. B. Ghaghada, S. A. Vasudevan, A. Kaay and A. Annapragada, PLoS One, 2017, 12, e0187563 CrossRef PubMed.
- S. Zhu, Q. Yang, A. L. Antaris, J. Yue, Z. Ma, H. Wang, W. Huang, H. Wan, J. Wang, S. Diao, B. Zhang, X. Li, Y. Zhong, K. Yu, G. Hong, J. Luo, Y. Liang and H. Dai, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 962–967 CrossRef CAS PubMed.
- B. Li, L. Lu, M. Zhao, Z. Lei and F. Zhang, Angew. Chem., Int. Ed., 2018, 57, 7483–7487 CrossRef CAS PubMed.
- G. Hong, J. C. Lee, J. T. Robinson, U. Raaz, L. Xie, N. F. Huang, J. P. Cooke and H. Dai, Nat. Med., 2012, 18, 1841–1846 CrossRef CAS PubMed.
- G. Hong, S. M. Tabakman, K. Welsher, H. Wang, X. Wang and H. Dai, J. Am. Chem. Soc., 2010, 132, 15920–15923 CrossRef CAS PubMed.
- H. Ashiba, Y. Iizumi, T. Okazaki, X. Wang and M. Fujimaki, Sensors, 2017, 17, 2569 CrossRef PubMed.
- C.-M. Tîlmaciu and M. C. Morris, Front. Chem., 2015, 3, 59 CrossRef PubMed.
- A. Jain, A. Homayoun, C. W. Bannister and K. Yum, Biotechnol. J., 2015, 10, 447–459 CrossRef CAS PubMed.
- S. Y. Ju, W. P. Kopcha and F. Papadimitrakopoulos, Science, 2009, 323, 1319–1323 CrossRef CAS PubMed.
- S. J. Rosenthal, J. C. Chang, O. Kovtun, J. R. McBride and I. D. Tomlinson, Chem. Biol., 2011, 18, 10–24 CrossRef CAS PubMed.
- Q. Ma and X. Su, Analyst, 2010, 135, 1867–1877 RSC.
- P. Zrazhevskiy, M. Sena and X. Gao, Chem. Soc. Rev., 2010, 39, 4326–4354 RSC.
- Y. Zhang, Y. Liu, C. Li, X. Chen and Q. Wang, J. Phys. Chem. C, 2014, 118, 4918–4923 CrossRef CAS.
- R. Tang, J. Xue, B. Xu, D. Shen, G. P. Sudlow and S. Achilefu, ACS Nano, 2015, 9, 220–230 CrossRef CAS PubMed.
- I. G. Theodorou, Z. A. R. Jawad, H. Qin, E. O. Aboagye, A. E. Porter, M. P. Ryan and F. Xie, Nanoscale, 2016, 8, 12869–12873 RSC.
- K. Wu, J. Zhang, S. Fan, J. Li, C. Zhang, K. Qiao, L. Qian, J. Han, J. Tang and S. Wang, Chem. Commun., 2015, 51, 141–144 RSC.
- S. Wilhelm, ACS Nano, 2017, 11, 10644–10653 CrossRef CAS PubMed.
- G. Chen, H. Qiu, P. N. Prasad and X. Chen, Chem. Rev., 2014, 114, 5161–5214 CrossRef CAS PubMed.
-
S. Wen, J. Zhou, K. Zheng, A. Bednarkiewicz, X. Liu and D. Jin, Nat. Commun., 2018, 9, 2415 Search PubMed.
- G. R. Tan, M. Wang, C. Y. Hsu, N. Chen and Y. Zhang, Adv. Opt. Mater., 2016, 4, 984–997 CrossRef CAS.
-
A. Ling Feng, M. Li You, L. Tian, S. Singamaneni, M. Liu, Z. Duan, T. Jian Lu, F. Xu and M. Lin, Nat. Sci. Rep., 2015, 5, 7779 Search PubMed.
- P. Alonso-Cristobal, P. Vilela, A. El-Sagheer, E. Lopez-Cabarcos, T. Brown, O. L. Muskens, J. Rubio-Retama and A. G. Kanaras, ACS Appl. Mater. Interfaces, 2015, 7, 33 Search PubMed.
- L. Zhou, Y. Fan, R. Wang, X. Li, L. Fan and F. Zhang, Angew. Chem., Int. Ed., 2018, 1–7 Search PubMed.
- D. Punj, P. Ghenuche, S. B. Moparthi, J. de Torres, V. Grigoriev, H. Rigneault and J. Wenger, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6, 268–282 CAS.
- A. Ono, M. Kikawada, R. Akimoto, W. Inami and Y. Kawata, Opt. Express, 2013, 21, 17447 CrossRef PubMed.
- N. Pourreza and M. Ghomi, Sens. Actuators, B, 2017, 251, 609–616 CrossRef CAS.
- S. Bharill, C. Chen, B. Stevens, J. Kaur, Z. Smilansky, W. Mandecki, I. Gryczynski, Z. Gryczynski, B. S. Cooperman and Y. E. Goldman, ACS Nano, 2011, 5, 399–407 CrossRef CAS PubMed.
- K. Aslan and T. A. J. Grell, Clin. Chem., 2011, 57, 746–752 CrossRef CAS PubMed.
- X. Ji, C. Xiao, W. F. Lau, J. Li and J. Fu, Biosens. Bioelectron., 2016, 82, 240–247 CrossRef CAS PubMed.
- K. Aslan, Z. Leonenko, J. R. Lakowicz and C. D. Geddes, J. Phys. Chem. B, 2005, 109, 3157–3162 CrossRef CAS PubMed.
- R. Bardhan, N. K. Grady, J. R. Cole, A. Joshi and N. J. Halas, ACS Nano, 2009, 3, 744–752 CrossRef CAS PubMed.
- A. M. Gabudean, M. Focsan and S. Astilean, J. Phys. Chem. C, 2012, 116, 12240–12249 CrossRef CAS.
- A. M. Gabudean, D. Biro and S. Astilean, Nanotechnology, 2012, 23, 485706 CrossRef CAS PubMed.
- S. R. Ahmed, M. A. Hossain, J. Y. Park, S. H. Kim, D. Lee, T. Suzuki, J. Lee and E. Y. Park, Biosens. Bioelectron., 2014, 58, 33–39 CrossRef CAS PubMed.
- Q. Zhang, L. Wu, T. I. Wong, J. Zhang, X. Liu, X. Zhou, P. Bai, B. Liedberg and Y. Wang, Int. J. Nanomed., 2017, 12, 2307–2314 CrossRef CAS PubMed.
- M. Kim, J. E. Kwon, K. Lee and W.-G. Koh, Biofabrication, 2018, 10, 035002 CrossRef PubMed.
- L. Liang, F. Lan, X. Yin, S. Ge, J. Yu and M. Yan, Biosens. Bioelectron., 2017, 95, 181–188 CrossRef CAS PubMed.
- K. Asian, J. R. Lakowicz, H. Szmacinski and C. D. Geddes, J. Fluoresc., 2004, 14, 677–679 CrossRef PubMed.
- J. Zhang, J. Malicka, I. Gryczynski and J. R. Lakowicz, Anal. Biochem., 2004, 330, 81–86 CrossRef CAS PubMed.
- O. G. Tovmachenko, C. Graf, D. J. Van Den Heuvel, A. Van Blaaderen and H. C. Gerritsen, Adv. Mater., 2006, 18, 91–95 CrossRef CAS.
- Z. Ren, X. Li, J. Guo, R. Wang, Y. Wu, M. Zhang, C. Li, Q. Han, J. Dong and H. Zheng, Opt. Commun., 2015, 357, 156–160 CrossRef CAS.
- J. Yang, F. Zhang, Y. Chen, S. Qian, P. Hu, W. Li, Y. Deng, Y. Fang, L. Han, M. Luqman and D. Zhao, Chem. Commun., 2011, 47, 11618 RSC.
- Y. Pang, Z. Rong, J. Wang, R. Xiao and S. Wang, Biosens. Bioelectron., 2015, 66, 527–532 CrossRef CAS PubMed.
- Y. Pang, Z. Rong, J. Wang, R. Xiao and S. Wang, Biosens. Bioelectron., 2015, 66, 527–532 CrossRef CAS PubMed.
- N. Sui, L. Wang, F. Xie, F. Liu, H. Xiao, M. Liu and W. W. Yu, Microchim. Acta, 2016, 183, 1563–1570 CrossRef CAS.
- L. Lu, Y. Qian, L. Wang, K. Ma and Y. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 1944–1950 CrossRef CAS PubMed.
- D. D. Xu, Y. L. Deng, C. Y. Li, Y. Lin and H. W. Tang, Biosens. Bioelectron., 2017, 87, 881–887 CrossRef CAS PubMed.
- N. Sui, L. Wang, T. Yan, F. Liu, J. Sui, Y. Jiang, J. Wan, M. Liu and W. W. Yu, Sens. Actuators, B, 2014, 202, 1148–1153 CrossRef CAS.
- J. Sun, J. Ji, Y. Sun, M. H. Abdalhai, Y. Zhang and X. Sun, Biosens. Bioelectron., 2015, 70, 239–245 CrossRef CAS PubMed.
- A. Gopalakrishnan, M. Chirumamilla, F. De Angelis, A. Toma, R. P. Zaccaria and R. Krahne, ACS Nano, 2014, 8, 7986–7994 CrossRef CAS PubMed.
- V. V. Thacker, L. O. Herrmann, D. O. Sigle, T. Zhang, T. Liedl, J. J. Baumberg and U. F. Keyser, Nat. Commun., 2014, 5, 3448 CrossRef PubMed.
- S. Sun, M. Li, Q. Du, C. E. Png and P. Bai, J. Phys. Chem. C, 2017, 121, 12871–12884 CrossRef CAS.
- M. Chirumamilla, A. Toma, A. Gopalakrishnan, G. Das, R. P. Zaccaria, R. Krahne, E. Rondanina, M. Leoncini, C. Liberale, F. De Angelis and E. Di Fabrizio, Adv. Mater., 2014, 26, 2353–2358 CrossRef CAS PubMed.
- B. Tang, J. Wang, J. A. Hutchison, L. Ma, N. Zhang, H. Guo, Z. Hu, M. Li and Y. Zhao, ACS Nano, 2016, 10, 871–879 CrossRef CAS PubMed.
- N. Banaei, A. Foley, J. M. Houghton, Y. Sun and B. Kim, Nanotechnology, 2017, 28, 0–11 CrossRef PubMed.
- A. L. Antaris, H. Chen, S. Diao, Z. Ma, Z. Zhang, S. Zhu, J. Wang, A. X. Lozano, Q. Fan, L. Chew, M. Zhu, K. Cheng, X. Hong, H. Dai and Z. Cheng, Nat. Commun., 2017, 8, 1–11 CrossRef PubMed.
-
P. Wang, Y. Fan, L. Lu, L. Liu, L. Fan, M. Zhao, Y. Xie, C. Xu and F. Zhang, Nat. Commun., 2018, 9, 2898 Search PubMed.
-
Y. Fan, P. Wang, Y. Lu, R. Wang, L. Zhou, X. Zheng, X. Li, J. A. Piper and F. Zhang, Nat. Nanotechnol., 2018, DOI:10.1038/s41565-018-0221-0.
- F. Xie, A. Centeno, M. R. Ryan, D. J. Riley and N. M. Alford, J. Mater. Chem. B, 2013, 1, 536–543 RSC.
- J. Zhang, Y. Fu, M. H. Chowdhury and J. R. Lakowicz, Nano Lett., 2007, 7, 2101–2107 CrossRef CAS PubMed.
- T. D. Corrigan, S. Guo, R. J. Phaneuf and H. Szmacinski, J. Fluoresc., 2005, 15, 777–784 CrossRef CAS PubMed.
- G. Zhang and D. Wang, Chem. – Asian J., 2009, 4, 236–245 CrossRef CAS PubMed.
- X. Wang, C. J. Summers and Z. L. Wang, Nano Lett., 2004, 4, 423–426 CrossRef CAS PubMed.
- J. Aizpurua, P. Hanarp, D. S. Sutherland, M. Käll, G. W. Bryant and F. J. García de Abajo, Phys. Rev. Lett., 2003, 90, 4 CrossRef PubMed.
- X. Huang, D. Ratchford, P. E. Pehrsson and J. Yeom, Nanotechnology, 2016, 27, 295302 CrossRef PubMed.
- Z. A. R. Jawad, I. G. Theodorou, L. R. Jiao and F. Xie, Sci. Rep., 2017, 7, 14309 CrossRef PubMed.
- Z. Mei and L. Tang, Anal. Chem., 2017, 89, 633–639 CrossRef CAS PubMed.
- W. Niu, Y. A. A. Chua, W. Zhang, H. Huang and X. Lu, J. Am. Chem. Soc., 2015, 137, 10460–10463 CrossRef CAS PubMed.
- Q. Su, X. Ma, J. Dong, C. Jiang and W. Qian, ACS Appl. Mater. Interfaces, 2011, 3, 1873–1879 CrossRef CAS PubMed.
- X. Li, L. Xing, K. Zheng, P. Wei, L. Du, M. Shen and X. Shi, ACS Appl. Mater. Interfaces, 2017, 9, 5817–5827 CrossRef CAS PubMed.
- Y. Il Park, H. Im, R. Weissleder and H. Lee, Bioconjugate Chem., 2015, 26, 1470–1474 CrossRef PubMed.
- M. F. Elshal and J. P. McCoy, Methods, 2006, 38, 317–323 CrossRef CAS PubMed.
- M. F. Templin, D. Stoll, M. Schrenk, P. C. Traub, C. F. Vöhringer and T. O. Joos, Drug Discovery Today, 2002, 7, 815–822 CrossRef CAS PubMed.
- M. Schena, D. Shalon, R. W. Davis and P. O. Brown, Adv. Sci., 1995, 270, 467–470 CAS.
- S. Spindel and K. E. Sapsford, Sensors, 2014, 14, 22313–22341 CrossRef CAS PubMed.
- J. Yao, M. Yang and Y. Duan, Chem. Rev., 2014, 114, 6130–6178 CrossRef CAS PubMed.
- A. Rapkiewicz, V. Espina, J. A. Zujewski, P. F. Lebowitz, A. Filie, J. Wulfkuhle, K. Camphausen, E. F. Petricoin, L. A. Liotta and A. Abati, Cancer, 2007, 111, 173–184 CrossRef CAS PubMed.
- Y. Wang, H. Li and D. Xu, Anal. Chim. Acta, 2016, 905, 149–155 CrossRef CAS PubMed.
- S. M. Tabakman, L. Lau, J. T. Robinson, J. Price, S. P. Sherlock, H. Wang, B. Zhang, Z. Chen, S. Tangsombatvisit, J. A. Jarrell, P. J. Utz and H. Dai, Nat. Commun., 2011, 2, 446 CrossRef PubMed.
- B. Liu, Y. Li, H. Wan, L. Wang, W. Xu, S. Zhu, Y. Liang, B. Zhang, J. Lou, H. Dai and K. Qian, Adv. Funct. Mater., 2016, 26, 7994–8002 CrossRef CAS.
- B. Zhang, R. B. Kumar, H. Dai and B. J. Feldman, Nat. Med., 2014, 20, 948–953 CrossRef CAS PubMed.
- B. Zhang, B. A. Pinsky, J. S. Ananta, S. Zhao, S. Arulkumar, H. Wan, M. K. Sahoo, J. Abeynayake, J. J. Waggoner, C. Hopes, M. Tang and H. Dai, Nat. Med., 2017, 23, 548–550 CrossRef CAS PubMed.
- X. Li, C. Pomares, G. Gonfrier, B. Koh, S. Zhu, M. Gong, J. G. Montoya and H. Dai, J. Clin. Microbiol., 2016, 54, 1726–1733 CrossRef CAS PubMed.
- B. Zhang, J. Yang, Y. Zou, M. Gong, H. Chen, G. Hong, A. L. Antaris, X. Li, C. L. Liu, C. Chen and H. Dai, Chem. Sci., 2014, 5, 4070–4075 RSC.
|
| This journal is © The Royal Society of Chemistry 2018 |
 Open Access Article
Open Access Article ,
Caoimhe
Joyce
,
Caoimhe
Joyce
 and
Fang
Xie
and
Fang
Xie
 *
*