Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ecology and evolution of metabolic cross-feeding interactions in bacteria†
Glen
D'Souza
 ab,
Shraddha
Shitut
ab,
Shraddha
Shitut
 ce,
Daniel
Preussger
c,
Ghada
Yousif
cde,
Silvio
Waschina
ce,
Daniel
Preussger
c,
Ghada
Yousif
cde,
Silvio
Waschina
 f and
Christian
Kost
f and
Christian
Kost
 *ce
*ce
aDepartment of Environmental Systems Sciences, ETH-Zürich, Zürich, Switzerland
bDepartment of Environmental Microbiology, Eawag: Swiss Federal Institute for Aquatic Sciences, Dübendorf, Switzerland
cExperimental Ecology and Evolution Research Group, Department of Bioorganic Chemistry, Max Planck Institute for Chemical Ecology, Jena, Germany. E-mail: christiankost@gmail.com
dDepartment of Botany and Microbiology, Faculty of Science, Beni-Suef University, Beni-Suef, Egypt
eDepartment of Ecology, School of Biology/Chemistry, Osnabrück University, Osnabrück, Germany
fResearch Group Medical Systems Biology, Institute for Experimental Medicine, Christian-Albrechts-University Kiel, Kiel, Germany
First published on 25th May 2018
Abstract
Literature covered: early 2000s to late 2017
Bacteria frequently exchange metabolites with other micro- and macro-organisms. In these often obligate cross-feeding interactions, primary metabolites such as vitamins, amino acids, nucleotides, or growth factors are exchanged. The widespread distribution of this type of metabolic interactions, however, is at odds with evolutionary theory: why should an organism invest costly resources to benefit other individuals rather than using these metabolites to maximize its own fitness? Recent empirical work has shown that bacterial genotypes can significantly benefit from trading metabolites with other bacteria relative to cells not engaging in such interactions. Here, we will provide a comprehensive overview over the ecological factors and evolutionary mechanisms that have been identified to explain the evolution and maintenance of metabolic mutualisms among microorganisms. Furthermore, we will highlight general principles that underlie the adaptive evolution of interconnected microbial metabolic networks as well as the evolutionary consequences that result for cells living in such communities.
1 Introduction
Bacteria are amongst the most ancient life forms on our planet.1,2 Even, the last common universal ancestor (LUCA) has been suggested to strongly resemble bacteria that dwell in extreme environments.3,4 During their evolutionary history of about 3.2 billion years, bacteria managed to colonize virtually every conceivable habitat on earth including air, soil, water, as well as other organisms such as animals and plants.5 Due to their widespread distribution and high abundance, bacteria play significant ecological roles in driving global biogeochemical cycles,6 determining homeostasis of the biosphere,7 and controlling the development, behaviour, and health of multicellular organisms.8In nature, bacteria usually exist within taxonomically and genotypically diverse communities.9–11 In these assemblages, bacteria compete for a wide variety of limiting resources such as favourable living spaces, nutrients, and minerals. Moreover, due to their metabolic activities, bacteria transform the environments they live in, thus drastically influencing the growth and metabolism of other co-occurring organisms.12 Strong selection pressures resulting from both of these factors have not only given rise to a plethora of ecological interactions, but also different bacterial strategies to survive and reproduce under these conditions.10 Accordingly, a large proportion of a bacterial cell's genetic material (between 17 and 42%) can encode traits that are involved in mediating ecological interactions.13
For heuristic purposes, ecological interactions between two individuals are typically classified based on the net fitness effects that result for the organisms involved. The typological spectrum of interactions resulting from this classification scheme ranges from antagonistic (i.e. negative fitness consequences) over neutral (i.e. no interaction) to beneficial interactions (i.e. positive fitness consequences).10 Examples of antagonistic behaviours displayed by bacteria include the active secretion of toxins such as colicins or antibiotics that kill or inhibit the growth of other bacteria,14,15 thus providing the toxin-producing bacteria with a competitive advantage. Evolutionary theory predicts that natural selection should favour such strategies that selfishly enhance the fitness of one organism at the expense of another one.16 Indeed, a large body of work has demonstrated the prevalence of antagonistic interactions in natural microbial communities.10,16,17
However, in recent years, awareness has grown that bacteria also show a range of cooperative behaviours, in which one individual helps another one at a cost to itself. A good example for this is so-called ‘public goods’. These are metabolites that are costly to produce, yet are released into the extracellular environment. As a consequence, these public goods do not only benefit the producing cell, but also other cells in the local group or population. Examples include antibiotic-degrading enzymes,18 motility-enhancing biosurfactants,19 matrix components for biofilms,20 or iron-scavenging molecules.21 Why would cells invest resources into behaviours that can be easily exploited by individuals that reap the benefits without bearing the costs for producing the public good? In most of the abovementioned cases, the individual producing the public good and the beneficiaries are genealogically related. Thus, by helping its relatives, the cooperative individual can increase the chance that its own genes are indirectly propagated. This so-called ‘kin-selection’ can explain altruistic cooperative behaviours among closely related individuals.22
The situation, however, is different for synergistic interactions that involve unrelated individuals or different species that reciprocally exchange metabolites such as sugars, growth factors, or amino acids with each other.23 A number of recent studies have suggested that these types of synergistic interactions might actually be common in the prokaryotic world.24–27 In many of these cases, the interactions are also obligatory for the individuals involved, meaning they can only exist when the required metabolite is externally supplied, for example by another bacterium.25,26 This type of metabolic interactions begs an evolutionary explanation: why should a bacterium give up its metabolic autonomy and rather rely on other organisms to provide essential metabolites? Moreover, why would a bacterial cell produce metabolites to benefit other, potentially unrelated individuals and not use these resources to maximize its own fitness?
In this article, we address these questions. By particularly focussing on metabolic interactions between two or more bacterial partners, we aim at developing a conceptual framework that allows not only to classify different types of metabolic interactions, but also to explain the evolution and maintenance of these relationships. In addition, we analyse how common metabolic cross-feeding interactions are in nature and what evolutionary consequences result for the organisms involved. The comprehensive picture that emerges from this analysis may provide an orientation to scientists that are new to this interesting field of study and identify avenues for future research.
2 Metabolic cross-feeding interactions
2.1 Historical account
A first and important step in understanding the origin of metabolic exchange in bacteria is to obtain a historical perspective on the discovery of this phenomenon. Early studies on what is now known as cross-feeding often discuss the phenomenon in the context of symbiosis.28,29 These studies mainly focussed on microbial interactions that impact plant growth (e.g. root nodule bacteria,30,31 mycorrhiza32) or play important roles for the fermentation of dairy products (i.e. lactic acid bacteria33–35). Back in 1887, Carl Garrè, a Swiss surgeon, was one of the first to mention that “one organism prepares food for another organism by changing the medium on which it grows”.29 Later in 1892, the British botanist Marshall Ward stumbled upon cross-feeding while trying to unravel the mystery of the Ginger-beer Plant. The substance in question is used to ferment ginger beer, a non-alcoholic, naturally sweetened beverage, from saccharine and ginger. Ward found out that this plant was, in effect, a symbiotic association between a yeast and bacteria that formed solid, semi-translucent, lumpy masses. More importantly, he found that an exchange of metabolites between both partners was an integral part of the fermentation process.36 Around this time, such mixed cultures of microbes were referred to as microbial associations.29 In 1897, Wilhelm Pfeffer, a German botanist and plant physiologist, introduced the terms conjunctive and disjunctive symbiosis to highlight the dependency of either partners for growth.37 Marshall Ward also proposed the use of terms like antibiosis and metabiosis to distinguish between negative and positive effects that result from an interaction for the partners involved.38The term ‘cross-feeding’ was coined by Hermann Reinheimer in 1921 – a British biologist who was interested in the evolutionary significance of cooperative symbiotic interactions.39 Reinheimer suggested two terms to differentiate the source of food or metabolite, namely ‘in-feeding’ for within-kingdom exchange and cross-feeding for between-kingdom exchange. This distinction, however, was not adopted by the scientific community at large. Instead, the term cross-feeding was subsequently used to describe interactions that involved an exchange of molecules and, thus, enhanced growth. Interestingly at this time, cross-feeding between auxotrophic strains was also used as a methodological tool to elucidate biochemical pathways.40,41 Notable work was done by Veikko Nurmikko, a Finnish microbiologist, who introduced the use of dialysis chambers to separate two auxotrophic strains of lactic acid bacteria such that they exchange metabolites via diffusion.42 In subsequent years, metabolic cross-feeding interactions were used to study the concerted degradation of herbicides43–45 or fatty acids,46 the enhanced production of amino acids,47 and to characterize auxotrophic strains.48–50 Interestingly, until today, mixed cultures of natural bacterial isolates are employed to identify novel pathways for the degradation of complex hydrocarbons like crude oil51–53 or toxic industrial dyes.54,55
Towards the end of the 1980's, microbiologists began to study bacterial interactions from an ecological and evolutionary perspective. Among them, Julian Adams and co-workers initiated long-term chemostat cultures of Escherichia coli in glucose-limited conditions.56,57 An intriguing observation from their continuous cultivation experiments was that bacterial strains repeatedly evolved mutations in the acetyl CoA synthetase enzyme. This mutation allowed the uptake of exogenous acetate, resulting in a stable coexistence of these mutants with wild type strains that secreted acetate as a by-product of glucose metabolism.58 Several subsequent studies analysed similar cases of diversifying selection in initially clonal populations that resulted from the evolution of metabolic cross-feeding interactions.59–62
In recent years, the phenomenon of metabolite exchange has gained momentum with an increasing number of working groups studying this type of ecological interactions from different perspectives and using different methodological approaches. However, depending on their research focus and scientific background, a number of different terms are used to describe qualitatively similar interactions. For example, terms like syntrophy,63 synergism,27 symbiosis,28 mutualism,28 or obligately mutualistic metabolism63 are often used interchangeably. Each of these terms describes a reciprocal exchange of molecules, yet in specific contexts. For instance, syntrophy denotes cases where the metabolism of two organisms are energetically coupled,63 while synergism simply refers to interactions from which both interacting partners benefit.27
2.2 Classification of cross-feeding interactions
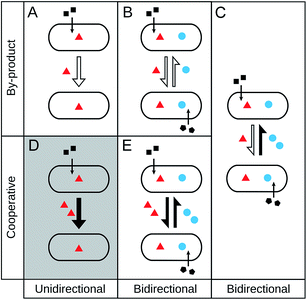
Given that a number of different terminologies are used to describe qualitatively similar ecological interactions, we begin by providing an unambiguous and comprehensive classification scheme to name different types of metabolic cross-feeding interactions. Due to the focus of this review, we only discuss interactions that involve an exchange of primary metabolites. In reality, however, bacteria often trade metabolites against other beneficial services such as detoxification of toxic metabolites or protection from predators.10 Even though we do not treat these interactions in detail, a similar nomenclature and conceptual logic can be applied to them as well.Our classification framework categorizes metabolic interactions along two main axes: (i) the degree of reciprocity (i.e. unidirectional versus bidirectional metabolite flow), and (ii) the investment by the involved partners (i.e. the cost to produce the exchanged metabolite) (Fig. 1). The first parameter, degree of reciprocity, categorizes cross-feeding interactions based on whether the metabolite exchange is unidirectional (one-way) or bidirectional (reciprocal) (Fig. 1). The second parameter, investment, divides cross-feeding interactions according to the cost of biosynthesis that the interacting partners bear during the interaction, resulting into two sub-categories (i) by-product cross-feeding (Fig. 1A and B) and (ii) cooperative cross-feeding (Fig. 1C and D). By-product cross-feeding is the exchange of metabolites that results from a selfish act of the producer.22 For example, by-products can be secreted due to the degradation of complex hydrocarbons,64 the accidental leakage of metabolites through the bacterial membrane,65 or overflow metabolism.66 In general, the production of metabolic by-products is independent of the presence of an interaction partner and positively correlated with producer's growth.
In contrast, cooperative cross-feeding occurs if one partner actively invests resources to produce metabolites that benefit an interaction partner (Fig. 1C and D). In this case, the cooperating cell is producing more of the metabolites than it would require for its own growth. Enhanced levels of metabolite production can be caused by an increased expression of the corresponding biosynthetic genes,67 a greater flux through the respective metabolic pathway,68 diverting resources into the production of a given metabolite,69,70 or harbouring a multi-copy plasmid that encodes the biosynthetic genes.71 In any case, a cell bearing this cost is significantly less fit than a cell that is not carrying the burden of increased metabolite production.69 Thus, an important difference between cooperative cross-feeding and an exchange of by-products is that cooperative cross-feeding must have been favoured by natural selection. In other words, a newly emerged mutant that produces increased amounts of a given metabolite found itself in an ecological setting, in which this cooperative trait was selectively favoured despite the concomitant fitness costs.
When cross-feeding interactions are classified in these two dimensions, it is possible to obtain five different outcomes.
2.3 Ways to study cross-feeding interactions
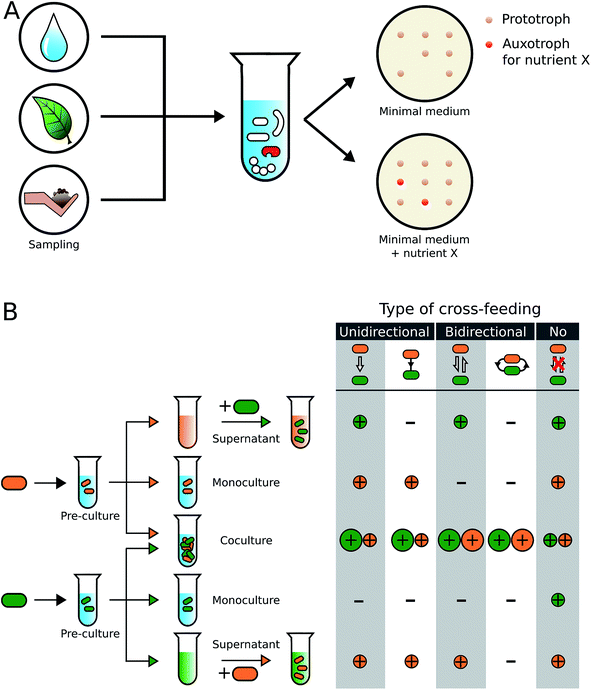
In the following two sections, we will provide an overview over different methodological approaches that have been employed to identify and characterize metabolic cross-feeding interactions in bacteria (Fig. 2).Sequencing, the whole genomes of the isolated strains and/or manipulating their genome (e.g. by mutagenesis) can shed further light on the molecular basis of the observed interaction. Intrinsic problems of this type of approaches are that only a fraction of the bacteria that were actually present in an environmental sample can be isolated and cultivated under laboratory conditions (see Section 4.5). Moreover, conditions that bacteria face in nature (e.g. spatial structure of soil particles, availability of specific nutrients, pH, etc.) are difficult to simulate under laboratory conditions. Moreover, mixing a certain number of different strains in all possible combinations of pairwise cocultures77,78 might bring together strains that would not meet in their natural habitat, thereby biasing the view on the true spectrum of existing interactions. Nevertheless, culture-dependent approaches have provided valuable insights into the rich diversity of metabolic interactions that exists within microbial communities (Fig. 2, Table S1†)79–82 and should be seen complementary to the so-called culture-independent approaches.
The major advantage of culture-independent approaches is that they provide hypotheses without the laborious and potentially biased isolation of environmental microorganisms. A downside, however, is that these techniques strongly depend on the quality of both the extraction process (i.e. DNA, RNA, or proteins) and the obtained reads. Furthermore, divergent sequences, the presence of metabolic enzyme homologs, and promiscuous enzymes with yet uncharacterized catalytic capabilities could lead to a potential overestimation of metabolic dependencies. As a consequence, the performed studies mainly provide hypotheses that need to be verified in subsequent experiments. Thus, many recent studies combine culture-dependent and independent approaches as complementary techniques to capture a more holistic picture of the microbial community.79,90,95
2.4 Distribution of cross-feeding interactions in nature
How prevalent is metabolite cross-feeding in nature? To address this question, we have screened the available literature for cases, which experimentally demonstrated cross-feeding of building block metabolites in natural bacterial isolates (Table S1†). In total, 77 studies were included that reported about 135 different interactions covering the period of 1952 to 2016. The metabolites identified in these studies were divided into the following six main categories: carbon source, nitrogen source, amino acids, nucleotides, vitamins, and others (i.e. phosphorus, iron, or organic compounds). Hormones, growth factors, or electron exchange were deliberately excluded from the analysis.The results of this meta-analysis indicated that metabolite cross-feeding is indeed very common both among different bacterial species and between bacteria and members of other kingdoms including archaea, fungi, animals, protists, and plants (Fig. 3). Moreover, cross-feeding of different molecules (Fig. 3B) was remarkably diverse with regards to the lifestyle of the involved partners and the habitats, in which the interaction occurred68,88,96–99 (Table S1†). Another insight that emerged from this comparative analysis was that in many cases, interacting bacterial cells tended to be localized in close spatial proximity, presumably to facilitate an exchange of metabolites.87,89 In general, photosynthetic and nitrogen-fixing organisms commonly traded carbon and nitrogen respectively against other commodities,94,100,101 which represents a major input of these fundamental elements into the global biochemical cycles.
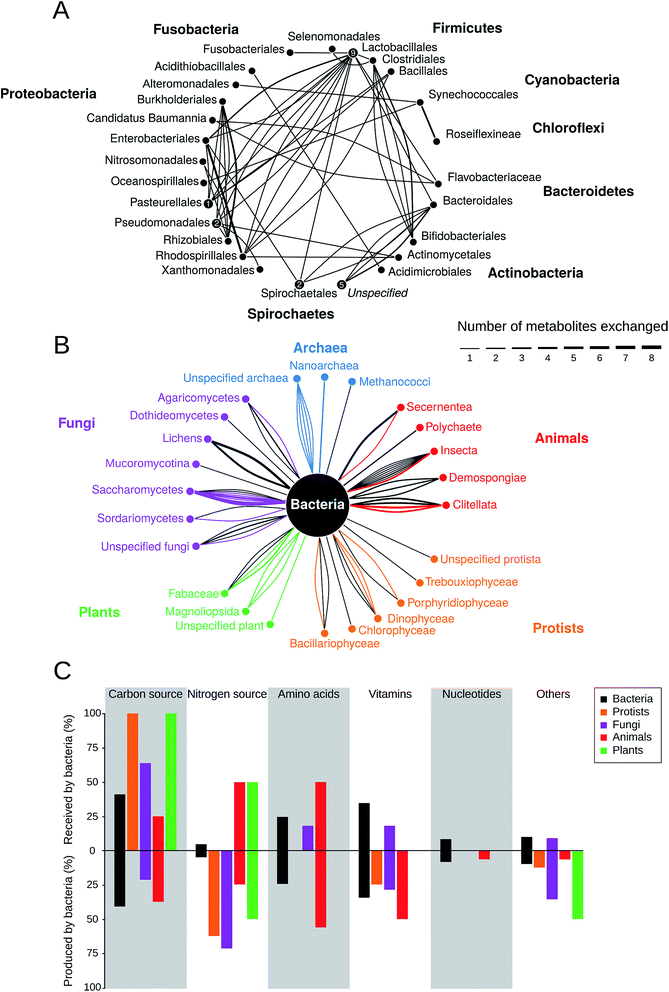 | ||
| Fig. 3 Prevalence of metabolic cross-feeding interactions. Data is the result of a meta-analysis of 78 studies that included 135 different cross-feeding interactions (Table S1†). (A) Metabolic cross-feeding interactions (edges) between bacteria (n = 68). Bacteria from the same order are summarized in nodes and nodes are grouped by the respective phylum. Numbers within nodes represent instances of within-order cross-feeding interactions. The thickness of edges indicates the number of different metabolites that are exchanged. (B) Interactions between bacteria and organisms from other kingdoms (n = 67). Edge thickness is scaled as in (A) and its colour corresponds to the partner that is producing the exchanged metabolite. (C) Percentage of specific metabolite classes that are either received (upper half) or produced (lower half) by bacteria in cross-feeding interactions relative to the total number of cases in each category (n = 135). | ||
Strikingly, the nature of the exchanged metabolites drastically depended on the corresponding partner, with which bacteria interacted (Fig. 3C). For example, plants and protists tended to mainly provide bacteria with (assimilated) carbon.102–104 In return, bacteria commonly supplied plants with nitrogen102 and algae with vitamins, which ∼50% of all algal species cannot produce autonomously.105 In general, bacteria are an important source of nitrogen for fungi, protists, plants, and animals. Animals commonly provide shelter and food to bacteria (e.g. in the gut99), while receiving a wide range of the metabolites including amino acids and vitamins in return. Interestingly, based on the collected data, bacteria are the only partner who cross-feed nucleotides either with other bacteria106 or with members of other kingdoms.91
Our literature survey also revealed that some specific types of cross-feeding interactions attracted more research attention than others. It is important to keep in mind that this pattern does not reflect an increased prevalence of these interactions in nature. For example, cross-feeding between Streptococcus and Lactobacillus has been extensively studied during the last decades, because of the biotechnological interest in these strains that are used in dairy production. On the other hand, many interactions remain likely undiscovered, because of a lack of scientific inquiry or because technical difficulties thwart the isolation and analysis of the partners involved.
2.5 Mechanisms of metabolite transfer
Given that cross-feeding is so common in the microbial world (Fig. 3, Table S1†), the question arises how metabolites are transferred between bacterial cells. Considering the large variety of bacterial lifestyles (e.g. biofilm growth versus planktonic cells, endosymbionts versus free-living bacteria) as well as the structural diversity of metabolites that can be exchanged, it is likely that bacteria use different mechanisms to transfer material from one cell to another one. Modes of metabolite exchange can be classified into contact-independent and contact-dependent. In this context, ‘contact’ refers to a direct physical connection between two or more interacting cells.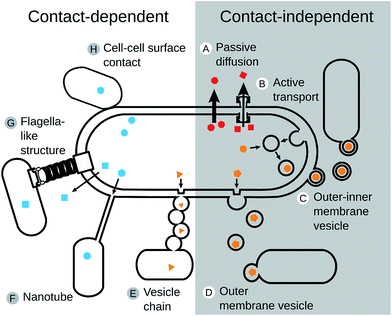 | ||
| Fig. 4 Mechanisms of metabolite transfer. Molecules can be transferred from one cell to another one using (A–D) contact-independent- or (E–H) contact-dependent means of cross-feeding in bacteria. Contact-independent mechanisms that are based on the diffusion through the extracellular environment require a release of the exchanged molecule by (A) a passive diffusion across the cellular membrane115,116,425 or (B) an active transport of molecules via membrane-based transporters.125,126 Alternatively, chemicals can be transferred via membrane vesicles with (C) a bilayer formed of the outer and inner membrane140,141 or (D) a single membrane.142 A contact-dependent exchange of metabolites between two cells can be mediated by (E) outer membrane vesicles that link to form a chain.143 or (F) intercellular nanotubes that allow an exchange of cytoplasmic contents.145,147 Moreover, also (G) flagella-like structures67,148 or (H) a direct surface contact149,150 can facilitate the exchange of metabolites between cells. | ||
2.5.1.1 Passive diffusion. The process of passive diffusion includes the passage of molecules through the cell membrane, often along concentration gradients and without the involvement of ATP (Fig. 4A). This type of exchange is commonly observed for small molecules like hydrogen, formate, potassium, volatile compounds like methanol,112 as well as metabolites like vitamins,103 acetate,113 amino acids, and intermediates of the TCA cycle (e.g. 2-ketoglutaric acid, gluconate).114 Metabolites that are transferred in this way are often released as a result of overflow metabolism,66 thus giving rise to interactions, in which by-products are being exchanged.
In this type of interactions, the speed of diffusion limits the exchange of metabolites and thus the growth of both interacting partners. For instance, Syntrophomonas wolfei and Methanobacterium formicicum exchange either hydrogen or formate as an electron carrier depending on the spatial distance between cells.115 Close proximity promotes electron transfer via hydrogen, because of its rapid diffusion through the medium, whereas formate is used for longer-distances.116
2.5.1.2 Active transport. Molecules that require an active transport are usually unable to cross the bacterial membrane due to their molecular weight, charge, or polarity. Examples include some amino acids,117 siderophores,118,119 enzymes,120 polymers,121,122 and vitamins.23 In these cases, the exchanged molecule needs to be exported into the extracellular environment, involving energy-dependent transport systems such as the ATP-binding cassette (ABC) transporter family123 or the phosphotransferase system.124 Also, corrinoids, a group of compounds, which consist of four pyrrole rings, fall into this category.23 Both Gram positive and Gram negative bacteria feature specific transporters for corrinoids (i.e. BtuFCD and BtuBFCD, respectively).125 Cobalamin (i.e. vitamin B12) is such a corrinoid, which is actively transported through the bacterial membrane. Cobalamin and its analogues have been identified in human faeces and are likely produced by members of the gut community. However, not all prokaryotes in the human gut can synthesize cobalamin. For instance, Bacteroides thetaiotaomicron contains multiple transporters for the uptake of externally available cobalamin,126 suggesting corrinoid cross-feeding in the gut.
2.5.1.3 Vesicle-mediated transport. Membrane vesicles (MVs) are small, spherical encapsulations that form via protrusion of the outer membrane and subsequent pinching off from the cell127–129 (Fig. 4C and D). For this reason, MVs consist primarily of outer membrane material (i.e. proteins, lipopolysaccharides, phospholipids) and encapsulate periplasmic components such as proteins,130–133 enzymes,18,134,135 nucleic acids,136 or signalling molecules.137,138 In addition, bacterial MVs are well-known shuttles for communication signals, which are especially common in pathogenic bacteria.128,131,139 As such, MVs provide an enclosed, protected environment for the exchanged molecules from the external chemical milieu.
Currently, two types of membrane vesicles are known from bacterial isolates. The first and most common type are outer membrane vesicles (OMVs). The second kind of vesicles are called outer–inner membrane vesicles (O-IMVs). These O-IMVs are formed by the protrusion of both the inner and outer membrane and contain cellular contents, especially nucleic acids.140 O-IMVs have been detected in Shewanella vesiculosa, Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Acinetobacter baumannii.141
Membrane vesicles are common and produced by a variety of different bacterial species. Until recently, research on MVs has mainly focussed on their role in transporting virulence factors from bacteria to host cells. Hence, little is known on whether MVs are also involved in transferring nutrients between bacterial cells. An exception is a study, in which the marine cyanobacterium Prochlorococcus was shown to release large quantities of OMVs.142 Besides DNA and RNA, OMVs also contained protein, which supported the growth of other marine bacteria such as Altermonas sp. and Halomonas sp., indicating cross-feeding of organic carbon. More work is necessary to fully evaluate the role of MVs as a means to shuttle nutrients between bacterial cells.
2.5.2.1 Vesicle chains. OMVs are not only used as transporting agents themselves, but also as building block materials to establish cell–cell conduits (Fig. 4E). For instance, predatory bacteria of the species Myxococcus xanthus link multiple individual membrane vesicles together to from so-called vesicle chains.143 The MVs within these chains contain lipids, sugars (fucose and mannose), carbohydrates (N-acetylglucosamine and N-acetylgalactoseamine), and certain proteins that are required for coordinated movement (CglB and Tgl).144 Vesicle chains provide an intercellular network for material transport. The molecular details, however, of how materials are transported within these interconnected vesicles, remain unknown.
2.5.2.2 Nanotubes. Advancement in imaging techniques to study cocultures of interacting bacteria has led to the discovery of several structures that might be used to transfer cytoplasmic materials between bacterial cells (Fig. 4F). For example, unshaken cells of Bacillus subtilis, for example, use nanotubes for shuttling cytoplasmic proteins and plasmid DNA to cells of the same or different bacterial species.145,146 These tubes were observed to connect neighbouring cells (intercellular nanotubes) as well as extend from cells into the surrounding (extending nanotubes).146 The membranous envelope of these structures was found to be constricted at certain points, giving the tube a sequential, bead-like appearance with a continuous lumen that is similar to the abovementioned vesicle chains.146 In another study, nanotubes were found to be used to transport essential amino acids between auxotrophic genotypes of E. coli that have been incubated under shaking conditions.147 Here, intercellular connections consisted of membrane-derived lipids, showed a continuous lumen, and were used to transport cytoplasmic materials between bacterial cells of the same or different species. In both nanotube-forming species (i.e. B. subtilis and E. coli) it remains unclear whether interaction partners are actively chosen (e.g. by receptors on the cell surface or chemotaxis) or if cell-attachment is unspecific (e.g. mediated by non-specific adhesins or sticky polymers).
2.5.2.3 Flagella-like filaments. A contact-dependent exchange of metabolites does not always rely on dedicated structures such as membrane vesicles or nanotubes, but can also be facilitated by already existing structures that are repurposed (Fig. 4G). The fermentative bacterium Pelotomaculum thermopropionicum was shown to form aggregates when cocultured with the methanogen Methanothermobacter thermautotrophicus to facilitate the transfer of hydrogen.148 Analysis of these aggregates indicated that flagellae were mediating this interaction.67 Gene expression analysis confirmed that binding of a flagellin protein (i.e. FliD) induced an up-regulation of genes for enzymes involved in methanogenesis. Thus, the flagellum is not only used to ensure physical proximity, but also to synchronize the metabolism of both interacting partners.
2.5.2.4 Cell–cell contact. The formation of extracellular appendages like nanotubes likely represents a significant cost to nutrient-limited cells, which should be avoided by cross-feeding bacteria. When cells are in close physical contact, such as within multicellular aggregates, the metabolite exchange is likely assisted by direct membrane contact (Fig. 4H). The green sulfur bacterium Prosthecochloris aestaurii for example is photoautotrophic, yet requires an electron donor to grow. The latter can be provided by a heterotrophic partner such as Geobacter sulfurreducens,149 which supports growth of P. aestaurii when both partners show an intimate cell contact. Additionally, a trans-outer membrane cytochrome complex in G. sulfurreducens was shown to be essential for cross-feeding of electrons. Another case of direct cell contact mediating an exchange of cytoplasmic materials was observed in a synthetic consortium of Clostridium acetobutylicum and Desulfovibrio vulgaris Hildenborough.150 In these cocultures, D. vulgaris could grow despite its inability to grow in monoculture. Differential labelling of cytoplasmic membrane and the peptidoglycan showed the absence of the peptidoglycan layer in the region of cell contact.150
3 The evolution and maintenance of metabolic cross-feeding interactions
The reported ubiquity of metabolic cross-feeding interactions in bacteria raises a fundamental question: why should bacterial cells start to actively invest resources to benefit other, potentially unrelated individuals? Natural selection predicts that organisms should maximize their fitness at the expense of others. How does this reconcile with an exchange of biosynthetic products that, in many cases, incurs significant fitness costs to the producing cell? For cooperative interactions, such as an exchange of costly metabolites, evolutionary theory predicts strategies should be favoured that reap cooperative benefits without reciprocating.22 These non-cooperating types, which utilize exchanged metabolites without contributing to their production, gain a significant fitness advantage over cells carrying this burden. Ultimately, the short-term advantage gained by such non-cooperators can, at least theoretically, result in an extinction of cooperating genotypes,151,152 thus representing a permanent threat to the existence of cooperative cross-feeding interactions. Consequently, a theory to solve this so-called tragedy of the commons needs to not only explain the emergence of reciprocal cross-feeding interactions, but also to provide mechanisms that can help explain the persistence of these cooperative relationships in the long-run.The problem, however, is multi-faceted, since different levels of biological organization can affect the dynamics of cross-feeding interactions in different ways. This is because cross-feeding interactions are strongly influenced by, for example, (i) the mode of function and regulation of the individual enzymes involved in the biosynthesis of the exchanged metabolites, (ii) the cellular allocation of limited resources (e.g. nutrients, expression machinery, space) to the cellular functions that are required for the cross-feeding interaction, as well as (iii) the biotic composition of the bacterial community that determines the frequency of potential producers and consumers of exchanged metabolites. Also features of the ecological environment like (i) the diffusibility of chemicals, (ii) the availability of nutrients, or (iii) the degree of spatial structuring will decide about the evolutionary fate of a metabolic cross-feeding interaction.
Hence, understanding the evolutionary trajectories that lead to obligate cross-feeding interactions requires the identification of the metabolic, physiological, and ecological factors enabling metabolite exchange as well as the evolutionary mechanism stabilising these interactions in the bacteria's natural environment.
3.1 Metabolic factors
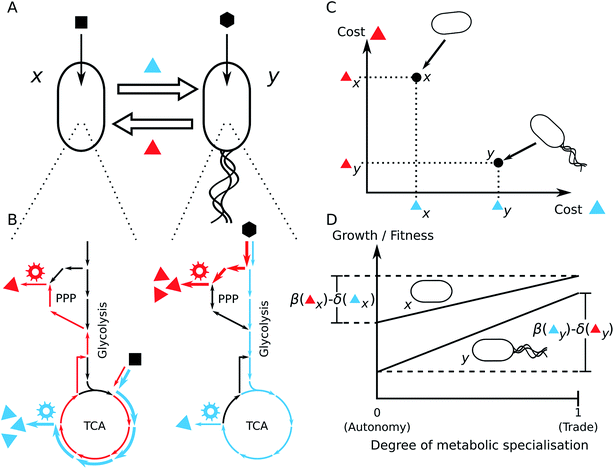
This situation strongly resembles trading interactions in human societies and there is growing appreciation in the scientific literature that the advantage of metabolic trade in bacterial communities can be assessed by applying economic models.155,156 One particular useful concept to investigate biochemical interdependencies between microorganisms is the theoretical framework of comparative advantages. In 1814, David Ricardo developed this economic theory to explain how two countries could benefit from international trade.157 Comparative advantages can quantitatively explain how the resource costs to produce required goods (e.g. metabolites) can translate into mutual benefits if two parties (e.g. different bacterial species) engage in trade of the respective goods (Fig. 5). Hence, such comparative advantages are likely important preconditions for the evolution of specialisation and cooperative biological trade.156
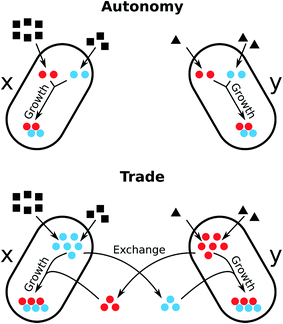 | ||
| Fig. 5 Economics of microbial metabolite trade and the role of comparative advantages. The scheme depicts the consequences on cell growth resulting from two opposing metabolic strategies, metabolic autonomy (above) and metabolite trade (below), in the presence of comparative advantages. Two bacteria (x and y) require two metabolites (red and blue) for cell growth. Each organism uses a different substrate from the environment and is able to produce each metabolite from the respective substrate. Both organisms differ in their metabolic costs to produce the two metabolites: bacterium x requires 3 units of its substrate to produce 1 unit of the red metabolite and 1.5 units substrate to produce the blue metabolite. In contrast, bacterium y requires 0.5 units of its substrate to produce the red metabolite and 1 unit substrate to synthesise the blue metabolite. Hence, organism y has an absolute advantage to produce both the red and the blue metabolite, as it requires less units of resources to synthesize them compared to bacterium x. However, organism x has a comparative advantage to produce the blue metabolite, because it can produce twice as many blue as red metabolites when it reallocates all resources from the production of red metabolites to the synthesis of blue. Analogously, organism y has a comparative advantage to produce the blue metabolite over the production of the red metabolite. Let us assume, that red and blue metabolites are required in equal quantities for cell growth. In case of metabolic autonomy, bacterium x requires 9 units of its resource to produce 2 units each of the red and blue metabolite. y requires 3 units of its resource to produce the same amounts. If each organism specialises for the biosynthesis of the metabolite for which it has the comparative advantage (x: blue, y: red) and trades half of the produced metabolites with the other organisms, each organism can dedicate 50% more of each metabolite to its growth, while consuming the same amount of resources. Thus, the trade of the red and blue metabolites can be mutually beneficial to both organisms. Adapted from ref. 155. | ||
Also, trade-offs in the cellular metabolic networks of single organisms could explain the benefits of metabolite exchange between different cells. Metabolic trade-offs occur if improving the metabolic cost efficiency of one metabolic process or pathway (e.g. due to adaptations) is coupled with increased costs for a different process. Such biochemical conflicts are known to play a central role in the evolution of specialisation158 and several trade-offs have been identified for a wide range of different metabolic processes in bacteria (for a recent review see Ferenci, 2010 (ref. 159)). Thus, trading metabolites may allow bacteria to increase resource efficiency by segregating conflicting metabolic pathways into separate cells.160
3.1.2.1 Molecular causes of comparative advantages. Differences in the architecture of metabolic networks and/or different resource preferences between bacterial species can entail different costs for metabolite production. This can cause reciprocal comparative advantages that can promote the evolution of cross-feeding interactions. Since different species often differ in the structure of their metabolic network, it is likely that these species also differ in their biosynthetic costs to produce different metabolites.166 Hence, reciprocal comparative advantages likely exist between phylogenetically distant species, which also differ in the structures of their metabolic networks.155
Another possibility of how the metabolic network structure can generate comparative advantages are different substrate preferences among bacteria. Coexisting bacterial species frequently utilise a distinct sub-set of carbon sources that are available in the environment.167,168 Different carbon sources often enter the metabolic network at different locations (Fig. 6B). It has been shown that the point, at which a carbon source enters the central metabolic network, strongly affects the distribution of metabolic fluxes169,170 and, in this way, also the production costs of individual amino acids.164 Hence, a consequence of diverse carbon source preferences is that one species of the bacterial community can have a comparative advantage in the biosynthetic cost efficiency of a specific set of metabolites over another species, which in turn has a comparative advantage in the production of another set of metabolites (Fig. 6C).
Comparative advantages can also arise in bacterial communities due to spatial structure. In a spatially structured bacterial population, cells may experience unequal access to different resources due to a heterogeneous distribution of chemicals on a micro-scale.171 For instance, in bacterial biofilms, nutrients are mainly accessible for cells residing close to the surface. In contrast, excreted metabolic by-products are likely enriched in the inner part of the biofilm and therefore more available to cells that dwell in the subsurface.171,172 In fact, it has been observed for a variety of different biofilm-forming bacteria that cells exhibit distinct metabolic phenotypes depending on their positioning within the biofilm and thus, the local environmental conditions to which cells are exposed.173 Such differences in metabolic phenotypes may cause reciprocal comparative advantages in the production of different metabolites that can promote cross-feeding interactions between different cells within the biofilm.
However, heterogeneity in metabolic phenotypes within bacterial populations is not limited to spatially structured environments or species with different genetically determined metabolic capabilities. Phenotypic heterogeneity can also arise in homogenous environments,174,175 which in turn can give rise to reciprocal comparative advantages between different phenotypes, thereby promoting a cooperative exchange of metabolites. Bacterial populations frequently display heterogeneity, where two essential metabolic functions are partitioned between two subpopulations. Prominent examples are nitrogen fixation and photosynthesis in cyanobacteria176 or acetate and acetoin production in Bacillus subtilis populations.177
3.1.2.2 Molecular causes of biochemical conflicts. Biochemical conflicts between two metabolic functions commonly arise due to resource allocation trade-offs.159 Different metabolic functions usually compete for the same cellular resources, e.g. the same precursor metabolites, ATP, or the use of the cellular transcription-/translation machinery.163 Importantly, resources that are consumed by one metabolic function are not available anymore to another one. Thus, the metabolic flux through one pathway might limit the activity of other metabolic processes.178,179 Segregating such antagonistic biochemical processes into different bacterial cells can resolve the biochemical conflict between them.158
The above-mentioned examples illustrate that comparative advantages, and biochemical conflicts in metabolite production between co-occurring organisms are prevalent in natural bacterial communities and are thus important determinants for the evolution of cooperative metabolite exchange. While biochemical conflicts and comparative advantages can explain the mutual fitness benefit that results from metabolic cross-feeding interactions, it is important to note that in isolation they are not sufficient to explain the evolution of cooperative interactions, since they do not provide a mechanism to prevent the exploitation of exchanged metabolites by non-cooperating cells (see Section 3.5).
3.2 Metabolite leakage: the first step towards the evolution of metabolic interactions
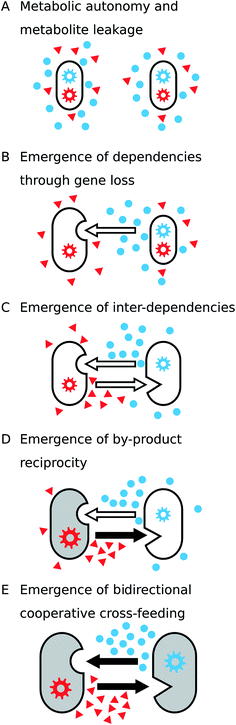
Many metabolic functions are leaky, which means that the products of these biochemical transformations are released into the extracellular environment, thus making them available to other cells180,181 (Fig. 7A and B). Metabolite leakage can facilitate the evolution of unidirectional by-product cross-feeding interactions as well as metabolic interdependence (Fig. 7A). This is because neighbouring cells can take advantage of the released resource, thus saving the costs of producing these metabolites by themselves (see Section 3.3). Cells, which use the metabolic by-products of other cells, adjust their metabolism by redistributing metabolic fluxes, which in-turn can cause leakage of other metabolites. The resulting mosaic of different metabolic strategies potentially provides the basis for the emergence of new metabolic dependencies (Fig. 7B and C).3.3 Emergence of by-product cross-feeding through gene loss
Loss or deactivation of a metabolic gene by mutation can render the survival of the resulting auxotrophic mutant contingent on an environmental supply of the focal metabolite. Potential sources for this metabolite are besides decaying organic matter, mainly other eukaryotic182–184 or prokaryotic organisms in the mutant's environment.185–187 Thus, the mutational loss of a conditionally-essential biosynthetic gene is a key step towards the establishment of an obligate metabolic cross-feeding interaction (Fig. 7B).3.3.2.1 Adaptive loss of biosynthetic functions in metabolite-rich environments. In principle, two evolutionary explanations can account for an adaptive loss of genes. First, selection is expected to remove a subset of genes from a bacterial genome that might not be essential in a given environment. Retaining genes that do not contribute to a bacterial cell's fitness is costly, because of the burden resulting from the functioning of the corresponding gene products within the cellular context.196 Moreover, the expression of unneeded proteins reduces the amount of resources that are available for other cellular processes.197,198 This is why mutants that lack these non-required functions may be adaptively favoured and thus increase in frequency relative to types that still carry these genes. This process, which is called genome-streamlining,199,200 can be considered as a way to cellular economization.199,201 This process should be particularly important in large bacterial populations, where the effect of natural selection is very strong.201–203 As a consequence, any fitness-enhancing mutation including loss-of-function mutations (e.g. deletions, frame-shifts) will be fixed in the population. Indeed, many free-living bacteria such as Prochlorococcus204 or Candidatus Pelagibacter ubique,203 which are oligotrophic and live in aquatic ecosystems that are relatively nutrient-deficient yet stable in terms of resource turnover,205 feature genomes of reduced sizes.
The second possibility is that selection favours a loss of biosynthetic functions in bacteria, when the resulting metabolic deficiency can be compensated by an environmental uptake of the corresponding compound. Indeed, several laboratory-based studies with Bacillus subtilis,206Escherichia coli,26,207 and Pseudomonas fluorescens208 clearly showed that amino acid auxotrophic bacterial strains gain a significant growth advantage (i.e. up to 20% relative to their prototrophic counterpart) when the metabolite they require for growth was sufficiently available in the environment. What can explain the strong fitness advantage observed in auxotrophic genotypes? Zamenhof and Eichhorn (1967), who first described this phenomenon, suggested that when the metabolite is present in the extracellular environment, bacteria that shut down their endogenous machinery to produce the metabolite gain a selective advantage over prototrophic cells, because they save the costs associated with producing the metabolite.206 Costs that could be saved by auxotrophic bacteria include (i) energetic costs that are required to drive biochemical reactions,163,165 (ii) ribosome costs that accrue for building the translational machinery,197,209 (iii) protein costs that stem from the need to produce the biosynthetic protein machinery,209 as well as (iv) carbon costs that result from the allocation of raw materials to produce the focal metabolite.164 All in all, a significant proportion of a bacterial cell's energy budget is allocated to amino acid biosynthesis.165 Given that many natural habitats of bacteria are rich in metabolites that bacterial cells require for growth (e.g. amino acids,210,211 vitamins,212 and nucleobases213), it appears plausible that natural selection may favour auxotrophic mutants that save the costs of metabolite production in these environments.
Compelling evidence for the importance of natural selection for driving gene loss in bacteria comes from several evolution experiments.196,214 Lee and Marx (2012) found that non-essential, accessory genes were frequently lost from almost 80% of evolving Methylobacterium extorquens AM1 populations that adapted to minimal medium.214 In this case, gene loss was accompanied by an increase in fitness, suggesting that selection favoured the loss of unneeded genes when adapting to a specific environment.214 In another study, Koskiniemi (2012) tested the fitness consequences196 of losing stretches of DNA from the genome of the bacterium Salmonella enterica and found that fitness-increasing deletions were rapidly fixed in populations that had been serially propagated in the same nutrient environment. Furthermore, E. coli populations that were selected in an amino acid-containing environment frequently lost the ability to autonomously biosynthesize these metabolites, with the evolved auxotrophies conferring an adaptive advantage.207
3.3.2.1.1 Mutational deactivation versus transcriptional down-regulation of metabolic genes. If gene loss is so beneficial, why then do bacteria not downregulate their biosynthetic machinery when the corresponding product is sufficiently available in the extracellular environment? In this way, cells could enjoy the benefits resulting from gene deactivation, yet retain the ability to grow autonomously when external metabolite pools are depleted. Two main reasons likely explain why a mutational gene loss or deactivation is probably more important in the context of metabolic cross-feeding interactions than a regulatory inactivation of the same biosynthetic pathways. First, the ability to sense environmental conditions in order to determine whether or not it is beneficial to switch from an autonomous metabolite production to an environmental uptake requires the maintenance of an extensive sensory and regulatory machinery. The production and maintenance of such a system likely requires a significant investment of resources and these costs would have to be outweighed by benefits resulting from it – even if the system remains in a certain configuration for extended periods of time. Second, a cell that is able to switch between an environmental uptake and an autonomous metabolite production has to be fitter than a cell, which specializes in just one strategy. A significant factor that works to the disadvantage of a regulation-based phenotype is the time and energy it takes to switch between both states. Indeed, a prototrophic genotype of E. coli that was cultivated in a minimal medium only rarely used environmentally supplied amino acids, while an auxotrophic loss-of-function mutant of the same genetic background gained a significant fitness advantage from tapping this resource.26 Even though it is not known at the moment whether the same pattern is true for other species as well, the above example clearly illustrates that auxotrophic and prototrophic cells are in different physiological states147,215,216 and that at least prototrophic E. coli cells do not downregulate their amino acid biosynthetic pathways to become functional auxotrophs when the corresponding metabolites are sufficiently available in the environment.26
3.3.2.2 Random genetic drift. The second main evolutionary mechanism that has been suggested to explain the loss of biosynthetic genes from bacterial genomes is random genetic drift. In populations of small size, random changes in allele frequencies can result in the fixation of maladaptive genes. Accordingly, genetic drift has been suggested to be the main cause for the extreme genome reduction that is commonly observed in endosymbiotic or endoparasitic bacteria.185,217,218 Several arguments seem to support this interpretation.
First, bacterial populations within host cells are usually small (103 to 104 cells per ml) and are subject to repeated reductions in their size (i.e. population bottlenecks) during transmission from parent host to its offspring.182,218,219 A reduction in effective population sizes (Ne) can greatly affect the impact of genetic drift.203,219Ne is the size of an idealized population that experiences the same magnitude of genetic drift as an existing population.203,219,220 However, this important parameter is difficult to estimate due to the stochasticity of genetic drift.201,203 Accordingly, the fixation probability of a mutant allele in a given populations depends on the product of Ne and s (i.e. the coefficient of selection).201,203 When Nes > 1, the fate of the mutant allele is primarily determined by selection, whereas when Nes <1, genetic drift determines the fixation probability of this allele.201,203,219,220 Thus, when Ne is low, mutations with deleterious effects can persist and even become fixed in the populations,219 because under these conditions, natural selection is less effective in eliminating deleterious mutation.201
Second, intracellular bacteria live in a nutrient-rich and rather constant environment. Thus, losing essential biosynthetic functions may not be penalized as strongly as is the case for bacteria living in nutrient-limiting conditions.
Third, reduced or absent levels of recombination in the intracellular environment of the host significantly restrict the opportunity to purge deleterious mutations.218,219 As a consequence, deleterious mutations even in key biosynthetic genes accumulate in the genome – a process, which has been termed Muller's Ratchet.201,218 Given that deletions of genes from bacterial genomes appear to be much more common than insertions,200,221 Muller's Ratchet is likely an important evolutionary force to account for gene loss in small bacterial populations. Indeed, Salmonella typhimurium populations that were repeatedly subjected to single-cell bottlenecks in rich medium revealed slow-growing phenotypes and auxotrophic loss of function mutants, which the authors interpret as evidence for an accumulation of deleterious mutations.222 Overwhelming evidence for the loss of biosynthetic genes from endosymbiotic or endoparasitic genomes stems from comparative genomic studies.184,185,223 Unfortunately, due to difficulties to cultivate the bacterial strains involved, it is often not possible to quantify the fitness consequences resulting from gene loss for the corresponding genotypes. Moreover, it often remains unclear whether drift or selection caused the observed pattern.
Moreover, the loss of a biosynthetic gene that causes metabolite starvation is well-known to trigger a stringent cellular response, which globally reorganizes the metabolism to economise available resources.215,224 Another consequence of auxotrophy-causing mutations, which has been observed in several species, is the formation of intercellular nanotubes145,147 (Fig. 4F). These physical intercellular connections allow the auxotrophic mutant to derive the focal metabolite from other cells that are still able to produce the required metabolite. Taken together, the abovementioned cellular responses to biosynthetic gene loss represent immediate physiological consequences that promote the evolution of cross-feeding interactions by either helping to establish unidirectional interactions or by adjusting their own metabolic processes to efficiently use the exchanged compounds.
3.4 The emergence of by-product reciprocity
A central step towards the evolution of cooperative cross-feeding interactions is the transition from pure by-product cross-feeding between two interacting organisms (Fig. 7C) to an interaction, in which costly metabolites are actively produced by one individual to benefit the corresponding partner (Fig. 7E). This transitional step includes the so-called by-product reciprocity interactions, where one organism performs a costly cooperative act such as an enhanced production of a metabolite, which is consumed by the corresponding partner. The actively overproduced metabolite benefits the recipient organism by fuelling its metabolic processes. As a consequence of the enhanced metabolic activity, the recipient organisms release higher amounts of metabolic by-products, thus benefiting the organisms that perform the cooperative act (Fig. 7D).225–227It is well-known that increasing the metabolic rate of a microorganism (i.e. population growth) also elevates the amount of metabolic by-products that are released.56,227 Thus, for two organisms that reciprocally exchange metabolic by-products, any mutation that will increase the production levels of the exchanged metabolite in one of the two partners, will be immediately rewarded by increased return levels. This automatic feed-back not only stabilizes the costly cooperative investment, but also paves the way for the evolution of reciprocal cooperative cross-feeding interactions.
3.5 Evolution of cooperative cross-feeding
The challenge with explaining the evolution of cooperative cross feeding lies in the fact that cooperation, such as the overproduction of metabolites, is costly. Thus, non-cooperating free-riders that do not contribute to the production of the traded metabolite may still benefit from the cooperative interaction. Consequently, whenever both cooperators and non-cooperators have an equal probability to gain access to the cooperative public good, natural selection predicts non-cooperative genotypes to increase in frequency, which may ultimately result in a collapse of the cooperative interaction.22,228–230 Another potential problem in complex microbial communities is that even if cooperative cross-feeding evolved, the respective genotypes need to ensure continued interaction with other, metabolically complementary genotypes.231 Hence, a key question in this context is which evolutionary mechanisms can stabilize metabolic cross-feeding within and between bacterial species.An answer to this question needs to take both the ecophysiological intricacies of the focal interaction as well as its eco-evolutionary context into account.232 For example, metabolite exchange via diffusion through the extracellular environment intrinsically imposes several limitations on conditions under which cross feeding can exist. In well-mixed environments, these so-called public goods should theoretically be equally available to both producing and non-producing cells167,231 and are thus prone to exploitation. Alternatively, metabolites may be transferred in a contact-dependent manner (e.g. via intercellular nanotubes147), which restricts the access to the exchanged metabolites. The ecological conditions under which cross-feeding between bacterial genotypes showing either means of metabolite transport can be maintained, is therefore drastically divergent, and will likely result in divergent evolutionary outcomes.
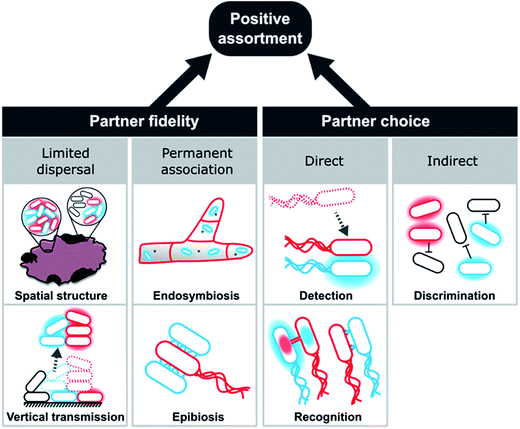
Partner choice can be subdivided into direct and indirect mechanisms (Fig. 8). Direct partner choice can be due to detection, which involves the active finding and subsequent interaction with specific genotypes or recognition that allows cross-feeding genotypes to identify compatible genotypes. Alternatively, indirect partner choice mechanisms operate via the exclusion or inactivation of non-cooperating genotypes. The resulting local enrichment of cooperating cells can also facilitate positive assortment.
These mechanisms should not be seen as mutually exclusive, but multiple processes can operate simultaneously. The contact-dependent inhibition (CDI) system, for instance, exemplifies the combination of partner choice by specific adhesion with non-partner discrimination via inactivation of targeted cells.242 Here, a two-component secretion system facilitates binding and toxin delivery into target cells that display specific receptors. However, carriers of immunity proteins, such as close relatives of the cell expressing the CDI system, are unharmed. This system is widely distributed in alpha-, beta-, and gamma-proteobacteria242 and can even mediate the specific assembly of multicellular biofilm communities.243,244 Below, we will explain each of these mechanisms in more detail and highlight a number of relevant examples.
3.5.2.1 Partner fidelity.
3.5.2.1.1 Limited dispersal. Limited dispersal refers to cases, in which groups of cells that exchange metabolites with each other, remain associated for extended periods of time. This type of increased population viscosity emerges in spatially structured communities that grow on surfaces or in multicellular aggregates.
In recent years, overwhelming evidence has accumulated that bacteria mainly grow attached to surfaces (i.e. a biofilm) or other bacteria (i.e. free-floating clusters), and thus prefer an aggregative lifestyle over a planktonic, free-living state.245–248 Simulations demonstrated the formation of such groups can not only facilitate cooperative interactions,249 but also conflicts among group members.250 Hence, the benefit of a newly established cooperation needs to outweigh the negative effect of conflicts and increased competition in local groups of cells. Importantly, the formation of biofilms and cell clusters can be caused by a number of non-social reasons, such as the protection from the abiotic (oxygen, pH, and drought) and biotic environment (predation,251,252 competition, and immune system), colonisation of favourable substrates (energy- and carbon-sources), or joint niche construction and exploitation (synergism).253
Once established, several important consequences result for cells that grow in spatially structured communities. First, metabolites that are released into the cell-external environment potentially accumulate254 and are thus increasingly available to resident cells. Second, local feedbacks increase the fitness of cell patches with a favourable combination of genotypes, while groups of incompatible types show weaker growth. This results in an interesting effect: the enhanced growth of cooperative groups leads to a spatial segregation of cooperating and non-cooperating cells and thus a spatial exclusion of non-cooperators from the shared goods.167,255–258 This pattern emerges due to a self-organization of initially well-mixed populations256,257,259–262 and can even prevent the invasion of non-cooperative, motile cells.263 This aspect of matrix-assisted population structure can be conceptualized as a passive means of non-cooperator exclusion. In combination, these effects strongly favour the evolution and maintenance of cooperative cross-feeding interactions as evidenced by numerous theoretical241,259,264 as well as experimental studies.255,256,258,261,265–267 Empirical evidence from syntrophic bacterial communities further corroborates this interpretation: the spatial distribution of metabolically interdependent members appears to be key to the functioning of these consortia.268,269 Taken together, various lines of evidence suggest that positive assortment in spatially structured populations can promote cooperation.
Bacterial cells that live in a spatially structured, aggregative community face the problem that with increasing cell density, competition for resources such as space and nutrients also increases. Thus, at one point, colonies need to disperse to populate new substrates. For cells that have started to engage in obligate cooperative cross-feeding, it is therefore crucial to remain associated with other, metabolically complementary cells. Since parts of biofilms are known to get detached,246 they likely function as a propagule to initiate a new biofilm. Notably, groups of cells that protrude from a biofilm or cell cluster are prone to be detached and dispersed more easily. In fact, this is the case for highly productive local groups.270 In this way, cells that are more cooperative, are more likely to transmit offspring to the next generation, thus enhancing selection for cooperative phenotypes. This type of group dispersal, which represents a type of vertical transmission, ensures that complementary and potentially coevolved genotypes interact for extended periods of time (Fig. 8). This is also the case for planktonic macrostructures and multicellular magnetotactic prokaryotes (MMPs) that exhibit propagule formation and fission of whole cell groups, respectively.253,271
3.5.2.1.2 Permanent associations. Extreme cases of staying together are permanent associations (Fig. 8), where prokaryotic cells either live on (epibiosis) or in (endosymbiosis) another organism. Examples include associations among archaea272 as well as interactions of bacteria with other bacteria,273–276 fungi,277–281 protists,282–286 or multicellular eukaryotes.284,287,288
A characteristic feature of these associations is the enormous potential of vertical transmission over evolutionary time when new generations of host–symbiont interactions are established. The tight fitness coupling of cells living in such close associations aligns their evolutionary interests, thereby limiting conflicts among interacting partners. This is particularly promoted by the fact that the fitness of these consortia is often not a property of individual cells any more, but an emergent feature that results from the interaction among host and symbiont. Selection acting on this level is therefore expected to increase complementarity and enhance metabolic cooperation among partners. Indeed, known cases of both epibiosis and endosymbiosis are frequently based on the reciprocal exchange of metabolites between both partners (Fig. 3B). Thus, to understand these systems, it is key to identify the causal mechanisms that initiate the assembly of these associations as well as the evolutionary forces that can tip the balance in favour of cooperative cross-feeding.
The first step in the evolution of a stable endosymbiosis is that a bacterial cell enters the intra- or intercellular environment of its host. Even though cases exist where environmental, commensal or already mutualistic bacteria successfully established as endosymbionts,289 parasitic bacterial strains have an advantage. Their intrinsic ability to enter host cells or tissues despite the presence of anti-bacterial defence strategies (e.g. immune system) enables them to persistently colonize the host.277 This then provides the opportunity for natural selection to transform the initially antagonistic interaction into a mutually beneficial one. This is corroborated by phylogenetic studies showing that proteobacterial mutualists were more frequently derived from parasitic than from free-living ancestors.290 Even if bacterial lineages do not feature strategies to repeatedly re-infect their host or be vertically transmitted, the host might strongly benefit from evolving means to transmit beneficial symbionts to its offspring itself.289 Unfortunately, direct experimental evidence to identify the factors that complete the transition to an obligately endosymbiotic association is lacking, thus complicating the exact assignment of cause and effect. Two key requirements that likely need to be fulfilled are (i) a strict vertical transfer of bacterial symbionts across host generations and (ii) a mutual benefit arising from this interaction. Whenever these criteria are fulfilled, natural selection can act on the symbiotic interaction. Common outcomes of this coevolutionary process are a drastic reduction in genome size of the symbiotic bacteria288,291 as well as the emergence of a metabolic complementarity of host and symbiont, where many different metabolites, precursors, or biochemical functions are reciprocally exchanged.292
The conditions favouring epibiotic associations are rather similar to the ones described previously for endosymbiotic interactions. The prospective epibiont needs to exhibit either features of a parasite or beneficial characteristics of a mutualist to allow repeated interactions with the respective host. In both cases, the ability to attach to the host is required, ultimately enabling the epibiont to exploit and adapt to the resources released by the host. This initially one-sided interaction opens the door to evolve a reciprocal exchange of metabolites, given that host and epibiont complement each other.282,293 Benefits that are associated with this interaction for the host and/or the epibiont favour a permanent association. Host–epibiont coevolution as well as competition with other groups of hosts and epibionts should intensify reciprocal interactions like cooperative cross-feeding.
An example for epibiosis is the TM7 phylotype (TM7x) that is obligately associated with Actinomyces odontolyticus in the oral cavity.273 The epibiont TM7 features a drastically reduced genome (∼700 genes)273 and derives all of its amino acids from its host A. odontolyticus. In phases of extreme starvation, TM7 can even kill its host, thus classifying as a parasitic interaction. Another well-studied case is the phototrophic consortium Chlorochromatium aggregatum, in which a flagellated β-proteobacterium within the family Comamonadaceae is surrounded by Chlorobium chlorochromatii, a green sulfur bacterium.294 In these physiologically highly intertwined consortia, the central bacterium provides motility and receives photosynthetically fixed carbon in return from its epibionts.
3.5.2.2 Partner choice. Partner choice is the second main principle that facilitates positive assortment among cooperative genotypes via either selecting preferred interaction partners (direct partner choice) or antagonizing undesirable or unsuitable cells (indirect partner choice).
Important to recognize for partner choice mechanisms is that they inherently rely on a mixing of interaction partners after every round of association and disassociation. A consequence of this is that these intermittent periods of mixing reduce the chance to repeatedly encounter the exact same genotype in subsequent rounds of interaction. Under these conditions, mutations, whose effects are highly partner-specific, are less likely to spread in the global gene pool, simply because they reduce the number of potential interaction partners. On the other hand, more favourable combinations of interaction partners could result from this process than would be expected to emerge in a regime exclusively relying on limited dispersal. However, at the moment, the above-mentioned population-genetic consequences are purely hypothetical and await experimental validation.
3.5.2.2.1 Direct partner choice. Detection as a partner choice mechanism involves the ability of bacterial strains, which are unable to produce certain metabolites autonomously, to find and identify other cells that are capable of providing these metabolites. This can be achieved in different ways. First, other cells or groups of cells release the required metabolite into the environment and the focal cell uses chemotaxis to trace the source of these metabolites. More cooperative cells will release more metabolites and are thus easier to find. Hence, the required metabolite is the most reliable indicator of a producing cell and should thus be the preferred cue for an auxotrophic cell. However, the focal cell can also use other chemicals to find suitable cells via positive chemotaxis. Here, in principle any metabolite, which can be sensed and followed along a concentration gradient, can serve as a cue. For example, quorum sensing, when used as a means of interspecific communication,295,296 can reveal the species identity of the signalling cell and with it also the potential of competition297 or metabolic compatibility.298 For interactions with higher organisms, bacteria are well-known to show a specific chemotactic response towards their hosts,299–302 while so far only few experimental examples of chemotaxis as a means of recognition among different bacterial species exist.248,303,304 Recently, Laganenka et al. (2016) reported that chemotactic accumulation followed by autoaggregation can also occur within species, which in this case, was mediated by the quorum sensing signal autoinducer-2 within populations of E. coli.305 In general, bacteria feature very sensitive molecular machineries to sense metabolites and quorum sensing molecules.306,307 Even though these systems likely evolved in a different ecological context, they can dramatically enhance the ability of cells to identify possible interaction partners, even in taxonomically diverse communities.
Even though the ability to find other cells can lead to positive assortment, this mechanism alone cannot explain the evolution or maintenance of cooperative cross-feeding. Positive feedback among interactions partners is again also necessary to operate in parallel: more cooperative cells that show positive chemotaxis have a fitness advantage over non-cooperative or non-chemotactic cells, thus increasing their frequency in the local community. This means that after detection of suitable cells, metabolites that are produced and exchanged must preferentially benefit cooperative interaction partners. This can be achieved by recognition mechanism (specific or unspecific) that allow attachment or group formation (see below). Indeed, the combination of chemical signalling with subsequent interspecies aggregation was recently observed in a mutually beneficial interaction between two bacterial species isolated from iron snow.248 More research, however, is necessary to fully evaluate the potential of this, at the moment rather theoretical possibility, to serve as a mechanism to identify suitable interaction partners.
The second main group of direct partner choice mechanisms is called recognition. Bacterial cells can recognize suitable interaction partners in two fundamentally different ways. First, recognition can be passive, by unspecifically attaching to other cells in the environment. In this case, growth of the focal cell will depend on the metabolic complementarity and cooperativity of the more or less randomly chosen interaction partner. Nevertheless, positive feed-back within interacting groups should increase the frequency of cooperators in a given population and thus raise the chance for a cooperative cell to encounter other cooperative cross-feeders in subsequent rounds of attachment and detachment (i.e. positive assortment).
Second, bacterial cells can be equipped with mechanisms, which allow them to specifically adhere to clonemates or members of other species that feature certain characteristics.263,308 In the context of biofilm formation, such behaviours are commonly referred to as co-aggregation.308,309 One evolutionary advantage of specific adhesion over random attachment is that specific adhesion minimizes the chance of associating with harmful, non-complementary, or non-cooperative genotypes. Moreover, adhesion can be key to the establishment of functional groups such as for the joint degradation of organic matter and the associated successful development of multispecies biofilms.308,309 Adhesion over multiple generations finally causes progeny to locally accumulate in clusters. Such a formation of interacting groups helps in directing resources to closely-related, cooperative genotypes. Thus, both mechanisms of unspecific adhesion and specific recognition help to solve the public goods dilemma by privatizing the exchanged metabolites within interacting groups.310
Recognition in microorganisms predominantly operates via structures on the outer membrane308,311,312 and is generally based on affinity between a receptor and an identification molecule.313 Accordingly, the mostly protein-based adhesins represent receptors that facilitate recognition via binding to specific structures on the surface of another cell.314 Other specific recognition systems can be pili,314 homotypic receptors for self-recognition,315–317 the contact-dependent inhibition system (CDI),242 and type VI secretion systems (T6SSs).318 These systems mediate a remarkable spectrum of partner choice ranging from interactions between different kingdoms such as the host choice of microbiota319 to specific autoaggregation with members of the same species.320
The molecular machinery that determines whether or not two cells can interact with each other basically functions like a greenbeard gene233 – i.e. a gene that allows cooperators to recognize other cooperators and thus to preferentially direct benefits towards them. Increasing evidence demonstrates greenbeards to be substantially more common in microorganisms than originally thought.311,321–323 For instance, the Flo1 flocculation gene of the yeast Saccharomyces cerevisiae and the csaA self-adhesion gene in the slime mould Dictyostelium discoideum were identified to represent such greenbeard genes that are key for the establishment of cooperative groups.311,324 Only carriers of the greenbeard genes are part of these groups and thus can enjoy cooperative benefits.228,230,321 In cooperative cross-feeding, however, recognition and cooperation are two separate functions. Hence, a non-cooperative carrier of the green beard allele could evolve and thus undermine the identification system. Nevertheless, recognition likely requires close physical contact, which automatically entails the abovementioned advantages of spatially structured metabolic interactions. Thus, a dedicated recognition system would still allow narrowing the spectrum of possible interaction partners to a subset of principally suitable cells. While research on microbial cooperation has focused more on traits like protection, reproduction, and siderophore production, cross-feeding of metabolites has received less attention so far. Thus, future work should evaluate how common such genetic recognitions systems are in natural microbial communities and whether they are also involved in stabilizing metabolic interactions.
Another attachment mechanism is the establishment of intercellular connections such as nanotubes, nanopods, or vesicle chains that are used to exchange materials between interacting cells (Fig. 4). In many of these cases, large clusters of interconnected cells emerge143,147 that should favour cooperative cross-feeding, because interactions within these clusters are highly localized. Analogous to cells growing on two-dimensional surfaces, also the self-organized growth of cells within three-dimensional networks should favour clusters of cooperative cells and penalize non-cooperators by spatial isolation.
3.5.2.2.2 Indirect partner choice. Indirect partner choice mechanisms lead to positive assortment of complementary cooperators by antagonizing non-complementary or non-cooperative genotypes. In general, this group of mechanisms either kills other genotypes or species or prevents them from invading a local population/community. Consequently, all cells within the social group need to be resistant to the harmful behaviour. By excluding other, potentially non-cooperating genotypes from cooperative benefits, indirect partner choice mechanisms can result in a local enrichment of cooperating genotypes.
Competition for space and other resources is common-place in microbial communities.325–329 Under these conditions, genotypes that can exclude or inactivate competitors are clearly at an advantage, thus explaining the widespread distribution of antagonistic behaviours among microorganisms.16,329,330 Whenever bacteria antagonize others, an almost automatic consequence is that these behaviours decrease the genetic diversity in the local community, thereby increasing the cohesiveness of the group displaying the harmful behaviour or being immune to its consequences. Antagonistic behaviours can facilitate the emergence and spread of cooperative behaviours within such groups, because metabolites that are produced as a cooperative act are more likely to benefit other group members. Discrimination against others can be achieved passively by generating a restrictive chemical environment, for instance by resource-depletion or pH-modification.331–333 The resulting competitive exclusion will reduce the number of genotypes present and possibly enrich close relatives of the strains causing the environmental modification. A property emerging from this process is often that already established bacterial communities, such as the ones colonizing corals,334 plant roots,320,335 or the intestine of animals,333,336,337 cannot be invaded by other bacteria. This phenomenon has been termed bacterial interference or colonization resistance.336,338,339 In the context of metabolic cooperation within a spatially structured bacterial community, the ability to prevent the establishment of foreign bacterial genotypes may help to promote the exclusion of exploitative individuals and thus to stabilize metabolic cooperative interactions.
Alternatively, bacteria can actively express phenotypes that inhibit or kill other bacteria in the vicinity. Besides the production of antimicrobial compounds such as antibiotics or bacteriocins,333 bacteria use a range of other strategies to eliminate competitors including harmful extracellular vesicles,340 contact-dependent inhibition systems (CDI),328 or type VI secretion systems (T6SS).328,341 Importantly, by killing genotypes that do not carry the gene that confers resistance against the antagonistic trait, cells that express the antagonistic trait and are resistant against it qualify as harming greenbeards.321,323 In such a scenario, the local elimination of susceptible genotypes generally increases genetic homogeneity for the greenbeard allele and promotes positive assortment. For example, the CDI-system of B. thailandensis was demonstrated to be capable of antagonizing carriers of different BcpA-CT and BcpI proteins, which were hence successfully excluded from a biofilm.323 Mutual antagonism between two bacterial species via the T6SS was demonstrated to cause segregation of different genotypes as well.342 Also in Bacillus species that were isolated from fresh water sediments, antagonistic interactions that were based on bacteriocin-like substances facilitated the assembly of cohesive communities, even in a homogeneous, aquatic environment.330 Under these conditions, positive assortment can be facilitated when multiple cooperative genotypes display the same antagonistic behaviour. This can even be orchestrated in a synchronized release of the antagonizing molecule in many individuals by quorum sensing.343–345
Both passive and active discrimination mechanisms differ drastically in the specificity, with which they affect other genotypes. For example, antibiotics usually kill or inhibit the growth of a broad range of bacterial species.346,347 In contrast, bacteriocins typically exhibit a narrow killing spectrum,346 enabling producers to specifically target other coexisting species or genotypes of the same species.333,348 Increasing evidence suggests that microorganisms are able to discriminate relatives with high selectivity: antagonistic interactions were identified to facilitate positive assortment among conspecific genotypes.233,329,342,349–351Proteus mirabilis, Myxococcus xanthus, and Bacillus subtilis are examples for bacterial species that can identify and antagonize other strains.352–354 When different populations of these species grow on two-dimensional surfaces, the formation of physical boundaries, so-called demarcation- or Dienes lines, indicate the presence of discrimination mechanisms.352–354 Strikingly, the underlying recognition mechanisms are usually highly selective and allow discrimination even within a species.313,352 Differences in a few loci already cause the formation of Dienes lines between swarming colonies of these bacteria.
The often very high specificity of antagonistic discrimination mechanisms combined with the fact that individual bacteria commonly use multiple discrimination mechanisms simultaneously355 suggests that bacteria aim at maximizing the chance to interact with desired genotypes of the same or different species. Here, the presence of antagonistic functions and the corresponding resistance genes serves as a system to discriminate bearers of these alleles (kin) from non-bearers (non-kin).355 Since these alleles can also be transferred via horizontal gene transfer,356 kinship does not necessarily imply affiliation to the same species. For this reason, the term kind discrimination is more applicable under these conditions.233 If members of the same kin/kind group engage in a mutualistic interaction, any kin/kind discrimination mechanism will help to protect cooperation from exploitation, as recently demonstrated for swarms of Bacillus subtilis.355 Moreover, positive assortment of genotypes in populations of Vibrio cholerae that was mediated via the T6SS correlated with increased cooperation.357 Accordingly, a phylogenetic comparison revealed that the extent of released public goods, in this case proteins, correlated with the total number of T6SS per strain, suggesting that positive assortment via killing promotes the evolution of public goods cooperation.357 Unfortunately, research in this field is just beginning to appreciate the potential significance of indirect partner choice mechanisms for the evolution of cooperative interactions. Thus, further research is necessary to fully evaluate to which extent positive assortment by specific discrimination mechanisms can favour cooperative cross-feeding among different genotypes or species.
4 Consequences of obligate metabolite cross-feeding
The evolutionary transition from a metabolically autonomous, free-living life-style to a state, in which the fitness of a bacterial cell hinges on the obligate cross-feeding of metabolites with other organisms has a number of significant evolutionary consequences for the focal genotype. Some of these effects are well-documented, while others are rather based on theoretical considerations. In the following, we will provide an overview over the manifold ramifications arising from obligate cooperative cross-feeding.4.1. Negative frequency-dependent selection
As outlined above, evolutionary theory predicts obligate cooperative interactions that are based on an exchange of public goods to be notoriously unstable.22,228–230 Particularly when cooperating and non-cooperating individuals have equal access to the cooperative public good, non-cooperators are expected to gain a significant fitness advantage relative to cooperating types, which ultimately should result in a collapse of the cooperative interaction. Surprisingly, this outcome is rarely observed in cases where essential metabolites are cooperatively exchanged (but see ref. 358). Instead, obligate cooperative cross-feeding interactions are commonly stabilized by negative frequency-dependent selection. Frequency dependence describes an evolutionary process, in which the fitness of a given genotype depends on the relative frequency of other genotypes in the total population. In the case of negative frequency-dependent selection, the fitness of a certain genotype decreases as it becomes more common in a given population. From this, results a stabilizing force that maintains interacting genotypes in the long-run.359–362Negative frequency-dependence has been shown to operate in both one-way by-product207,363 and two-way cooperative cross-feeding interactions.74 In both cases, frequencies of the two interacting cell types oscillated around a stable equilibrium point that most likely was determined by rates of metabolite production and consumption. Interestingly, the same pattern prevails when non-cooperators are included: also consortia consisting of amino acid cross-feeding E. coli cells and non-cooperating auxotrophs74 or cocultures between producers of a public good and the corresponding non-cooperators258,360,364–367 were stabilized by negative frequency-dependent selection. Observing negative frequency-dependent selection for different types of metabolic interactions in both spatially structured and unstructured environments suggests this pattern is a common principle emerging in synergistic microbial communities.
However, cooperators and non-cooperating individuals of the abovementioned examples are unlikely to have equal access to the cooperatively produced metabolite. Instead, mechanisms of positive assortment, which have either evolved or result as a by-product from other features of the interaction, ensure the cooperatively produced metabolite is predominantly benefitting other producing genotypes. Examples of such mechanisms include a privatization of metabolites or biosynthetic functions due to contact-dependent exchange mechanisms74,147 or the localization of public goods in spatially structured environments.255,256,258,261,265–267
As a consequence, no genotype can take over in these obligate interactions. Instead, negative frequency-dependence maintains genotypic diversity.167,259,368 A prediction that follows from this is that the evolution of cooperative cross-feeding should promote a metabolic and genotypic diversification within microbial communities. Indeed, this pattern has been repeatedly observed in theoretical models167 and experimentally evolved microbial communities.56,207
4.2 Coevolutionary dynamics
Unfortunately, very little is known on the coevolutionary consequences resulting from cooperative cross-feeding.369 An important aspect is certainly the positive feed-back loop that results when positive assortment assures repeated interactions among complementary partners across generations (i.e. partner fidelity feedback):76 in reciprocal cross-feeding interactions, an increased metabolic investment on one side automatically enhances growth and thus also metabolite production on the other side.69 Thus, continued coevolution is expected to increase productivity in cross-feeding communities, which is corroborated by experimental evidence.370For antagonistic interactions such as host–parasite interactions, both empirical and theoretical work suggest increased rates of molecular evolution (i.e. red-queen effect),371 mainly affecting loci involved in virulence and resistance. In contrast, much less research has been devoted to the question how mutualistic interactions – such as cooperative cross-feeding – affect rates of evolution. Theoretical work implies that in mutualistic interactions, species generally evolve more slowly to increase their share of the benefits (i.e. red-king effect372). Experimental tests whether horizontally or vertically transmitted bacterial symbionts indeed evolve to some point of evolutionary stasis,373 however, revealed inconclusive patterns.374 Thus, further work should examine in more detail how synergistic coevolution affects the rate of evolution.
Finally, also the genomic signature that results from synergistic coevolution of cross-feeding genotypes is not very well understood. Given the transient nature of cross-feeding interactions, it is for example unclear, whether a pattern of reciprocal coevolutionary change, in which mutations in one interaction partner favour a specific set of mutations in the other partner, can be expected. Coevolution experiments with syntrophic consortia consisting of a sulfate reducer (Desulfovibrio vulgaris) and a hydrogenotrophic methanogen (Methanococcus maripaludis) for 1000 generations documented an extremely rapid loss of functional independence of Desulfovibrio vulgaris, which was driven by a mutational inactivation of genes involved in sulfate respiration.375 Also pairs of lactic acid bacteria (i.e. Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus) that have been serially propagated in coculture to ferment yoghurt reciprocally exchange a larger number of metabolites in a mutualistic manner.376 This striking metabolic complementarity, which likely evolved in response to the ecological interaction between both species, is largely due to gene loss.377,378 Given that consortia of co-occurring bacterial endosymbionts89,223,379 display a similar pattern on a genomic level, it is tempting to speculate that metabolic complementarity is a common outcome of a synergistic metabolic coevolution.
4.3 Formation of intercellular metabolic networks
The frequent loss of metabolic genes from microbial genomes along with intricate patterns of cross-feeding, where other coexisting genotypes compensate the functional deficiency, suggests that within bacterial consortia, metabolism is often a community-level property and not a feature of an individual cell any more. If true, this implies that a bacterial communities' metabolism is in essence a super-metabolic network, where many of its constituents perform specialized biosynthetic or catabolic tasks that also benefit other members of the community. Indeed, recent metagenomic and empirical surveys of environmental bacterial communities have revealed that individual genotypes in a community can have distinct patterns of amino acid auxotrophies, such that some members lack multiple biosynthesis pathways, and, at the same time, specialize in the production of another set of amino acids.23,378,380,381 Also specialized genotypes have been found, which provide biosynthetically expensive amino acids to other community members.380 Moreover, coevolved symbiotic interactions between bacteria and higher organisms often show signatures of a functional fusion, in which interacting parties operate as one integrated metabolic unit. For instance, the partitioning of biosynthetic pathways between host and endosymbionts219,382 or between multiple, co-occurring endosymbionts187,383 illustrates that only the sum of the organismal metabolic pathways can satisfy the nutritional needs of all interacting parties.187Metabolic cross-feeding interactions can strongly determine community structure and function.380 As discussed above, the establishment of such interactions is driven by a complex interplay between fitness-advantages of individual mutants and the eco-evolutionary dynamics between multiple genotypes. Ultimately, the highly-conserved structure of core metabolic pathways in otherwise rather divergent bacterial lineages could guide the evolutionary self-organization of metabolic exchange even between very different bacterial species.384 Whether and to which extent microbial communities act as one integrated metabolic unit that maximizes both productivity and stability of the entire consortium, however, needs to be addressed in future studies. Here, targeted analyses of the metabolic abilities and activities of individual strains living in natural bacterial communities are necessary to identify whether the distribution of metabolic functions within these communities is indeed determined by global constraints that are dictated by the structure of the underlying metabolic network.385,386
4.4 Bacterial unculturability
In 1932, Razumov was isolating freshwater bacteria and noticed that the majority of bacteria, which he observed under the microscope, resisted cultivation on agar plates.387 Since then, microbiologists have repeatedly corroborated that less than 1% of all archaea and bacteria can be grown in vitro388 – a problem that has been popularized as the great plate count anomaly.389 Accordingly, recent advances in metagenome sequencing of environmental samples revealed not only the presence of a tremendous microbial diversity, but also discovered completely novel genera and even phyla, which have not been isolated yet.390–392Many potential explanations have been proposed to account for the unculturability of most bacterial species. These can be grouped into four main categories:
(1) Niche mismatch: Mismatch between the physiological and nutritional requirements of a bacterial strain and the conditions provided (e.g. pH, temperature, salts, minerals, and nutrient levels).393,394
(2) Dormancy: Bacterial cells might be viable, but unculturable.395,396
(3) Antagonistic effects: Competitively superior strains outcompete others and/or produce toxic compounds (e.g. antibiotics) to kill or inhibit other strains.14,397
(4) Obligate metabolic interactions: Bacterial strains have evolved obligate metabolic relationships with other neighbours in their environment. Thus, attempts to isolate a single species must fail, because of the lacking nutrients or biochemical functions.398,399
Although all of the above-mentioned reasons likely contribute to explaining the unculturability of many bacterial isolates, accumulating experimental evidence suggests that an obligate exchange of metabolites or biochemical functions among bacterial strains is particularly important in this context. For example, attempts to preserve ecological interaction within microbial communities has significantly increased bacterial recovery. In 2002, Kaeberlein et al. designed a diffusion chamber, which allowed an exchange of metabolites between cells and their environment, but prevented a mixing of cells.400 Using this system, the authors managed to isolate novel cultures, which increased bacterial culturability to up to 50%.401 Interestingly, some of the isolated strains did not grow on synthetic media afterwards, indicating that a direct contact with the native microbial community was essentially required for growth.400 Other related isolation techniques are enrichment cultures, in which environmental conditions are tailored to favour certain genotypes,402 or dilution cultures, where environmental samples are diluted to low, but known cell numbers.403 Both approaches frequently do not result in the isolation of individual strains, but mixtures of strains that can only grow together, but not in isolation.85 Thus, this method provides the opportunity to isolate coexisting genotypes that cross-feed essential metabolites and study their interaction in more detail. Taken together, the available evidence suggests that metabolic interdependencies within natural microbial communities are an important determinant of the commonly observed unculturability of natural bacterial isolates.
4.5 Levels of selection and transitions in individuality
When auxotrophic bacteria engage in cooperative cross-feeding with other bacterial cells, the question arises whether natural selection acts on individual bacterial cells or if groups of cells are the unit of selection. Before addressing this question, some relevant key terms need to be defined: an ‘evolutionary individual’ is the unit, whose frequencies are adjusted by natural selection.404,405 Traits that determine the Darwinian fitness of such an individual must be heritable, variable, and give rise to differential reproduction among competing individuals that differ in the respective trait.404,406 As a consequence, natural selection does not only act on, for example, populations of bacterial cells, but can simultaneously operate on lower (e.g. plasmids inside of bacterial cells) or higher levels (e.g. multicellular prokaryotes such as cyanobacteria). A transition in individuality is now observed when the level of selection is shifted from lower level units that are units of selection themselves, to a higher-level entity, which is composed of several lower-level units.406 The fitness of the collective that emerges during this transition is not proportional to the average fitness of the assembled lower-level units, but an emergent property of the higher-level entity.407–409 Moreover, after the transition, “genetic information is transmitted between generations” such that “entities that were capable of independent replication before the transition can replicate only as part of a larger whole of it”.410 Major leaps in biological complexity have resulted from evolutionary transitions, during which previously independent organisms were functionally integrated to form a new, higher-level entity.411–413 Eminent examples of such transformative events include the origin of the eukaryotic cell414,415 or the emergence of plastids from a cyanobacterial progenitor.416How does natural selection now act in bacterial communities that engage in obligate cooperative cross-feeding of metabolites? First, it is important to recognize that under these conditions, fitness is not only determined by the traits of the individual cells, but is a property that emerges from interactions among cells.74,417 Even if cells that do not invest in the cooperative production of shared metabolites may save the cost of metabolite production,418,419 their lack of investment in cooperative cross-feeding likely curtails their own fitness. This is largely due to the fact that multi-level selection acts on both the level of individual cells and on groups of cells (e.g. sub-populations, different parts of the same biofilm, cell clusters, and so on). Auxotrophic cells of a local community assemble in groups to facilitate the exchange of metabolites.147,420 Growth of cells within such a group depends on the genotypic composition of the whole group: clusters that contain more cooperative cells grow more than clusters consisting mainly of non-cooperators (Fig. 9). Even if non-cooperators gain an advantage in their local group, they are selected against on a global level, if more cooperative groups export their productivity in subsequent rounds of assembly and disassembly. Different mechanisms of positive assortment (Fig. 8) and principles of self-organization,256,260 ensure that cooperative cells remain associated with other cooperators over time.
 | ||
| Fig. 9 Multi-level selection favours cooperative cross-feeding. Cells in a global pool assemble into clusters consisting of non-cooperative (white) and cooperative (grey) cells. On a cluster-level, non-cooperating cells gain a selective advantage over cooperators, because they save the costs of metabolite production. However, the productivity of clusters depends on the relative proportion of cooperative cells within a cluster. As a consequence, differential growth of clusters selects against non-cooperation and favours cooperation on a global level. Modified after ref. 426 and 427. | ||
A second important consequence of living in a close metabolic entanglement with other bacterial cells is that the mutational landscape, which is available to a cell to improve its fitness, will likely dependent on the current interaction partner. Thus, the spectrum of mutations that is expected to be favoured within cross feeding interactions should be radically different from those that might be beneficial in a metabolically autonomous bacterium. Hence, mutations that arise from within these interactions and which improve the performance of the cell group (e.g. mutations increasing among-cell adhesion or levels of metabolite production), are likely maladaptive outside these interactions and thus, should be interpreted as group-level adaptations.
Finally, the question remains: are groups of bacteria, whose survival depends on obligate cross-feeding of metabolites, evolutionary individuals? Put differently: what is the unit that is most relevant to evolution – the individual cell that is unable to survive in isolation, or the group of cells, in which auxotrophic bacteria can thrive? If groups were the relevant evolutionary individual, cell groups would have undergone a transition in individuality. To answer this question, it is useful to consider cases, in which a new individual has been formed by natural selection upon the merging of previously independent lower-level units and identify hallmarks that characterize these cases (Table 1).
| Characteristic | Cooperative cross-feeding bacteria | Example |
|---|---|---|
| a Reproduction only as part of a multicellular consortium. b Synergistic fitness benefit arises upon combination of functions. c Groups of cross-feeding bacteria beget new groups. | ||
| Mutual dependencea | Yes | 428 |
| Functional specialisation of cells/division of labourb | Yes | 429 and 430 |
| Cooperation | Yes | 151 and 431 |
| Group-level reproductionc | Likely yes, to some extent | 432 |
| Cell-attachment | Yes, but likely transient | 147 and 419 |
| Strict vertical transmission | Likely not | |
| Conflict resolution strategies | Yes | 256 and 433 |
| Coordination of cellular activities | Yes | 417 and 426 |
Evaluating whether consortia of cross-feeding bacteria fulfil these criteria indicates that even though important features such as mutual dependence, functional specialisation, cooperation, and cell-attachment result almost automatically from cooperative cross-feeding, a striking difference is that these interactions are often transient (Table 1). Due to the often non-permanent nature of association between cells, heritability of group-traits is likely low. Moreover, cross-feeding consortia do not form a cohesive unit that is clearly delimitable from other cells in the environment, but rather a delicate network of transiently interacting cells. Nevertheless, selection is expected to favour extended associations between compatible genotypes. Moreover, frequency-dependent selection and spatial self-organization within clusters should adjust the mixture and the positioning of cells within clusters, thus maximizing the supply of limiting metabolites for cooperative cells.420 Mutations that were favourable in the context of one group, might work equally well when the focal mutant is combined with other genotypes, thus compensating for the lack of a strict vertical transmission. Finally, repeated bouts of association and disassociation allow to purge detrimental mutations on a cluster-level, thus accelerating molecular evolution. Taken together, consortia of bacteria that engage in obligate cooperative cross-feeding do not form a coherent, multicellular organism. Still, their performance results from complex metabolic interactions among the constituent cells, which is more than the sum of its parts. Future work is necessary to determine how durable cross-feeding interactions are and how this affects coevolution of interacting cells.
5 Concluding remarks
Our comprehensive analysis of the available literature revealed how commonly bacteria are involved in metabolic cross-feeding interactions with other bacteria, archaea, or eukaryotic organisms (Fig. 3). Particularly striking was the tremendous diversity in terms of mode of metabolite exchange that characterized the analysed interactions (Fig. 4). What also became obvious from screening the available literature, however, is that despite intensive efforts to study the molecular details of numerous metabolic cross-feeding interactions during the past decades,23,63,376 ecological or evolutionary aspects of these interactions are still notoriously understudied. Moreover, research attention so far has been mainly directed towards a relatively small set of model systems that have been studied in more detail.23,63,376 Thus, more empirical work is required to systematically compare cross-feeding interactions of different environmental origins and taxonomic compositions. In addition, the prevalence and ecological significance of metabolite cross-feeding should be increasingly analysed in different natural microbial communities, especially focussing on its importance for structuring these communities. Finally, the analysis of isolated consortia should be complemented by studies of synthetically engineered or experimentally evolved interactions, in which the causal molecular and evolutionary factors can be identified much easier.With the growing realization that metabolic interactions within microbial communities and populations are key for determining human health,8 global biogeochemical cycles6 or the yield in biotechnological production processes,421 the need to understand the rules that govern the emergence and evolution of these interactions is becoming particularly urgent.
Undoubtedly, the development of new technologies to chemically identify and characterize exchanged metabolites, to derive transcriptional and proteomic information of individual genotypes in a coculture context, as well as to differentially label and image interacting cells under controlled conditions, will significantly advance the study of microbial metabolite exchange.13 Especially the possibility to combine different methods will provide exciting opportunities, such as to image living cocultures on a single-cell level and simultaneously visualize the distribution of metabolites with a high spatial resolution. Moreover, current computational advances in simulating metabolic processes of cells that are embedded in complex communities hold the potential to predict bacterial metabolite exchange interactions based on the genome sequence of the organisms present in the community.422,423
A wealth of exciting research opportunities is waiting in this rapidly emerging field. Interesting questions that should be addressed in the future include (i) which ecological factors determine the assembly of metabolically interacting consortia in natural microbial communities (e.g. motility, chemotaxis, antagonistic interactions)? (ii) How stable/transient are obligate metabolic interactions in natural environments? (iii) Which evolutionary consequences result for genotypes that transition into an obligate metabolic dependency with other genotypes (e.g. local adaptation, genome streamlining, improved efficiency)? (iv) Which rules govern the division of metabolic functions within microbial communities? or (v) Are clusters of metabolite cross-feeding cells evolutionary individuals?
Evolution does not only proceed by giving rise to new species, but also by merging previously independent organisms into new life-forms.424 Consequently, intricate metabolic interdependencies between two or more individuals are a general feature of life. Answering the abovementioned questions using metabolite cross-feeding within microbial communities as a tractable model therefore holds the potential to help resolve the fundamental evolutionary problem of how biological complexity can emerge from the establishment of cooperative interactions among simpler units.
6 Conflicts of interest
There are no conflicts of interest to declare.7 Acknowledgements
We thank Wilhelm Boland and Martin Ackermann for support as well as Christoph Kaleta, Martin Kaltenpoth, Paul Rainey, Will Ratcliff as well as the whole Research Group Insect Symbiosis for fruitful discussions. Funding from the Jena School of Microbial Communication (SW, DP, CK), the Volkswagen Foundation (I/85 290 to GD, SS, CK), the DAAD GERLS program (GY, CK), the German Research Foundation (SFB 944-P19 and KO 3909/2-1 to DP, SS, CK), the ETH-Zürich postdoctoral fellowship program (GD), the Marie Curie Actions for People COFUND Program (GD), the Simons Collaboration on the Principles of Microbial Ecosystems (GD), and the Cluster of Excellence ‘Inflammation at Interfaces’ (ExC 306 to SW) is gratefully acknowledged.8 References
- W. B. Whitman, D. C. Coleman and W. J. Wiebe, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6578–6583 CrossRef.
- M. C. Horner-Devine, K. M. Carney and B. J. Bohannan, Proc. R. Soc. London, Ser. B, 2004, 271, 113–122 CrossRef PubMed.
- J. O. McInerney, Nat. Microbiol., 2016, 1, 16139 CrossRef PubMed.
- M. C. Weiss, F. L. Sousa, N. Mrnjavac, S. Neukirchen, M. Roettger, S. Nelson-Sathi and W. F. Martin, Nat. Microbiol., 2016, 1, 16116 CrossRef PubMed.
- F. U. Battistuzzi, A. Feijao and S. B. Hedges, BMC Evol. Biol., 2004, 4, 44 CrossRef PubMed.
- P. G. Falkowski, T. Fenchel and E. F. Delong, Science, 2008, 320, 1034–1039 CrossRef PubMed.
- J. E. Lovelock and L. Margulis, Tellus, 1974, 26, 2–10 CrossRef.
- M. McFall-Ngai, M. G. Hadfield, T. C. Bosch, H. V. Carey, T. Domazet-Lošo, A. E. Douglas, N. Dubilier, G. Eberl, T. Fukami and S. F. Gilbert, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3229–3236 CrossRef PubMed.
- S. Mitri and K. R. Foster, Annu. Rev. Genet., 2013, 47, 247–273 CrossRef PubMed.
- A. E. Little, C. J. Robinson, S. B. Peterson, K. F. Raffa and J. Handelsman, Annu. Rev. Microbiol., 2008, 62, 375–401 CrossRef PubMed.
- J. L. Sachs and A. C. Hollowell, mBio, 2012, 3(3), e00099-12 CrossRef PubMed.
- R. González-Cabaleiro, I. D. Ofiţeru, J. M. Lema and J. Rodríguez, ISME J., 2015, 9, 2630–2641 CrossRef PubMed.
- V. V. Phelan, W.-T. Liu, K. Pogliano and P. C. Dorrestein, Nat. Chem. Biol., 2012, 8, 26–35 CrossRef PubMed.
- B. Kerr, M. A. Riley, M. W. Feldman and B. J. Bohannan, Nature, 2002, 418, 171–174 CrossRef PubMed.
- B. C. Kirkup and M. A. Riley, Nature, 2004, 428, 412–414 CrossRef PubMed.
- K. R. Foster and T. Bell, Curr. Biol., 2012, 22, 1845–1850 CrossRef PubMed.
- M. E. Hibbing, C. Fuqua, M. R. Parsek and S. B. Peterson, Nat. Rev. Microbiol., 2009, 8, nrmicro2259 Search PubMed.
- O. Ciofu, T. J. Beveridge, J. Kadurugamuwa, J. Walther-Rasmussen and N. Høiby, J. Antimicrob. Chemother., 2000, 45, 9–13 CrossRef PubMed.
- R. Daniels, J. Vanderleyden and J. Michiels, FEMS Microbiol. Rev., 2004, 28, 261–289 CrossRef PubMed.
- D. Davies and G. Geesey, Appl. Environ. Microbiol., 1995, 61, 860–867 Search PubMed.
- A. S. Griffin, S. A. West and A. Buckling, Nature, 2004, 430, 1024–1027 CrossRef PubMed.
- S. A. West, A. S. Griffin, A. Gardner and S. P. Diggle, Nat. Rev. Microbiol., 2006, 4, 597–607 CrossRef PubMed.
- E. C. Seth and M. E. Taga, Front. Microbiol., 2014, 5, 350 Search PubMed.
- A. Zelezniak, S. Andrejev, O. Ponomarova, D. R. Mende, P. Bork and K. R. Patil, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 6449–6454 CrossRef PubMed.
- M. T. Mee, J. J. Collins, G. M. Church and H. H. Wang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E2149–E2156 CrossRef PubMed.
- G. D'Souza, S. Waschina, S. Pande, K. Bohl, C. Kaleta and C. Kost, Evolution, 2014, 68, 2559–2570 CrossRef PubMed.
- B. Schink, Antonie van Leeuwenhoek, 2002, 81, 257–261 CrossRef PubMed.
- R. Pound, Am. Nat., 1893, 27, 509–520 CrossRef.
- C. E. Marshall, Science, 1915, 306–312 Search PubMed.
- M. Dawson, Proc. R. Soc. London, Ser. B, 1899, 64, 167–168 Search PubMed.
- D. T. MacDougal, Bull. Torrey Bot. Club, 1899, 26, 511–530 CrossRef.
- T. P. Ferguson and G. Bond, Ann. Bot., 1954, 18, 385–396 CrossRef.
- M. S. Marshall, J. Dairy Sci., 1920, 3, 406–413 CrossRef.
- V. Graham, J. Bacteriol., 1943, 45, 51 Search PubMed.
- D. A. Soulides, Appl. Microbiol., 1955, 3, 129–131 Search PubMed.
- H. M. Ward, Philos. Trans. R. Soc., B, 1892, 183, 125–197 CrossRef.
- W. Pfeffer, Pflanzenphysiologie: ein Handbuch der Lehre vom Stoffwechsel und Kraftwechsel in der Pflanze, Engelmann, Leipzig, 1897 Search PubMed.
- H. Marshall Ward D.Sc, Ann. Bot., 1899, 549–562 Search PubMed.
- H. Reinheimer, Symbiogenesis, the universal law of progressive evolution, Knapp, Drewett, 1915 Search PubMed.
- F. Gibson and M. Jones, J. Sci. Comput., 1954, 17, 33–34 Search PubMed.
- V. Nurmikko, Experientia, 1956, 12, 245–249 CrossRef PubMed.
- V. Nurmikko, Acta Chem. Scand., 1955, 9, 1317–1322 CrossRef.
- S. R. Sørensen, Z. Ronen and J. Aamand, Appl. Environ. Microbiol., 2002, 68, 3478–3485 CrossRef.
- M. L. de Souza, D. Newcombe, S. Alvey, D. E. Crowley, A. Hay, M. J. Sadowsky and L. P. Wackett, Appl. Environ. Microbiol., 1998, 64, 178–184 Search PubMed.
- S. J. Roberts, A. Walker, N. R. Parekh, S. J. Welch and M. J. Waddington, Pest Manage. Sci., 1993, 39, 71–78 CrossRef.
- M. J. McInerney, M. P. Bryant and N. Pfennig, Arch. Microbiol., 1979, 122, 129–135 CrossRef.
- P. Fildes, Microbiology, 1956, 15, 636–642 CrossRef PubMed.
- W. B. Dempsey and P. F. Pachler, J. Bacteriol., 1966, 91, 642–645 Search PubMed.
- S. Pomper, J. Bacteriol., 1952, 63, 707 Search PubMed.
- E. Glanville and M. Demerec, Genetics, 1960, 45, 1359 Search PubMed.
- K. Rahman, J. Thahira-Rahman, P. Lakshmanaperumalsamy and I. Banat, Bioresour. Technol., 2002, 85, 257–261 CrossRef PubMed.
- A. Haritash and C. Kaushik, J. Hazard. Mater., 2009, 169, 1–15 CrossRef PubMed.
- N. Das and P. Chandran, Biotechnol. Res. Int., 2011, 2011, 941810 Search PubMed.
- H. Ali, Water, Air, Soil Pollut., 2010, 213, 251–273 CrossRef.
- R. G. Saratale, G. D. Saratale, J.-S. Chang and S. Govindwar, J. Taiwan Inst. Chem. Eng., 2011, 42, 138–157 CrossRef.
- D. S. Treves, S. Manning and J. Adams, Mol. Biol. Evol., 1998, 15, 789–797 CrossRef PubMed.
- R. B. Helling, T. Kinney and J. Adams, Microbiology, 1981, 123, 129–141 Search PubMed.
- R. B. Helling, C. N. Vargas and J. Adams, Genetics, 1987, 116, 349–358 Search PubMed.
- R. F. Rosenzweig, R. Sharp, D. S. Treves and J. Adams, Genetics, 1994, 137, 903–917 Search PubMed.
- P. E. Turner, V. Souza and R. E. Lenski, Ecology, 1996, 77, 2119–2129 CrossRef.
- M. L. Friesen, G. Saxer, M. Travisano and M. Doebeli, Evolution, 2004, 58, 245–260 CrossRef PubMed.
- T. Pfeiffer and S. Bonhoeffer, Am. Nat., 2004, 163, E126–E135 CrossRef PubMed.
- B. E. Morris, R. Henneberger, H. Huber and C. Moissl-Eichinger, FEMS Microbiol. Rev., 2013, 37, 384–406 CrossRef PubMed.
- E. Doyle, L. Muckian, A. M. Hickey and N. Clipson, Adv. Appl. Microbiol., 2008, 65, 27–66 Search PubMed.
- I. Shiio, S.-I. Ôtsuka and M. Takahashi, J. Biochem., 1962, 51, 56–62 CrossRef PubMed.
- N. Paczia, A. Nilgen, T. Lehmann, J. Gätgens, W. Wiechert and S. Noack, Microb. Cell Fact., 2012, 11, 122 CrossRef PubMed.
- T. Shimoyama, S. Kato, S. i. Ishii and K. Watanabe, Science, 2009, 323, 1574 CrossRef PubMed.
- A. Belenguer, S. H. Duncan, A. G. Calder, G. Holtrop, P. Louis, G. E. Lobley and H. J. Flint, Appl. Environ. Microbiol., 2006, 72, 3593–3599 CrossRef PubMed.
- W. Harcombe, Evolution, 2010, 64, 2166–2172 Search PubMed.
- E. H. Wintermute and P. A. Silver, Mol. Syst. Biol., 2010, 6, 407 CrossRef PubMed.
- L. Baumann, P. Baumann, N. A. Moran, J. Sandström and M. L. Thao, J. Mol. Evol., 1999, 48, 77–85 CrossRef PubMed.
- F. Lipschultz, O. Zafiriou, S. Wofsy, M. McElroy, F. Valois and S. Watson, Nature, 1981, 294, 641–643 CrossRef.
- H. Koch, S. Lücker, M. Albertsen, K. Kitzinger, C. Herbold, E. Spieck, P. H. Nielsen, M. Wagner and H. Daims, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11371–11376 CrossRef PubMed.
- S. Pande, H. Merker, K. Bohl, M. Reichelt, S. Schuster, L. F. de Figueiredo, C. Kaleta and C. Kost, ISME J., 2014, 8, 953–962 CrossRef PubMed.
- O. Leimar and R. C. Connor, Genetic and cultural evolution of cooperation, 2003, pp. 203–222 Search PubMed.
- J. L. Sachs, U. G. Mueller, T. P. Wilcox and J. J. Bull, Q. Rev. Biol., 2004, 79, 135–160 CrossRef PubMed.
- J. Russel, H. L. Røder, J. S. Madsen, M. Burmølle and S. J. Sørensen, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10684–10688 CrossRef PubMed.
- J. S. Madsen, H. L. Røder, J. Russel, H. Sørensen, M. Burmølle and S. J. Sørensen, Environ. Microbiol., 2016, 18, 2565–2574 CrossRef PubMed.
- A. Z. Rosenthal, E. G. Matson, A. Eldar and J. R. Leadbetter, ISME J., 2011, 5, 1133–1142 CrossRef PubMed.
- J. R. Graber and J. A. Breznak, Appl. Environ. Microbiol., 2005, 71, 1883–1889 CrossRef PubMed.
- P. Kaiser, in Azospirillum VI and Related Microorganisms: Genetics-Physiology-Ecology, ed. I. Fendrik, M. del Gallo, J. Vanderleyden and M. de Zamaroczy, Springer Berlin Heidelberg, Berlin, Heidelberg, 1995, pp. 207–212, DOI:10.1007/978-3-642-79906-8_21.
- J. Kives, D. Guadarrama, B. Orgaz, A. Rivera-Sen, J. Vazquez and C. SanJose, J. Dairy Sci., 2005, 88, 4165–4171 CrossRef PubMed.
- T. Woyke, H. Teeling, N. N. Ivanova, M. Huntemann, M. Richter, F. O. Gloeckner, D. Boffelli, I. J. Anderson, K. W. Barry, H. J. Shapiro, E. Szeto, N. C. Kyrpides, M. Mussmann, R. Amann, C. Bergin, C. Ruehland, E. M. Rubin and N. Dubilier, Nature, 2006, 443, 950–955 CrossRef PubMed.
- J. Foster, M. Ganatra, I. Kamal, J. Ware, K. Makarova, N. Ivanova, A. Bhattacharyya, V. Kapatral, S. Kumar, J. Posfai, T. Vincze, J. Ingram, L. Moran, A. Lapidus, M. Omelchenko, N. Kyrpides, E. Ghedin, S. Wang, E. Goltsman, V. Joukov, O. Ostrovskaya, K. Tsukerman, M. Mazur, D. Comb, E. Koonin and B. Slatko, PLoS Biol., 2005, 3(4), e121 Search PubMed.
- S. L. Garcia, M. Buck, K. D. McMahon, H.-P. Grossart, A. Eiler and F. Warnecke, Mol. Ecol., 2015, 24, 4449–4459 CrossRef PubMed.
- S. Ghignone, A. Salvioli, I. Anca, E. Lumini, G. Ortu, L. Petiti, S. Cruveiller, V. Bianciotto, P. Piffanelli, L. Lanfranco and P. Bonfante, ISME J., 2012, 6, 136–145 CrossRef PubMed.
- J. L. Mark Welch, B. J. Rossetti, C. W. Rieken, F. E. Dewhirst and G. G. Borisy, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E791–E800 CrossRef PubMed.
- A. E. Dekas, R. S. Poretsky and V. J. Orphan, Science, 2009, 326, 422–426 CrossRef PubMed.
- D. Wu, S. C. Daugherty, S. E. Van Aken, G. H. Pai, K. L. Watkins, H. Khouri, L. J. Tallon, J. M. Zaborsky, H. E. Dunbar, P. L. Tran, N. A. Moran and J. A. Eisen, PLoS Biol., 2006, 4, e188 Search PubMed.
- C. B. Walker, A. M. Redding-Johanson, E. E. Baidoo, L. Rajeev, Z. L. He, E. L. Hendrickson, M. P. Joachimiak, S. Stolyar, A. P. Arkin, J. A. Leigh, J. Z. Zhou, J. D. Keasling, A. Mukhopadhyay and D. A. Stahl, ISME J., 2012, 6, 2045–2055 CrossRef PubMed.
- S. Sieuwerts, D. Molenaar, S. van Hijum, M. Beerthuyzen, M. J. A. Stevens, P. W. M. Janssen, C. J. Ingham, F. A. M. de Bok, W. M. de Vos and J. Vlieg, Appl. Environ. Microbiol., 2010, 76, 7775–7784 CrossRef PubMed.
- U. Jahn, R. Summons, H. Sturt, E. Grosjean and H. Huber, Arch. Microbiol., 2004, 182, 404–413 CrossRef PubMed.
- S. Sawanon and Y. Kobayashi, Anim. Sci. J., 2006, 77, 208–214 CrossRef.
- R. A. Foster, M. M. M. Kuypers, T. Vagner, R. W. Paerl, N. Musat and J. P. Zehr, ISME J., 2011, 5, 1484–1493 CrossRef PubMed.
- A. Rivière, M. Gagnon, S. Weckx, D. Roy and L. De Vuyst, Appl. Environ. Microbiol., 2015, 81, 7767–7781 CrossRef PubMed.
- A. Deveau, C. Brulé, B. Palin, D. Champmartin, P. Rubini, J. Garbaye, A. Sarniguet and P. Frey-Klett, Environ. Microbiol. Rep., 2010, 2, 560–568 CrossRef PubMed.
- M. Gobbetti, A. Corsetti and J. Rossi, Appl. Microbiol. Biotechnol., 1994, 41, 456–460 CrossRef.
- M. M. Ramsey, K. P. Rumbaugh and M. Whiteley, PLoS Pathog., 2011, 7, e1002012 Search PubMed.
- P. Engel and N. A. Moran, FEMS Microbiol. Rev., 2013, 37, 699–735 CrossRef PubMed.
- C. Kaiser, M. R. Kilburn, P. L. Clode, L. Fuchslueger, M. Koranda, J. B. Cliff, Z. M. Solaiman and D. V. Murphy, New Phytol., 2015, 205, 1537–1551 CrossRef PubMed.
- R. L. Chapman, Mitig. Adapt. Strategies Glob. Change, 2013, 18, 5–12 CrossRef.
- M. K. Bach, W. E. Magee and R. H. Burris, Plant Physiol., 1958, 33, 118–124 CrossRef PubMed.
- M. T. Croft, A. D. Lawrence, E. Raux-Deery, M. J. Warren and A. G. Smith, Nature, 2005, 438, 90–93 CrossRef PubMed.
- S. A. Amin, D. H. Green, M. C. Hart, F. C. Küpper, W. G. Sunda and C. J. Carrano, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17071–17076 CrossRef PubMed.
- M. T. Croft, M. J. Warren and A. G. Smith, Eukaryotic Cell, 2006, 5, 1175–1183 CrossRef PubMed.
- J. Foster, M. Ganatra, I. Kamal, J. Ware, K. Makarova, N. Ivanova, A. Bhattacharyya, V. Kapatral, S. Kumar, J. Posfai, T. Vincze, J. Ingram, L. Moran, A. Lapidus, M. Omelchenko, N. Kyrpides, E. Ghedin, S. Wang, E. Goltsman, V. Joukov, O. Ostrovskaya, K. Tsukerman, M. Mazur, D. Comb, E. Koonin and B. Slatko, PLoS Biol., 2005, 3, e121 Search PubMed.
- M. E. Hibbing, C. Fuqua, M. R. Parsek and S. B. Peterson, Nat. Rev. Microbiol., 2010, 8, 15–25 CrossRef PubMed.
- K. B. Xavier and B. L. Bassler, Nature, 2005, 437, 750–753 CrossRef PubMed.
- S. Uroz, S. R. Chhabra, M. Camara, P. Williams, P. Oger and Y. Dessaux, Microbiology, 2005, 151, 3313–3322 CrossRef PubMed.
- B.-Y. Wang and H. K. Kuramitsu, Appl. Environ. Microbiol., 2005, 71, 354–362 CrossRef PubMed.
- L. M. Mashburn and M. Whiteley, Nature, 2005, 437, 422 CrossRef PubMed.
- H. Nikaido, Microbiol. Mol. Biol. Rev., 2003, 67, 593–656 CrossRef PubMed.
- Z. Ren, T. E. Ward and J. M. Regan, Environ. Sci. Technol., 2007, 41, 4781–4786 CrossRef PubMed.
- J. Zhou, Q. Ma, H. Yi, L. Wang, H. Song and Y.-J. Yuan, Appl. Environ. Microbiol., 2011, 77, 7023–7030 CrossRef PubMed.
- D. R. Boone, R. L. Johnson and Y. Liu, Appl. Environ. Microbiol., 1989, 55, 1735–1741 Search PubMed.
- B. Schink and A. J. M. Stams, in The Prokaryotes: Prokaryotic Communities and Ecophysiology, ed. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt and F. Thompson, Springer Berlin Heidelberg, Berlin, Heidelberg, 2013, pp. 471–493, DOI:10.1007/978-3-642-30123-0_59.
- R. Krämer, FEMS Microbiol. Rev., 1994, 13, 75–93 CrossRef.
- J. H. Crosa and C. T. Walsh, Microbiol. Mol. Biol. Rev., 2002, 66, 223–249 CrossRef PubMed.
- J. L. Furrer, D. N. Sanders, I. G. Hook-Barnard and M. A. McIntosh, Mol. Microbiol., 2002, 44, 1225–1234 CrossRef PubMed.
- C. Arnosti, Ann. Rev. Mar. Sci., 2011, 3, 401–425 CrossRef PubMed.
- H.-C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef PubMed.
- J. Wingender, T. R. Neu and H.-C. Flemming, in Microbial extracellular polymeric substances, Springer, 1999, pp. 1–19 Search PubMed.
- D. C. Rees, E. Johnson and O. Lewinson, Nat. Rev. Mol. Cell Biol., 2009, 10, 218–227 CrossRef PubMed.
- P. Postma, J. Lengeler and G. Jacobson, Microbiol. Rev., 1993, 57, 543–594 Search PubMed.
- Y. Zhang, D. A. Rodionov, M. S. Gelfand and V. N. Gladyshev, BMC Genom., 2009, 10, 78 CrossRef PubMed.
- P. H. Degnan, N. A. Barry, K. C. Mok, M. E. Taga and A. L. Goodman, Cell Host Microbe, 2014, 15, 47–57 Search PubMed.
- A. Nasarabadi, J. E. Berleman and M. Auer, Biogenesis of Fatty Acids, Lipids and Membranes, 2017, pp. 1–15 Search PubMed.
- A. Kulp and M. J. Kuehn, Annu. Rev. Microbiol., 2010, 64, 163–184 CrossRef PubMed.
- T. J. Beveridge, J. Bacteriol., 1999, 181, 4725–4733 Search PubMed.
- R. Fiocca, V. Necchi, P. Sommi, V. Ricci, J. Telford, T. L. Cover and E. Solcia, J. Pathol., 1999, 188, 220–226 CrossRef PubMed.
- S. Kato, Y. Kowashi and D. R. Demuth, Microb. Pathog., 2002, 32, 1–13 CrossRef PubMed.
- S. N. Wai, B. Lindmark, T. Söderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe and B. E. Uhlin, Cell, 2003, 115, 25–35 CrossRef PubMed.
- K. Yokoyama, T. Horii, T. Yamashino, S. Hashikawa, S. Barua, T. Hasegawa, H. Watanabe and M. Ohta, FEMS Microbiol. Lett., 2000, 192, 139–144 CrossRef PubMed.
- S. Patrick, J. P. McKenna, O. Seamus and E. Dermott, Microb. Pathog., 1996, 20, 191–202 CrossRef PubMed.
- J. L. Kadurugamuwa and T. J. Beveridge, J. Bacteriol., 1995, 177, 3998–4008 CrossRef PubMed.
- D. W. Dorward and C. F. Garon, Appl. Environ. Microbiol., 1990, 56, 1960–1962 Search PubMed.
- L. Mashburn-Warren, R. Mclean and M. Whiteley, Geobiology, 2008, 6, 214–219 CrossRef PubMed.
- J. P. Remis, J. W. Costerton and M. Auer, ISME J., 2010, 4, 1085–1087 CrossRef PubMed.
- H. Yonezawa, T. Osaki, S. Kurata, M. Fukuda, H. Kawakami, K. Ochiai, T. Hanawa and S. Kamiya, BMC Microbiol., 2009, 9, 197 CrossRef PubMed.
- C. Pérez-Cruz, O. Carrión, L. Delgado, G. Martinez, C. López-Iglesias and E. Mercade, Appl. Environ. Microbiol., 2013, 79, 1874–1881 CrossRef PubMed.
- C. Pérez-Cruz, L. Delgado, C. López-Iglesias and E. Mercade, PLoS One, 2015, 10, e0116896 Search PubMed.
- S. J. Biller, F. Schubotz, S. E. Roggensack, A. W. Thompson, R. E. Summons and S. W. Chisholm, Science, 2014, 343, 183–186 CrossRef PubMed.
- J. P. Remis, D. Wei, A. Gorur, M. Zemla, J. Haraga, S. Allen, H. E. Witkowska, J. W. Costerton, J. E. Berleman and M. Auer, Environ. Microbiol., 2014, 16, 598–610 CrossRef PubMed.
- J. E. Berleman, S. Allen, M. A. Danielewicz, J. P. Remis, A. Gorur, J. Cunha, M. Z. Hadi, D. R. Zusman, T. R. Northen and H. E. Witkowska, Front. Microbiol., 2014, 5, 474 Search PubMed.
- G. P. Dubey and S. Ben-Yehuda, Cell, 2011, 144, 590–600 CrossRef PubMed.
- G. P. Dubey, G. B. M. Mohan, A. Dubrovsky, T. Amen, S. Tsipshtein, A. Rouvinski, A. Rosenberg, D. Kaganovich, E. Sherman and O. Medalia, Dev. Cell, 2016, 36, 453–461 CrossRef PubMed.
- S. Pande, S. Shitut, L. Freund, M. Westermann, F. Bertels, C. Colesie, I. B. Bischofs and C. Kost, Nat. Commun., 2015, 6, 6238 CrossRef PubMed.
- S. i. Ishii, T. Kosaka, K. Hori, Y. Hotta and K. Watanabe, Appl. Environ. Microbiol., 2005, 71, 7838–7845 CrossRef PubMed.
- P. T. Ha, S. R. Lindemann, L. Shi, A. C. Dohnalkova, J. K. Fredrickson, M. T. Madigan and H. Beyenal, Nat. Commun., 2017, 8, 13924 CrossRef PubMed.
- S. Benomar, D. Ranava, M. L. Cárdenas, E. Trably, Y. Rafrafi, A. Ducret, J. Hamelin, E. Lojou, J.-P. Steyer and M.-T. Giudici-Orticoni, Nat. Commun., 2015, 6, 6283 CrossRef PubMed.
- M. A. Nowak, Science, 2006, 314, 1560–1563 CrossRef PubMed.
- G. Hardin, J. Nat. Resour. Pol. Res., 2009, 1, 243–253 CrossRef.
- A. Goelzer and V. Fromion, Biochim. Biophys. Acta Gen. Subj., 2011, 1810, 978–988 CrossRef PubMed.
- A. Varma and B. O. Palsson, Appl. Environ. Microbiol., 1994, 60, 3724–3731 Search PubMed.
- J. Tasoff, M. T. Mee and H. H. Wang, PLoS One, 2015, 10, e0132907 Search PubMed.
- P. Hammerstein and R. Noë, Philos. Trans. R. Soc., B, 2016, 371, 20150101 CrossRef PubMed.
- D. Ricardo, The works and correspondence of David Ricardo Vol. 1: On the principles of political economy and taxation, 1817 Search PubMed.
- D. R. Johnson, F. Goldschmidt, E. E. Lilja and M. Ackermann, ISME J., 2012, 6, 1985–1991 CrossRef PubMed.
- T. Ferenci, Trends Microbiol., 2016, 24, 209–223 CrossRef PubMed.
- E. E. Lilja and D. R. Johnson, ISME J., 2016, 10, 1568–1578 CrossRef PubMed.
- O. Ponomarova and K. R. Patil, Curr. Opin. Microbiol., 2015, 27, 37–44 CrossRef PubMed.
- G. D. Werner, J. E. Strassmann, A. B. Ivens, D. J. Engelmoer, E. Verbruggen, D. C. Queller, R. Noë, N. C. Johnson, P. Hammerstein and E. T. Kiers, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1237–1244 CrossRef PubMed.
- C. Kaleta, S. Schäuble, U. Rinas and S. Schuster, Biotechnol. J., 2013, 8, 1105–1114 CrossRef PubMed.
- S. Waschina, G. D'Souza, C. Kost and C. Kaleta, FEBS J., 2016, 283, 2149–2163 CrossRef PubMed.
- H. Akashi and T. Gojobori, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 3695–3700 CrossRef PubMed.
- E. M. Heizer Jr, D. W. Raiford, M. L. Raymer, T. E. Doom, R. V. Miller and D. E. Krane, Mol. Biol. Evol., 2006, 23, 1670–1680 CrossRef PubMed.
- S. Germerodt, K. Bohl, A. Lück, S. Pande, A. Schröter, C. Kaleta, S. Schuster and C. Kost, PLoS Comput. Biol., 2016, 12, e1004986 Search PubMed.
- P. Hu, E. A. Dubinsky, A. J. Probst, J. Wang, C. M. Sieber, L. M. Tom, P. R. Gardinali, J. F. Banfield, R. M. Atlas and G. L. Andersen, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 7432–7437 CrossRef PubMed.
- J. Zhao and K. Shimizu, J. Biotechnol., 2003, 101, 101–117 CrossRef PubMed.
- V. Sabarly, C. Aubron, J. Glodt, T. Balliau, O. Langella, D. Chevret, O. Rigal, A. Bourgais, B. Picard and D. Vienne, Environ. Microbiol., 2016, 18, 100–117 CrossRef PubMed.
- J. L. M. Welch, B. J. Rossetti, C. W. Rieken, F. E. Dewhirst and G. G. Borisy, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E791–E800 CrossRef PubMed.
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef PubMed.
- P. S. Stewart and M. J. Franklin, Nat. Rev. Microbiol., 2008, 6, 199–210 CrossRef PubMed.
- M. Ackermann, Nat. Rev. Microbiol., 2015, 13, 497–508 CrossRef PubMed.
- D. Healey, K. Axelrod and J. Gore, Mol. Syst. Biol., 2016, 12, 877 CrossRef PubMed.
- K. Kumar, R. A. Mella-Herrera and J. W. Golden, Cold Spring Harbor Perspect. Biol., 2010, 2, a000315 CrossRef PubMed.
- A. Z. Rosenthal, Y. Qi, S. Hormoz, J. Park, C. H.-J. Li and M. Elowitz, bioRxiv, 2017, 208686 Search PubMed.
- M. T. Wortel, E. Noor, M. Ferris, F. J. Bruggeman and W. Liebermeister, bioRxiv, 2017, 111161 Search PubMed.
- S. M. Carroll, M.-C. Lee and C. J. Marx, Evolution, 2014, 68, 760–771 CrossRef PubMed.
- J. J. Morris, Trends Genet., 2015, 31, 475–482 CrossRef PubMed.
- Y. Kallus, J. H. Miller and E. Libby, Nat. Commun., 2017, 8, 1361 CrossRef PubMed.
- J. J. Wernegreen, Nat. Rev. Genet., 2002, 3, 850–861 CrossRef PubMed.
- G. H. Thomas, J. Zucker, S. J. Macdonald, A. Sorokin, I. Goryanin and A. E. Douglas, BMC Syst. Biol., 2009, 3, 24 CrossRef PubMed.
- N. A. Moran, J. P. McCutcheon and A. Nakabachi, Annu. Rev. Genet., 2008, 42, 165–190 CrossRef PubMed.
- J. P. McCutcheon and N. A. Moran, Nat. Rev. Microbiol., 2012, 10, 13–26 CrossRef PubMed.
- J. P. McCutcheon and N. A. Moran, Genome Biol. Evol., 2010, 2, 708–718 Search PubMed.
- J. P. McCutcheon and N. A. Moran, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19392–19397 CrossRef PubMed.
- A. Mira, L. Klasson and S. G. Andersson, Curr. Opin. Microbiol., 2002, 5, 506–512 CrossRef PubMed.
- K. Han, Z.-f. Li, R. Peng, L.-p. Zhu, T. Zhou, L.-g. Wang, S.-g. Li, X.-b. Zhang, W. Hu and Z.-h. Wu, Sci. Rep., 2013, 3, 2101 CrossRef PubMed.
- A. Nakabachi, A. Yamashita, H. Toh, H. Ishikawa, H. E. Dunbar, N. A. Moran and M. Hattori, Science, 2006, 314, 267 CrossRef PubMed.
- P. Puigbò, A. Lobkovsky, D. Kristensen, Y. Wolf and E. Koonin, BMC Biol., 2014, 12, 1–19 CrossRef PubMed.
- E. N. Gordienko, M. D. Kazanov and M. S. Gelfand, J. Bacteriol., 2013, 195, 2786–2792 CrossRef PubMed.
- H. Tettelin, D. Riley, C. Cattuto and D. Medini, Curr. Opin. Microbiol., 2008, 11, 472–477 CrossRef PubMed.
- D. Medini, C. Donati, H. Tettelin, V. Masignani and R. Rappuoli, Curr. Opin. Genet. Dev., 2005, 15, 589–594 CrossRef PubMed.
- S. J. Biller, P. M. Berube, D. Lindell and S. W. Chisholm, Nat. Rev. Microbiol., 2015, 13, 13–27 CrossRef PubMed.
- S. Koskiniemi, S. Sun, O. G. Berg and D. I. Andersson, PLoS Genet., 2012, 8(6), e1002787 Search PubMed.
- I. Shachrai, A. Zaslaver, U. Alon and E. Dekel, Mol. Cell, 2010, 38, 758–767 CrossRef PubMed.
- E. Dekel and U. Alon, Nature, 2005, 436, 588–592 CrossRef PubMed.
- A. Dufresne, L. Garczarek and F. Partensky, Genome Biol., 2005, 6, R14 CrossRef PubMed.
- A. Mira, H. Ochman and N. A. Moran, Trends Genet., 2001, 17, 589–596 CrossRef PubMed.
- D. J. Martínez-Cano, M. Reyes-Prieto, E. Martínez-Romero, L. P. Partida-Martínez, A. Latorre, A. Moya and L. Delaye, Front. Microbiol., 2014, 5, 742 Search PubMed.
- B. K. Swan, B. Tupper, A. Sczyrba, F. M. Lauro, M. Martinez-Garcia, J. M. Gonzalez, H. Luo, J. J. Wright, Z. C. Landry, N. W. Hanson, B. P. Thompson, N. J. Poulton, P. Schwientek, S. G. Acinas, S. J. Giovannoni, M. A. Moran, S. J. Hallam, R. Cavicchioli, T. Woyke and R. Stepanauskas, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11463–11468 CrossRef PubMed.
- B. Batut, C. Knibbe, G. Marais and V. Daubin, Nat. Rev. Microbiol., 2014, 12, 841–850 CrossRef PubMed.
- J. Grote, J. C. Thrash, M. J. Huggett, Z. C. Landry, P. Carini, S. J. Giovannoni and M. S. Rappe, mBio, 2012, 3(5), e00252-12 CrossRef PubMed.
- L. A. Bristow, W. Mohr, S. Ahmerkamp and M. M. Kuypers, Curr. Biol., 2017, 27, R474–R478 CrossRef PubMed.
- S. Zamenhof and H. H. Eichhorn, Nature, 1967, 216, 456–458 CrossRef PubMed.
- G. D'Souza and C. Kost, PLoS Genet., 2016, 12, e1006364 Search PubMed.
- W. Kim and S. B. Levy, Appl. Environ. Microbiol., 2008, 74, 3644–3651 CrossRef PubMed.
- F. Wessely, M. Bartl, R. Guthke, P. Li, S. Schuster and C. Kaleta, Mol. Syst. Biol., 2011, 7, 515 CrossRef PubMed.
- A. G. Lochhead, Bacteriol. Rev., 1958, 22, 145–153 Search PubMed.
- A. Moura, M. A. Savageau and R. Alves, PLoS One, 2013, 8, e77319 Search PubMed.
- E. Schmidt and R. L. Starkey, Soil Sci., 1951, 71, 221–232 CrossRef.
- D. J. Levy-Booth, R. G. Campbell, R. H. Gulden, M. M. Hart, J. R. Powell, J. N. Klironomos, K. P. Pauls, C. J. Swanton, J. T. Trevors and K. E. Dunfield, Soil Biol. Biochem., 2007, 39, 2977–2991 CrossRef.
- M. C. Lee and C. J. Marx, PLoS Genet., 2012, 8, e1002651 Search PubMed.
- B. J. Paul, M. B. Berkmen and R. L. Gourse, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 7823–7828 CrossRef PubMed.
- V. Santos and I. Hirshfield, PLoS One, 2016, 11, e0157578 Search PubMed.
- D. J. Martínez-Cano, M. Reyes-Prieto, E. Martinez-Romero, L. P. Partida-Martinez, A. Latorre, A. Moya and L. Delaye, Front. Microbiol., 2015, 5, 742 Search PubMed.
- N. A. Moran, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 2873–2878 CrossRef.
- J. J. Wernegreen, Ann. N. Y. Acad. Sci., 2015, 1360, 16–35 CrossRef PubMed.
- J. H. Gillespie, Population genetics: a concise guide, JHU Press, 2010 Search PubMed.
- C.-H. Kuo and H. Ochman, Genome Biol. Evol., 2009, 1, 145–152 CrossRef PubMed.
- A. I. Nilsson, S. Koskiniemi, S. Eriksson, E. Kugelberg, J. C. D. Hinton and D. I. Andersson, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 12112–12116 CrossRef PubMed.
- P. Łukasik, K. Nazario, J. T. Van Leuven, M. A. Campbell, M. Meyer, A. Michalik, P. Pessacq, C. Simon, C. Veloso and J. P. McCutcheon, Proc. Natl. Acad. Sci. U. S. A., 2017, 201712321 Search PubMed.
- M. F. Traxler, S. M. Summers, H. T. Nguyen, V. M. Zacharia, G. A. Hightower, J. T. Smith and T. Conway, Mol. Microbiol., 2008, 68, 1128–1148 CrossRef PubMed.
- D. K. Aanen, H. Henrik, A. J. Debets, N. A. Kerstes, R. F. Hoekstra and J. J. Boomsma, Science, 2009, 326, 1103–1106 CrossRef PubMed.
- R. C. Connor, Anim. Behav., 1986, 34, 1562–1566 CrossRef.
- J. Sachs and A. Hollowell, mBio, 2012, 3, e00099-12 CrossRef PubMed.
- W. D. Hamilton, J. Theor. Biol., 1964, 7, 1–52 CrossRef PubMed.
- J. M. Smith, Nature, 1964, 201, 1145–1147 CrossRef.
- R. Dawkins, The selfish gene/Richard Dawkins, Oxford University Press, New York, 1976 Search PubMed.
- N. M. Oliveira, R. Niehus and K. R. Foster, Proc. Natl. Acad. Sci. U.S.A., 2014, 111, 17941–17946 CrossRef PubMed.
- C. E. Tarnita, J. Exp. Biol., 2017, 220, 18–24 CrossRef PubMed.
- J. E. Strassmann, O. M. Gilbert and D. C. Queller, Annu. Rev. Microbiol., 2011, 65, 349–367 CrossRef PubMed.
- R. C. MacLean, Heredity, 2008, 100, 233–239 CrossRef PubMed.
- J. A. Fletcher and M. Doebeli, Proc. R. Soc. B, 2009, 276, 13–19 CrossRef PubMed.
- D. S. Wilson, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 143–146 CrossRef.
- K. R. Foster and T. Wenseleers, J. Evol. Biol., 2006, 19, 1283–1293 CrossRef PubMed.
- A. Traulsen and M. A. Nowak, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10952–10955 CrossRef PubMed.
- T. Antal, H. Ohtsuki, J. Wakeley, P. D. Taylor and M. A. Nowak, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8597–8600 CrossRef PubMed.
- C. D. Nadell, K. R. Foster and J. B. Xavier, PLoS Comput. Biol., 2010, 6, e1000716 Search PubMed.
- M. Doebeli and N. Knowlton, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 8676–8680 CrossRef.
- S. K. Aoki, E. J. Diner, C. T. de Roodenbeke, B. R. Burgess, S. J. Poole, B. A. Braaten, A. M. Jones, J. S. Webb, C. S. Hayes, P. A. Cotter and D. A. Low, Nature, 2010, 468, 439–442 CrossRef PubMed.
- E. C. Garcia, M. S. Anderson, J. A. Hagar and P. A. Cotter, Mol. Microbiol., 2013, 89, 1213–1225 CrossRef PubMed.
- M. S. Anderson, E. C. Garcia and P. A. Cotter, PLoS Pathog., 2014, 10, 14 Search PubMed.
- C. E. Zobell, J. Bacteriol., 1943, 46, 39–56 Search PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef PubMed.
- O. X. Cordero and M. S. Datta, Curr. Opin. Microbiol., 2016, 31, 227–234 CrossRef PubMed.
- J. F. Mori, N. Ueberschaar, S. Lu, R. E. Cooper, G. Pohnert and K. Kusel, ISME J., 2017, 11, 1075–1086 CrossRef PubMed.
- M. van Veelen, J. Garcia and L. Aviles, J. Theor. Biol., 2010, 264, 1240–1253 CrossRef PubMed.
- P. B. Rainey and S. De Monte, Annu. Rev. Ecol. Evol. Syst., 2014, 45, 599–620 CrossRef.
- G. Corno and K. Jürgens, Appl. Environ. Microbiol., 2006, 72, 78–86 CrossRef PubMed.
- R. Liébana, L. Arregui, A. Santos, A. Murciano, D. Marquina and S. Serrano, FEMS Microbiol. Ecol., 2016, 92, fiw134 CrossRef PubMed.
- N. A. Lyons and R. Kolter, Curr. Opin. Microbiol., 2015, 24, 21–28 CrossRef PubMed.
- H.-C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef PubMed.
- J. D. Van Dyken, M. J. I. Müller, K. M. L. Mack and M. M. Desai, Curr. Biol., 2013, 23, 919–923 CrossRef PubMed.
- S. Pande, F. Kaftan, S. Lang, A. Svatos, S. Germerodt and C. Kost, ISME J., 2016, 10, 1413–1423 CrossRef PubMed.
- M. S. Datta, K. S. Korolev, I. Cvijovic, C. Dudley and J. Gore, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7354–7359 CrossRef PubMed.
- K. Drescher, C. D. Nadell, H. A. Stone, N. S. Wingreen and B. L. Bassler, Curr. Biol., 2014, 24, 50–55 CrossRef PubMed.
- S. Estrela and S. P. Brown, PLoS Comput. Biol., 2013, 9, e1003398 Search PubMed.
- J. van Gestel, F. J. Weissing, O. P. Kuipers and A. T. Kovacs, ISME J., 2014, 8, 2069–2079 CrossRef PubMed.
- B. Momeni, A. J. Waite and W. Shou, eLife, 2013, 2, e00960 Search PubMed.
- R. Tecon and D. Or, Sci. Rep., 2017, 7, 43726 CrossRef PubMed.
- C. D. Nadell, K. Drescher, N. S. Wingreen and B. L. Bassler, ISME J., 2015, 9, 1700–1709 CrossRef PubMed.
- M. A. Nowak, S. Bonhoeffer and R. M. May, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 4877–4881 CrossRef.
- M. Burmølle, J. S. Webb, D. Rao, L. H. Hansen, S. J. Sørensen and S. Kjelleberg, Appl. Environ. Microbiol., 2006, 72, 3916–3923 CrossRef PubMed.
- J. E. Strassmann and D. C. Queller, Evolution, 2016, 70, 848–859 CrossRef PubMed.
- L. M. Wahl, Am. Nat., 2002, 160(1), 135–145 CrossRef PubMed.
- A. Pernthaler, A. E. Dekas, C. T. Brown, S. K. Goffredi, T. Embaye and V. J. Orphan, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 7052–7057 CrossRef PubMed.
- M. F. Haroon, S. Hu, Y. Shi, M. Imelfort, J. Keller, P. Hugenholtz, Z. Yuan and G. W. Tyson, Nature, 2013, 500, 567–570 CrossRef PubMed.
- Á. T. Kovács, Front. Microbiol., 2014, 5, 649 Search PubMed.
- K. L. I. Norlund, G. Southam, T. Tyliszczak, Y. Hu, C. Karunakaran, M. Obst, A. P. Hitchcock and L. A. Warren, Environ. Sci. Technol., 2009, 43, 8781–8786 CrossRef PubMed.
- U. Jahn, M. Gallenberger, W. Paper, B. Junglas, W. Eisenreich, K. O. Stetter, R. Rachel and H. Huber, J. Bacteriol., 2008, 190, 1743–1750 CrossRef PubMed.
- X. S. He, J. S. McLean, A. Edlund, S. Yooseph, A. P. Hall, S. Y. Liu, P. C. Dorrestein, E. Esquenazi, R. C. Hunter, G. H. Cheng, K. E. Nelson, R. Lux and W. Y. Shi, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 244–249 CrossRef PubMed.
- F. Husnik and J. P. McCutcheon, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E5416–E5424 CrossRef PubMed.
- J. Overmann, in Symbiosis: Mechanisms and Model Systems, ed. J. Seckbach, Springer, Netherlands, Dordrecht, 2002, pp. 239–255, DOI:10.1007/0-306-48173-1_15.
- Y. Xia, Y. Kong, T. R. Thomsen and P. Halkjær Nielsen, Appl. Environ. Microbiol., 2008, 74, 2229–2238 CrossRef PubMed.
- D. Y. Kobayashi and J. A. Crouch, Annu. Rev. Phytopathol., 2009, 47, 63–82 CrossRef PubMed.
- R. Honegger, in Fungal Associations, ed. B. Hock, Springer Berlin Heidelberg, Berlin, Heidelberg, 2012, pp. 287–339, DOI:10.1007/978-3-642-30826-0_15.
- L. P. Partida-Martinez, S. Monajembashi, K.-O. Greulich and C. Hertweck, Curr. Biol., 2007, 17, 773–777 CrossRef PubMed.
- Z. Li, Q. Yao, S. P. Dearth, M. R. Entler, H. F. Castro Gonzalez, J. K. Uehling, R. J. Vilgalys, G. B. Hurst, S. R. Campagna, J. L. Labbé and C. Pan, Environ. Microbiol., 2017, 19, 1041–1053 CrossRef PubMed.
- H. Guo, S. P. Glaeser, I. Alabid, J. Imani, H. Haghighi, P. Kämpfer and K.-H. Kogel, Front. Microbiol., 2017, 8, 629 Search PubMed.
- V. P. Edgcomb, S. A. Breglia, N. Yubuki, D. Beaudoin, D. J. Patterson, B. S. Leander and J. M. Bernhard, ISME J., 2011, 5, 231–243 CrossRef PubMed.
- S. S. Epstein, D. A. Bazylinski and W. H. Fowle, J. Eukaryot. Microbiol., 1998, 45, 64–70 CrossRef.
- J. M. Bernhard, K. R. Buck, M. A. Farmer and S. S. Bowser, Nature, 2000, 403, 77–80 CrossRef PubMed.
- E. C. M. Nowack and M. Melkonian, Philos. Trans. R. Soc., B, 2010, 365, 699–712 CrossRef PubMed.
- R. Ramanan, B.-H. Kim, D.-H. Cho, H.-M. Oh and H.-S. Kim, Biotechnol. Adv., 2016, 34, 14–29 CrossRef PubMed.
- L. Cai, G. Zhou, R.-M. Tian, H. Tong, W. Zhang, J. Sun, W. Ding, Y. H. Wong, J. Y. Xie, J.-W. Qiu, S. Liu, H. Huang and P.-Y. Qian, Sci. Rep., 2017, 7, 9320 CrossRef PubMed.
- N. A. Moran, J. P. McCutcheon and A. Nakabachi, Annu. Rev. Genet., 2008, 42, 165–190 CrossRef PubMed.
- J. L. Sachs, R. G. Skophammer and J. U. Regus, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10800–10807 CrossRef PubMed.
- J. L. Sachs, R. G. Skophammer, N. Bansal and J. E. Stajich, Proc. R. Soc. Lond. B Biol. Sci., 2014, 281, 20132146 CrossRef PubMed.
- C. Toft and S. G. E. Andersson, Nat. Rev. Genet., 2010, 11, 465–475 CrossRef PubMed.
- E. Zientz, T. Dandekar and R. Gross, Microbiol. Mol. Biol. Rev., 2004, 68, 745–770 CrossRef PubMed.
- Z. Liu, J. Müller, T. Li, R. M. Alvey, K. Vogl, N.-U. Frigaard, N. C. Rockwell, E. S. Boyd, L. P. Tomsho, S. C. Schuster, P. Henke, M. Rohde, J. Overmann and D. A. Bryant, Genome Biol., 2013, 14, R127 CrossRef PubMed.
- G. Wanner, K. Vogl and J. Overmann, J. Bacteriol., 2008, 190, 3721–3730 CrossRef PubMed.
- B.-A. Zohar and I. Kolodkin-Gal, in Quorum Sensing vs. Quorum Quenching: A Battle with No End in Sight, ed. V. C. Kalia, Springer, India, New Delhi, 2015, pp. 85–99, DOI:10.1007/978-81-322-1982-8_9.
- A. Clinton and K. P. Rumbaugh, Isr. J. Chem., 2016, 56, 265–272 CrossRef.
- R. Levy and E. Borenstein, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12804–12809 CrossRef PubMed.
- S. Bernhardsson, P. Gerlee and L. Lizana, BMC Evol. Biol., 2011, 11, 20 CrossRef PubMed.
- S. de Weert, H. Vermeiren, I. H. M. Mulders, I. Kuiper, N. Hendrickx, G. V. Bloemberg, J. Vanderleyden, R. De Mot and B. J. J. Lugtenberg, Mol. Plant-Microbe Interact., 2002, 15, 1173–1180 CrossRef PubMed.
- G. H. Wadhams and J. P. Armitage, Nat. Rev. Mol. Cell Biol., 2004, 5, 1024–1037 CrossRef PubMed.
- G. Bay, N. Nahar, M. Oubre, M. J. Whitehouse, D. A. Wardle, O. Zackrisson, M.-C. Nilsson and U. Rasmussen, New Phytol., 2013, 200, 54–60 CrossRef PubMed.
- J. Tout, T. C. Jeffries, K. Petrou, G. W. Tyson, N. S. Webster, M. Garren, R. Stocker, P. J. Ralph and J. R. Seymour, ISME J., 2015, 9, 1764–1777 CrossRef PubMed.
- T. R. Miller, K. Hnilicka, A. Dziedzic, P. Desplats and R. Belas, Appl. Environ. Microbiol., 2004, 70, 4692–4701 CrossRef PubMed.
- R. D. Sieg, K. L. Poulson-Ellestad and J. Kubanek, Nat. Prod. Rep., 2011, 28, 388–399 RSC.
- L. Laganenka, R. Colin and V. Sourjik, Nat. Commun., 2016, 7, 12984 CrossRef PubMed.
- V. Sourjik and H. C. Berg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 123–127 CrossRef PubMed.
- J. Müller, C. Kuttler and B. A. Hense, Biosystems, 2008, 92, 76–81 CrossRef PubMed.
- A. H. Rickard, P. Gilbert, N. J. High, P. E. Kolenbrander and P. S. Handley, Trends Microbiol., 2003, 11, 94–100 CrossRef PubMed.
- S. Katharios-Lanwermeyer, C. Xi, N. S. Jakubovics and A. H. Rickard, Biofouling, 2014, 30, 1235–1251 CrossRef PubMed.
- T. Julou, T. Mora, L. Guillon, V. Croquette, I. J. Schalk, D. Bensimon and N. Desprat, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12577–12582 CrossRef PubMed.
- S. Smukalla, M. Caldara, N. Pochet, A. Beauvais, S. Guadagnini, C. Yan, M. D. Vinces, A. Jansen, M. C. Prevost, J.-P. Latgé, G. R. Fink, K. R. Foster and K. J. Verstrepen, Cell, 2008, 135, 726–737 CrossRef PubMed.
- A. Kouzuma, S. Kato and K. Watanabe, Front. Microbiol., 2015, 6, 477 Search PubMed.
- D. Wall, Annu. Rev. Microbiol., 2016, 70, 143–160 CrossRef PubMed.
- K. Vengadesan and S. V. L. Narayana, Protein Sci., 2011, 20, 759–772 CrossRef PubMed.
- N. Dautin and H. D. Bernstein, Annu. Rev. Microbiol., 2007, 61, 89–112 CrossRef PubMed.
- B. Heras, M. Totsika, K. M. Peters, J. J. Paxman, C. L. Gee, R. J. Jarrott, M. A. Perugini, A. E. Whitten and M. A. Schembri, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 457–462 CrossRef PubMed.
- O. Sherlock, R. M. Vejborg and P. Klemm, Infect. Immun., 2005, 73, 1954–1963 CrossRef PubMed.
- L. Liu, S. Hao, R. Lan, G. Wang, D. Xiao, H. Sun and J. Xu, Infect. Immun., 2015, 83, 2596–2604 CrossRef PubMed.
- K. McLoughlin, J. Schluter, S. Rakoff-Nahoum, A. L. Smith and K. R. Foster, Cell Host Microbe, 2016, 19, 550–559 Search PubMed.
- P. C. Bogino, M. de las Mercedes Oliva, F. G. Sorroche and W. Giordano, Int. J. Mol. Sci., 2013, 14, 15838–15859 CrossRef PubMed.
- A. Gardner and S. A. West, Evolution, 2010, 64, 25–38 CrossRef PubMed.
- J. Heller, J. Zhao, G. Rosenfield, D. J. Kowbel, P. Gladieux and N. L. Glass, PLoS Biol., 2016, 14, e1002431 Search PubMed.
- E. S. Danka, E. C. Garcia and P. A. Cotter, Trends Microbiol., 2017, 25, 391–401 CrossRef PubMed.
- D. C. Queller, E. Ponte, S. Bozzaro and J. E. Strassmann, Science, 2003, 299, 105–106 CrossRef PubMed.
- S. Freilich, R. Zarecki, O. Eilam, E. S. Segal, C. S. Henry, M. Kupiec, U. Gophna, R. Sharan and E. Ruppin, Nat. Commun., 2011, 2, 589 CrossRef PubMed.
- X. Liu, M. M. Ramsey, X. Chen, D. Koley, M. Whiteley and A. J. Bard, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 2668–2673 CrossRef PubMed.
- M. E. Hibbing, C. Fuqua, M. R. Parsek and S. B. Peterson, Nat. Rev. Microbiol., 2010, 8, 15–25 CrossRef PubMed.
- R. M. Stubbendieck and P. D. Straight, J. Bacteriol., 2016, 198, 2145–2155 CrossRef PubMed.
- M. Ghoul and S. Mitri, Trends Microbiol., 2016, 24, 833–845 CrossRef PubMed.
- R.-A. Pérez-Gutiérrez, V. López-Ramírez, Á. Islas, L. D. Alcaraz, I. Hernández-González, B. C. L. Olivera, M. Santillán, L. E. Eguiarte, V. Souza, M. Travisano and G. Olmedo-Alvarez, ISME J., 2013, 7, 487–497 CrossRef PubMed.
- M. Sipiczki, Appl. Environ. Microbiol., 2006, 72, 6716–6724 CrossRef PubMed.
- S. E. Lindgren and W. J. Dobrogosz, FEMS Microbiol. Lett., 1990, 87, 149–164 CrossRef.
- A. G. T. K. M. Fons, Microb. Ecol. Health Dis., 2000, 12, 240–246 CrossRef.
- R. S. Peixoto, P. M. Rosado, D. C. d. A. Leite, A. S. Rosado and D. G. Bourne, Front. Microbiol., 2017, 8, 341 Search PubMed.
- T. Rudrappa, M. L. Biedrzycki and H. P. Bais, FEMS Microbiol. Ecol., 2008, 64, 153–166 CrossRef PubMed.
- C. G. Buffie and E. G. Pamer, Nat. Rev. Immunol., 2013, 13, 790–801 CrossRef PubMed.
- B. Stecher, D. Berry and A. Loy, FEMS Microbiol. Rev., 2013, 37, 793–829 CrossRef PubMed.
- X. He, J. S. McLean, L. Guo, R. Lux and W. Shi, ISME J., 2014, 8, 564–574 CrossRef PubMed.
- S. Caballero, S. Kim, R. A. Carter, I. M. Leiner, B. Susac, L. Miller, G. J. Kim, L. L. Ling and E. G. Pamer, Cell Host Microbe, 2017, 21, 592–602 Search PubMed.
- Z. Li, A. J. Clarke and T. J. Beveridge, J. Bacteriol., 1998, 180, 5478–5483 Search PubMed.
- A. B. Russell, S. B. Peterson and J. D. Mougous, Nat. Rev. Microbiol., 2014, 12, 137 CrossRef PubMed.
- M. J. Q. Wong, X. Liang, M. Smart, L. Tang, R. Moore, B. Ingalls and T. G. Dong, Appl. Environ. Microbiol., 2016, 82, 6881–6888 CrossRef PubMed.
- L. E. N. Quadri, Antonie van Leeuwenhoek, 2002, 82, 133–145 CrossRef PubMed.
- E. Shanker and M. J. Federle, Genes, 2017, 8, 15 CrossRef PubMed.
- J. R. van der Ploeg, J. Bacteriol., 2005, 187, 3980–3989 CrossRef PubMed.
- P. D. Cotter, R. P. Ross and C. Hill, Nat. Rev. Microbiol., 2013, 11, 95–105 CrossRef PubMed.
- M. Bassetti, M. Merelli, C. Temperoni and A. Astilean, Ann. Clin. Microbiol. Antimicrob., 2013, 12, 22 CrossRef PubMed.
- M. P. Zacharof and R. W. Lovitt, APCBEE Proc., 2012, 2, 50–56 CrossRef.
- C. D. Nadell, K. Drescher and K. R. Foster, Nat. Rev. Microbiol., 2016, 14, 589–600 CrossRef PubMed.
- L. Chao and B. R. Levin, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 6324–6328 CrossRef.
- O. Rendueles, P. C. Zee, I. Dinkelacker, M. Amherd, S. Wielgoss and G. J. Velicer, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9076–9081 CrossRef PubMed.
- K. A. Gibbs and E. P. Greenberg, Chem. Rev., 2011, 111, 188–194 CrossRef PubMed.
- M. Vos and G. J. Velicer, Curr. Biol., 2009, 19, 1763–1767 CrossRef PubMed.
- P. Stefanic, B. Kraigher, N. A. Lyons, R. Kolter and I. Mandic-Mulec, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14042–14047 CrossRef PubMed.
- N. A. Lyons, B. Kraigher, P. Stefanic, I. Mandic-Mulec and R. Kolter, Curr. Biol., 2016, 26, 733–742 CrossRef PubMed.
- S. Borgeaud, L. C. Metzger, T. Scrignari and M. Blokesch, Science, 2015, 347, 63–67 CrossRef PubMed.
- L. McNally, E. Bernardy, J. Thomas, A. Kalziqi, J. Pentz, S. P. Brown, B. K. Hammer, P. J. Yunker and W. C. Ratcliff, Nat. Commun., 2017, 8, 11 CrossRef PubMed.
- A. J. Waite and W. Shou, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19079–19086 CrossRef PubMed.
- J. B. Xavier and K. R. Foster, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 876–881 CrossRef PubMed.
- J. Gore, H. Youk and A. van Oudenaarden, Nature, 2009, 459, 253–256 CrossRef PubMed.
- A. Sanchez and J. Gore, PLoS Biol., 2013, 11, e1001547 Search PubMed.
- O. X. Cordero and M. F. Polz, Nat. Rev. Microbiol., 2014, 12, 263–273 CrossRef PubMed.
- N. Ribeck and R. E. Lenski, Evolution, 2015, 69, 1313–1320 CrossRef PubMed.
- D. Greig and M. Travisano, Proc. R. Soc. London, Ser. B, 2004, 271, S25–S26 CrossRef PubMed.
- R. C. MacLean, A. Fuentes-Hernandez, D. Greig, L. D. Hurst and I. Gudelj, PLoS Biol., 2010, 8, e1000486 Search PubMed.
- A. Ross-Gillespie, A. Gardner, S. A. West and A. S. Griffin, Am. Nat., 2007, 170, 331–342 CrossRef PubMed.
- D. B. Borenstein, Y. Meir, J. W. Shaevitz and N. S. Wingreen, PLoS One, 2013, 8, e63304 Search PubMed.
- M. Doebeli and I. Ispolatov, Science, 2010, 328, 494–497 CrossRef PubMed.
- K. L. Hillesland, Ann. N. Y. Acad. Sci., 2017 DOI:10.1111/nyas.13515.
- K. L. Hillesland and D. A. Stahl, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2124–2129 CrossRef PubMed.
- S. Paterson, T. Vogwill, A. Buckling, R. Benmayor, A. J. Spiers, N. R. Thomson, M. Quail, F. Smith, D. Walker and B. Libberton, Nature, 2010, 464, 275–278 CrossRef PubMed.
- C. T. Bergstrom and M. Lachmann, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 593–598 CrossRef PubMed.
- R. Law, The biology of mutualism: Ecology and evolution, 1985, pp. 145–170 Search PubMed.
- J. L. Sachs, C. J. Essenberg and M. M. Turcotte, Trends Ecol. Evol., 2011, 26, 202–209 CrossRef PubMed.
- K. L. Hillesland, S. Lim, J. J. Flowers, S. Turkarslan, N. Pinel, G. M. Zane, N. Elliott, Y. Qin, L. Wu and N. S. Baliga, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 14822–14827 CrossRef PubMed.
- S. Sieuwerts, F. A. M. de Bok, J. Hugenholtz and J. E. T. van Hylckama Vlieg, Appl. Environ. Microbiol., 2008, 74, 4997–5007 CrossRef PubMed.
- A. Bolotin, B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau and P. Hols, Nat. Biotechnol., 2004, 22, 1554–1558 CrossRef PubMed.
- M. van de Guchte, S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessieres, J. Weissenbach, S. D. Ehrlich and E. Maguin, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9274–9279 CrossRef PubMed.
- N. Y. Ankrah, J. Luan and A. E. Douglas, J. Bacteriol., 2017, 199, e00872-16 CrossRef PubMed.
- M. Embree, J. K. Liu, M. M. Al-Bassam and K. Zengler, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(50), 15450 CrossRef PubMed.
- S. L. Garcia, K. D. McMahon, M. Martinez-Garcia, A. Srivastava, A. Sczyrba, R. Stepanauskas, H.-P. Grossart, T. Woyke and F. Warnecke, ISME J., 2013, 7, 137–147 CrossRef PubMed.
- S. Shigenobu, H. Watanabe, M. Hattori, Y. Sakaki and H. Ishikawa, Nature, 2000, 407, 81–86 CrossRef PubMed.
- J. P. McCutcheon and C. D. von Dohlen, Curr. Biol., 2011, 21, 1366–1372 CrossRef PubMed.
- R. Braakman, M. J. Follows and S. W. Chisholm, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E3091–E3100 CrossRef PubMed.
- P. D. Schloss and J. Handelsman, Genome Biol., 2005, 6, 229 CrossRef PubMed.
- H.-C. Chiu, R. Levy and E. Borenstein, PLOS Comput. Biol, 2014, 10(7), e1003695 Search PubMed.
- A. Razumov, Mikrobiologiya, 1932, 1, 131–146 Search PubMed.
- R. I. Amann, W. Ludwig and K. H. Schleifer, Microbiol. Rev., 1995, 59, 143–169 Search PubMed.
- A. J. T. Staley and A. Konopka, Annu. Rev. Microbiol., 1985, 39, 321–346 CrossRef PubMed.
- P. Hugenholtz, B. M. Goebel and N. R. Pace, J. Bacteriol., 1998, 180, 4765–4774 Search PubMed.
- K. C. Wrighton, B. C. Thomas, I. Sharon, C. S. Miller, C. J. Castelle, N. C. VerBerkmoes, M. J. Wilkins, R. L. Hettich, M. S. Lipton, K. H. Williams, P. E. Long and J. F. Banfield, Science, 2012, 337, 1661–1665 CrossRef PubMed.
- M. S. Rappé and S. J. Giovannoni, Annu. Rev. Microbiol., 2003, 57, 369–394 CrossRef PubMed.
- H. Tamaki, Y. Sekiguchi, S. Hanada, K. Nakamura, N. Nomura, M. Matsumura and Y. Kamagata, Appl. Environ. Microbiol., 2005, 71, 2162–2169 CrossRef PubMed.
- E. G. Biosca, R. Flores, R. D. Santander, J. L. Díez-Gil and E. Barreno, PLoS One, 2016, 11, e0160328 Search PubMed.
- X. Huai-Shu, N. Roberts, F. L. Singleton, R. W. Attwell, D. J. Grimes and R. R. Colwell, Microb. Ecol., 1982, 8, 313–323 CrossRef PubMed.
- J. T. Lennon and S. E. Jones, Nat. Rev. Microbiol., 2011, 9, 119–130 CrossRef PubMed.
- G. J. Velicer and H. Mendes-Soares, Curr. Biol., 2009, 19, R55–R56 CrossRef PubMed.
- S. Pande and C. Kost, Trends Microbiol., 2017, 25, 349–361 CrossRef PubMed.
- A. D'Onofrio, J. M. Crawford, E. J. Stewart, K. Witt, E. Gavrish, S. Epstein, J. Clardy and K. Lewis, Chem. Biol., 2010, 17, 254–264 CrossRef PubMed.
- T. Kaeberlein, K. Lewis and S. S. Epstein, Science, 2002, 296, 1127–1129 CrossRef PubMed.
- D. Nichols, N. Cahoon, E. M. Trakhtenberg, L. Pham, A. Mehta, A. Belanger, T. Kanigan, K. Lewis and S. S. Epstein, Appl. Environ. Microbiol., 2010, 76, 2445–2450 CrossRef PubMed.
- S. L. Garcia, K. D. McMahon, H.-P. Grossart and F. Warnecke, Environ. Microbiol. Rep., 2014, 6, 21–27 CrossRef PubMed.
- D. K. Button, F. Schut, P. Quang, R. Martin and B. R. Robertson, Appl. Environ. Microbiol., 1993, 59, 881–891 Search PubMed.
- R. C. Lewontin, Annu. Rev. Ecol. Systemat., 1970, 1, 1–18 CrossRef.
- D. L. Hull, Annu. Rev. Ecol. Systemat., 1980, 11, 311–332 CrossRef.
- R. E. Michod, Am. Nat., 1997, 150, S5–S21 CrossRef PubMed.
- S. Okasha, Evolution and the levels of selection, Oxford University Press, Oxford, 2006 Search PubMed.
- J. Damuth and I. L. Heisler, Biol. Philos., 1988, 3, 407–430 CrossRef.
- R. E. Michod, Biol. Philos., 2005, 20, 967–987 CrossRef.
- J. Maynard Smith and E. Szathmáry, The major transitions in evolution, Oxford University Press, Oxford, 1995 Search PubMed.
- S. A. West, R. M. Fisher, A. Gardner and E. T. Kiers, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 10112–10119 CrossRef PubMed.
- E. Clarke, J. Biosci., 2014, 39, 303–317 CrossRef PubMed.
- C. M. Fraser, J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison and J. C. Venter, Science, 1995, 270, 397–403 Search PubMed.
- A. E. Douglas, Cold Spring Harbor Perspect. Biol., 2014, 6, a016113 CrossRef PubMed.
- S. Estrela, B. Kerr and J. J. Morris, Curr. Opin. Microbiol., 2016, 31, 191–198 CrossRef PubMed.
- D. Bhattacharya, J. M. Archibald, A. P. M. Weber and A. Reyes-Prieto, BioEssays, 2007, 29, 1239–1246 CrossRef PubMed.
- S. Shitut, T. Ahsendorf, S. Pande, M. Egbert and C. Kost, bioRxiv, 2017, 114462 Search PubMed.
- G. M. Dunny, T. J. Brickman and M. Dworkin, Bioessays, 2008, 30, 296–298 CrossRef PubMed.
- M. Marchal, F. Goldschmidt, S. N. Derksen-Müller, S. Panke, M. Ackermann and D. R. Johnson, BMC Evol. Biol., 2017, 17, 106 CrossRef PubMed.
- W. Shou, S. Ram and J. M. Vilar, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 1877–1882 CrossRef PubMed.
- M. Cavaliere, S. Feng, O. S. Soyer and J. I. Jiménez, Environ. Microbiol., 2017, 2949–2963 CrossRef PubMed.
- E. Bauer, J. Zimmermann, F. Baldini, I. Thiele and C. Kaleta, PLoS Comput. Biol., 2017, 13, e1005544 Search PubMed.
- W. R. Harcombe, W. J. Riehl, I. Dukovski, B. R. Granger, A. Betts, A. H. Lang, G. Bonilla, A. Kar, N. Leiby and P. Mehta, Cell Rep., 2014, 7, 1104–1115 CrossRef PubMed.
- L. Margulis, Origin of eukaryotic cells: Evidence and research implications for a theory of the origin and evolution of microbial, plant and animal cells on the precambrian Earth, Yale University Press, 1970 Search PubMed.
- S. M. B. Krause, T. Johnson, Y. Samadhi Karunaratne, Y. Fu, D. A. C. Beck, L. Chistoserdova and M. E. Lidstrom, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 358–363 CrossRef PubMed.
- J. B. Xavier, Mol. Syst. Biol., 2011, 7, 483 CrossRef PubMed.
- E. Sober and D. S. Wilson, Unto others: The evolution and psychology of unselfish behavior, Harvard University Press, 1999 Search PubMed.
- J. Overmann, in Recent Advances in Phototrophic Prokaryotes, Springer, 2010, pp. 15–29 Search PubMed.
- M. Doebeli, Popul. Ecol., 2002, 44, 59–70 CrossRef.
- S. Estrela, C. H. Trisos and S. P. Brown, Am. Nat., 2012, 180, 566–576 CrossRef PubMed.
- J. M. Biernaskie and S. A. West, Proc. R. Soc. Lond. B Biol. Sci., 2015, 282(1813), 20151075 CrossRef PubMed.
- K. Brenner and F. H. Arnold, PLoS One, 2011, 6, e16791 Search PubMed.
- J. B. Bruce, G. A. Cooper, H. Chabas, S. A. West and A. S. Griffin, Evolution, 2017, 71, 2484–2495 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8np00009c |
| This journal is © The Royal Society of Chemistry 2018 |