Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rational design of multi-functional gold nanoparticles with controlled biomolecule adsorption: a multi-method approach for in-depth characterization†
Isaac
Ojea-Jiménez‡
*a,
Robin
Capomaccio‡
a,
Inês
Osório
 a,
Dora
Mehn
a,
Dora
Mehn
 a,
Giacomo
Ceccone
a,
Rohanah
Hussain
a,
Giacomo
Ceccone
a,
Rohanah
Hussain
 b,
Giuliano
Siligardi
b,
Giuliano
Siligardi
 b,
Pascal
Colpo
a,
François
Rossi
c,
Douglas
Gilliland
a and
Luigi
Calzolai
b,
Pascal
Colpo
a,
François
Rossi
c,
Douglas
Gilliland
a and
Luigi
Calzolai
 *a
*a
aEuropean Commission, DG-Joint Research Centre, Via E. Fermi 2749, 21027 Ispra, VA, Italy. E-mail: Isaac.OJEA-JIMENEZ@ec.europa.eu; Luigi.CALZOLAI@ec.europa.eu
bDiamond Light Source, Chilton, Didcot, OX11 0DE, UK
cEuropean Commission, DG-Joint Research Centre, Westerduinweg 3, 1755ZG Petten, Netherlands
First published on 22nd May 2018
Abstract
Multi-functionalized nanoparticles are of great interest in biotechnology and biomedicine, especially for diagnostic and therapeutic purposes. However, at the moment the characterization of complex, multi-functional nanoparticles is still challenging and this hampers the development of advanced nanomaterials for biological applications. In this work, we have designed a model system consisting of gold nanoparticles functionalized with two differentially-terminated poly(ethylene oxide) ligands, providing both “stealth” properties and protein-binding capabilities to the nanoparticles. We use a combination of techniques (Centrifugal Liquid Sedimentation, Dynamic Light Scattering, Flow Field Flow Fractionation, Transmission Electron Microscopy, and Circular Dichroism) to: (i) monitor and quantify the ratios of ligand molecules per nanoparticle; (ii) determine the effect of coating density on non-specific protein adsorption; (iii) to assess the number and structure of the covalently-bound proteins. This article aims at comparing the complementary outcomes from typical and orthogonal techniques used in nanoparticle characterization by employing a versatile nanoparticle-ligands-biomolecule model system.
1. Introduction
Diagnostic and therapeutic applications of functionalized nanoparticles are highly attractive due to the inherent multivalent nature of their surfaces, since the binding affinity of a nanoparticle (NP) conjugate is enhanced proportionally to the density of its binding sites.1,2 In this context a plethora of nanoparticles have been formulated with multiple drugs and targeting agents in order to simultaneously tackle several points of the biological pathways.3–5 Within the past decades researchers have therefore been struggling to create multifunctional nanoparticles that may contain specific drugs together with different compounds designed to guide the particles towards specific targets for treatment and/or diagnosis. In these multifunctional vectors each component should be present in the appropriate ratio to fulfill its function. This requirement imposes a difficult challenge for the techniques used to characterize these complex systems, but their availability is a pre-requisite for the optimization of the synthesis of multifunctional NPs. Thus, the NP-platform should be able to accommodate multiple binding chemistries sharing the same surface. Besides, the design of the final NP conjugate has important effects on pharmacokinetics, biodistribution and toxicity.6 In addition, small modifications of its physico-chemical nature may have strong influences on the aggregation, degradation, biological interactions and evolution during the full life-cycle.7,8In particular, gold nanoparticles (AuNPs) have been proven to be excellent candidates for such biotechnological and biomedical breakthroughs.9–11 Their tunable plasmonic properties, readily preparation in a wide range of sizes,12 relatively high chemical stability and biocompatibility,13 and well established protocols for the binding of various ligands14 qualify AuNPs as an excellent system for both fundamental studies and applied research. The most straightforward way to prepare biomolecule-functionalized AuNPs is by direct immobilization of thiol residues on the nanoparticle surface. However, in order to ensure sufficient stabilization of the colloidal solution and avoid undesired displacement in complex physiological milieus, a valuable approach consists in the deliberate separation of the functionalization and passivation processes, in which coupling of biomolecules is made to carboxyl-terminated poly(ethylene glycol) (PEG)-coated AuNPs.15,16 The most widely applied strategy is to coat first nanoparticles with linear chains of PEG, which suppresses non-specific protein absorption by blocking protein binding sites and by creating a thermodynamic barrier to protein diffusion.17–19 In addition, PEGylation is also effective at slowing the rate of uptake by the mononuclear phagocyte system and prolonging the circulation in blood.20 In the literature the use of PEGylated nanomaterials has led to varying performance between formulations with noticeable differences in blood half-lives (from more than 24 hours to a few minutes).21
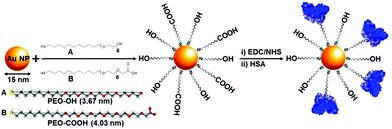
It seems that there is currently an incomplete understanding and connection between the physico-chemical properties of a nanomaterial and its real functionalization state, and as a result it remains unclear how to optimize the synthesis of functionalized nanomaterials for biological applications. Here, we use a AuNPs model system to investigate first the effect of coating density of a hydroxy-terminated poly(ethylene oxide) (PEO-OH) ligand on non-specific protein adsorption and to quantify the ratios of ligand molecules per NP (Fig. 1). Then, we monitor the grafting density of covalently bound proteins per nanoparticle by controlling the ratio between hydroxy and carboxylic acid-terminated ligands (PEO-OH and PEO-COOH, respectively) on the surface of AuNPs (Fig. 1). Proper characterization of NP functionalization is supported by a robust set of complementary characterization techniques (Centrifugal Liquid Sedimentation, CLS; Dynamic Light Scattering, DLS; Asymmetric Flow Field Flow Fractionation, AF4; Transmission Electron Microscopy, TEM; Z-potential; sodium dodecyl sulfate polyacrylamide gel electrophoresis, SDS-PAGE; and Circular Dichroism, CD) after systematic modification of the surface characteristics via fine-tuning of the reaction conditions. By understanding the influence of key parameters such as the density of particles, grafting density and arrangement of PEO-ligands as well as conformation of biomolecules, this study establishes principles for the rational design of multi-functionalized NPs with controlled biomolecule adsorption and/or binding.
2. Results and discussion
A. Determination of the PEO-ligand coating density
We synthesized AuNPs using established protocols,22 and functionalized them with variable amounts of thiolated PEO-OH ligands. We achieved various coverage densities of PEO-OH functionalization by mixing pre-formed AuNPs with variable concentrations of thiolated-PEO-OH, starting from 10% to 100% of the theoretical saturation density. Two additional samples with an excess of ligand (10-fold and 50-fold the theoretical saturating density) were also prepared. The theoretical saturation density was calculated based on electron diffraction studies of monolayers of alkanethiolates on Au(111) surface, which show a calculated area per molecule of 21.4 Å2.23 Based on a calculated surface area of 616 nm2 for 14 nm AuNPs (see below for NP characterization), 100% of ligand binding (or theoretical maximum saturation density) would correspond to approximately 2800 ligand molecules per AuNP.We chose 14 nm as a typical nominal diameter of AuNPs considering the compromise of accommodating a significant biological load while still having a small size.24,25 The choice of thiol-PEO-ligand of approximately 1 kDa was based on: (i) the thiol group at one terminus to provide a strong affinity for gold, (ii) a non-polar alkyl chain to give to the structure the ability to self-assemble into a dense layer that excludes water due to the hydrophobic core, (iii) the polyethylene oxide sequence to enhance colloidal stability in water, and (iv) the ability of PEO ligands to reduce plasma protein binding and to delay opsonization. Coating densities were controlled by varying the PEO-to-NP starting stoichiometry in the reaction mixture in order to obtain samples below saturation (10% saturation, corresponding to 280 molecules per NP; 50% saturation, corresponding to 1400 molecules per NP), at 100% saturation (corresponding to 2800 molecules per NP) and with 10 and 50-fold excess of PEO-ligand above the saturating density (28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 140
000 and 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 PEO molecules per NP, respectively).23 Specific details of the starting reaction mixtures are reported in the materials and methods section.
000 PEO molecules per NP, respectively).23 Specific details of the starting reaction mixtures are reported in the materials and methods section.
After synthesis, the size, and morphology of pristine AuNPs were analyzed by TEM (ESI, Fig. S1†) and the hydrodynamic diameter by batch-mode DLS (ESI, Fig. S2†). The data indicate well monodispersed sample of 13.9 ± 1.1 nm by TEM (Fig. S1†) and a hydrodynamic diameter of 16.1 nm by DLS. While it is not possible to detect a change in size upon functionalization by using TEM, DLS measurements in batch mode indicate an increase in hydrodynamic diameter for the functionalized AuNP (Fig. S2†), up to 22.6 nm for the AuNP-PEO at 100% saturation.
Functionalization of the AuNP surface was additionally proven by a red-shift of the Surface Plasmon Resonance (SPR) band in the absorbance spectra (ESI, Fig. S3†) and a change in surface charge by Z-potential from −36.5 mV (pristine AuNPs) to −19.4 mV (100% of PEO-coverage). The Z-potential changes of the different samples (Table 1) indicate that the increase in the amount of PEO ligand on the AuNP cause a progressive change towards more neutral potential of the AuNPs. While this reduction in Z-potential could lead to a lower colloidal stability, the PEO-ligands provide steric stabilization to the AuNPs.26
| Sample | Starting ratio free PEO-OH/NP | Diametera (nm) | Densityb (g cm−3) | Z-potential (mV) |
|---|---|---|---|---|
| a Hydrodynamic diameter measured by AF4-DLS online. b Apparent density calculated from the AF4-DLS-CLS combined approach, considering for pristine AuNPs a hydrodynamic diameter of 18.5 nm with a density of 10.9 g cm−3. | ||||
| Pristine AuNP | 0 | 18.5 | 10.9 | −36.5 |
| AuNP-PEO-OH 10% | 280 | 18.5 | 10.9 | −27.9 |
| AuNP-PEO-OH 50% | 1400 | 21.5 | 7.4 | −22.5 |
| AuNP-PEO-OH 100% | 2800 | 22.4 | 6.8 | −19.4 |
| AuNP-PEO-OH 10× excess | 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
23.3 | 6.2 | −20.1 |
| AuNP-PEO-OH 50× excess | 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
22.7 | 6.5 | −19.8 |
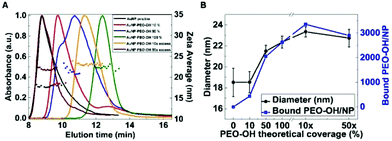
To better understand the amount of PEO-coating in the various AuNP-PEO samples, we used CLS and AF4 coupled to DLS (AF4-DLS), two separative characterization techniques, to determine the nanoparticle size distribution. Using CLS, the time needed by each particle to reach the detector under the centrifugal field is a function of the hydrodynamic diameter and the density of the particle. Fig. S4† shows that the time to reach the detector gradually increases for the various AuNP-PEO complexes when the amount of PEO-ligand increases. AuNP-PEO complexes with higher PEO-coating densities have larger hydrodynamic size (see Table 1), thus the increase in sedimentation time in CLS could only be explained by a decrease in particle density. In previous studies, we have used AF4-DLS to measure the accurate value of the hydrodynamic diameter of the various AuNPs complexes.22 The hydrodynamic values obtained with AF4-DLS are more accurate than those obtained by DLS in batch-mode because the size-separation step performed by AF4 ensures that online DLS measures a monodispersed sample. Fig. 2 shows the AF4-DLS results for the various AuNP samples at increasing PEO coverage densities. Results indicate that for the majority of samples an increase in exit time from the AF4 channel corresponds to an increase in size (from 18.0 nm for pristine AuNPs to 22.4 nm at 100% PEO coverage). The samples with 10× and 50× PEO excess have a size of 23.3 nm and 22.7 nm, respectively, but give somewhat anomalous AF4 fractograms. Such anomalies have been documented before by our (and others) previous work,27 and it can be explained by the presence of the large excess of ligand in the sample that could interact with the AF4 semipermeable membrane, changing its properties and causing an anomalous AF4 separation process. These results confirm our previous conclusions27 that sizing using AF4 exit time alone should be used with extreme care and that the AF4-DLS combination is a much more robust method for determination of particle size distributions.
We have previously shown that combining hydrodynamic diameters obtained by AF4-DLS with the CLS data allows determining the density of AuNP-protein complexes.22 In this case the same approach allowed us to determine the densities of the AuNP-PEO complexes, which showed a consistent decrease when the coverage of PEO-ligand on the surface of AuNPs increased, starting from 10.9 g cm−3 for pristine AuNPs and lowering to 6.8 g cm−3 for 100% coverage (Table 1). Using the density of the AuNP-PEO complexes, together with their hydrodynamic diameter and the diameter of the AuNP core, it is possible to determine the mass of the organic layer surrounding the metallic core of the complex and estimate the average number of PEO-ligand molecules present in each NP (Fig. 2B), corresponding to around 470, 2080, 2660, 3390 and 2930 PEO-OH molecules for the different AuNP-PEO samples.
As it can be seen in Table 1, the calculated number of PEO-ligands per each NP correlates well with the theoretical values and it indicates that at saturation there are approximately 3000 PEO-OH molecules on the surface of each NP. However, in some cases such as for 10% PEO coverage, the experimental error is high, probably due to the fact that there are still citrate molecules present on the NP surface and thus they contribute to overestimate the amount of PEO ligand bound to each NP.
B. Effect of PEO-OH ligand coating density on protein adsorption
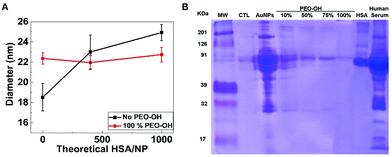
In order to directly study the effect of coating density on protein adsorption, we incubated samples of pristine AuNPs and AuNP-PEO-OH 100% with different amounts (400 and 1000 proteins per NP) of human serum albumin (HSA), then separated and measured the size of the samples with AF4-DLS (Fig. 3A). Non-coated AuNPs adsorb HSA proteins increasing the size of the resulting nanoparticle-protein complex. On the contrary, the AuNP-PEO-OH 100% does not show any size increase when incubated even with a large excess of HSA, suggesting that protein does not bind to the functionalized NPs.8To assess the functional properties of AuNP-PEO-OH constructs, we incubated the different samples with human serum plasma and analysed the formation of the protein corona on the AuNP-PEO-OH construct using SDS-PAGE. Fig. 3B indicates that pristine AuNPs adsorbed quite a large amount of proteins of different molecular weight, with human serum albumin, not surprisingly, being the most abundant. The data shows that increasing the density of the PEO-functionalization (10, 50, 75 and 100%) consistently reduces the amount of protein absorbed on the NPs. At 100% functionalization, only a very weak band (probably due to HSA, as it fits with its molecular weight) is barely visible, thus indicating that the AuNP-PEO-OH 100% sample is almost completely shielded from protein binding in plasma serum.
C. Bi-functionalization of AuNPs surfaces with PEO-ligands
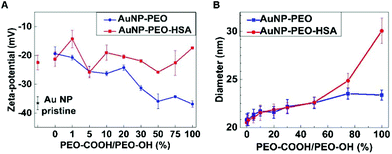
The development of next generation of targeted AuNP-based drug delivery systems requires nanoparticles with multifunctionalization and thus the availability of at least two different functional groups on the surface of AuNPs. To assess the synthetic route and requirements of bi-functionalized AuNPs we introduced a terminal PEO-COOH functionalization in combination with the PEO-OH ligand previously described. In particular, citrate-stabilized AuNPs reaction was carried out with different ratios of PEO-OH and PEO-COOH ligands at 100% total PEO theoretical coverage (Fig. 1).Starting with PEO-OH 100% ligand we systematically introduced increasing amounts of PEO-COOH (1, 5, 10, 20, 30, 50, 75 and 100%), thus leading to a variety of mixed monolayers. Although all the AuNP-PEO samples were prepared at the theoretical saturating density (100% PEO-coverage), AuNP-PEO complexes were separated from the free, unreacted PEO ligands by AF4 (Fig. S5A†). Fig. 4A (blue data plot) shows the Z-potential of the different bi-functional AuNPs. Starting from a value of around −20 mV for the PEO-OH 100% sample the Z-potential decreases with increasing amounts of PEO-COOH ligands to a value of −38 mV for the PEO-COOH 100% sample. The size of the bi-functional complexes (measured by AF4-DLS) increases slightly with increasing amounts of PEO-COOH (from 22.1 nm for PEO-COOH 1%, to 23.9 nm for PEO-COOH 100%). This small increase in hydrodynamic diameter could be explained by a combination of two factors: the longer length of PEO-COOH compared to PEO-OH (see Fig. 1) and the more negative total surface charge for PEO-COOH (Fig. 4A) leading to an increase of the electrodynamic layer thickness.
D. Covalent attachment of HSA to PEO-bifunctionalized AuNP complexes
The availability of bi-functional AuNPs containing PEO-COOH groups should allow the controlled binding of proteins and polypeptides. The coupling of proteins to inorganic colloidal NPs is quite challenging because one needs to consider not only the several chemical parameters that affect the kinetics of the reaction but also the stability of the final NP-protein complex. In our case, the presence of terminal –COOH groups allows using the well-established EDC/NHS chemistry. In fact, EDC/NHS coupling is one of the most commonly used strategies due to the high solubility of the reagents in water and the easy removal of byproducts. [1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride] (EDC) catalyzes the formation of amide bonds between carboxyl and amine groups, while (N-hydroxysulfosuccinimide) (NHS) is used to increase the stability of active intermediates via the formation of ester functional groups with carboxylates. Several reports have demonstrated the successful coupling of biomolecules to colloidal AuNPs in water using EDC/NHS cross-linking reactions.15,16,28Fig. S5B† shows selected parts of the AF4 fractograms of AuNP-PEO samples with different ratios of PEO-COOH/PEO-OH ligands (from 0% to 100%), followed by pre-activation with EDC/NHS, and subsequent coupling to HSA protein. On one hand, it is apparent that the size of the particles increases when the number of terminal COOH-groups become more abundant, as longer elution times correspond to larger particle diameters and from the hydrodynamic diameters measured by AF4-DLS (Fig. 4B, blue data plot). On the other hand, the AF4 fractograms indicates that multiple cross-linking between proteins and particles was mainly avoided during the coupling reaction. Besides, any remaining HSA protein that has not coupled to the NHS-activated particles appears as a separate peak at an elution time ∼10 min (data not shown). Fig. 4B (red data dots) shows the size (measured by AF4-DLS, see also Table S1†) of the AuNP-PEO-COOH-HSA complexes. The results show an increase in size from 23.4 nm to 30.0 nm in the case of HSA covalently bound to AuNP-PEO-COOH 100%, while the increase in size is smaller or not significant for AuNP-PEO with lower amounts of available COOH groups.
We also measured the Z-potential before and after covalent attachment of HSA to AuNP-PEO samples with different ratios of PEO-COOH/PEO-OH ligands (from 0% to 100% of PEO-COOH). The Z-potential of the bi-functional AuNPs changes from −19.4 mV for the PEO-OH 100% to −36.9 mV for the PEO-COOH 100%, suggesting an increase in the colloidal stability of the AuNP for PEO-COOH 100% compared to PEO-OH (Fig. 4A, blue plot). In contrast, after EDC/NHS pre-activation, reaction with HSA molecules, and proper purification steps the Z-potential showed only minimal variations with values fluctuating around −21.1 ± 3.6 mV for the bi-functional AuNP-PEO-HSA complexes (Fig. 4A, red plot).
The average number of proteins present in each AuNP-PEO-HSA complex can be estimated from the mass of the protein layer surrounding the AuNP-PEO complex, which was calculated combining the densities of the AuNP-PEO and AuNP-PEO-HSA complexes together with their corresponding hydrodynamic diameters (ESI, Table S1†). In the case of the maximal loading capacity (i.e. for AuNP-PEO-COOH 100%), we can estimate approximately 50 HSA molecules per NP. This value compares well with a maximum predicted number of 54 HSA protein molecules based on geometrical considerations and the area occupied by a single HSA protein in its native conformation. The experimental number of 50 HSA proteins covalently-bound to AuNP-PEO is significantly larger than the 20 HSA proteins adsorbed on pristine AuNP that we measured in our previous study.22 Such a larger number of bounded proteins per particle is explained by the increase in the hydrodynamic diameter of the NPs (i.e. AuNP-PEO of 23.3 nm in the present study vs. pristine AuNPs of 18.5 nm in our previous work), leading to a larger surface area available for protein binding.
AuNPs of around 10–30 nm can cause endothelial leakiness (the so called “nanoparticle induced endothelial leakiness”, NanoEL36). As density seems to play an important role in inducing endothelial permeability37 (with denser particle inducing higher leakiness), the quite substantial density decrease for AuNP-PEO-COOH 100% covalently bound to HSA (from 10.9 g cm−3 for pristine particles to 2.6 g cm−3) could determine a reduction of the NanoEL effect, even if the critical density for NanoEL effect in silica nanoparticles was found to be around 1.6 g cm−3.
TEM analysis of bi-functional AuNP-PEO samples was also performed before and after the covalent attachment of HSA and it is shown in Fig. 5. Despite the sample preparation step requiring the drying of specimens and observation under high vacuum conditions, the increase of interparticle distance in the case of AuNP-PEO-COOH 100% coating becomes clearly visible. The interparticle distance is enhanced in all cases after the coupling of HSA. Analysis of the averaged minimal interparticle distance in a hexagonal packing arrangement of the particles resulted in values of 1.5, 3.9 and 4.4 nm before HSA coupling and of 1.7, 3.5 and 7.1 nm after HSA coupling, for bi-functional AuNPs with PEO-COOH/PEO-OH ligand ratios of 0%, 50% and 100% respectively.
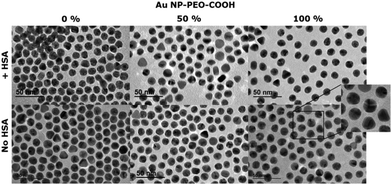 | ||
| Fig. 5 TEM analysis of bi-functional AuNP-PEO complexes at different ratios of PEO-COOH/PEO-OH ligands (0%, 50% and 100%) before and after the covalent attachment of HSA molecules. | ||
The AuNP-PEO-COOH system could also be used for conjugation to targeting antibodies. Compared to DNA-antibody nanostructures that have been recently developed for cancer cell targeting38 and for detection of pathogens39 the AuNP-PEO-COOH-antibody system would be less stoichiometrically and spatially controlled, while it could probably achieve the binding of more antibody molecules per nanoparticle.
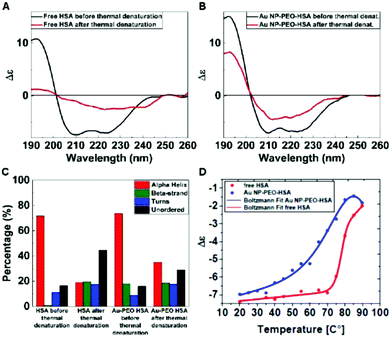
By means of Circular Dichroism, we were able to study the changes in the secondary structure of the HSA molecules after the covalent attachment to AuNP-PEO-COOH 100% particles. Free HSA molecules were removed from AuNP-PEO-HSA complexes by repeated centrifugation steps, and purification was monitored by acquiring the CD spectra of either the supernatants and of the resuspended pellets (ESI, Fig. S10†). These experiments confirmed that after 2 purification steps the unreacted HSA molecules were removed. Synchrotron radiation circular dichroism (SRCD) analysis was used to monitor the thermal unfolding of free HSA and HSA covalently bound to the AuNP-PEO-COOH particles. The CD spectrum of AuNP-PEO-HSA is very similar to that of free HSA (Fig. 6A and B). This qualitative analysis is confirmed by a more detailed analysis: the deconvolution of the CD spectra show that free HSA contains around 72% of α-helical secondary structure, while HSA covalently bound to AuNP-PEO-COOH is 74% α-helical (Fig. 6C). This confirms that the overall secondary structure of HSA covalently bound to the particles is very similar to that of the free HSA. Using SRCD we studied the thermal unfolding of both samples, by following the change in intensity of the α-helical peak at 209 nm. Fig. 6D shows the melting unfolding curves for the two samples. Fitting the data to the equation below:
| y = A2 + (A1 − A2)/(1 + exp(x − x0)/dx) |
3. Experimental
A. Materials and methods
Gold(III) chloride trihydrate (>99.9%), trisodium citrate dihydrate (>99.9), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS) and deuterium oxide (D2O, >99.9 atom% D) were purchased from Sigma-Aldrich and used as received without further purification. Ligands (2-(2-{{2-[2-(1-mercaptoundec-11-yloxy)-ethoxy]-ethoxy}-ethoxy)-ethoxy}-ethoxy)-ethanol (PEO-OH) and 2-(2-[2-(11-mercapto-undecyloxy)-ethoxy]-ethoxy)-ethoxy-ethoxy-ethoxy-ethoxy-acetic acid (PEO-COOH) were purchased from ProChimia and kept under N2 and in the freezer at −20 °C. Human serum albumin (fatty acid free and globulin free, HSA) was purchased from Sigma Aldrich and used at the concentration of 2 mg mL−1 in phosphate buffer (1 mM, pH 7.4). Human plasma was purchased from Sigma-Aldrich and stored in aliquots at −20 °C.NPs were visualized using a transmission electron microscope TEM (JEOL 2100, Japan) at an accelerating voltage of 200 kV. The samples were prepared by placing a drop (4 μL) onto ultrathin Formvar-coated 200-mesh copper grids (Tedpella Inc.) and left to dry in air at 4 °C. For each sample, the size of at least 100 particles was measured to obtain the average and the size distribution. Digital images were analyzed with the ImageJ software and a custom macro performing smoothing (3 × 3 or 5 × 5 median filter), manual global threshold and automatic particle analysis provided by the ImageJ. The macro can be downloaded from http://code.google.com/p/psa-macro. The circularity filter of 0.8 was used to exclude agglomerates formed during drying. UV-vis absorption spectra were recorded with an Evolution 300 UV-Vis Spectrophotometer (Thermo Scientific, USA) at room temperature. DLS and Z-potential measurements were obtained using a Zetasizer Nano ZS instrument (Malvern Instruments, UK). Hydrodynamic diameters were calculated using the internal software analysis from the DLS intensity-weighted particle size distribution. The Z-potential was measured for independent samples just after adjusting the pH of the dispersions with either 1 mM NaOH or 1 mM HCl and at a total NaCl concentration of 1 mM to keep the conductivity at approximately 1 mS cm−1. The size distribution of the NPs was determined by CLS in a sucrose density gradient using a CPS Disc Centrifuge model DC24000UHR (CPS Instruments Europe, Netherlands). The instrument was operated in line-start mode at a disc rotation speeed of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm using a sucrose gradient (8–24% w/w) capped with dodecane to prevent solvent evaporation. Each reference was precedented by a calibration step done using an aqueous reference standard of 239 nm diameter polyvinyl chloride (PVC) spheres. All samples were vortexed for 30 seconds prior to injection.
000 rpm using a sucrose gradient (8–24% w/w) capped with dodecane to prevent solvent evaporation. Each reference was precedented by a calibration step done using an aqueous reference standard of 239 nm diameter polyvinyl chloride (PVC) spheres. All samples were vortexed for 30 seconds prior to injection.
B. AF4 purification
The asymmetrical flow field-flow fractionation system used was a Postnova AF2000 (Postnova Analytics, Landsberg, Germany). 1 mM Phospate buffer pH 7.4 has been used as eluent. The channel was set up with a 350 μm spacer and with a regenerated membrane with a 10 kDa cut-off. Detector flow was set at 0.5 mL min−1. The injection step has been done during 5 min at 0.25 mL per minute. The elution time came after a transition step of 1 minute with a cross flow of 2 mL min−1. The UV detector is a PN3212 spectrophotometer (Postnova Analytics, Landsberg, Germany) tuned at 525 nm and 500 nm. The fluorescence detector is a PN3412 fluorescence detector (Postnova Analytics, Landsberg, Germany) with an excitation wavelength tuned at 280 nm and emission wavelength tuned at 340 nm and the MALS is a seven-angle multi-angle light scattering detector (PN 3070) from Postnova (Postnova Analytics, Landsberg, Germany). DLS from Malvern Zetasizer was coupled online with AF4. 5 runs have been performed and 700 μL of each peak have been collected and mixed together. Data analysis was performed by filtering out AF4-DLS data points with polydispersity values greater than 0.1.C. Circular dichroism
Circular dichroism spectra were collected using a Chirascan CD (Applied Biophysics) instrument. A 1 cm pathlength low volume quartz cell was used to analyze low concentration of AuNP-PEO-HSA complexes. For each sample 4 scans were acquired and averaged according to the quality of the spectra and the concentration of protein. The CD spectra of free HSA and AuNP-PEO-HSA samples were baseline-corrected by substracting the CD spectrum of the buffer (for free HSA) or the CD spectrum of AuNPs at the same concentration than in AuNP-PEO-HSA samples (for AuNP-PEO-HSA complexes). The baseline-corrected spectra were smoothed with Savitzky–Golay smoothing function of six points. CD spectra were analyzed with the DICHROWEB analysis software using the CONTINLL algorithm and the protein dataset 7 (http://dichroweb.cryst.bbk.ac.uk). SRCD experiments were performed using a nitrogen flushed module B end-station spectrophotometer at the B23 synchrotron radiation CD beamline at the Diamond Light Source (Oxfordshire, UK).30–33 Measurements were carried out at 23 °C unless otherwise stated with wavelength set between 185 and 260 nm, 0.5 mm slit equivalent to 1 nm bandwidth, increment of 1 nm, and interval time of 1 s. The cell used has a 10 cm pathlength and a volume of 820 μl. Data were collected on protein samples as repeated scans, without any averaging over 30 scans. Protein secondary structure was estimated with the B23 software, CDAPPS,34 available for in-house and remote processing CDAPPS using the CONTIN-LL algorithm and the dataset CLSTR.35 All samples for CD and SRCD were obtained by centrifuge separation (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf, 20 min, 4 °C) of two fractions: the free, unbound HSA (supernatant) and the AuNP-PEO-HSA complex (pellet, followed by resuspension in the original volume of milliQ water). Thermal unfolding studies were carried out with free HSA (0.1 mg mL−1) diluted in milliQ water and AuNP-PEO-HSA complex after 2 centrifuge separation steps. Spectra were recorded every 5 °C over a 20–90 °C temperature range with 5 minutes equilibration time for each temperature. The reversibility of changes was monitored by recording the spectrum at 20 °C after cooling the samples from 90 °C in 20 min.
000 rcf, 20 min, 4 °C) of two fractions: the free, unbound HSA (supernatant) and the AuNP-PEO-HSA complex (pellet, followed by resuspension in the original volume of milliQ water). Thermal unfolding studies were carried out with free HSA (0.1 mg mL−1) diluted in milliQ water and AuNP-PEO-HSA complex after 2 centrifuge separation steps. Spectra were recorded every 5 °C over a 20–90 °C temperature range with 5 minutes equilibration time for each temperature. The reversibility of changes was monitored by recording the spectrum at 20 °C after cooling the samples from 90 °C in 20 min.
D. Synthesis of AuNPs of 14 nm
AuNPs of 14 nm nominal diameter were synthesized using a modified Turkevich method using a specialized microwave apparatus (Discover S by CEM corporation). Briefly, 5 mL of aqueous gold(III) chloride trihydrate (10 mM) were added to 95 mL of milliQ-water in a single necked 100 mL round bottom flask equipped with magnetic stirrer and a glass distilling column. The flask was mounted in the microwave reactor, heated rapidly (<1 min) to 97 °C while stirring and then hold for 5 min using a maximum microwave power of 150 W. Under vigorous stirring, 2.5 mL of sodium citrate (100 mM) were injected and the reaction mixture was maintained at 97 °C for 20 min after which the reaction vessel was rapidly cooled to 60 °C with compressed air and then allowed to cool to room temperature. Z-potential value of the as synthesized AuNPs was −36.5 mV at a pH = 6.5. The approximate concentration was calculated to be 0.5 mM (0.096 mg mL−1) of gold stabilized with 2.4 mM of sodium citrate and at pH = 6.5.E. Mono-functionalization of citrate AuNPs with different percentages of PEO-OH
The AuNPs of 14 nm were functionalized with different percentages of polyethylene oxide (PEO) ligand in order to produce different degrees of coating densities on the surface. Samples were prepared by mixing a volume (either 0.2, 1, 2, 20 and 100 μL to obtain 10%, 50%, 100% saturation and 10 and 50-fold excess of PEO ligand above the saturation density on the AuNP surface, respectively) of a stock solution of PEO-ligand (100 mM) in milliQ H2O with a AuNPs suspension (10 mL) at room temperature under stirring overnight. Successful functionalization was proven by UV-vis spectroscopy and the change in surface charge by Z-potential from −36.5 mV (pristine AuNPs) to −19.4 mV (100% of PEO-coverage). Samples were fully characterized by batch-mode DLS, CPS and AF4-DLS online.F. SDS-PAGE gel electrophoresis
The AuNPs of 14 nm, both pristine and the variously PEO-functionalized NPs were incubated with human serum (Sigma-Aldrich) for 24 h at 37 °C. The NPs mix was centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf, 5 min, 4 °C), the supernatant was carefully removed and the NPs pellet was subsequently washed with 1× PBS (Gibco). This washing procedure was repeated three times. The final pellet was suspended in 20 μL Pierce Lane Marker reducing sample buffer (ThermoFisher) and incubated at 95 °C for 5 min. After a short spin down, the supernatant was loaded in 12% SDS Polyacrylamide gel and run at 110 V, 25 mA in 1× SDS Running Buffer. After electrophoresis, the gel was Coomassie stained.
000 rcf, 5 min, 4 °C), the supernatant was carefully removed and the NPs pellet was subsequently washed with 1× PBS (Gibco). This washing procedure was repeated three times. The final pellet was suspended in 20 μL Pierce Lane Marker reducing sample buffer (ThermoFisher) and incubated at 95 °C for 5 min. After a short spin down, the supernatant was loaded in 12% SDS Polyacrylamide gel and run at 110 V, 25 mA in 1× SDS Running Buffer. After electrophoresis, the gel was Coomassie stained.
G. Bi-functionalization of citrate AuNPs with different ratios of PEO-COOH/PEO-OH ligands (from 0% to 100%)
The degree of coating of the AuNPs was controlled by adding a fixed final ratio of 2 PEO-molecules per total AuNPs surface in order to obtain 100% coverage of the total available surface. Samples were prepared by mixing an aqueous volume of 100 mM PEO-OH (20, 19.8, 19, 18, 16, 14, 10, 5 and 0 μL) with an aqueous volume of 100 mM PEO-COOH (0, 0.2, 1, 2, 4, 6, 10, 15 and 20 μL, respectively) in a glass vial containing milliQ H2O (100 μL), to obtain 0, 1, 5, 10, 20, 30, 50, 75 and 100% of COOH-groups available on the surface of AuNPs, respectively. A suspension of pristine AuNPs (10 mL) was quickly added and the reaction mixture was allowed to stir at RT overnight. Successful functionalization was proven by UV-vis spectroscopy and by the change in surface charge by Z-potential from −19.4 mV (100% PEO-OH coverage) to −36.9 mV (100% PEO-COOH coverage). The pH value of all the samples ranged between 6.0–6.2. All the samples were fully characterized by batch-mode DLS and AF4-DLS online and the change in surface charge determined by Z-potential at a pH value of 6.1.H. Covalent attachment of HSA to PEO-functionalized AuNPs
Freshly prepared aqueous solutions of EDC (0.25 mL, 0.4 M) and NHS (0.25 mL, 0.1 M) were mixed in 5 mL eppendorf tubes and the corresponding AuNP-PEO suspensions with different ratios of PEO-COOH/PEO-OH ligands (from 0% to 100%) (5 mL) were added. Samples were allowed to mix gently for 1 h at 18 °C before being centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf, 10 min, 4 °C) and the resulting pellets were resuspended in the same volume of milliQ H2O. Stock solutions of HSA of 1 and 10 mg mL−1 were prepared with milliQ H2O from the mother solution of HSA at 2 mg mL−1 in phosphate buffer 1 mM pH 7.4. An aliquot of HSA 1 mg mL−1 (131 μL) was added to each of the NHS-activated AuNP-PEO samples and the reaction mixtures were allowed to mix gently overnight. Finally, ethanolamine (6 μL, 100 mmol) was added in order to block any unreacted NHS-active groups. The same procedure was repeated with the stock solution of HSA 10 mg mL−1 and the NHS-activated AuNP-PEO sample containing 100% of COOH-terminal groups. Materials were fully characterized by DLS, TEM, CLS, CD and the change in surface charge determined by Z-potential at a pH value of 6.1.
000 rcf, 10 min, 4 °C) and the resulting pellets were resuspended in the same volume of milliQ H2O. Stock solutions of HSA of 1 and 10 mg mL−1 were prepared with milliQ H2O from the mother solution of HSA at 2 mg mL−1 in phosphate buffer 1 mM pH 7.4. An aliquot of HSA 1 mg mL−1 (131 μL) was added to each of the NHS-activated AuNP-PEO samples and the reaction mixtures were allowed to mix gently overnight. Finally, ethanolamine (6 μL, 100 mmol) was added in order to block any unreacted NHS-active groups. The same procedure was repeated with the stock solution of HSA 10 mg mL−1 and the NHS-activated AuNP-PEO sample containing 100% of COOH-terminal groups. Materials were fully characterized by DLS, TEM, CLS, CD and the change in surface charge determined by Z-potential at a pH value of 6.1.
4. Conclusions
In this work, we employ a simple but robust AuNP-PEO-protein model as a platform to control variations on the NP surface, and systematically interrogate the system with a variety of physico-chemical characterization techniques. On one hand, PEO-coating can reduce protein adsorption and thereby confer a stealth effect, while on the other hand the number of proteins covalently attached can be controlled by the available –COOH groups in a bi-functional mixture of PEO-COOH/PEO-OH on the NP surface. Such detailed control is essential for the development and reproducibility of high quality targeted nanoparticle systems (for either therapy or diagnostic applications) able to reach their intended target while avoiding non-specific absorption of serum proteins.Overall, we have demonstrated that it is possible to reliably characterize multi-functionalized AuNPs by combining the separation ability of AF4 with sizing measurements from online DLS. These findings are strongly supported by complementary characterization techniques, such as batch-mode DLS, Z-potential, CLS, SDS-PAGE, and TEM. The outcome of this detailed study provides important information such as the scope of non-specific protein adsorption or valuable quantification of ratios of ligands or covalently-bound protein molecules per NP. SRCD spectroscopy showed that the secondary structure of the covalently-bound HSA protein was conserved and that it could be partially recovered after thermal unfolding. The rationale behind these experiments focuses on establishing an unambiguous connection between the outcome from physico-chemical characterizations and real composition of engineered nanomaterials, which is of key relevance for the development of multi-functional NP systems for biomedical applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We like to thank Diamond Light Source for beamtime allocation on B23 beamline (SM11373).Notes and references
- J. E. Gestwicki, C. W. Cairo, L. E. Strong, K. A. Oetjen and L. L. Kiessling, J. Am. Chem. Soc., 2002, 124, 14922–14933 CrossRef PubMed.
- R. Weissleder, K. Kelly, E. Y. Sun, T. Shtatland and L. Josephson, Nat. Biotechnol., 2005, 23, 1418–1423 CrossRef PubMed.
- L. Hosta-Rigau, I. Olmedo, J. Arbiol, L. J. Cruz, M. J. Kogan and F. Albericio, Bioconjugate Chem., 2010, 21, 1070–1078 CrossRef PubMed.
- G. F. Paciotti, D. G. I. Kingston and L. Tamarkin, Drug Dev. Res., 2006, 67, 47–54 CrossRef.
- G. F. Paciotti, L. Myer, D. Weinreich, D. Goia, N. Pavel, R. E. McLaughlin and L. Tamarkin, Drug Delivery, 2004, 11, 169–183 CrossRef PubMed.
- P. Szabo and R. Zelko, Curr. Pharm. Des., 2015, 21, 3148–3157 CrossRef PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. W. Chan, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef PubMed.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- R. A. Sperling, P. Rivera Gil, F. Zhang, M. Zanella and W. J. Parak, Chem. Soc. Rev., 2008, 37, 1896–1908 RSC.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, Chem. Soc. Rev., 2012, 41, 2740–2779 RSC.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- E. E. Connor, J. Mwamuka, A. Gole, C. J. Murphy and M. D. Wyatt, Small, 2005, 1, 325–327 CrossRef PubMed.
- A. C. Templeton, W. P. Wuelfing and R. W. Murray, Acc. Chem. Res., 2000, 33, 27–36 CrossRef PubMed.
- D. Bartczak and A. G. Kanaras, Langmuir, 2011, 27, 10119–10123 CrossRef PubMed.
- W. Eck, G. Craig, A. Sigdel, G. Ritter, L. J. Old, L. Tang, M. F. Brennan, P. J. Allen and M. D. Mason, ACS Nano, 2008, 2, 2263–2272 CrossRef PubMed.
- S. I. Jeon, J. H. Lee, J. D. Andrade and P. G. De Gennes, J. Colloid Interface Sci., 1991, 142, 149–158 CrossRef.
- J. Satulovsky, M. A. Carignano and I. Szleifer, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 9037–9041 CrossRef PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Nat. Nanotechnol., 2016, 11, 372–377 CrossRef PubMed.
- N. J. Butcher, G. M. Mortimer and R. F. Minchin, Nat. Nanotechnol., 2016, 11, 310–311 CrossRef PubMed.
- S. D. Perrault, C. Walkey, T. Jennings, H. C. Fischer and W. C. W. Chan, Nano Lett., 2009, 9, 1909–1915 CrossRef PubMed.
- R. Capomaccio, I. Ojea Jimenez, P. Colpo, D. Gilliland, G. Ceccone, F. Rossi and L. Calzolai, Nanoscale, 2015, 7, 17653–17657 RSC.
- H. Sellers, A. Ulman, Y. Shnidman and J. E. Eilers, J. Am. Chem. Soc., 1993, 115, 9389–9401 CrossRef.
- Ž. Krpetić, P. Nativo, I. A. Prior and M. Brust, Small, 2011, 7, 1982–1986 CrossRef PubMed.
- P. Nativo, I. A. Prior and M. Brust, ACS Nano, 2008, 2, 1639–1644 CrossRef PubMed.
- I. Ojea-Jiménez, L. García-Fernández, J. Lorenzo and V. F. Puntes, ACS Nano, 2012, 6, 7692–7702 CrossRef PubMed.
- D. C. António, C. Cascio, D. Gilliland, A. J. A. Nogueira, F. Rossi and L. Calzolai, Biointerphases, 2016, 11, 04B309 CrossRef PubMed.
- C. Rocker, M. Potzl, F. Zhang, W. J. Parak and G. U. Nienhaus, Nat. Nanotechnol., 2009, 4, 577–580 CrossRef PubMed.
- H. D. Young and F. W. Sears, University physics, Addison-Wesley Pub. Co., Reading, Mass., 1992 Search PubMed.
- R. Hussain, T. Javorfi and G. Siligardi, J. Synchrotron Radiat., 2012, 19, 132–135 CrossRef PubMed.
- R. Hussain, T. Jávorfi and G. Siligardi, in Comprehensive Chirality, ed. E. M. Carreira and H. Yamamoto, Elsevier, Amsterdam, 2012, pp. 438–448 Search PubMed.
- T. Jávorfi, R. Hussain, D. Myatt and G. Siligardi, Chirality, 2010, 22, E149–E153 CrossRef PubMed.
- G. Siligardi and R. Hussain, in Structural Proteomics: High-Throughput Methods, ed. R. J. Owens, Springer New York, New York, NY, 2015, pp. 255–276 Search PubMed.
- R. Hussain, K. Benning, T. Javorfi, E. Longo, T. R. Rudd, B. Pulford and G. Siligardi, J. Synchrotron Radiat., 2015, 22, 465–468 CrossRef PubMed.
- S. W. Provencher and J. Gloeckner, Biochemistry, 1981, 20, 33–37 CrossRef PubMed.
- M. I. Setyawati, C. Y. Tay, B. H. Bay and D. T. Leong, ACS Nano, 2017, 11, 5020–5030 CrossRef PubMed.
- C.Y. Tay, M. I. Setyawati and D.T. Leong, ACS Nano, 2017, 11, 2764–2772 CrossRef PubMed.
- M. I. Setyawati, R. V. Kutty and D. T. Leong, Small, 2016, 12, 5601–5611 CrossRef PubMed.
- J. Wang, M. C. Leong, E. Z. Leong, W. S. Kuan and D. T. Leong, Anal. Chem., 2017, 89, 6900–6906 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8nr00973b |
| ‡ Both authors have equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2018 |