None of us is the same as all of us: resolving the heterogeneity of extracellular vesicles using single-vesicle, nanoscale characterization with resonance enhanced atomic force microscope infrared spectroscopy (AFM-IR)
Sally Yunsun
Kim†
 ab,
Dipesh
Khanal†
ab,
Dipesh
Khanal†
 ab,
Priyanka
Tharkar
ab,
Bill
Kalionis
ab,
Priyanka
Tharkar
ab,
Bill
Kalionis
 c and
Wojciech
Chrzanowski
c and
Wojciech
Chrzanowski
 *ab
*ab
aFaculty of Pharmacy, The University of Sydney, NSW, Australia. E-mail: wojciech.chrzanowski@sydney.edu.au
bThe University of Sydney Nano Institute, NSW, Australia
cDepartment of Maternal-Fetal Medicine Pregnancy Research Centre and University of Melbourne, Department of Obstetrics and Gynaecology, Royal Women's Hospital, Parkville, Victoria, Australia
First published on 4th April 2018
Abstract
Extracellular vesicles (EVs) are highly specialized, nanoscale messengers that deliver biological signals and in doing so mediate intercellular communication. Increasing evidence shows that within populations of EVs, important properties including morphology, membrane composition, and content vary substantially. This heterogeneity arises in response to the nature, state, and environmental conditions of the cell source. However, currently there are no effective approaches, which unequivocally discriminate differences between individual EVs, which critically hampers progress in this emerging scientific area. Measuring EV heterogeneity is paramount to our understanding of how EVs influence the physiological and pathological functions of their target cells. Moreover, understanding EV heterogeneity is essential for their application as diagnostics and therapeutics. We propose an innovative approach using resonance enhanced atomic force microscope infrared spectroscopy (AFM-IR) to identify the nanoscale structural composition of EVs, as demonstrated and validated using EVs derived from two types of placenta stem cells. The particular strength of this approach is that it is a label-free and ultra-high sensitivity technique that has the power to measure individual EV heterogeneity. New insights gained by this method into EV heterogeneity will have a profound impact not only on our basic understanding of EV biology but also on disease diagnostics and the emerging area of EV-therapies.
Conceptual insightsExtracellular vesicles (EVs) emerge as key messenger molecules that regulate cellular function and have become recognized for their therapeutic and diagnostic potential. Despite significant advances in analytical techniques, multifaceted characterization of individual vesicles (of sizes below 200 nm) and EV subpopulations has not been achieved until now. This critical limitation hinders progress in the fundamental science of EVs and in clinical translation, for which nanoscale characterization of the biological signals packaged into EVs and their heterogeneity is essential. To address this unmet need, we present, for the first time, a study of the molecular structure and composition of individual EVs using AFM-IR. We demonstrate that AFM-IR enables characterization of EVs at ultra-high resolution to define subtle differences between individual vesicles and their subpopulations that affect downstream applications in the emerging area of EV-therapies. The rigorous study of the structural and molecular differences of individual EVs at nanoscale resolution may also provide robust evidence of pathological processes, which may pave the way for a new approach for a global challenge of early disease detection. Our approach outperforms other methods for EV characterization with unmatched resolution (single vesicle) and is ‘probe free’, avoiding the bias and major resolution limitations of molecular probes. |
Introduction
Extracellular vesicles (EVs) are highly specialized biological nanoscale messengers that orchestrate both physiological and pathophysiological processes in recipient cells. EVs are classified based on their cellular origin and biological function, or on their distinct biogenesis pathways. Intense research focuses on understanding EV biogenesis; developing methods for their isolation, purification and characterization; defining their package of bioactive macromolecules (e.g. proteins, lipids, nucleic acids); and understanding how their uptake by recipient cells results in cellular reprogramming. Currently, key global challenges in EV research are:– establishing the heterogeneity of EV populations,
– high resolution detection of differences in the molecular structures of individual vesicles, and
– validation of isolation and purification protocols.
All these challenges hinge on our ability to probe individual vesicles at a significantly improved resolution than what is currently achieved with conventional protocols.
The study of EVs is fundamental to our understanding of cell signaling in normal and pathological tissues. EVs are also in the limelight because of their potential as biomarkers for cancer, cardiovascular and kidney diseases,1–4 their promise as ‘natural’ drug delivery systems, but primarily for their intrinsic therapeutic properties. This is most notable in the field of cell-based therapies for regenerative medicine where EVs deliver biological signals that direct a cascade of protective biological responses that ultimately restore cellular function in damaged or diseased tissues.5–7 Therefore, to unlock the true potential of EVs for their versatile applications, what is needed is a new approach to unequivocally define nanoscale differences at a single EV level. Ironically, currently used methods for EV characterization are only suitable for bulk EV populations and fall short of pinpointing differences between individual EVs and there are no current methods to discriminate the structural and molecular differences of individual vesicles of nanosize range.
Nanoscale understanding of the biological signals packaged by cells into EVs is essential for application in the diagnosis and therapy of diseases. However, while there have been advances in characterizing intracellular lipid droplets and other organelles, which have no direct correlation with EVs and have different size ranges, there are no reported methods for analyzing individual EV composition within a population of EVs. While conventional spectroscopy techniques may probe larger EVs, e.g. apoptotic bodies, it has not been possible to probe individual EVs, e.g. exosomes (20–120 nm). Comparing subpopulations of EVs has only been possible by separating them from the total population of EVs with little control in which a portion is selected for characterization. The heterogeneity assessment of EV populations has only recently been recognized as critically important for advancing the field. We present an innovative approach using resonance enhanced atomic force microscope infrared spectroscopy (AFM-IR) to assess the heterogeneity and identify the nanoscale structural composition of EVs. AFM-IR not only provides ultra-high resolution but also overcomes limitations of conventional methods suited to bulk EV populations, which include limited resolution and sensitivity. Since the individual EVs can be probed within a bulk EV sample, this technique is a useful high-resolution screening tool for identifying the protein, lipid and nucleic acid contents of EVs simultaneously prior to conducting molecular assays to interrogate the exact molecular make-up. Within the context of EVs being highlighted as early biomarkers of a number of diseases and stem cell-derived EVs being developed as therapeutics to replace cell therapies, nanoscale examination of individual EVs using AFM-IR has the potential for profound significance in disease diagnostics and in the emerging area of EV-therapies. Thus, the development of the AFM-IR technique for individual EV profiling is pushing the forefront of regenerative medicine closer to the creation of truly bioactive therapies, the demand for which is rapidly increasing.
Our interest is human mesenchymal stromal cells (MSCs), which have stem cell-like properties and are derived from tissues and fluids. MSCs have therapeutic potential for many applications,8 including enhancing recovery from cellular injuries caused by myocardial infarction,9,10 and they have additional beneficial properties of attenuating inflammation and immune responses.11–13 The therapeutic action of MSCs is primarily through a paracrine mechanism mediated by EVs. EVs secreted by mesenchymal stromal cells have been extensively researched for their regenerative medicine applications.8
A major problem that besets all EV research is that current isolation methods produce heterogeneous populations of EVs with respect to their size and structure. These problems arise in part due the nanoscale size of EVs and the paucity of specific markers for isolating and identifying the different classes of EVs. The composition of EVs is another confounding factor. The EV composition of bioactive macromolecules in several classes of EVs is the result of selective packaging. The secreting cell uses a regulated mechanism that responds to various stimuli (e.g. growth conditions such as hypoxia,5 the cellular microenvironment, and pathological changes within the cell of origin14), which determines the final specific composition of EVs. Thus, the physical structure and composition of the EV population, and of individual EVs, are heterogeneous. The heterogeneity of EVs is a substantial problem for EV research even where EVs are isolated from a source of uniform cell populations. However, for MSC-based therapeutics a further confounding factor is that MSCs that produce EVs are themselves a heterogeneous population, and this heterogeneity is maintained during ex vivo cell expansion. Methods to monitor EVs during MSC expansion would therefore be highly beneficial to this area of research.
The absence of available techniques to interrogate the heterogeneity of EVs restricts our understanding of EV function.15–17 Conventional technologies to probe individual EVs are limited to determining the particle size, morphology and number of individual EVs. The overall molecular composition of a population of EVs is primarily determined on the entire EV population using protein-, lipid- or nucleic acid-based analyses. There are no currently available methods for analyzing subpopulations of EVs without isolating them, using capture antibodies for example, from the total population of EVs. Moreover, there are no methods for determining individual EV composition within a population. Despite the recent progress in the development of IR-based characterization methods, many conventional methods have limited resolution and are only suitable for bulk materials without enabling direct comparison of individual EVs within a sample. We present an innovative approach using AFM-IR, to study the structure, composition and dynamics of whole populations, subpopulations and individual EVs at an ultra-high resolution of less than 20 nm (Scheme 1).
 | ||
| Scheme 1 Schematic diagram of AFM-IR: (a) probing individual EVs within a population and (b) probing multiple points on an individual EV particle. | ||
AFM-IR has recently been used for determining conformational changes in intra- and extracellular protein structures, which indicate altered biological function,18 for probing chemical and mechanical nanodomains within nanoparticles,19 and for studying intracellular lipid droplets, vesicles and cytoskeletal filaments of cells at the subcellular level.20 However, AFM-IR for complex biological nanoparticles such as EVs has not yet been reported. The advantages of incorporating AFM-IR for EV characterization are based primarily on ultra-high sensitivity and resolution and represent a leap forward in a quest to unequivocally define the molecular composition of individual EVs and their subpopulations. These allow the user to profile the composition and structure of single EVs within a population to identify nanoscale heterogeneity, which is critical for assessing the effects of EV isolation and processing methods, or for identifying alterations in protein to lipid ratios. The latter application may potentially add significant value in the diagnosis and prognosis of diseases in the field of biomarkers. This fast, label-free technique requires only a few microliters of samples for mapping IR-absorbing species of individual EVs, and is a convenient tool for comparing individual EV compositions and structures. With the outstanding resolution of less than 20 nm, we can probe multiple points on individual EVs to interrogate differences in composition and structure within single EVs. Gaining a better understanding of EV composition and heterogeneity will lead to advances in the precise development of EV therapeutics and biomarkers, as well as reveal the purity of EV populations.
We demonstrate the feasibility of probing individual EVs and identifying differences in the molecular composition of EV populations at nanoscale resolution. We present the comparison of EV samples isolated using ultrafiltration and after further purification using the additional size-exclusion chromatography (SEC) column. For comparison, we also present EV samples isolated using the conventional ultracentrifugation method and demonstrate AFM-IR to pinpoint differences between two stem cell lines. The nanoscale structural and compositional differences within individual EVs of the same EV population are presented to highlight the importance of defining the differences in potentially maximizing the use of EVs in regenerative medicine.
Experimental
Cell culture for EV isolation
EVs were isolated from a well-characterized, telomerase (hTERT)-transformed human chorionic mesenchymal stromal cell line (CMSC29) and decidual mesenchymal stromal cell line (DMSC23).21 CMSC29 and DMSC23 retain many of the important characteristics of the primary mesenchymal stromal/stem cells21,22 while ensuring reproducibility by avoiding the confounder of patient-to-patient variation between primary mesenchymal stromal cell preparations. CMSC29 and DMSC23 were cultured in AmnioMAX™ (Life Technologies) and MesenCult™ (StemCell Technologies) media, respectively, with their supplements until 80% confluent, then cells were washed twice in HBSS(−) (Sigma Aldrich) and incubated in EV isolation media (bovine serum albumin 0.5%, GlutaMAX™ 1%, penicillin 100 U mL−1, streptomycin 100 μg mL−1 (Life Technologies) in AmnioMAX™ basal media) for 48 h. Cells were counted to ensure comparable numbers of cells between replicates and cell viability >90% was confirmed using trypan blue assay prior to the isolation of EVs from the conditioned media. To ensure reproducibility, biological triplicates were used for the isolation of EVs.EV isolation and preparation for AFM-IR
EVs were isolated from conditioned media by the ultrafiltration method using an Amicon® Ultra centrifugal filter (10K MWCO, Merck Millipore), following the manufacturer's protocol.23 Enriched EV populations were achieved by passing through the qEVsingle SEC column (Izon Science) following the manufacturer's protocol,24 to remove background proteins, lipids, and other contaminating particulates.25 For EV isolation by ultracentrifugation, initially cells and debris were removed by centrifuging media at 500 × g for 5 min followed by further centrifugation at 2000 × g for 10 min. The supernatant was transferred to thick-wall polycarbonate ultracentrifuge tubes (Seton Scientific Inc.) and centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 60 min at 4 °C using rotor Ti-70 in an Optima LE-80K Ultracentrifuge (Beckman Coulter). The supernatant was removed and the EV pellet was resuspended in 1 mL of RNase-free PBS (Lonza) using RNase-free pipette tips. Then ultracentrifugation was repeated at 31
000 × g for 60 min at 4 °C using rotor Ti-70 in an Optima LE-80K Ultracentrifuge (Beckman Coulter). The supernatant was removed and the EV pellet was resuspended in 1 mL of RNase-free PBS (Lonza) using RNase-free pipette tips. Then ultracentrifugation was repeated at 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 rpm (100
200 rpm (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g) for 60 min at 4 °C, and the supernatant was removed. The pellet was resuspended in 200 μL of RNase-free PBS.
000 × g) for 60 min at 4 °C, and the supernatant was removed. The pellet was resuspended in 200 μL of RNase-free PBS.
Five microliters each of fraction 7 containing EVs, and the original batch of EV samples prior to SEC, were placed on a zinc selenide prism to dry at room temperature for AFM-IR.
Measurement of the molecular composition of EVs using AFM-IR
Nanoscale infrared biospectroscopy was conducted using an AFM-IR instrument (nanoIR™, Anasys Instruments) as per a previously published protocol.18 Briefly, four IR background spectra were collected from 1000 to 1800 cm−1; these spectra were then averaged and normalized for calibration of the signal intensity prior to measurement. The cantilever ringdown signal was optimized at its frequency center of above 75 kHz and frequency window of 50 kHz. After optimization of the laser signal, AFM-IR spectra were collected from 1000 to 1800 cm−1 at 1 cm−1 intervals with a scan rate of 0.1 Hz with a coverage of 16. A silicon nitride cantilever (EXC450 tips, AppNano, CA, USA) with a nominal spring constant of 0.5 N m−1 was used for all measurements. The scan sizes used were 10 μm × 10 μm for probing one point per EV and 1 μm × 1 μm for probing multiple points on individual EVs. Data analysis was done using the Analysis Studio™ software. Smoothing of the spectra using the ‘Savitzky–Golay’ function was achieved using the polynomial function of 2 and 15 numbers of points. All data from AFM-IR are representative of at least triplicate samples and positions.Results and discussion
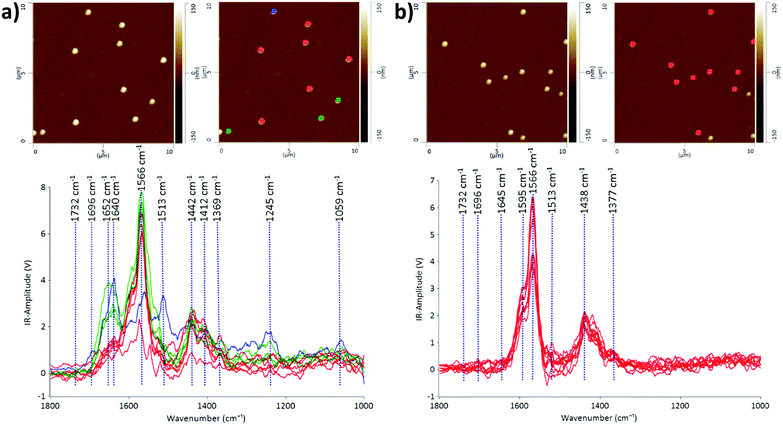
We demonstrate that the AFM-IR technique is capable of probing the heterogeneity of EV populations and interrogating nanoscale structural and compositional differences in individual EVs in two different EV populations: CMSC29 EVs isolated using ultrafiltration and CMSC29 EVs isolated by ultrafiltration followed by additional purification through SEC. Ultrafiltration is a faster method than ultracentrifugation and produces higher yields in the isolation of EVs from cell culture conditioned media.26 However, various methods of EV isolation produce heterogeneous populations of EVs, and identifying this heterogeneity in EV populations is critical in advancing this field of research.27The AFM-IR spectra of EVs give information on the molecular constituents and their structures, enabling the identification of heterogeneity in the structure and composition of individual EVs at nanoscale resolution. The IR peaks with the highest intensities at 1566 cm−1 and 1595 cm−1 are common in both samples before and after passing through the SEC column (Fig. 1). Both peaks indicate the presence of amide II, arising from the N–H bending vibrations of the peptide groups within individual EVs.28,29 The EV samples obtained by ultrafiltration prior to passing through the SEC column were more heterogeneous, as grouped in red, green and blue subgroups according to the similarities of IR peak profiles (Fig. 1a). The prominent absorption peaks at 1640 cm−1 and 1652 cm−1 for the EVs marked green or blue in Fig. 1a indicate the presence of the amide I structure of proteins, originating mainly from the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the protein peptide backbone.30 The intensities of these peaks decreased dramatically or these peaks were absent in the EV samples after passing through the SEC column (Fig. 1b), demonstrating the effectiveness of SEC in purifying EV populations. The absorption bands at 1438 cm−1 and 1442 cm−1 are due to the CH2 and CH3 of the protein or the bending (scissoring) vibration of lipid acyl CH2 groups, found commonly in both EV populations before and after passing through SEC. The peaks at 1412 cm−1 are present in both samples at similar intensities, which refer to the phosphatidylcholine head group.30 The low intensity peaks at 1369 cm−1 and 1377 cm−1 represent thymine and this reveals that RNAs are contained inside CMSC29 EVs obtained by ultrafiltration, regardless of a further SEC purification step.30 The low intensity peaks at 1732 cm−1 in both EV populations (Fig. 1) are attributed to the ester groups of phospholipids, triglycerides and cholesterol esters.31
O stretching vibrations of the protein peptide backbone.30 The intensities of these peaks decreased dramatically or these peaks were absent in the EV samples after passing through the SEC column (Fig. 1b), demonstrating the effectiveness of SEC in purifying EV populations. The absorption bands at 1438 cm−1 and 1442 cm−1 are due to the CH2 and CH3 of the protein or the bending (scissoring) vibration of lipid acyl CH2 groups, found commonly in both EV populations before and after passing through SEC. The peaks at 1412 cm−1 are present in both samples at similar intensities, which refer to the phosphatidylcholine head group.30 The low intensity peaks at 1369 cm−1 and 1377 cm−1 represent thymine and this reveals that RNAs are contained inside CMSC29 EVs obtained by ultrafiltration, regardless of a further SEC purification step.30 The low intensity peaks at 1732 cm−1 in both EV populations (Fig. 1) are attributed to the ester groups of phospholipids, triglycerides and cholesterol esters.31
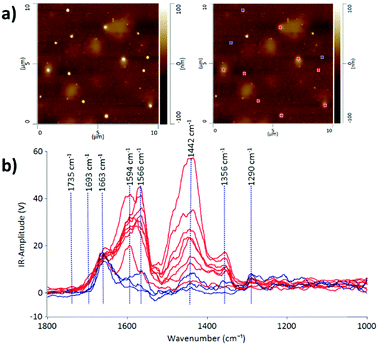
Some protein-rich regions were found within the EV sample in fraction 7 after passing through the SEC column as can be observed in the lighter patchy regions on the AFM height image (Fig. 2). The IR profiles of the EVs embedded inside the protein-rich regions (indicated in red) are different from those of the individual EVs on their own (indicated in blue). This is evidence that the detection of heterogeneity in EV populations can arise from the surrounding contaminating molecules at the nanoscale level. With the precise technique of AFM-IR we can identify that the EVs embedded within the protein structures have a stronger intensity peak at 1442 cm−1. As discussed above, this peak is attributed to the CH2 and CH3 of a protein assignment or the bending (scissoring) vibration of lipid acyl CH2 groups. This reveals the presence of free proteins and lipids in the EV samples; however, it is more likely referring to the presence of proteins, since the lipid peak at 1735 cm−1 is of low intensity. An important aspect of EV samples for all applications is not merely detecting the presence of contaminating molecules but also identifying the composition of the contamination in order to assess suitability for downstream applications. Thus, we demonstrate that AFM-IR is capable of measuring the purity of EV samples quantitatively and with ultrahigh resolution. AFM-IR also has the power to identify the composition of the contaminating molecules, which is critical for optimizing EV purification protocols.
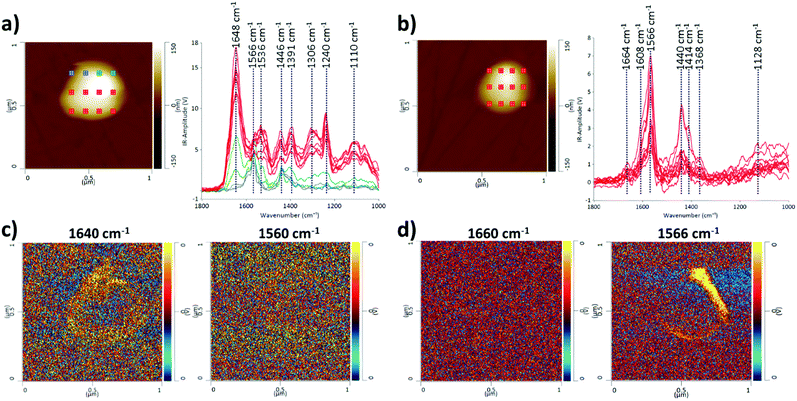
We also present the capability of AFM-IR to probe multiple points on a single EV as an array, for obtaining IR spectra and maps (Fig. 3), allowing visualization and comparison of structural and compositional differences within a single EV. Multipoint analysis (nanoscale mapping) of single EVs is a particular strength of AFM-IR, and it is not achievable with any other currently used techniques that characterize EVs. Mapping at the nanoscale level is critically important to probe the heterogeneity of individual EVs and their populations. Although it is possible to compare EVs within a single population, in Fig. 3 we compare single EVs in the two EV populations consistent with the above examples; CMSC29 EVs isolated using ultrafiltration, before and after SEC. Although the majority of CMSC29 EVs retain the same AFM-IR spectra before and after SEC as observed in Fig. 1, we identified that the EV population before SEC is more heterogeneous and there were some minor subpopulations with different AFM-IR profiles. An example of a single EV that represents this minor subpopulation is presented in Fig. 3a, for comparison with Fig. 3b, which represents the majority of other EVs in the same sample before SEC as well as the overall population of EVs after SEC. The peak at 1640–60 cm−1 in Fig. 3a is characteristic of the amide I band in the protein, while the peak at 1566 cm−1 in Fig. 3b is linked to the –N–H bending vibration of the amide II band.28,30 The peaks at 1500 to 1700 cm−1 in Fig. 3a represent a conventional protein band with distinct peaks for amides I and II composed of overlapping bands that belong to the different secondary structures of proteins such as α-helices, β-sheets, random coils.30,31 In contrast, the AFM-IR spectra of CMSC29 EVs after SEC (Fig. 3b), which are also representative of the majority of other EVs in the sample before SEC, display unresolved bands with overlapping amide I and II peaks, as observed in the conventional IR characterization of microvesicles.30 The AFM-IR profile of a minor single EV presented in Fig. 3a corresponds to the IR profile of the EV marked in blue in Fig. 1a representing a minor subpopulation. Thus, we demonstrate that AFM-IR is a sensitive method to identify heterogeneity and is capable of interrogating structural differences between the subpopulations within the same EV sample. As multiple points are probed on a single EV, the area with the highest amide I concentration can be viewed at a nanoscale level.
Furthermore, AFM-IR allows high resolution mapping of individual EVs (IR-absorbing species). The different IR peaks were clearly identifiable and the regions rich in amide I and II structures were visualized in EVs before and after SEC, as demonstrated by maximum absorption in yellow at 1640 cm−1 and 1566 cm−1, respectively (Fig. 3c and d). Hence, we demonstrated that AFM-IR is a highly sensitive method to detect nanoscale compositional and structural differences between EV populations and individual EVs within a sample.
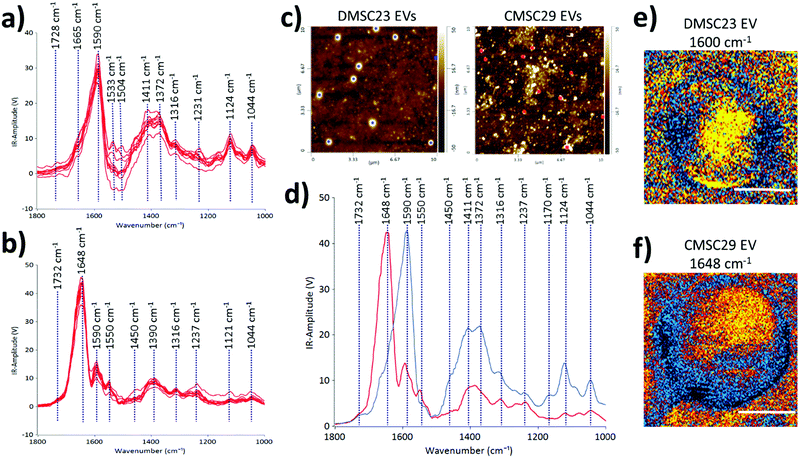
To demonstrate the use of AFM-IR in identifying optimal EV formulations for potential EV-therapies, the differences in the molecular compositions of CMSC29 EVs and DMSC23 EVs were interrogated. Despite that CMSC29 and DMSC23 cell lines are derived from the same source (human placenta), there are clear differences in their EV composition as determined by AFM-IR (Fig. 4). The AFM-IR spectra collected from multiple individual DMSC23 EVs (Fig. 4a) and CMSC29 EVs (Fig. 4b) show a peak within the amide stretching region of peptides (1500 to 1700 cm−1), which corresponds to the bulk protein composition.29 The amide I peak in proteins (1590 to 1700 cm−1) is composed of overlapping bands that belong to different secondary structures of proteins such as α-helices, β-sheets, random coils.30 Therefore, using AFM-IR, changes to the individual protein components can be resolved. For CMSC29, the amide I peak was in the typical wavenumber of 1648 cm−1 (Fig. 4b), which suggests a normal conformation of the protein that contains a highly α-helical structure. On the other hand, for DMSC23 EVs, the main amide I peak was broadened and shifted to a lower wavenumber with the main peak present at 1590 cm−1 (Fig. 4a). In addition, the amide I band peak with a weaker intensity is also seen at 1665 cm−1. Cumulatively, broadening of the amide I peak with the shift to lower frequencies and appearance of a weak intensity peak at 1665 cm−1 indicated changes to the protein conformation in the DMSC23 EV sample.
AFM-IR also enables a fast screening of the differences in the nucleic acid composition between the samples. In the DMSC23 EV spectra, we observed the presence of two well-defined peaks at 1590 cm−1 and 1533 cm−1, which correspond to adenine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond) and guanine (C
N bond) and guanine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond).29 Both peaks were detected only at a very low level in CMSC29 EVs (Fig. 4b). For both DMSC23 EVs and CMSC29 EVs a low intensity peak at 1730 cm−1 that corresponds to purine base (C
N bond).29 Both peaks were detected only at a very low level in CMSC29 EVs (Fig. 4b). For both DMSC23 EVs and CMSC29 EVs a low intensity peak at 1730 cm−1 that corresponds to purine base (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond) was observed. However, the intensity of this peak varied across the EV populations, which demonstrated the heterogeneity of nucleic acid content in the samples. Overall, this study confirmed differences in protein structure and nucleic acid content between both types of EVs, as well as differences in the composition in single EVs between the two populations.
O bond) was observed. However, the intensity of this peak varied across the EV populations, which demonstrated the heterogeneity of nucleic acid content in the samples. Overall, this study confirmed differences in protein structure and nucleic acid content between both types of EVs, as well as differences in the composition in single EVs between the two populations.
The AFM height images (Fig. 4c), and the AFM-IR averaged spectra (Fig. 4d) obtained from two different EV populations allow direct comparison of DMSC23 EVs and CMSC29 EVs. The AFM height images were used to select the EVs of the same size for AFM-IR measurements. The peaks at 1411 cm−1 and 1372 cm−1 had higher intensities in the spectra of DMSC23 EVs, which refer to the phosphatidylcholine head group and thymine, respectively.29 These data indicate larger amounts of those components in lipids and nucleic acids packaged in DMSC23 EVs compared to CMSC29 EVs. The IR maps in the maximum absorption region at 1590–1600 cm−1 for DMSC23 EVs (Fig. 4e) and at 1648 cm−1 for CMSC29 EVs (Fig. 4f) demonstrate the capacity of AFM-IR to generate a fingerprint for individual EVs. The AFM-IR analyses of EVs enable subpopulations of EVs to be analysed at the level of individual EVs and also distinguish differences in the compositions and structures of various EV populations.
Taken together, AFM-IR has the potential to be utilized for highly sensitive, precise and relatively fast measurement of alterations in EV structure and composition to verify EV purification protocols as well as to establish the fundamental science of EVs and their diagnostic and therapeutic potential.
Conclusions
One of the critical challenges in the EV area, which curbs the translation of EVs to clinical applications is the capability to characterize individual vesicles and their heterogeneity. Whilst the ideal scenario would be to have a full profile of RNAs, DNA, proteins and lipids in each individual nanosize vesicle, it is technically unachievable. For molecular biological analyses, large quantities of EVs are required (hundreds of thousands of EVs) and the typical yield of the molecules from such preparation does not exceed a few picograms. These quantities are currently the lower detection limit of existing techniques and thus the final result merely represents the averaged profile of the entire, large vesicle population. These significant technological constraints are unlikely to be resolved in the near future. Therefore, alternative approaches which can determine the heterogeneity and composition of individual vesicles are highly sought after.Indeed, we have demonstrated for the first time that the AFM-IR technique is capable of probing the molecular constituents and structures of individual vesicles. We showed that using this approach we can validate purification protocols and clearly distinguish protein aggregates from vesicles, which is not possible with other approaches due their similar sizes. The data presented in this study suggest that AFM-IR can transform existing protocols for interrogating EV compositions and structures, and assessing EV purity. Despite that this technique does not enable sequencing of RNAs it provides robust data on protein, DNA and lipid contents and their structure that point to the molecular structure of each of the individual vesicles. We demonstrated the strength of the technique to determine structural differences of vesicles from different cells by probing vesicles isolated from two subtypes of placenta stem cells, which are characterized by different molecular compositions.
We know that variations in IR spectra can be used to identify pathologies, which has been previously demonstrated for microvesicles at the micron scale. However, due to the resolution of the conventional techniques, a very high concentration of the sample is required and perhaps only advanced stages of the diseases can be identified. Therefore, our ability to probe individual vesicles is a leap forward in diagnosis and is likely to enable early disease detection from a few vesicles. This advance may transform the way we diagnose (earlier detection with smaller sample quantities) and treat diseases in particular for diseases which are characterized by high mortalities (cancer, multiple sclerosis or dementia).
The fact that only a few microliters of samples are required is an added benefit for the use of this technique in the diagnosis or prognosis of diseases. For therapeutic applications, AFM-IR will allow researchers to not only assess EV purity and identify the contaminating molecules in EV preparations but furthermore can be developed to:
(i) enable the development of protocols for effective EV preparation,
(ii) assist fundamental studies of biological principles that govern the production and export of the vesicles via a better understanding of the packaging and assembly of EVs, and
(iii) identify the most effective EV formulations or optimal EV subtypes that are most therapeutically relevant, manipulated via pre-conditioning of secreting cells.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
WCh acknowledges the University of Sydney for a SOAR Fellowship.References
- L. Guo and B. He, Extracellular vesicles and their diagnostic and prognostic potential in cancer, Transl. Cancer Res., 2017, 6(3), 599–612 CrossRef.
- W. Zhang, X. Zhou, H. Zhang, Q. Yao, Y. Liu and Z. Dong, Extracellular vesicles in diagnosis and therapy of kidney diseases, Am. J. Physiol.: Renal. Physiol., 2016, 311(5), F844–F851 CrossRef CAS PubMed.
- F. Jansen, G. Nickenig and N. Werner, Extracellular Vesicles in Cardiovascular Disease: Potential Applications in Diagnosis, Prognosis, and Epidemiology, Circ. Res., 2017, 120(10), 1649–1657 CrossRef CAS PubMed.
- M. Verma, T. K. Lam, E. Hebert and R. L. Divi, Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology, BMC Clin. Pathol., 2015, 15, 6 CrossRef PubMed.
- M. J. Shurtleff, M. M. Temoche-Diaz, K. V. Karfilis, S. Ri and R. Schekman, Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction, eLife, 2016, 5, e19276 Search PubMed.
- E. van der Pol, A. N. Böing, P. Harrison, A. Sturk and R. Nieuwland, Classification, Functions, and Clinical Relevance of Extracellular Vesicles, Pharmacol. Rev., 2012, 64(3), 676–705 CrossRef CAS PubMed.
- A. M. Silva, J. H. Teixeira, M. I. Almeida, R. M. Goncalves, M. A. Barbosa and S. G. Santos, Extracellular Vesicles: Immunomodulatory messengers in the context of tissue repair/regeneration, Eur. J. Pharm. Sci., 2017, 98, 86–95 CrossRef CAS PubMed.
- I. M. Bjorge, S. Y. Kim, J. F. Mano, B. Kalionis and W. Chrzanowski, Extracellular vesicles, exosomes and shedding vesicles in regenerative medicine – a new paradigm for tissue repair, Biomater. Sci., 2018, 6, 60–78 RSC.
- S. Bian, L. Zhang, L. Duan, X. Wang, Y. Min and H. Yu, Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model, J. Mol. Med., 2014, 92(4), 387–397 CrossRef CAS PubMed.
- F. Arslan, R. C. Lai, M. B. Smeets, L. Akeroyd, A. Choo, E. N. Aguor, L. Timmers, H. V. van Rijen, P. A. Doevendans, G. Pasterkamp, S. K. Lim and D. P. de Kleijn, Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury, Stem Cell Res., 2013, 10(3), 301–312 CrossRef CAS PubMed.
- K. Drommelschmidt, M. Serdar, I. Bendix, J. Herz, F. Bertling, S. Prager, M. Keller, A.-K. Ludwig, V. Duhan, S. Radtke, K. de Miroschedji, P. A. Horn, Y. van de Looij, B. Giebel and U. Felderhoff-Müser, Mesenchymal stem cell-derived extracellular vesicles ameliorate inflammation-induced preterm brain injury, Brain, Behav., Immun., 2017, 60, 220–232 CrossRef CAS PubMed.
- C. Lo Sicco, D. Reverberi, C. Balbi, V. Ulivi, E. Principi, L. Pascucci, P. Becherini, M. C. Bosco, L. Varesio, C. Franzin, M. Pozzobon, R. Cancedda and R. Tasso, Mesenchymal Stem Cell-Derived Extracellular Vesicles as Mediators of Anti-Inflammatory Effects: Endorsement of Macrophage Polarization, Stem Cells Transl. Med., 2017, 6(3), 1018–1028 CrossRef CAS PubMed.
- T. Shigemoto-Kuroda, J. Y. Oh, D.-K. Kim, H. J. Jeong, S. Y. Park, H. J. Lee, J. W. Park, T. W. Kim, S. Y. An, D. J. Prockop and R. H. Lee, MSC-derived Extracellular Vesicles Attenuate Immune Responses in Two Autoimmune Murine Models: Type 1 Diabetes and Uveoretinitis, Stem Cell Rep., 2017, 8(5), 1214–1225 CrossRef CAS PubMed.
- Y. Yuana, A. Sturk and R. Nieuwland, Extracellular vesicles in physiological and pathological conditions, Blood Rev., 2013, 27(1), 31–39 CrossRef CAS PubMed.
- F. Fatima and M. Nawaz, Nexus between extracellular vesicles, immunomodulation and tissue remodeling: for good or for bad?, Ann. Transl. Med., 2017, 5(6), 139 CrossRef PubMed.
- B. Giebel, On the function and heterogeneity of extracellular vesicles, Ann. Transl. Med., 2017, 5(6), 150 CrossRef PubMed.
- E. Willms, H. J. Johansson, I. Mäger, Y. Lee, K. E. M. Blomberg, M. Sadik, A. Alaarg, C. I. E. Smith, J. Lehtiö, S. El Andaloussi, M. J. A. Wood and P. Vader, Cells release subpopulations of exosomes with distinct molecular and biological properties, Sci. Rep., 2016, 6, 22519 CrossRef CAS PubMed.
- D. Khanal, A. Kondyurin, H. Hau, J. C. Knowles, O. Levinson, I. Ramzan, D. Fu, C. Marcott and W. Chrzanowski, Biospectroscopy of Nanodiamond-Induced Alterations in Conformation of Intra- and Extracellular Proteins: A Nanoscale IR Study, Anal. Chem., 2016, 88(15), 7530–7538 CrossRef CAS PubMed.
- D. Khanal, B. Zhang, I. Ramzan, C. Marcott, Q. Li and W. Chrzanowski, Probing Chemical and Mechanical Nanodomains in Co-Polymer Nanorods with Correlative Atomic Force Microscopy – Nano-correscopy, Part. Part. Syst. Charact., 2018 DOI:10.1002/ppsc.201700409.
- L. Quaroni, K. Pogoda, J.-W. Zuber and W. Kwiatek, Mid-Infrared Spectroscopy and Microscopy of Subcellular Structures in Eukaryotic Cells with Atomic Force Microscopy-Infrared Spectroscopy, 2017, arXiv:1704.01395, arXiv preprint.
- S. Q. Qin, G. D. Kusuma, B. Al-Sowayan, R. A. Pace, S. Isenmann, M. D. Pertile, S. Gronthos, M. H. Abumaree, S. P. Brennecke and B. Kalionis, Establishment and characterization of fetal and maternal mesenchymal stem/stromal cell lines from the human term placenta, Placenta, 2016, 39, 134–146 CrossRef CAS PubMed.
- G. D. Kusuma, M. H. Abumaree, M. D. Pertile, A. V. Perkins, S. P. Brennecke and B. Kalionis, Mesenchymal Stem/Stromal Cells Derived From a Reproductive Tissue Niche Under Oxidative Stress Have High Aldehyde Dehydrogenase Activity, Stem Cell Rev. Rep., 2016, 12(3), 285–297 CrossRef CAS PubMed.
- Merck, Extracellular Vesicle Protocol, 2017.
- I. Science, qEV Application Note: qEV Size Exclusion Column, Izon Science Ltd, Oxford, UK, 2016 Search PubMed.
- A. N. Böing, E. van der Pol, A. E. Grootemaat, F. A. W. Coumans, A. Sturk and R. Nieuwland, Single-step isolation of extracellular vesicles by size-exclusion chromatography, J. Extracell. Vesicles, 2014, 3(1), 23430 CrossRef PubMed.
- R. J. Lobb, M. Becker, S. W. Wen, C. S. F. Wong, A. P. Wiegmans, A. Leimgruber and A. Möller, Optimized exosome isolation protocol for cell culture supernatant and human plasma, J. Extracell. Vesicles, 2015, 4(1), 27031 CrossRef PubMed.
- A. Bobrie, M. Colombo, S. Krumeich, G. Raposo and C. Thery, Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation, J. Extracell. Vesicles, 2012, 1(1), 18397 CrossRef CAS PubMed.
- E. S. Guang Choo, X. Tang, Y. Sheng, B. Shuter and J. Xue, Controlled loading of superparamagnetic nanoparticles in fluorescent nanogels as effective T2-weighted MRI contrast agents, J. Mater. Chem., 2011, 21(7), 2310–2319 RSC.
- S.-Y. Lin, M.-J. Li and W.-T. Cheng, FT-IR and Raman vibrational microspectroscopies used for spectral biodiagnosis of human tissues, Spectroscopy, 2007, 21(1), 1–30 CrossRef CAS.
- J. Lee, B. Wen, E. A. Carter, V. Combes, G. E. R. Grau and P. A. Lay, Infrared spectroscopic characterization of monocytic microvesicles (microparticles) released upon lipopolysaccharide stimulation, FASEB J., 2017, 31(7), 2817–2827 CrossRef CAS PubMed.
- J. Mihály, R. Deák, I. C. Szigyártó, A. Bóta, T. Beke-Somfai and Z. Varga, Characterization of extracellular vesicles by IR spectroscopy: fast and simple classification based on amide and CH stretching vibrations, Biochim. Biophys. Acta, 2017, 1859(3), 459–466 CrossRef PubMed.
Footnote |
| † S. Y. K. and D. K. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |