Multifunctional nanozymes: enzyme-like catalytic activity combined with magnetism and surface plasmon resonance
Jiangjiexing
Wu
ab,
Sirong
Li
a and
Hui
Wei
 *abc
*abc
aDepartment of Biomedical Engineering, College of Engineering and Applied Sciences, Nanjing National Laboratory of Microstructures, Nanjing University, Nanjing 210093, China. E-mail: weihui@nju.edu.cn; Web: http://weilab.nju.edu.cn Fax: +86-25-83594648; Tel: +86-25-83593272
bState Key Laboratory of Coordination Chemistry, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210093, China
cState Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210093, China
First published on 15th May 2018
Abstract
Over decades, as alternatives to natural enzymes, highly-stable and low-cost artificial enzymes have been widely explored for various applications. In the field of artificial enzymes, functional nanomaterials with enzyme-like characteristics, termed as nanozymes, are currently attracting immense attention. Significant progress has been made in nanozyme research due to the exquisite control and impressive development of nanomaterials. Since nanozymes are endowed with unique properties from nanomaterials, an interesting investigation is multifunctionality, which opens up new potential applications for biomedical sensing and sustainable chemistry due to the combination of two or more distinct functions of high-performance nanozymes. To highlight the progress, in this review, we discuss two representative types of multifunctional nanozymes, including iron oxide nanomaterials with magnetic properties and metal nanomaterials with surface plasmon resonance. The applications are also covered to show the great promise of such multifunctional nanozymes. Future challenges and prospects are discussed at the end of this review.
Introduction
Recently, nanozymes have emerged as a new kind of artificial enzymes. They are functional nanomaterials with enzyme-like characteristics.1–15 Compared with natural enzymes, nanozymes have various advantages, such as robustness to harsh environments, low cost, and ease of mass production. Moreover, nanozymes are endowed with some unique properties in terms of their responses towards external stimuli, size- (shape-, composition-) dependent catalytic activities, large surface area, multiple functionalities besides catalysis, etc. By combining the enzyme mimicking activities with the other properties (such as optical, electrical, and magnetic properties) of nanomaterials, the designed multifunctional nanozymes would not only integrate multiple processes (e.g., separation and catalysis) more efficiently and economically but also provide high-performance platforms for practical applications (e.g., in situ monitoring and ultra-sensitive sensing) (Scheme 1). Multifunctional nanozymes have been extensively investigated for diverse applications in biosensor development, environmental remediation, diagnosis and therapeutics, etc. To highlight the recent progress, two types of multifunctional nanozymes are summarized in this review as they are the most widely studied. One is iron oxide nanozymes with magnetic properties and the other is metal nanozymes with surface plasmon resonance (SPR). The advantages of the combination of magnetic properties or SPR with enzyme mimicking activities are illustrated by discussing representative applications. Finally, the current challenges facing multifunctional nanozymes and future directions are discussed.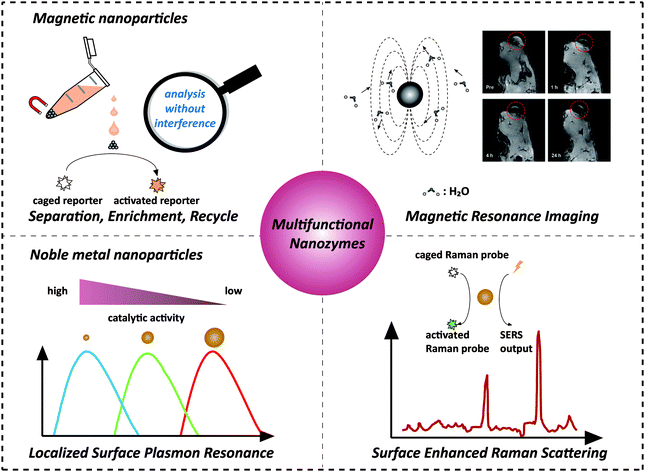 | ||
| Scheme 1 Combination of magnetic properties and surface plasmon resonance with enzyme-like activities for constructing multifunctional nanozymes. The magnetic resonance image was adapted with permission from ref. 43. Copyright (2013) American Chemical Society. | ||
Iron oxide nanozymes with magnetic properties
Magnetic iron oxide nanomaterials are unique because of their interactions with magnetic fields and field gradients. These enable magnetic separation, enrichment, and recycling of magnetic nanoparticles (MNPs), as well as tracking and visualization of the local environment of MNP labelled cells through magnetic resonance imaging (MRI).16–18 Previously, MNPs were considered to be catalytically inert, thus they had been widely used as a support to bind catalysts or enzymes for further functionalization.19 Unexpectedly, Yan et al. found that Fe3O4 MNPs exhibited an intrinsic peroxidase mimicking activity.20 Later, Wang et al. reported the application of Fe3O4 MNPs in hydrogen peroxide (H2O2) and glucose detection based on their peroxidase mimicking activity.21 Since then, ever-growing interest has been shown to exploring the potential applications of magnetic nanozymes.22,23 In this section, we discuss the combination of magnetic properties and enzyme mimicking properties of iron oxide nanomaterials and their applications.Separation, enrichment and reusability
One obvious advantage of MNPs is that they can be easily separated and controlled in an external magnetic field due to their inherent magnetic properties. Further modifications (such as coating,24–29 doping,30 and hybridization31–41) were made to improve their enzyme-like catalytic activities and stabilities. Moreover, the inherent magnetic properties of the modified magnetic nanozymes could still be retained. For example, Gu et al. modified Fe2O3 MNPs with varied amounts of Prussian blue (PB). They found that the catalytic activities of the Fe2O3 MNPs were enhanced as the PB content increased and simultaneously the magnetism was still retained.24 On the other hand, the magnetic properties of MNPs could be tuned through changing the synthetic conditions while the high catalytic activities were retained.34,42Taking advantage of the easy separation of MNPs, the reaction solution can be separated into two parts; one is the supernatant part and the other is the enriched MNPs. This greatly helps to measure the supernatant solution without interference.30,36,39,44–46 For example, Tang and co-workers developed a reverse colorimetric immunoassay for sensitive detection of prostate specific antigen (PSA) which utilized the peroxidase-like activity of magnetic beads and natural catalase carried by gold nanoparticles (AuNPs) (Fig. 1a).47 The magnetic beads and AuNPs were first functionalized with an anti-PSA capture antibody and a catalase/anti-PSA detection antibody, respectively. Then in the presence of PSA, a sandwich-type immunoassay was constructed. More PSA would provide more catalase. The catalase decomposed the hydrogen peroxide in the detection solution, leading to a reduced catalytic efficiency of magnetic bead based peroxidase mimics. Besides the peroxidase-like activities, magnetic beads also provided an easy method of separation by applying a local magnetic field, as shown in Fig. 1b. The separation eliminated the potential interference for the absorbance measurement of the supernatant solution. A dynamic range of 0.05–20 ng mL−1 with a detection limit of 0.03 ng mL−1 for PSA detection was achieved with this reverse enzyme colorimetric immunoassay.
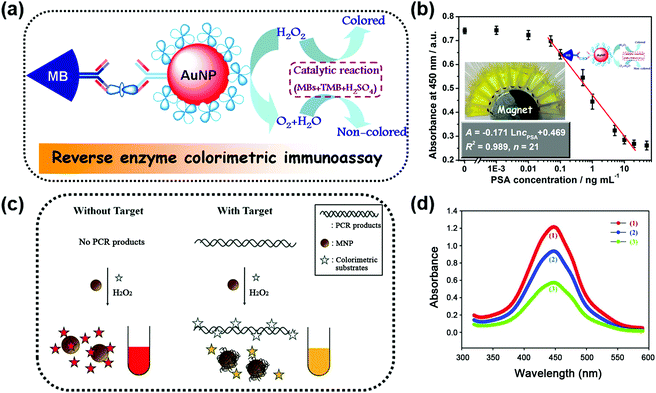 | ||
| Fig. 1 (a) Scheme for the magnetocontrolled enzyme-mediated reverse colorimetric immunosensing strategy. (b) Calibration plots between the absorbance of oxidized TMB at 450 nm and the logarithm of PSA concentrations. (c) Schematic illustration of the MNP-based, label-free, colorimetric detection of target DNA. (d) Reduction of the peroxidase activity of MNPs induced by target DNA: a control sample without target DNA (1, red), a test sample with target DNA after removal of the DNA in the solution (2, blue), and a test sample with target DNA before removal of the DNA in the solution (3, green). (a and b) Adapted with permission from ref. 47. Copyright (2013) American Chemical Society; (c and d) adapted with permission from ref. 50. Copyright (2011) John Wiley and Sons. | ||
Interference-free measurement of the supernatant solution not only ensured the accuracy of the final signal but could also be used to elucidate the role of leached iron ions. According to previous studies, the optimal condition for performing peroxidase-like catalytic reaction of MNPs was at low pH.20,21 However, the iron ions might be leached from MNPs in this condition, which would then have some effects on the peroxidase-like catalytic activity. To figure out whether the peroxidase-like catalytic activity came from the MNPs or the leached iron ions, the magnetic properties of MNPs were utilized to remove the MNPs from the solution with a magnet. Then, the catalytic activity of the supernatant solution was tested, and the results confirmed that the high peroxidase mimicking activity was attributed to MNPs rather than to the leached iron ions.20,36,37,48
Similarly, another interesting application benefiting from interference free measurement was DNA detection. Gao and co-workers reported a simple and cheap colorimetric assay for single-nucleotide polymorphism (SNP) genotyping, which combined the peroxidase mimicking activities and magnetic properties of ferrofluidic nano-particulate probes (FNPs) with a ligase. The oligonucleotide-coated FNPs were first hybridized with target SNP strands and then an aggregate of the FNPs was formed through a ligase chain reaction. After magnetic separation of the FNP aggregate from the unreacted FNPs under an external magnetic field, the FNPs left in the supernatant produced the colorimetric signal through the catalytic oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) in the presence of H2O2. A high sensitivity with a concentration as low as 3 pM and excellent selectivity to discriminate one copy of the SNP target from 1000 copies of the wild type targets were achieved.49
Besides interference-free measurement of the supernatant solution, the potential interference from the supernatant solution could also be eliminated when performing the test with MNPs. A colorimetric DNA detection based on the shielding of double-stranded DNA (dsDNA) against the peroxidase-like activity of MNPs is shown in Fig. 1c. On the one hand, the polymerase chain reaction-amplified target DNA would be adsorbed directly onto the surface of MNPs, inhibiting the binding of the colorimetric substrates to MNPs. On the other hand, the negatively charged target DNA remaining in the solution would interact with the positively charged substrates, leading to a reduced amount of the substrates for binding. Thus, the catalytic rate of the MNPs was lower than that of the system without target DNA (see the red and green lines in Fig. 1d). However, a quantity of unbound nucleic acids remained in the solution and the contribution of their inhibition effect was unclear. To clarify these issues, the MNPs were separated from the solution with an external magnetic field. First, through interference-free fluorescent measurement of the supernatant solution, nearly 60% of the target DNA remaining in the solution was determined according to the fluorescent signal generated by Eva-green dye intercalated into dsDNA. Then the separated MNPs were re-dispersed in the solution and the colorimetric assays were performed again. Without the interference of the unbound DNA, the absorption measurement results still showed a significant decrease of the colorimetric signal (red and blue lines in Fig. 1d) but only a 40% reduction of the signal obtained before removal of unbound DNA (blue and green lines in Fig. 1d). This result illustrated that the inhibition effect of unbound DNA on the peroxidase-like activity of MNPs was nearly the same as that of the target DNA adsorbed on the MNPs.50
Notably, the separated MNPs also played a role in capture and enrichment during the magnetic separation, which would amplify the readout signal.20,51–59 For example, Yan and co-workers reported two immunoassays based on MNPs. The first one was similar to a conventional enzyme-linked immunosorbent assay, except using Fe3O4 MNPs in place of enzymes. As shown in Fig. 2a, hepatitis B virus surface antigen (preS1) was first adsorbed onto a plate, followed by adding anti-preS1 antibodies for specific target recognition, and then protein A-Fe3O4 MNP conjugates were added for signal development. The utilization of peroxidase mimicking Fe3O4 MNPs made this process more robust, easier, and more economical. In the second case, a modified capture-detection sandwich immunoassay format was developed with Fe3O4 MNPs to detect myocardial infarction biomarker troponin I (TnI) (Fig. 2b). Different from the first one, the MNPs here were used to capture and enrich the antigens from a blood sample, as well as to generate the colorimetric signal through the catalytic oxidation of TMB.20 This enrichment could provide accurate measurement without any interference from the complex biological system as well as greater detection sensitivity. Further, they developed a MNP-based nanozyme-strip for detecting the glycoprotein of Ebola virus (EBOV-GP) (Fig. 3b). The nanozyme catalyzed colorimetric reaction amplified the final signal, which was 100-fold more sensitive than that of a standard colloidal gold strip (Fig. 3c and d). Moreover, a 10-fold enhancement of sensitivity could be achieved through the magnetic separation and enrichment of targets (Fig. 3e).52
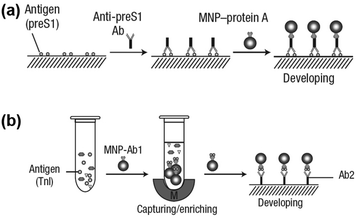 | ||
| Fig. 2 (a) Fe3O4 MNP based antigen-down immunoassay. (b) A capture-detection sandwich immunoassay format with Fe3O4 MNPs as nanozymes. Adapted with permission from ref. 20. Copyright (2007) Nature Publishing Group. | ||
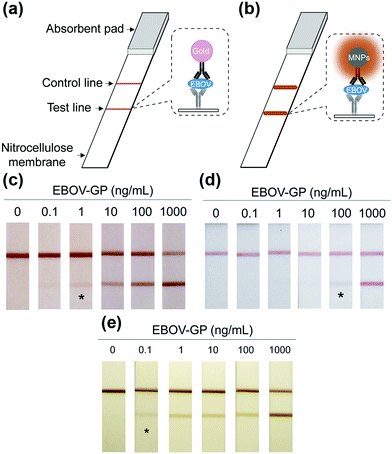 | ||
| Fig. 3 (a) Standard AuNPs-based strip. (b) Nanozyme-strip employing Fe3O4 MNPs in place of AuNPs. (c) Nanozyme-strip, (d) standard colloidal gold strip and (e) nanozyme-strip combined with magnetic enrichment for EBOV-GP detection. The asterisk (*) indicates the limit of visual detection of the test line in strips. Adapted with permission from ref. 52. Copyright (2015) Elsevier. | ||
A strategy for chemiluminescent (CL) detection of pesticides was reported by Han, Zhang and co-workers (Fig. 4). They demonstrated that the dissolved oxygen would be converted to superoxide anions by the peroxidase-like Fe3O4 MNPs, and then the CL signal of luminol would be generated. In the presence of the radical scavenger ethanol, the CL signal would be quenched. However, the specific binding of pesticide could inhibit this quenching effect through protecting superoxide anions from interacting with ethanol, leading to a “turn-on” signal. Meanwhile, the superparamagnetic properties of Fe3O4 MNPs provided a simple magnetic enrichment and separation approach to attain interference-free measurement for detection of pesticide ethoprophos (EP). Obvious CL enhancement was observed with an enhancement coefficient of 2.11.53 In this direction, Zheng et al. reported a sensitive detection of arsenic and antimony in fish based on Fe3O4 MNPs combined with microwave digestion. Magnetic separation and effective catalytic reaction based on Fe3O4 MNPs helped to eliminate potential interferences from the sample matrices and also raised the sensitivity by one order of magnitude compared with conventional techniques without MNPs.54
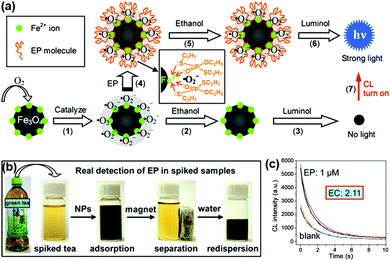 | ||
| Fig. 4 (a) Mechanism of CL switching at the surface of Fe3O4 MNPs. (b) Schematic illustration of the magnetic separation/re-dispersion process for overcoming the interfering effect in green tea. (c) CL response resulting from EP molecules in green tea after carrying out the separation/re-dispersion process two times. The enhancement coefficient (EC) is the ratio of the CL intensity of the sample with pesticide to that of the sample without pesticide. Adapted with permission from ref. 53. Copyright (2012) American Chemical Society. | ||
Furthermore, magnetic enrichment of MNPs was also reported for other interesting therapeutic applications. Murohara et al. found that magnetic separation and enrichment of Fe3O4 MNPs with an external magnet helped to construct induced pluripotent stem (iPS) cell-derived fetal liver kinase-1 positive (Flk-1+) cell sheets with an extracellular matrix (ECM) precursor. The iPS cell sheet was comprised of pile-ups of 15–20 layered cells with a thickness of nearly 300 μm. The obtained iPS cell sheet was easy to handle. As shown in Fig. 5d, the iPS cell sheets were placed on the adductor muscles of nude mice using the alnico magnet. If using ECM precursor only, the fluctuating cellular density would affect the forming process, thus it was hard to have a truly sheet-like structure. Later, they investigated the effect of iPS cell sheet transplantation on blood flow recovery of the ischemic muscle. The results in Fig. 5e showed that the iPS cell sheets promoted angiogenesis more effectively than the conventional direct-injection of Flk-1+ cell-MNPs-ECM, Flk-1+ cell-ECM, and control buffer groups. The anti-oxidative activities of the Fe3O4 MNPs were the reason for enhancing angiogenesis. Therefore, the combination of magnetic enrichment and separation as well as the enzyme-like catalytic activities of the MNPs provided a promising method of tissue engineering for therapeutic angiogenesis (Fig. 5).60
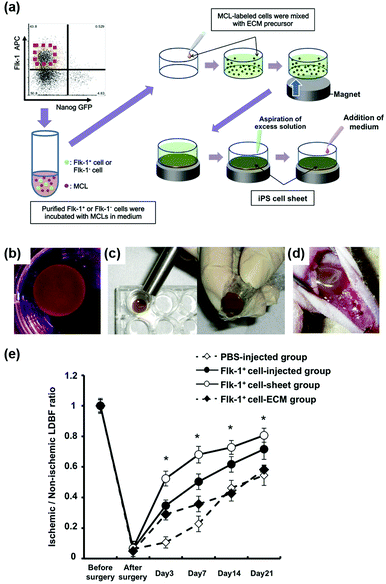 | ||
| Fig. 5 Fe3O4 MNP-facilitated formation of iPS cell sheets for reparative angiogenesis. (a) Procedure for construction of the iPS cell-derived cell sheet. (b) An alnico magnet was positioned on the surface of the culture medium. The Flk-1 cell sheet floated up to the surface of the culture medium without disruption. (c) The magnetized Flk-1 cell sheet attached to an alnico magnet covered with polyvinylidene difluoride membrane via a magnetic force. (d) Flk-1 cell sheets were placed on the adductor muscles of nude mice using the alnico magnet. (e) Quantitative analysis of the ischemic/normal laser Doppler blood flow (LDBF) ratios in the Flk-1+ cell-sheet, Flk-1+ cell-injected, Flk-1+ cell-ECM and control phosphate buffer saline (PBS) groups. Adapted with permission from ref. 60. Copyright (2013) Nature Publishing Group. | ||
In addition to enrichment, the separated MNPs can be recycled. Thus enormous interest and efforts have been focused on the reusability of magnetic nanozymes, which would help the development of green chemistry or sustainable chemistry. For example, Fe3O4 MNPs were reported to remove phenol and aniline from aqueous solution through MNPs based peroxidase-like activity. The separation of MNPs by applying an external magnetic field made the catalysts reusable, and the reused MNPs still retained good activities to remove over 80% of phenol even after 8 recycles. The slightly reduced removal efficiency was due to the conglomeration or loss of MNPs during the recycle and rinse process.61 More applications and details of reused nanozymes based on Fe3O4 MNPs or the iron oxide composites are listed in Table 1.
| Nanomaterials | Enzyme activity | Application | Cycle number | Comments | Ref. |
|---|---|---|---|---|---|
| Fe3O4 | Peroxidase | DNA detection | 4 | 50 | |
| Degradation of phenol and aniline | 8 | 61 | |||
| Glucose or cholesterol detection | 30 | Immobilizing MNPs and glucose oxidase (GOx) or cholesterol oxidase within mesocellular silica | 62 | ||
| Removal of sulfathiazole | 3 | 63 | |||
| Degradation of xylenol orange | 7 | 64 | |||
| Galactose detection | 20 | Entrapping MNPs and galactose oxidase in mesocellular silica | 65 | ||
| Degradation of methylene blue | 5 | 66 | |||
| Degradation of bisphenol A | 5 | 67 | |||
| Human chorionic gonadotropin detection through sandwich immunoassay | 8 | Amino-functionalized Fe3O4 | 68 | ||
| H2O2 and glucose detection | 5 | 5,10,15,20-Tetrakis(4-carboxyphenyl)-porphyrin-functionalized Fe3O4 | 69 | ||
| H2O2 and glucose detection | 5 | Casein incorporated on MNPs | 70 | ||
| Lipid peroxidation in liposomes | 5 | 71 | |||
| Degradation of phenol | 3 | Hydrogel coated Fe3O4 magnetic composite nanospheres | 72 | ||
| Degradation of organic dye pollutants crystal violet | 3 | 73 | |||
| Degradation of 4-chlorophenol | 3 | 74 | |||
| Oxidation of benzyl alcohol | 5 | Epichlorohydrin-modified Fe3O4 | 75 | ||
| Removal of organic pollutants Rhodamine B | 5 | 76 | |||
| Zn2+ detection | 10 | 77 | |||
| H2O2 and glucose detection as well as degradation of phenol and Congo red dye | 7 | 78 | |||
| Fe3O4@MSN | Peroxidase | H2O2 and glucose detection | 5 | 79 | |
| Carboxyl cellulose nanospheres@Fe3O4 | Peroxidase | Removal of textile dye Navy blue | 5 | 80 | |
| Fe3O4/β-cyclodextrin polymer composite | Peroxidase | Glucose detection | 10 | 81 | |
| Fe3O4/Al pillared bentonite | Peroxidase | Degradation of Rhodamine B | 4 | 82 | |
| Fe3O4@mSiO2@hydroxypropyl β-cyclodextrin | Peroxidase | H2O2 and β-estradiol detection | 10 | 83 | |
| Fe3O4/CeO2 composite | Peroxidase | Degradation of 4-chlorophenol | 6 | 84 | |
| Fe3O4–Au nanocomposites | Peroxidase | N/A | 9 | 85 | |
| Grapheme quantum dots/Fe3O4 | Peroxidase | Removal of phenolic compound | 10 | 86 | |
| Yolk-like Fe3O4@Fe3O4/C | Peroxidase | Degradation of 4-chlorophenol | 4 | 87 | |
| Au/Fe3O4/graphene oxide (GO) hybrid material | Peroxidase | Hg2+ detection | 15 | 88 | |
| Fe3O4/Au NPs | Peroxidase | Degradation of methyl orange | 10 | 89 | |
| Au–Pd–Fe3O4/reduced graphene oxide (rGO) | Peroxidase | N/A | 5 | 90 | |
| Fe3O4–Au@mesoporous SiO2 | Peroxidase and oxidase | Glucose detection | 6 | 91 | |
| Magnetite/rGO nanocomposite | Peroxidase | Degradation of methylene blue | 5 | 92 | |
| GOx/Fe3O4/GO magnetic nanocomposite | Peroxidase | H2O2 and glucose detection | 4 | 93 | |
| Three-dimensional (3D) rGO_Fe3O4–Pd | Peroxidase and oxidase | Reduced glutathione and glucose detection | 10 | Self-assembled 3D graphene nanohybrids with in situ-formed Fe3O4 and Pd NPs | 94 |
| Magnetic zeolitic imidazolate framework 8 (ZIF-8)@GOx | Peroxidase | Glucose detection | 12 | Using Fe3O4 to grow core–shell magnetic ZIF-8 and then embedding GOx to form magnetic ZIF-8@GOx | 95 |
| Poly(diallyldimethylammonium chloride)–Fe3O4/GOx composite | Peroxidase | Glucose detection | 6 | GOx was assembled onto the Fe3O4 MNPs through electrostatic attraction | 96 |
| Poly(diallyldimethylammonium chloride)–Fe3O4/GOx composite | Peroxidase | Glucose detection | 7 | GOx was assembled onto the Fe3O4 MNPs through electrostatic attraction | 97 |
| Fe2O3 | Peroxidase | Degradation of methylene blue | 5 | Prussian-blue modified Fe2O3 | 98 |
| Janus γ-Fe2O3/SiO2 | Peroxidase | Glucose detection | 5 | 99 | |
| FeVO4 | Peroxidase | H2O2 detection | 10 | 100 | |
| MnFe2O4 | Oxidase | N/A | 6 | 101 |
Magnetic resonance imaging
MNPs can also be employed as contrast agents for MRI.102 Chen, Wang and co-workers used folic acid, Eu complexes, fluorescein isothiocyanate, and Au–Fe3O4 NPs to fabricate a multifunctional nanoprobe for fluorescence/MR dual-modal imaging and TMB-based colorimetric/calcium dipicolinate-based fluorigenic detection of cancer cells (Fig. 6). They found that the catalytic activity of this nanoprobe was enhanced due to the synergistic effect between Au and Fe3O4. Thus the nanoprobe conjugated cancer cells would catalyze the oxidation of TMB into blue oxidized product more easily and quickly, which could be judged by the naked eye and quantitatively determined through absorption measurement at 653 nm. The detection limit of the TMB based colorimetric assay was as low as 100 cancer cells, the same as the fluorigenic detection results. Therefore, such a multimodal nanoprobe would be a promising and reliable candidate for early diagnosis of cancers in biomedical application.103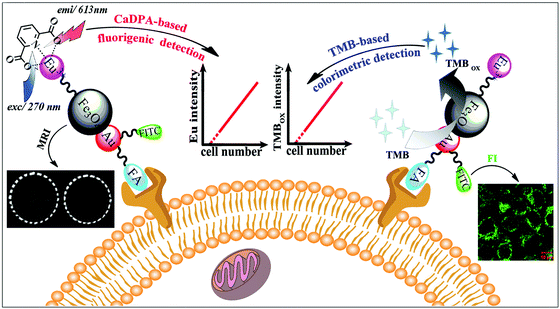 | ||
| Fig. 6 Fluorescence and colorimetric response of the multifunctional probe as well as fluorescence/T2-weighted MRI of cells. Reprinted with permission from ref. 103. Copyright (2013) Royal Society of Chemistry. | ||
Besides colorimetric detection of cancer cells, MNPs can also inhibit tumor growth through reactive oxygen species (ROS) generated by peroxidase-like catalytic decomposition of H2O2. In Wang's work, the T2-weighted MRI and peroxidase mimicking properties of MNPs were combined for tumor targeting and inhibiting in vivo. Both the MRI in vivo and inductively coupled plasma-atomic emission spectroscopy analysis on ex vivo tissues demonstrated that the PEGylated MNPs preferentially accumulated in tumor tissue, which was due to the enhanced permeability and retention (EPR) effect. Later, anti-tumor studies were conducted by intravenously injecting MNPs and H2O2 into mice. The results showed that the combination of MNPs and H2O2 gave better anti-tumor efficacy than MNPs/H2O2 alone or the control, and nearly 99% of tumors were inhibited after 17 days.104
Recently, Hyeon et al. reported that enhanced photodynamic therapy (PDT) was realized by simultaneously using manganese ferrite nanoparticles (MFNs) and the photosensitizer molecule chlorin e6 (Ce6). The MFNs could continuously produce a sufficient amount of O2via Fenton reaction in a H2O2-rich cancer microenvironment, overcoming the cancer hypoxic limitation of PDT. The MFNs were combined with Ce6, which was loaded in mesoporous silica nanoparticles, to form nanocomposites of manganese ferrite mesoporous silica nanoparticles (MFMSNs). ROS could be continuously generated from the MFMSNs under laser irradiation, thus the therapeutic efficiency of PDT would be consequently enhanced (Fig. 7b). Moreover, the T2 contrast effect in MRI provided a tracking method to check the accumulation of MFMSNs in vivo (Fig. 7c). The selective accumulation at tumor sites and continuous supply of O2 for PDT made the MFMSNs a potential candidate in cancer therapy.43 Likewise, a multifunctional theranostic nanoagent based on Fe-doped polydiaminopyridine nanofusiforms combined with dihydroartemisinin and methylene blue was constructed for chemo/photodynamic/photothermal therapy. The Fe-doped polydiaminopyridine nanofusiforms not only overcame tumor hypoxia to enhance the efficiency of PDT as catalase-like enzymes but also validated the accumulation of this nanoagent in tumors through T2-weighted MRI.105
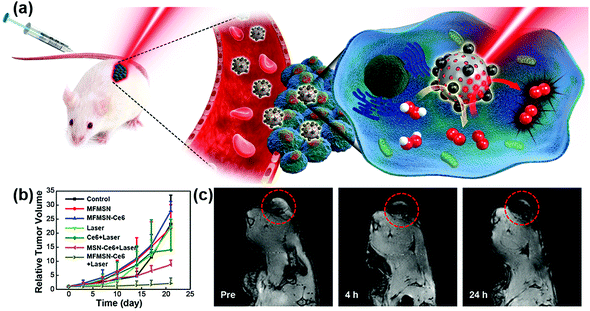 | ||
| Fig. 7 (a) Schematic illustration of MFMSNs for efficient PDT in hypoxic cancer. (b) Tumor volume changes of each group after 3 weeks. (c) In vivo T2-weighted MRI tracking of a tumor-bearing mouse at various time periods. Tumors are circled with red dashed lines. Adapted with permission from ref. 43. Copyright (2017) American Chemical Society. | ||
In addition, cell tracking provided by MRI of MNPs could benefit the study of biosafety issues associated with MNPs’ use. According to Huang's study, commercialized superparamagnetic iron oxide (SPIO) NPs were not toxic but instead promoted stem cell growth. The reason was likely due to the ability of SPIO NPs to remove intracellular H2O2 through peroxidase mimicking properties.106 Meanwhile, with the degradation of SPIO NPs within lysosome, free iron ions would be generated and released to accelerate the cell cycle progression (Fig. 8).
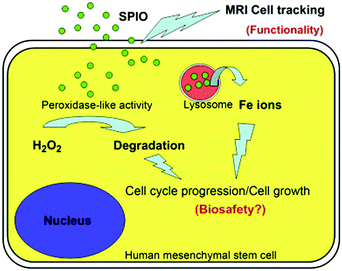 | ||
| Fig. 8 SPIO NPs could promote cell growth due to their ability to diminish intracellular H2O2 through intrinsic peroxidase-like activity. Also, SPIO could accelerate cell cycle progression, which may be mediated by the free iron released from lysosomal degradation. Reprinted with permission from ref. 106. Copyright (2009) Elsevier. | ||
Metal nanozymes with surface plasmon resonance
Metal nanomaterials (such as AuNPs, silver nanoparticles (AgNPs), and copper nanoparticles (CuNPs)) possess distinct optical properties including localized surface plasmon resonance (LSPR) and surface enhanced Raman scattering (SERS).107–114 These properties are strongly influenced by the size, morphology, and inter-particle distance. Thus great interest has been shown to develop sensitive assays with AuNPs as colorimetric signals or AgNPs as SERS substrates.111,115,116 Although metal nanomaterials had also been used as catalysts, it was still surprising when Rossi et al. found “hidden talents” of AuNPs in that citrate-coated AuNPs exhibited GOx mimicking activity.117 Since then, a significant amount of effort has been made to explore metal NPs based enzyme mimicking activities and their applications.118–121 For instance, the generation of fluorescence signals catalyzed by metal nanozymes was reported for biomedical sensing.122–128 Here, we focus on the combination of LSPR/SERS properties and enzyme mimicking properties of metal nanomaterials and their applications.Localized surface plasmon resonance
LSPR is a result of the interaction between the local plasmons across the surface of nanomaterials and incident light. The LSPR spectrum would have a dramatic shift when the size or morphology of nanomaterials changes. Fan, Li and co-workers found that small (13 nm) AuNPs with GOx-like catalytic activities would produce H2O2in situ to initiate the seeded growth of small AuNPs, and then the growth process would be self-limited by the size-dependent catalytic activity decrease and the product-caused surface passivation (Fig. 9a).129 Inspired by this phenomenon, they further constructed a sensing platform for DNA based on the modulation of AuNPs’ GOx-like catalytic activities with DNA. As shown in Fig. 9b, different binding affinities of single-stranded DNA (ssDNA) and dsDNA to the AuNPs’ surface resulted in different catalytic activities of AuNPs, thereby affecting the final seeded growth of AuNPs. The strong adsorption of the probe ssDNA to the surface of AuNPs inhibited the intrinsic oxidase-like catalytic activities, and thus the AuNPs would not grow larger. After forming dsDNA by hybridizing with complementary DNA, the catalytic activities of AuNPs would be recovered, which then could be used for DNA or microRNA detection. For monitoring the nanozymes’ activities, they chose different suitable output signals. For example, either a colorimetric or chemiluminescent signal would be produced when the nanozymes were coupled with the horseradish peroxidase (HRP)-based catalytic reaction. Without HRP, the SPR-enhanced light scattering spectra of the individual enlarged AuNPs could also be used as a signal.130 Similarly, an aptasensor for adenosine triphosphate (ATP) was developed through changing the probe DNA to an anti-ATP aptamer. As shown in Fig. 10, the plasmonic resonance Rayleigh scattering (PRRS) spectra peak shift was used as a readout signal. The more ATP, the larger the peak shift, and the detection limit was calculated to be as low as 4.0 nM.131 The dark field microscopy (DFM) study with catalytic AuNPs as the probe brought out a new method for biomolecular sensing. However, the limited optical resolution of DFM made this method only suitable for uniformly large AuNPs (>50 nm), which had a low catalytic activity.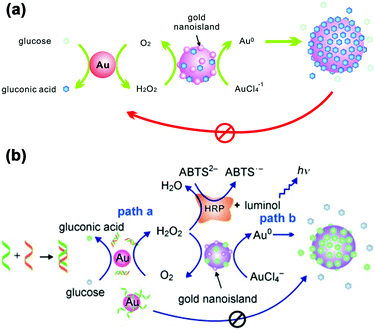 | ||
| Fig. 9 (a) Schematic demonstration of the AuNP-based self-limiting growth system. The size, shape, and catalytic activity of AuNPs were self-limited by the integrated influence from the catalytic reaction, seeded enlargement, and surface passivation of AuNPs. Reprinted with permission from ref. 129. Copyright (2010) American Chemical Society. (b) Illustration of the GOx-like catalytic activity of AuNPs regulated by DNA hybridization, which could be either amplified by HRP cascaded color or chemiluminescence variations (path a) or lead to nanoplasmonic changes owing to size enlargement (path b). Reprinted with permission from ref. 130. Copyright (2011) John Wiley and Sons. | ||
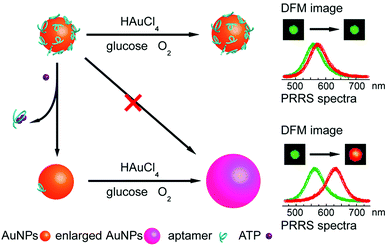 | ||
| Fig. 10 Schematic illustration of the aptasensor. The plasmonic signal was generated by the ATP-induced conformational change of surface adsorbed anti-ATP aptamer that recovered the self-catalytic activities of AuNPs. Adapted with permission from ref. 131. Copyright (2012) Royal Society of Chemistry. | ||
To overcome this limitation, they further combined plasmonic large AuNPs (50 nm) with catalytically active small AuNPs (13 nm) to fabricate a multicomponent halo-like structure (nanohalo) based on DNA-directed self-assembly. The Au nanohalo showed higher electromagnetic field intensity with almost 15-fold enhancement compared to that on the surface of large AuNPs, which would benefit the sensing of dielectric changes near the small AuNPs. Due to the high sensitivity of LSPR to the refractive index or dielectric constant of the surroundings, the small change in the permittivity of small AuNPs would lead to a significant change of the LSPR of large AuNPs, thereby enabling monitoring of the catalytic reactions and the states of the nanocatalyst. As shown in Fig. 11, the LSPR spectra of the Au nanohalo were totally different from those of the large AuNPs. The adsorption of glucose, charging–discharging of small AuNPs, and the adsorption of gluconic acid were reflected as an initial red-shift of 2.52 nm, a rapid blue-shift of 6.88 nm, then a slow red-shift of 3.53 nm, and a final position at 3.53 nm of red-shift, respectively (Fig. 11b). While for the large AuNPs, there was only an initial red-shift of 1.16 nm attributed to the adsorption of glucose, thereby reflecting the inert or slow catalytic activity of large AuNPs.132
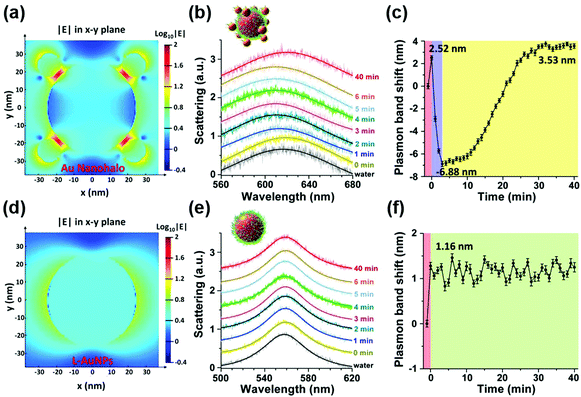 | ||
| Fig. 11 Electromagnetic finite-difference time-domain simulation of the local electric field enhancement of the Au nanohalo (a) and large AuNPs (d) at the X–Y plane. (b) LSPR spectra and (c) the corresponding plasmonic band peak shifts of one Au nanohalo during the catalytic reaction. (e) LSPR spectra and (f) the corresponding plasmon band peak shifts of one large AuNP along the catalytic process under the same experimental conditions. LSPR spectra were collected using DFM with a time interval of 1 min and then fitted by a Lorentz curve to determine the peak position. Error bars in panels c and f represented the deviations coming from the Lorentzian fitting procedure. Adapted with permission from ref. 132. Copyright (2015) American Chemical Society. | ||
Surface enhanced Raman scattering
SERS is a surface-sensitive technique to enhance the Raman scattering of molecules bound to or near plasmonic surfaces, thereby providing highly sensitive detection of adsorbed molecules. In 2013, Faulds and co-workers combined AgNPs as SERS substrates with their peroxidase-like activities for a highly sensitive detection of H2O2 as low as 100 nM.133 Moreover, SERS can acquire the unique fingerprints of the molecules up to single-molecule sensitivity and reflect the real catalytic process. For example, Song and co-workers reported in situ super-sensitive monitoring of the catalytic oxidation of TMB through Ag@carbon dots (CDs) based peroxidase mimicking activity (Fig. 12).134 The synthesized Ag@CDs showed an enhanced catalytic activity due to the synergistic effect between the AgNPs’ core and the CDs’ shell. Besides excellent catalytic activity, the Ag@CDs also worked as an ultrasensitive SERS substrate. As shown in Fig. 12, the catalytic oxidation of TMB in the presence of H2O2 was monitored in situ. The product oxidized TMB showed three characteristic bands at 1190, 1336, and 1605 cm−1, which were assigned to the CH3 bending, inter-ring C–C stretching, and C–H bending + ring stretching modes, respectively. The intensities of these bands gradually increased over the whole reaction time from 0 to 20 min and stayed invariant after 20 min, indicating that almost all the TMB molecules had been consumed. The results were consistent with their observation of the adsorption spectra of oxidized TMB at 650 nm. As the peroxidase-like catalytic reaction was H2O2-dependent, a H2O2 sensor was accordingly developed with a detection limit of 16 nM.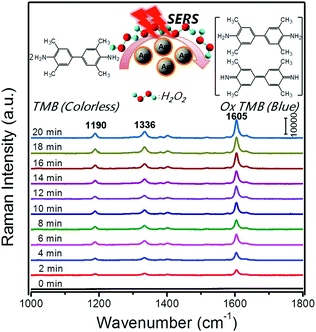 | ||
| Fig. 12 Time-dependent SERS spectra for the oxidation of TMB by H2O2 (3 mM) catalyzed by Ag@CDs NPs. The time between consecutive curves is 2 min. Reprinted with permission from ref. 134. Copyright (2016) American Chemical Society. | ||
Besides AgNPs, AuNPs were also demonstrated as good SERS substrates and even more stable than AgNPs. Our group recently developed SERS active AuNPs with peroxidase-mimicking properties for versatile bioassays,135 and we also combined an oxidase with the peroxidase mimic AuNPs; the corresponding oxidase substrate was determined subsequently. As shown in Fig. 13a, the AuNPs were grown in situ in a porous metal–organic framework MIL-101, then an oxidase (i.e., GOx or lactate oxidase (LOx)) was assembled onto AuNPs@MIL-101 to construct integrated nanozymes AuNPs@MIL-101@GOx and AuNPs@MIL-101@LOx for glucose and lactate sensing via SERS, respectively. Further, this assay was explored for monitoring the glucose and lactate in living brain and tumor samples. If an ischemic stroke happened, the striatum glucose level would decrease while the lactate level would increase. When an anti-oxidation drug like astaxanthin (ATX) was used for treatment, the dynamic change of both the glucose and lactate would be suppressed as shown in Fig. 13c, which verified the effectiveness of ATX as a drug for alleviating cerebral ischemic injuries. We also measured the glucose and lactate metabolism in both normal and tumor tissues. Therefore, the developed integrative nanozyme not only provided a general method for monitoring the important biological events evolving glucose and lactate, but could also evaluate the therapeutic efficacy of potential drugs.
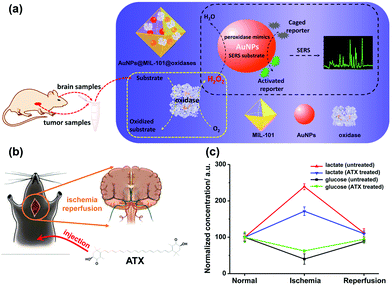 | ||
| Fig. 13 (a) Schematic illustration of AuNPs@MIL-101@oxidases for efficient enzymatic cascade reactions. First, oxidases catalyzed the oxidation of a substrate target (i.e., glucose or lactate) to produce H2O2; peroxidase-like AuNPs in AuNPs@MIL-101@oxidases then catalytically oxidized caged Raman reporters with H2O2 to produce active Raman reporters and simultaneously enhanced the reporters’ Raman signals for SERS measurements. (b) Schematic illustration of global cerebral ischemia/reperfusion and the treatment with ATX. (c) Dynamic changes of glucose and lactate following ischemia and reperfusion with and without ATX pretreatment. The glucose and lactate levels before ischemia were normalized as 100. Adapted with permission from ref. 135. Copyright (2017) American Chemical Society. | ||
However, the low catalytic activity of AuNPs made the SERS detection need half an hour's incubation, which in turn tremendously limited their further applications. To overcome this limitation, we further fabricated multifunctional Au@Pt nanozymes with simultaneous plasmonic and peroxidase-mimicking activities (Fig. 14). The experimental and computational studies indicated that due to the weak electric effect and the decreased electric field, neither a lower nor higher amount of Pt would give cooperatively enhanced performance of Au@Pt nanozymes. Only a suitable amount (2.5%) of Pt decorated onto the AuNPs produced the highest Raman signal within 2 min. The high performance of the Au@Pt2.5% bifunctional nanozyme benefited not only from the significantly enhanced catalytic oxidation of TMB with the help of the Pt shell, but also from the high electric field retained from the AuNP core for SERS.136 Other systems based on SERS active nanozymes are summarized in Table 2.
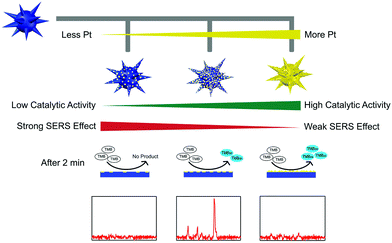 | ||
| Fig. 14 Rational design of high-performance Au@Pt NPs bifunctional nanozymes by controlling the Pt amount. Reprinted with permission from ref. 136. Copyright (2018) American Chemical Society. | ||
| Nanomaterials | Enzyme activity | Targets | Linear range | LOD | Comments | Ref. |
|---|---|---|---|---|---|---|
| AgNPs | Peroxidase | H2O2 | 4–10 μM | 100 nM | 133 | |
| AgNPs | Peroxidase | Human C-reactive protein | 1.56–25 ng mL−1 | 1.09 ng mL−1 | Silver-linked immunosorbent assay | 137 |
| Ag@CDs | Peroxidase | H2O2 | 30 nM–3 mM | 16 nM | 134 | |
| AgNPs/CDs | Peroxidase | H2O2 | 10 nM–10 μM | 10 nM | 138 | |
| Ag–Cu2O/rGO nanocomposites | Peroxidase | H2O2 | 10 nM–10 mM | 10 nM | 139 | |
| Glucose | 10 nM–10 mM | 10 nM | ||||
| AuNPs@MIL-101 | Peroxidase | Glucose | 10–200 μM | 4.2 μM | Assembly of GOx and LOx onto AuNPs@MIL-101 to form AuNPs@MIL-101@GOx and AuNPs@MIL-101@LOx integrative nanozymes | 135 |
| Lactate | 10–200 μM | 5.0 μM | ||||
| Au@Pt multibranched NPs | Peroxidase | H2O2 | 1–30 μM | 0.38 μM | 136 | |
| Cu/Au complex | Peroxidase | H2O2 | 10 μM–10 mM | 1.5 μM | Fabrication of a novel dual-biomimetic functional (superhydrophobic and peroxidase-like activity) material for H2O2 analysis based on SERS | 140 |
| rGO/CuS/Au composite nanosheets | Peroxidase | H2O2 | 3.05–50 μM | 2.1 μM | 141 | |
| Au@Ag@Pt NPs | Peroxidase | H2O2 | 10 pM–100 mM | 10 pM | 142 | |
| Glucose | 1 nM–200 mM | 1 nM | ||||
| Chitosan-modified popcorn-like Au–Ag NPs | Peroxidase | Melamine | 10 nM–50 μM | 8.51 nM | Melamine can react with H2O2 and make the available amount of H2O2 for the oxidation of TMB reduced | 143 |
| Au/CuS composite | Peroxidase | Rhodamine 6G | 100 nM–20 μM | N/A | Real-time monitoring of the degradation process | 144 |
| CoS2/Au (CoS/Au) composite | Peroxidase | Rhodamine 6G | 100 nM–100 μM | N/A | Real-time monitoring of the degradation process | 145 |
| Au/FeS (Au/Co3O4) composite | Peroxidase | Rhodamine 6G | 100 nM–100 μM | N/A | Real-time monitoring of the degradation process | 146 |
Conclusions and perspective
This review highlights recent exciting developments of multifunctional nanozymes, especially the utilization of magnetic properties of iron oxide nanozymes and SPR of metal nanozymes and their applications. The benefits—such as easy separation, enrichment, recycling, and MRI from iron oxide, as well as LSPR based colorimetric sensors, ultrasensitive SERS based detection, and in situ monitoring of the catalytic process—have been discussed in detail. While the development of multifunctional nanozymes has been evidenced from the above examples, there are still some challenges remaining to be tackled. Here, we speculate on a few such directions which need to be addressed in future studies.(a) Though many nanomaterials have been explored to mimic natural enzymes, currently mainly two major types of multifunctional nanozymes (i.e., magnetic iron nanoparticle based and plasmonic metal nanoparticle based) have been developed. Therefore, new multifunctional nanozymes from other well-developed nanomaterials should be explored. For example, other nanomaterials such as iron chalcogenides (e.g., FeS,147–149 Fe3S4,150 FeSe,147,151,152 and FeTe153), PtCo bimetallic NPs,154 CePO4:Tb,Gd hollow nanospheres,155 and α-MnSe156 also showed both excellent magnetic properties and high enzyme mimicking activities. Magnetic α-MnSe was reported to be a robust peroxidase mimic and can be easily recovered and reused for 10 cycles.156
(b) For the current applications of multifunctional nanozymes, most experiments were performed based on peroxidase mimics or a combination with natural oxidases for detection of H2O2 and glucose, as well as degradation of organic pollutants. Given the fact that there are six major types of natural enzymes, we expect to see more multifunctional nanozymes with new catalytic activities beyond redox reactions.
(c) Taking advantage of multifunctional nanozymes to contribute more in bionanotechnology fields should be an interesting and challenging direction. More applications such as cancer diagnostics103 and therapy43,104,157–160 as well as stem cell promotion60,106 should be investigated. However, various problems should be carefully studied and tested before the clinical use of the multifunctional nanozymes, which include clinical toxicity, stability, and catalytic mechanisms.
(d) For MRI applications, besides magnetic iron oxide T2 agents, Gd-based nanomaterials are another type of MR contrast agents, which exhibit an increase in signal intensity and appear bright in T1-weighted images. T1-weighted imaging can be used for identifying fatty tissue while T2-weighted is useful for detecting inflammation. Thus, Gd-based nanomaterials as a peroxidase mimic155,161 should be considered and explored to broaden the MRI applications of multifunctional nanozymes.
(e) For SERS-based active nanozymes, usually their catalytic activities are relatively low, which are limited by the metal nanoparticles (Au, Ag) themselves. To solve this issue, one nanomaterial with high catalytic activity could be deposited on the surface of Au/Ag, as most catalytic reactions happen on the surface of current nanozymes. However, the deposition would dampen the SERS properties, thus better experimental strategies and detailed theoretical studies are required to balance these two aspects for designing high-performance nanozymes. For example, Au@Pt NPs were rationally designed with enhanced enzyme-like catalytic activities while retaining their excellent SERS properties.136
(f) Besides magnetism and SPR, other functions (e.g., fluorescence162–165 and thermal166) should also be investigated to broaden the research and applications of multifunctional nanozymes. Cai and co-workers constructed a nanoprobe of fluorescence enzyme mimicking Au nanoclusters, which can be used for tumor tissue detection based on simultaneous fluorescent and peroxidase staining.162 This front has not been explored to a great extent, which could be a new important direction of future studies.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Natural Science Foundation of China (21722503), Natural Science Foundation of Jiangsu Province (BK20160615), 973 Program (2015CB659400), PAPD program, Shuangchuang Program of Jiangsu Province, Open Funds of the State Key Laboratory of Coordination Chemistry (SKLCC1619 and SKLCC1819), Open Funds of the State Key Laboratory of Analytical Chemistry for Life Science (SKLACLS1704), China Postdoctoral Science Foundation (2016M590437), Fundamental Research Funds for the Central Universities (021314380046) and Thousand Talents Program for Young Researchers.References
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- X. Wang, Y. Hu and H. Wei, Inorg. Chem. Front., 2016, 3, 41–60 RSC.
- Y. Zhou, B. Liu, R. Yang and J. Liu, Bioconjugate Chem., 2017, 28, 2903–2909 CrossRef PubMed.
- X. Wang, W. Guo, Y. Hu, J. Wu and H. Wei, Nanozymes: Next Wave of Artificial Enzymes, Springer, 2016 Search PubMed.
- L. Gao and X. Yan, Sci. China: Life Sci., 2016, 59, 400–402 CrossRef PubMed.
- Y. Lin, J. Ren and X. Qu, Acc. Chem. Res., 2014, 47, 1097–1105 CrossRef PubMed.
- R. Ragg, M. N. Tahir and W. Tremel, Eur. J. Inorg. Chem., 2016, 1906–1915 CrossRef.
- A. Karakoti, S. Singh, J. M. Dowding, S. Seal and W. T. Self, Chem. Soc. Rev., 2010, 39, 4422–4432 RSC.
- H. Cheng, S. Lin, F. Muhammad, Y.-W. Lin and H. Wei, ACS Sens., 2016, 1, 1336–1343 CrossRef.
- H. Cheng, Y. Liu, Y. Hu, Y. Ding, S. Lin, W. Cao, Q. Wang, J. Wu, F. Muhammad, X. Zhao, D. Zhao, Z. Li, H. Xing and H. Wei, Anal. Chem., 2017, 89, 11552–11559 CrossRef PubMed.
- X. Wang, W. Cao, L. Qin, T. Lin, W. Chen, S. Lin, J. Yao, X. Zhao, M. Zhou, C. Hang and H. Wei, Theranostics, 2017, 7, 2277–2286 CrossRef PubMed.
- S. Li, Y. Huang, J. Liu, E. Wang and H. Wei, Prog. Biochem. Biophys., 2018, 45, 129–147 Search PubMed.
- H. Cheng, L. Zhang, J. He, W. Guo, Z. Zhou, X. Zhang, S. Nie and H. Wei, Anal. Chem., 2016, 88, 5489–5497 CrossRef PubMed.
- J. Yao, Y. Cheng, M. Zhou, S. Zhao, S. Lin, X. Wang, J. Wu, S. Li and H. Wei, Chem. Sci., 2018, 9, 2927–2933 RSC.
- S. Lin, J. Wu, J. Yao, W. Cao, F. Muhammad and H. Wei, Nanozymes for Biomedical Sensing Applications: from in vitro Sensing to Living Systems, Elsevier, 2018 Search PubMed.
- V. Polshettiwar, R. Luque, A. Fihri, H. B. Zhu, M. Bouhrara and J. M. Bassett, Chem. Rev., 2011, 111, 3036–3075 CrossRef PubMed.
- N. Lee, D. Yoo, D. Ling, M. H. Cho, T. Hyeon and J. Cheon, Chem. Rev., 2015, 115, 10637–10689 CrossRef PubMed.
- M. Colombo, S. Carregal-Romero, M. F. Casula, L. Gutierrez, M. P. Morales, I. B. Bohm, J. T. Heverhagen, D. Prosperi and W. J. Parak, Chem. Soc. Rev., 2012, 41, 4306–4334 RSC.
- M. B. Gawande, P. S. Branco and R. S. Varma, Chem. Soc. Rev., 2013, 42, 3371–3393 RSC.
- L. Z. Gao, J. Zhuang, L. Nie, J. B. Zhang, Y. Zhang, N. Gu, T. H. Wang, J. Feng, D. L. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef PubMed.
- H. Wei and E. Wang, Anal. Chem., 2008, 80, 2250–2254 CrossRef PubMed.
- L. Gao, K. Fan and X. Yan, Theranostics, 2017, 7, 3207–3227 CrossRef PubMed.
- K. Fan, C. Cao, Y. Pan, D. Lu, D. Yang, J. Feng, L. Song, M. Liang and X. Yan, Nat. Nanotechnol., 2012, 7, 459 CrossRef PubMed.
- X. Q. Zhang, S. W. Gong, Y. Zhang, T. Yang, C. Y. Wang and N. Gu, J. Mater. Chem., 2010, 20, 5110–5116 RSC.
- Y. H. Wu, M. J. Song, Z. A. Xin, X. Q. Zhang, Y. Zhang, C. Y. Wang, S. Y. Li and N. Gu, Nanotechnology, 2011, 22, 8 Search PubMed.
- Q. Y. Liu, L. Y. Zhang, H. Li, Q. Y. Jia, Y. L. Jiang, Y. T. Yang and R. R. Zhu, Mater. Sci. Eng., C, 2015, 55, 193–200 CrossRef PubMed.
- J. L. Sang, R. L. Wu, P. P. Guo, J. Du, S. M. Xu and J. D. Wang, J. Appl. Polym. Sci., 2016, 133, 43065 CrossRef.
- J. A. R. Guivar, E. G. R. Fernandes and V. Zucolotto, Talanta, 2015, 141, 307–314 CrossRef PubMed.
- B. Liu and J. Liu, Nano Res., 2017, 10, 1125–1148 CrossRef.
- X. Y. Niu, Y. Y. Xu, Y. L. Dong, L. Y. Qi, S. D. Qi, H. L. Chen and X. G. Chen, J. Alloys Compd., 2014, 587, 74–81 CrossRef.
- H. Zhang, L. Ma, P. Li and J. Zheng, Biosens. Bioelectron., 2016, 85, 343–350 CrossRef PubMed.
- L. Song, C. Huang, W. Zhang, M. Ma, Z. Chen, N. Gu and Y. Zhang, Colloids Surf., A, 2016, 506, 747–755 CrossRef.
- S. Li, H. Li, F. Chen, J. Liu, H. Zhang, Z. Yang and B. Wang, Dyes Pigm., 2016, 125, 64–71 CrossRef.
- P. Janoš, P. Kuráň, V. Pilařová, J. Trögl, M. Šťastný, O. Pelant, J. Henych, S. Bakardjieva, O. Životský, M. Kormunda, K. Mazanec and M. Skoumal, Chem. Eng. J., 2015, 262, 747–755 CrossRef.
- J. Chun, H. Lee, S. H. Lee, S. W. Hong, J. Lee, C. Lee and J. Lee, Chemosphere, 2012, 89, 1230–1237 CrossRef PubMed.
- Y. L. Dong, H. G. Zhang, Z. U. Rahman, L. Su, X. J. Chen, J. Hu and X. G. Chen, Nanoscale, 2012, 4, 3969–3976 RSC.
- J. W. Lee, H. J. Jeon, H. J. Shin and J. K. Kang, Chem. Commun., 2012, 48, 422–424 RSC.
- Y. Song, K. Qu, C. Xu, J. Ren and X. Qu, Chem. Commun., 2010, 46, 6572–6574 RSC.
- C. Q. Wang, J. Qian, K. Wang, X. W. Yang, Q. Liu, N. Hao, C. K. Wang, X. Y. Dong and X. Y. Huang, Biosens. Bioelectron., 2016, 77, 1183–1191 CrossRef PubMed.
- W. Zhang, C. Chen, D. Yang, G. Dong, S. Jia, B. Zhao, L. Yan, Q. Yao, A. Sunna and Y. Liu, Adv. Mater. Interfaces, 2016, 3, 1600590 CrossRef.
- N. Salarizadeh, M. Sadri and R. H. Sajedi, Appl. Organomet. Chem., 2018, 32, e4018 CrossRef.
- A. Roy, R. Sahoo, C. Ray, S. Dutta and T. Pal, RSC Adv., 2016, 6, 32308–32318 RSC.
- J. Kim, H. R. Cho, H. Jeon, D. Kim, C. Song, N. Lee, S. H. Choi and T. Hyeon, J. Am. Chem. Soc., 2017, 139, 10992–10995 CrossRef PubMed.
- L. Su, W. J. Qin, H. G. Zhang, Z. U. Rahman, C. L. Ren, S. D. Ma and X. G. Chen, Biosens. Bioelectron., 2015, 63, 384–391 CrossRef PubMed.
- R. Cheng, C. Cheng, G. H. Liu, X. Zheng, G. Q. Li and J. Li, Chemosphere, 2015, 141, 138–143 CrossRef PubMed.
- J. Qian, X. W. Yang, L. Jiang, C. D. Zhu, H. P. Mao and K. Wang, Sens. Actuators, B, 2014, 201, 160–166 CrossRef.
- Z. Q. Gao, M. D. Xu, L. Hou, G. N. Chen and D. P. Tang, Anal. Chem., 2013, 85, 6945–6952 CrossRef PubMed.
- N. Puvvada, P. K. Panigrahi, D. Mandal and A. Pathak, RSC Adv., 2012, 2, 3270–3273 RSC.
- W. Shen, C. L. Lim and Z. Q. Gao, Chem. Commun., 2013, 49, 8114–8116 RSC.
- K. S. Park, M. I. Kim, D. Y. Cho and H. G. Park, Small, 2011, 7, 1521–1525 CrossRef PubMed.
- X. Ai, L. Wu, M. N. Zhang, X. D. Hou, L. Yang and C. B. Zheng, J. Agric. Food Chem., 2014, 62, 8586–8593 CrossRef PubMed.
- D. Duan, K. Fan, D. Zhang, S. Tan, M. Liang, Y. Liu, J. Zhang, P. Zhang, W. Liu, X. Qiu, G. P. Kobinger, G. F. Gao and X. Yan, Biosens. Bioelectron., 2015, 74, 134–141 CrossRef PubMed.
- G. J. Guan, L. Yang, Q. S. Mei, K. Zhang, Z. P. Zhang and M. Y. Han, Anal. Chem., 2012, 84, 9492–9497 CrossRef PubMed.
- Y. Jia, H. M. Yu, L. Wu, X. D. Hou, L. Yang and C. B. Zheng, Anal. Chem., 2015, 87, 5866–5871 CrossRef PubMed.
- W. S. Zou, J. Yang, T. T. Yang, X. Hu and H. Z. Lian, J. Mater. Chem., 2012, 22, 4720–4727 RSC.
- R. Thiramanas, K. Jangpatarapongsa, P. Tangboriboonrat and D. Polpanich, Anal. Chem., 2013, 85, 5996–6002 CrossRef PubMed.
- J. M. Perez, Nat. Nanotechnol., 2007, 2, 535–536 CrossRef PubMed.
- L. Gao, J. Wu, S. Lyle, K. Zehr, L. Cao and D. Gao, J. Phys. Chem. C, 2008, 112, 17357–17361 Search PubMed.
- M. S. Kim, S. H. Kweon, S. Cho, S. S. A. An, M. I. Kim, J. Doh and J. Lee, ACS Appl. Mater. Interfaces, 2017, 9, 35133–35140 Search PubMed.
- T. Kito, R. Shibata, M. Ishii, H. Suzuki, T. Himeno, Y. Kataoka, Y. Yamamura, T. Yamamoto, N. Nishio, S. Ito, Y. Numaguchi, T. Tanigawa, J. K. Yamashita, N. Ouchi, H. Honda, K. Isobe and T. Murohara, Sci. Rep., 2013, 3, 1418 CrossRef PubMed.
- S. X. Zhang, X. L. Zhao, H. Y. Niu, Y. L. Shi, Y. Q. Cai and G. B. Jiang, J. Hazard. Mater., 2009, 167, 560–566 CrossRef PubMed.
- M. I. Kim, J. Shim, T. Li, J. Lee and H. G. Park, Chem. – Eur. J., 2011, 17, 10700–10707 CrossRef PubMed.
- H. Y. Niu, D. Zhang, S. X. Zhang, X. L. Zhang, Z. F. Meng and Y. Q. Cai, J. Hazard. Mater., 2011, 190, 559–565 CrossRef PubMed.
- M. Y. Zhu and G. W. Diao, J. Phys. Chem. C, 2011, 115, 18923–18934 Search PubMed.
- M. I. Kim, J. Shim, T. Li, M. A. Woo, D. Cho, J. Lee and H. G. Park, Analyst, 2012, 137, 1137–1143 RSC.
- X. L. Cheng, J. S. Jiang, D. M. Jiang and Z. J. Zhao, J. Phys. Chem. C, 2014, 118, 12588–12598 Search PubMed.
- R. X. Huang, Z. Q. Fang, X. B. Fang and E. P. Tsang, J. Colloid Interface Sci., 2014, 436, 258–266 CrossRef PubMed.
- M. Z. Yang, Y. P. Guan, Y. Yang, T. T. Xia, W. B. Xiong, N. Wang and C. Guo, J. Colloid Interface Sci., 2013, 405, 291–295 CrossRef PubMed.
- Q. Y. Liu, H. Li, Q. R. Zhao, R. R. Zhu, Y. T. Yang, Q. Y. Jia, B. Bian and L. H. Zhuo, Mater. Sci. Eng., C, 2014, 41, 142–151 CrossRef PubMed.
- Y. Liu, M. Yuan, L. J. Qiao and R. Guo, Biosens. Bioelectron., 2014, 52, 391–396 CrossRef PubMed.
- L. J. Wang, Y. Min, D. D. Xu, F. J. Yu, W. Z. Zhou and A. Cuschieri, Chem. Commun., 2014, 50, 11147–11150 RSC.
- W. Wang, Y. Liu, T. L. Li and M. H. Zhou, Chem. Eng. J., 2014, 242, 1–9 CrossRef.
- D. Bhuyan, S. S. Arbuj and L. Saikia, New J. Chem., 2015, 39, 7759–7762 RSC.
- R. Cheng, G. Q. Li, C. Cheng, L. Shi, X. Zheng and Z. Ma, RSC Adv., 2015, 5, 66927–66933 RSC.
- S. W. Xiao, C. T. Zhang, R. Chen and F. X. Chen, New J. Chem., 2015, 39, 4924–4932 RSC.
- D. Wan, W. Li, G. Wang and X. Wei, J. Mater. Eng. Perform., 2016, 25, 4333–4340 CrossRef.
- B. Liu, X. Han and J. Liu, Nanoscale, 2016, 8, 13620–13626 RSC.
- Y. Pan, N. Li, J. S. Mu, R. H. Zhou, Y. Xu, D. Z. Cui, Y. Wang and M. Zhao, Appl. Microbiol. Biotechnol., 2015, 99, 703–715 CrossRef PubMed.
- Y. H. Wang, B. Zhou, S. Wu, K. M. Wang and X. X. He, Talanta, 2015, 134, 712–717 CrossRef PubMed.
- Y. F. Qin, Z. Y. Qin, Y. N. Liu, M. Cheng, P. F. Qian, Q. Wang and M. F. Zhu, Appl. Surf. Sci., 2015, 357, 2103–2111 CrossRef.
- Y. Shi, J. Huang, J. N. Wang, P. Su and Y. Yang, Talanta, 2015, 143, 457–463 CrossRef PubMed.
- D. Wan, W. B. Li, G. H. Wang, K. Chen, L. L. Lu and Q. Hu, Appl. Surf. Sci., 2015, 349, 988–996 CrossRef.
- S. L. Wei, J. W. Li and Y. Liu, RSC Adv., 2015, 5, 107670–107679 RSC.
- L. J. Xu and J. L. Wang, Environ. Sci. Technol., 2012, 46, 10145–10153 CrossRef PubMed.
- H. Y. Sun, X. L. Jiao, Y. Y. Han, Z. Jiang and D. R. Chen, Eur. J. Inorg. Chem., 2013, 109–114 CrossRef.
- X. C. Wu, Y. Zhang, T. Han, H. X. Wu, S. W. Guo and J. Y. Zhang, RSC Adv., 2014, 4, 3299–3305 RSC.
- T. Zeng, X. L. Zhang, S. H. Wang, Y. R. Ma, H. Y. Niu and Y. Q. Cai, Chem. – Eur. J., 2014, 20, 6474–6481 CrossRef PubMed.
- S. Zhang, H. Li, Z. Wang, J. Liu, H. Zhang, B. Wang and Z. Yang, Nanoscale, 2015, 7, 8495–8502 RSC.
- Q. Gao, Y. Xing, M. L. Peng, Y. S. Liu, Z. Y. Luo, Y. Y. Jin, H. M. Fan, K. B. Li, C. Chen and Y. L. Cui, Chin. J. Chem., 2017, 35, 1431–1436 CrossRef.
- J. Zhang, J. Ma, X. Fan, W. Peng, G. Zhang, F. Zhang and Y. Li, Catal. Commun., 2017, 89, 148–151 CrossRef.
- X. He, L. Tan, D. Chen, X. Wu, X. Ren, Y. Zhang, X. Meng and F. Tang, Chem. Commun., 2013, 49, 4643–4645 RSC.
- G. F. Liu, N. Wang, J. T. Zhou, A. J. Wang, J. Wang, R. F. Jin and H. Lv, RSC Adv., 2015, 5, 95857–95865 RSC.
- Q. Chang and H. Q. Tang, Microchim. Acta, 2014, 181, 527–534 CrossRef.
- X. J. Zheng, Q. Zhu, H. Q. Song, X. R. Zhao, T. Yi, H. L. Chen and X. G. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 3480–3491 Search PubMed.
- C. Hou, Y. Wang, Q. H. Ding, L. Jiang, M. Li, W. W. Zhu, D. Pan, H. Zhu and M. Z. Liu, Nanoscale, 2015, 7, 18770–18779 RSC.
- C.-H. Liu and W.-L. Tseng, Anal. Chim. Acta, 2011, 703, 87–93 CrossRef PubMed.
- C.-J. Yu, C.-Y. Lin, C.-H. Liu, T.-L. Cheng and W.-L. Tseng, Biosens. Bioelectron., 2010, 26, 913–917 CrossRef PubMed.
- H. Wang and Y. M. Huang, J. Hazard. Mater., 2011, 191, 163–169 CrossRef PubMed.
- C. Lu, X. Liu, Y. Li, F. Yu, L. Tang, Y. Hu and Y. Yine, ACS Appl. Mater. Interfaces, 2015, 7, 15395–15402 Search PubMed.
- Y. Yu, P. Ju, D. Zhang, X. Han, X. Yin, L. Zheng and C. Sun, Sens. Actuators, B, 2016, 233, 162–172 CrossRef.
- A. A. Vernekar, T. Das, S. Ghosh and G. Mugesh, Chem. – Asian J., 2016, 11, 72–76 CrossRef PubMed.
- S. Laurent, D. Forge, M. Port, A. Roch, C. Robic, L. Vander Elst and R. N. Muller, Chem. Rev., 2008, 108, 2064–2110 CrossRef PubMed.
- J. Liu, W. Zhang, H. L. Zhang, Z. Y. Yang, T. R. Li, B. D. Wang, X. Huo, R. Wang and H. T. Chen, Chem. Commun., 2013, 49, 4938–4940 RSC.
- D. Zhang, Y. X. Zhao, Y. J. Gao, F. P. Gao, Y. S. Fan, X. J. Li, Z. Y. Duan and H. Wang, J. Mater. Chem. B, 2013, 1, 5100–5107 RSC.
- J. Bai, X. Jia, W. Zhen, W. Cheng and X. Jiang, J. Am. Chem. Soc., 2018, 140, 106–109 CrossRef PubMed.
- D.-M. Huang, J.-K. Hsiao, Y.-C. Chen, L.-Y. Chien, M. Yao, Y.-K. Chen, B.-S. Ko, S.-C. Hsu, L.-A. Tai, H.-Y. Cheng, S.-W. Wang, C.-S. Yang and Y.-C. Chen, Biomaterials, 2009, 30, 3645–3651 CrossRef PubMed.
- H. Wei, C. Chen, B. Han, E. Wang and A. Chem, Anal. Chem., 2008, 80, 7051–7055 CrossRef PubMed.
- W. Guo, Y. Hu and H. Wei, Analyst, 2017, 142, 2322–2326 RSC.
- Y. Wang, H. Wei, B. Li, W. Ren, S. Guo, S. Dong and E. Wang, Chem. Commun., 2007, 5220–5222 RSC.
- H. Wei, B. Li, J. Li, E. Wang and S. Dong, Chem. Commun., 2007, 3735–3737 RSC.
- J. Wu, L. H. Tan, K. Hwang, H. Xing, P. Wu, W. Li and Y. Lu, J. Am. Chem. Soc., 2014, 136, 15195–15202 CrossRef PubMed.
- H. Wei, B. Li, J. Li, S. Dong and E. Wang, Nanotechnology, 2008, 19, 095501 CrossRef PubMed.
- H. Wei, J. Li, Y. Wang and E. Wang, Nanotechnology, 2007, 18, 175610 CrossRef.
- H. Wei, Z. Wang, J. Zhang, S. House, Y. G. Gao, L. Yang, H. Robinson, L. H. Tan, H. Xing and C. Hou, Nat. Nanotechnol., 2011, 6, 93–97 CrossRef PubMed.
- S. Guo and E. Wang, Nano Today, 2011, 6, 240–264 CrossRef.
- L. H. Tan, H. Xing and Y. Lu, Acc. Chem. Res., 2014, 47, 1881–1890 CrossRef PubMed.
- M. Comotti, C. Della Pina, R. Matarrese and M. Rossi, Angew. Chem., Int. Ed., 2004, 43, 5812–5815 CrossRef PubMed.
- Y. Lin, J. Ren and X. Qu, Adv. Mater., 2014, 26, 4200–4217 CrossRef PubMed.
- L. N. Zhang, H. H. Deng, F. L. Lin, X. W. Xu, S. H. Weng, A. L. Liu, X. H. Lin, X. H. Xia and W. Chen, Anal. Chem., 2014, 86, 2711–2718 CrossRef PubMed.
- C. Wang, Y. Shi, Y. Y. Dan, X. G. Nie, J. Li and X. H. Xia, Chem. – Eur. J., 2017, 23, 6717–6723 CrossRef PubMed.
- T. Yu, M. Li, B. Kim and D. T. Auguste, Theranostics, 2017, 7, 899–911 CrossRef PubMed.
- Y.-S. Wu, F.-F. Huang and Y.-W. Lin, ACS Appl. Mater. Interfaces, 2013, 5, 1503–1509 Search PubMed.
- C.-I. Wang, W.-T. Chen and H.-T. Chang, Anal. Chem., 2012, 84, 9706–9712 CrossRef PubMed.
- P.-C. Kuo, C.-W. Lien, J.-Y. Mao, B. Unnikrishnan, H.-T. Chang, H.-J. Lin and C.-C. Huang, Anal. Chim. Acta, 2018, 1009, 89–97 CrossRef PubMed.
- C.-I. Wang, C.-C. Huang, Y.-W. Lin, W.-T. Chen and H.-T. Chang, Anal. Chim. Acta, 2012, 745, 124–130 CrossRef PubMed.
- C.-L. Li, C.-C. Huang, W.-H. Chen, C.-K. Chiang and H.-T. Chang, Analyst, 2012, 137, 5222–5228 RSC.
- C.-W. Lien, C.-C. Huang and H.-T. Chang, Chem. Commun., 2012, 48, 7952–7954 RSC.
- R. Gill, R. Polsky and I. Willner, Small, 2006, 2, 1037–1041 CrossRef PubMed.
- W. Luo, C. Zhu, S. Su, D. Li, Y. He, Q. Huang and C. Fan, ACS Nano, 2010, 4, 7451–7458 CrossRef PubMed.
- X. Zheng, Q. Liu, C. Jing, Y. Li, D. Li, W. Luo, Y. Wen, Y. He, Q. Huang, Y.-T. Long and C. Fan, Angew. Chem., Int. Ed., 2011, 50, 11994–11998 CrossRef PubMed.
- Q. Liu, C. Jing, X. Zheng, Z. Gu, D. Li, D.-W. Li, Q. Huang, Y.-T. Long and C. Fan, Chem. Commun., 2012, 48, 9574–9576 RSC.
- K. Li, K. Wang, W. Qin, S. Deng, D. Li, J. Shi, Q. Huang and C. Fan, J. Am. Chem. Soc., 2015, 137, 4292–4295 CrossRef PubMed.
- K. S. McKeating, S. Sloan-Dennison, D. Graham and K. Faulds, Analyst, 2013, 138, 6347–6353 RSC.
- J. Jin, S. Zhu, Y. Song, H. Zhao, Z. Zhang, Y. Guo, J. Li, W. Song, B. Yang and B. Zhao, ACS Appl. Mater. Interfaces, 2016, 8, 27956–27965 Search PubMed.
- Y. Hu, H. Cheng, X. Zhao, J. Wu, F. Muhammad, S. Lin, J. He, L. Zhou, C. Zhang, Y. Deng, P. Wang, Z. Zhou, S. Nie and H. Wei, ACS Nano, 2017, 11, 5558–5566 CrossRef PubMed.
- J. Wu, K. Qin, D. Yuan, J. Tan, L. Qin, X. Zhang and H. Wei, ACS Appl. Mater. Interfaces, 2018, 10, 12954–12959 Search PubMed.
- S. Sloan-Dennison, S. Laing, N. C. Shand, D. Graham and K. Faulds, Analyst, 2017, 142, 2484–2490 RSC.
- H. Zhao, Y. Guo, S. Zhu, Y. Song, J. Jin, W. Ji, W. Song, B. Zhao, B. Yang and Y. Ozaki, Appl. Surf. Sci., 2017, 410, 42–50 CrossRef.
- Y. Guo, H. Wang, X. Ma, J. Jin, W. Ji, X. Wang, W. Song, B. Zhao and C. He, ACS Appl. Mater. Interfaces, 2017, 9, 19074–19081 Search PubMed.
- Z. Yu, Y. Park, L. Chen, B. Zhao, Y. M. Jung and Q. Cong, ACS Appl. Mater. Interfaces, 2015, 7, 23472–23480 Search PubMed.
- W. Song, G. Nie, W. Ji, Y. Jiang, X. Lu, B. Zhao and Y. Ozaki, RSC Adv., 2016, 6, 54456–54462 RSC.
- J. Li, L. Lv, G. Zhang, X. Zhou, A. Shen and J. Hu, Anal. Methods, 2016, 8, 2097–2105 RSC.
- J. Li, G. Zhang, L. Wang, A. Shen and J. Hu, Talanta, 2015, 140, 204–211 CrossRef PubMed.
- Q. Cai, S. Lu, F. Liao, Y. Li, S. Ma and M. Shao, Nanoscale, 2014, 6, 8117–8123 RSC.
- S. Lu, S. Ma, H. Wang and M. Shao, RSC Adv., 2016, 6, 78852–78857 RSC.
- S. Ma, Q. Cai, K. Lu, F. Liao and M. Shao, J. Nanopart. Res., 2016, 18, 26 CrossRef.
- A. K. Dutta, S. K. Maji, D. N. Srivastava, A. Mondal, P. Biswas, P. Paul and B. Adhikary, ACS Appl. Mater. Interfaces, 2012, 4, 1919–1927 Search PubMed.
- S. K. Maji, A. K. Dutta, P. Biswas, D. N. Srivastava, P. Paul, A. Mondal and B. Adhikary, Appl. Catal., A, 2012, 419–420, 170–177 CrossRef.
- Z. Dai, S. Liu, J. Bao and H. Ju, Chem. – Eur. J., 2009, 15, 4321–4326 CrossRef PubMed.
- C. Ding, Y. Yan, D. Xiang, C. Zhang and Y. Xian, Microchim. Acta, 2016, 183, 625–631 CrossRef.
- F. Qiao, Z. Wang, K. Xu and S. Ai, Analyst, 2015, 140, 6684–6691 RSC.
- A. K. Dutta, S. K. Maji, A. Mondal, B. Karmakar, P. Biswas and B. Adhikary, Sens. Actuators, B, 2012, 173, 724–731 CrossRef.
- P. Roy, Z.-H. Lin, C.-T. Liang and H.-T. Chang, Chem. Commun., 2012, 48, 4079–4081 RSC.
- S. Cai, C. Qi, Y. Li, Q. Han, R. Yang and C. Wang, J. Mater. Chem. B, 2016, 4, 1869–1877 RSC.
- W. Wang, X. P. Jiang and K. Z. Chen, Chem. Commun., 2012, 48, 6839–6841 RSC.
- Y. Wang, D. Zhang and Z. B. Xiang, Mater. Res. Bull., 2015, 67, 152–157 CrossRef.
- Q. Han, S. Cai, L. Yang, X. Wang, C. Qi, R. Yang and C. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 21116–21123 Search PubMed.
- Z. Ma, M. Zhang, X. Jia, J. Bai, Y. Ruan, C. Wang, X. Sun and X. Jiang, Small, 2016, 12, 5477–5487 CrossRef PubMed.
- Y. Guan, M. Li, K. Dong, N. Gao, J. Ren, Y. Zheng and X. Qu, Biomaterials, 2016, 98, 92–102 CrossRef PubMed.
- T. Lin, X. Zhao, S. Zhao, H. Yu, W. Cao, W. Chen, H. Wei and H. Guo, Theranostics, 2018, 8, 990–1004 CrossRef PubMed.
- M. Singh, P. Weerathunge, P. D. Liyanage, E. Mayes, R. Ramanathan and V. Bansal, Langmuir, 2017, 33, 10006–10015 CrossRef PubMed.
- D. Hu, Z. Sheng, S. Fang, Y. Wang, D. Gao, P. Zhang, P. Gong, Y. Ma and L. Cai, Theranostics, 2014, 4, 142–153 CrossRef PubMed.
- X. Ren, J. Liu, J. Ren, F. Tang and X. Meng, Nanoscale, 2015, 7, 19641–19646 RSC.
- Y. Li, Q. Ma, Z. Liu, X. Wang and X. Su, Anal. Chim. Acta, 2014, 840, 68–74 CrossRef PubMed.
- A. Pratsinis, G. A. Kelesidis, S. Zuercher, F. Krumeich, S. Bolisetty, R. Mezzenga, J. C. Leroux and G. A. Sotiriou, ACS Nano, 2017, 11, 12210–12218 CrossRef PubMed.
- Y. Song, Q. Shi, C. Zhu, Y. Luo, Q. Lu, H. Li, R. Ye, D. Du and Y. Lin, Nanoscale, 2017, 9, 15813–15824 RSC.
| This journal is © The Royal Society of Chemistry 2018 |



