Genetic, structural, and functional diversity of low and high-affinity siderophores in strains of nitrogen fixing Azotobacter chroococcum†
Xinning
Zhang‡
 *ab,
Oliver
Baars‡
*ab,
Oliver
Baars‡
 *ac and
François M. M.
Morel
a
*ac and
François M. M.
Morel
a
aDepartment of Geosciences, Princeton University, USA. E-mail: xinningz@princeton.edu
bPrinceton Environmental Institute, Princeton University, USA
cDepartment of Entomology & Plant Pathology, North Carolina State University, USA. E-mail: obaars@ncsu.edu
First published on 16th November 2018
Abstract
To increase iron (Fe) bioavailability in surface soils, microbes secrete siderophores, chelators with widely varying Fe affinities. Strains of the soil bacterium Azotobacter chroococcum (AC), plant-growth promoting rhizobacteria used as agricultural inoculants, require high Fe concentrations for aerobic respiration and nitrogen fixation. Recently, A. chroococcum str. NCIMB 8003 was shown to synthesize three siderophore classes: (1) vibrioferrin, a low-affinity α-hydroxy carboxylate (pFe = 18.4), (2) amphibactins, high-affinity tris-hydroxamates, and (3) crochelin A, a high-affinity siderophore with mixed Fe-chelating groups (pFe = 23.9). The relevance and specific functions of these siderophores in AC strains remain unclear. We analyzed the genome and siderophores of a second AC strain, A. chroococcum str. B3, and found that it also produces vibrioferrin and amphibactins, but not crochelin A. Genome comparisons indicate that vibrioferrin production is a vertically inherited, conserved strategy for Fe uptake in A. chroococcum and other species of Azotobacter. Amphibactin and crochelin biosynthesis reflects a more complex evolutionary history, shaped by vertical gene transfer, gene gain and loss through recombination at a genomic hotspot. We found conserved patterns of low vs. high-affinity siderophore production across strains: the low-affinity vibrioferrin was produced by mildly Fe limited cultures. As cells became more severely Fe starved, vibrioferrin production decreased in favor of high-affinity amphibactins (str. B3, NCIMB 8003) and crochelin A (str. NCIMB 8003). Our results show the evolution of low and high-affinity siderophore families and conserved patterns for their production in response to Fe bioavailability in a common soil diazotroph.
Significance to metallomicsBacteria employ diverse iron-binding structures to access the micronutrient iron, yet the relevance and specific function of different structures is not well understood. We show that the widespread soil N2-fixer, Azotobacter chroococcum, employs the low-affinity siderophore vibrioferrin under mild Fe limitation and high-affinity siderophores, amphibactins and crochelin, under moderate to strong Fe limitation. Comparative analyses indicate that vibrioferrin production is a conserved, vertically inherited feature of Azotobacter physiology, whereas production of the high-affinity siderophores is lineage specific, shaped to a greater degree by genetic recombination. The results provide insight on why many bacteria have the capacity to produce multiple siderophore families. |
Introduction
Strains of the γ-proteobacterium Azotobacter chroococcum are free-living, aerobic, N2-fixing (diazotrophic) heterotrophs commonly found in soil, water, rhizosphere, and phyllosphere environments.1–3A. chroococcum is the predominant species recovered in soil surveys of Azotobacter species and has been extensively studied as a plant-growth promoting rhizobacterium and agricultural inoculant.1,3,4A. chroococcum strains may stimulate plant growth through N2 fixation, and the release of plant-growth promoting compounds,4–6 including the phytohormones indole-acetic acid, gibberellins, auxins as well as antifungal compounds.5,7 In addition to its roles in soil fertility and plant nutrition, A. chroococcum (hereafter AC) has also been a model bacterium in studies of biological nitrogen (N2) fixation and H2 metabolism.8–13 An important but understudied area of AC physiology is its production of siderophores, iron-chelating secondary metabolites that aid in the acquisition of iron (Fe), an essential micronutrient for respiratory metabolism and diazotrophy that has low bioavailability in oxic environments. Our previous analyses of Azotobacter species showed that siderophore concentrations can reach hundreds of micromolar in batch cultures14 and represent the major nitrogen containing secreted metabolites under N2-fixing conditions.15A recent study of A. chroococcum str. NCIMB 8003 (hereafter AC-8003) characterized the first siderophore structures made by an AC strain.16 Three types of siderophores were identified: vibrioferrin (an α-hydroxy carboxylate), amphibactins (tris-hydroxamates), and crochelin A, a siderophore with mixed chelating groups including a new Fe chelating moiety.16 These structures exhibit a range of Fe-binding affinities indicated by their pFe values (where pFe = −log[Feaq3+] at defined standardized concentrations of siderophore, iron, and pH; higher pFe values indicate high Fe binding affinities). Vibrioferrin has one of the lowest known pFe values for siderophores (pFe = 18.417) and we refer to vibrioferrin as a low-affinity siderophore. Amphibactins are tris-hydroxamates and can be expected to be orders of magnitude stronger. No pFe value has been reported for amphibactins but other known tris-hydroxamate siderophores range from pFe = 25–27 (exochelin MS: pFe = 25.0,18 desferrichrome: pFe = 25.8,16,19 desferrioxamine B: pFe = 26.616,19). The pFe value for crochelin A is 23.9.16 In the following we refer to amphibactins and crochelin A as high affinity siderophores. Chemical assays suggest that the production of hydroxamate-type siderophores may be a common strategy for Fe acquisition among AC strains.20–22 In comparison, the well-studied model organism Azotobacter vinelandii also makes the low-affinity siderophore vibrioferrin14 but not hydroxamates or crochelins. Instead it produces a number of high-affinity catecholate siderophores and azotobactins14,23–27 (azotochelin pFe = 23.1;28 protochelin pFe = 28.4;29 azotobactin pFe = 27.830).
How AC-8003 uses vibrioferrin, amphibactin, and crochelin A to acquire Fe and whether these siderophore classes form a general strategy to combat Fe stress by other AC strains is unknown. Analyses of the genomes and Fe-chelating metabolites of additional AC strains will help to identify conserved and lineage-specific accessory siderophores for AC. Here, we first analyze the genome of a second strain of A. chroococcum (str. B3, hereafter AC-B3) to identify shared and lineage specific siderophores of A. chroococcum. We then perform a detailed structural characterization of all siderophores produced by AC-8003 and AC-B3 using an Fe-isotope guided, high-resolution liquid chromatography-mass spectrometry (LC-MS) approach (ChelomEx).31 We tracked the concentrations of the siderophores at different degrees of Fe limitation to better understand how the siderophores are used. Together with previous analyses of Azotobacter vinelandii,14,15 our results show that the low-affinity Fe-binding siderophore vibrioferrin is conserved among all Azotobacter species and is produced in response to mild Fe limitation. Higher affinity siderophores, which have more complex evolutionary histories, are produced for Fe acquisition at strong Fe limitation. Our study provides insight into why and how AC uses multiple siderophore structures, thus shedding light on the common bacterial strategy to harbor gene clusters for both low- and high-affinity siderophores.
Results and discussion
Bioinformatic analyses of siderophore genes
To examine siderophore genes in AC-B3 for comparison with AC-8003,2,16 we used PacBio single-molecule, real time sequencing to obtain a draft genome of 5.0 Mb (Table S1, ESI†), consisting of a single, closed 4.6 Mb chromosome (Genbank accession CP011835) and three incomplete plasmids ranging from ∼66 kb to 306 kb (Genbank accessions CP011836–CP011838). The genome size and structure of AC-B3 is roughly comparable that of AC-8003 (5.2 Mb total genome comprising 4.6 Mb chromosome and six plasmids ranging in size from ∼10 to 312 kb).Bioinformatic techniques can help identify putative siderophore gene clusters and also provide preliminary structural information related to amino acid content.32 The AC-8003 genome2 includes clusters that are associated with the biosynthesis of vibrioferrin, amphibactins, and crochelins (Fig. 1A–C and Table S2, ESI†). AC-B3 which we sequenced for this study, also contains gene clusters for vibrioferrin (ACG10_16855 to ACG10_16880) and amphibactin (ACG10_19975 to ACG10_19940) (Fig. 1A, B and Table S2, ESI†). Vibrioferrin clusters from AC-8003 and AC-B3 are 96% identical, while amphibactin clusters are 92% identical based on amino acid sequence analysis of all proteins encoded within the cluster. The mixed NRPS-polyketide synthase cro cluster of AC-8003 encoding crochelin2,16 (Achr_38950 to Achr_38800, Fig. 1C) was not identified in the AC-B3 genome. We then searched for the presence of AC siderophore genes in other completed and draft Azotobacter genomes within Genbank (Table S3, ESI†), constructed phylogenies for siderophore biosynthetic genes (Fig. 2A and B) and their host organisms (Fig. 2D), and analyzed siderophore gene context and distribution (Fig. 2C and D) to better understand the evolutionary history of Azotobacter siderophores (Fig. 3).
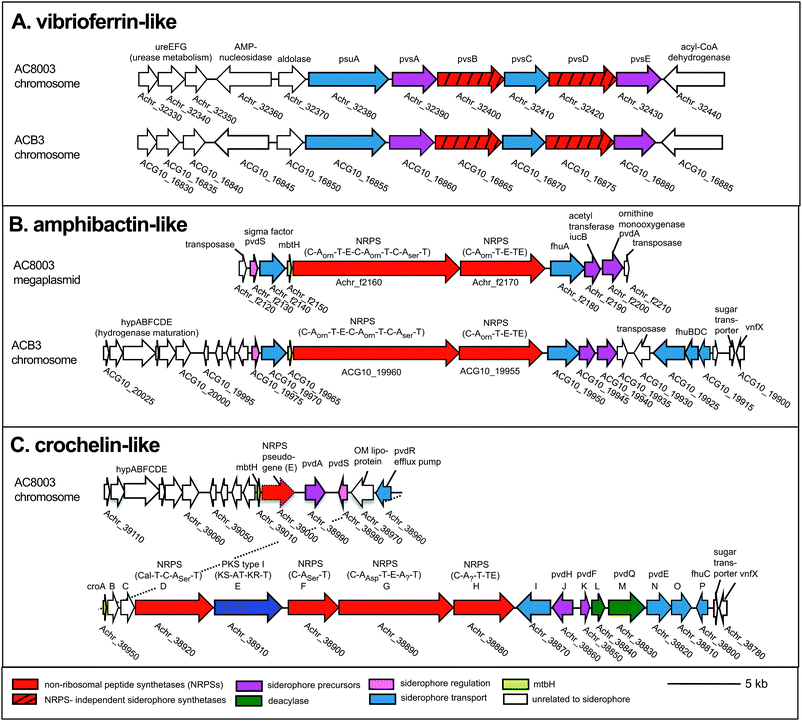 | ||
| Fig. 1 Siderophore gene clusters and their gene context in A. chroococcum str. NCIMB 8003 and str. B3. NRPS domains: C, condensation, A, adenylation, T, thiolation, E, epimerization, TE termination. Arrows with the same color represent similar genes. Accession numbers are found in Table S2 (ESI†). | ||
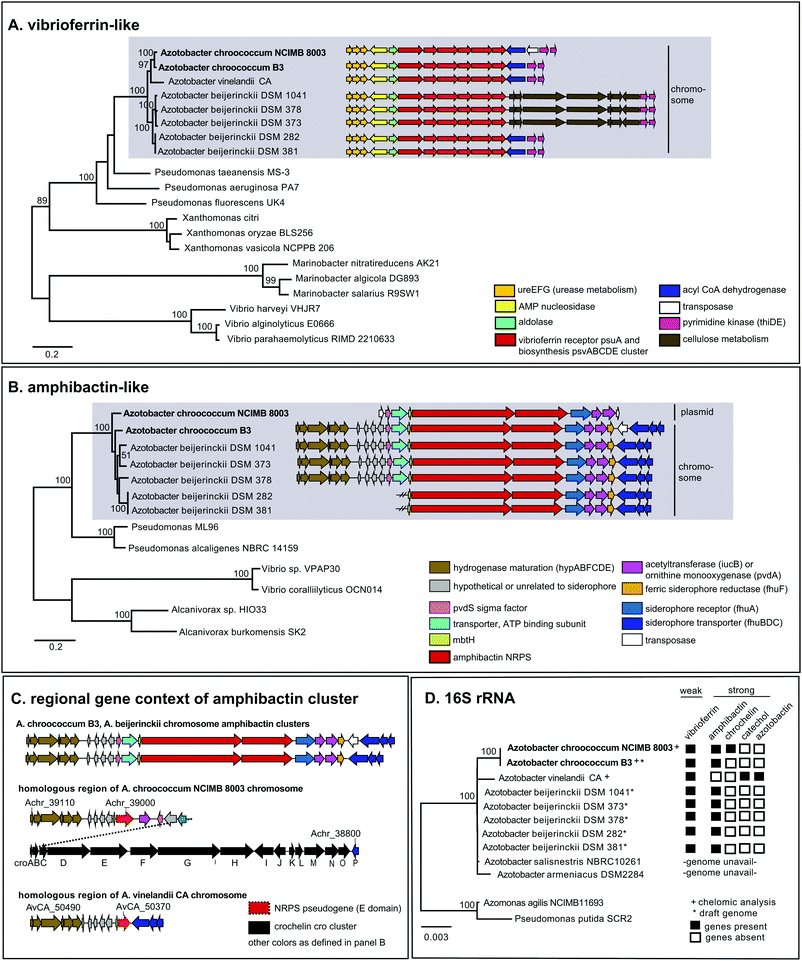 | ||
| Fig. 2 Phylogeny and distribution of siderophore biosynthetic proteins and their host organisms. (A) Maximum likelihood tree of concatenated vibrioferrin-like biosynthetic proteins, PvsABCDE. (B) Maximum likelihood tree of concatenated amphibactin-like NRPS biosynthetic proteins (A. borkumensis ABO2093, ABO2092 homologs). (C) Gene context of amphibactin cluster and related regions in Azotobacter genomes. (D) Neighbor joining distance tree of Azotobacter species and siderophore gene distribution. PhyML and WAG amino acid substitution models were used for protein phylogenies; neighbor joining and HKY nucleotide substitution models for the 16s rRNA tree. Numbers indicate clustering based on 100 bootstrap calculations for protein trees, 1000 bootstraps for 16s rRNA tree. Accession and loci information are found in Tables S3–S6 (ESI†). | ||
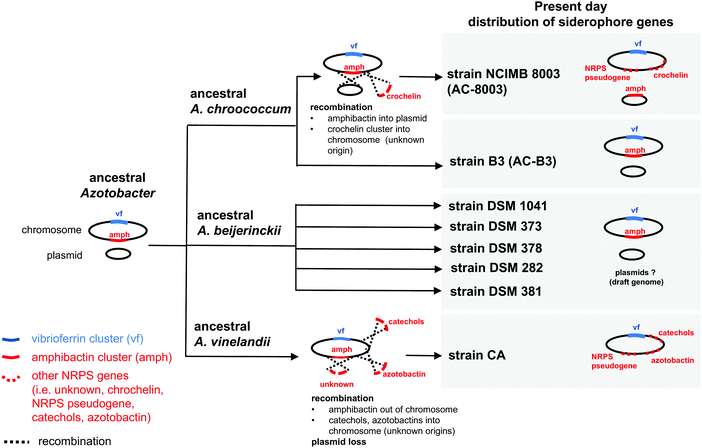 | ||
| Fig. 3 Possible evolutionary scenario for present day distribution of siderophore gene clusters in the genus Azotobacter. | ||
Phylogenetic analysis of concatenated amphibactin NRPS proteins shows clustering patterns that suggest a more complex evolutionary history for amphibactins compared to vibrioferrins, as sequences from the same Azotobacter species do not consistently group together (i.e. AC-8003 and AC-B3 sequences do not form a clade to the exclusion of A. beijerinckii, Fig. 2B). While not as clear as for the phylogeny of vibrioferrin biosynthesis, signals for vertical inheritance of amphibactin production genes are present. Sequences from various Azotobacter species group together separate from sequences from other genera, forming a greater Azotobacter amphibactin clade (Fig. 2B sequences in grey box). This phylogenetic pattern implies that some vertical gene transfer of the amphibactin cluster must have occurred during Azotobacter speciation. Moreover, within this large cluster, subclusters that are well-supported (nodes with high bootstrap values) only comprise strains from the same species (A. beijerincki DSM 1041 and 373; DSM 282 and 381). Finally, vertical inheritance is reflected by the conserved gene context across different species (Fig. 2B, AC-B3 and A. beijerinckii strains), wherein amphibactin cluster genes on the chromosome are flanked by hydrogenase maturation and siderophore transport genes (hypABFCDE and fhuBDC, respectively, Fig. 2B).
Interestingly, the AC-8003 amphibactin cluster is nested between two transposase genes (Achr_f2120 and Achr_f2210, Fig. 1) on a ∼300 kb mega plasmid (pAcX50f) in contrast to amphibactin genes in other Azotobacter species. The homologous section of the AC-8003 chromosome that encodes amphibactins in AC-B3 and A. beijerinckii strains, contains, instead, a NRPS pseudogene with a single epimerization (E) domain followed by the large cro cluster for crochelin A synthesis (Fig. 2C). A NRPS pseudogene is also present in the homologous region of the A. vinelandii CA chromosome (Fig. 2C).
Based on gene context and phylogenetic data, we propose an evolutionary scenario in which amphibactin genes were originally located on the chromosome of an ancestral Azotobacter species, whose genome comprised chromosome and plasmid replicons (Fig. 3). Vertical inheritance during organism speciation could explain the continued presence of amphibactin genes on the chromosome of AC-B3 and A. beijerinckii strains. Chromosome located amphibactin genes, facilitated by flanking transposases (Fig. 2B, Achr_f2120, Achr_f2210), could have then recombined into the plasmid in the AC-8003 lineage, explaining the plasmid-encoded amphibactins on AC-8003 (Fig. 3). The recombination of novel NRPS genes (encoding crochelin) into a region near the NRPS-rich amphibactin chromosome locus in the AC-8003 lineage (Fig. 3) would have also had to occur to explain the proximate location of crochelin and amphibactin chromosome gene clusters (Fig. 2C). Further evidence consistent with the chromosome to plasmid movement of amphibactin genes (Fig. 2D) is the absence of both amphibactin genes and plasmids in A. vinelandii. This can be explained by plasmid loss in an A. vinelandii ancestor following amphibactin gene recombination into the plasmid (as in AC-8003) (Fig. 3). Finally, the occurrence of an amphibactin biosynthetic locus on the ancestral chromosome of all Azotobacter species is also suggested by the presence of the NRPS pseudogene in AC-8003 and A. vinelandii chromosomes. While recombination in an NRPS-rich region of the chromosome helps to explain amphibactin and crochelin biosynthesis, horizontal gene transfer is also likely to have introduced siderophore biosynthetic genes in the other locations on the chromosome (e.g. for catechols and azotobactin in the A. vinelandii lineage, Fig. 3).
Alternatively, the amphibactin cluster might have originally been encoded on the plasmid of an ancestral Azotobacter species, with the current distribution of genes shaped by a mixture of vertical inheritance of the plasmid (with plasmid based amphibactin genes of AC-8003 representing the ancient locus), plasmid/chromosome recombination, and plasmid loss from different lineages. However, two separate recombination events (each moving the amphibactin genes from the plasmid to the exact same location of the chromosome) would have needed to occur to explain why AC-B3 and A. beijerinckii amphibactin genes do not form a cluster to the exclusion of AC-8003, but still feature conserved gene contexts. Finally, in this alternative scenario, plasmid loss from the A. vinelandii lineage would result in the absence of amphibactins in A. vinelandii CA. Based on the more rapid evolution of plasmid sequences compared to chromosomal genes,39 we believe this scenario to be less likely. Regardless, plausible scenarii for amphibactin evolution must involve vertical gene transfer as well as gene gain and loss through homologous recombination.
Genetic recombination within NRPS-rich areas of bacterial genomes, aided by mobile genetic elements and resulting in gene loss and gain, can lead to movement of entire siderophore biosynthetic loci between replicons40–42 (e.g. onto the plasmid in AC-8003) and explain new loci that encode for entirely new siderophores,43 such as crochelin A. Given the proximity of chromosomal loci for amphibactins, crochelins, and transposases, siderophore diversity in Azotobacter appears to be strongly facilitated by “mixing and matching” between natural product gene clusters.43 Indeed, analyses of the AC-8003 genome2 has indicated a high number of transposases, which would facilitate DNA movement within and between genomes.
Analysis of siderophore structures
The result of siderophore gene evolution is the conservation of genes for the low-affinity siderophore vibrioferrin across all Azotobacter species. Genes encoding for amphibactin-like siderophores are only found in AC strains but not in AV strains, which instead utilize a group of catechol siderophores and azotobactins (Fig. 2D). Crochelin appears to be only produced by AC-8003. To study the siderophore products from these gene clusters, we used high-resolution LC-MS to screen for characteristic 54Fe–56Fe stable isotope pattern associated with Fe chelates and the exact mass difference between apo- and Fe-bound forms as described previously (Fig. S2, ESI†).16 We also searched high-resolution LC-MS/MS data for siderophore-characteristic fragmentation patterns.31 We have previously reported siderophores produced by AC-8003 focusing on crochelins as a new siderophore family.18 Here, we provide a detailed analysis of a suite of new hydrophilic amphibactins in the AC-8003 supernatants and present the first analysis of AC-B3 siderophores. We then target the identified sideorphores to quantify their production under high, intermediate, and low Fe availability conditions (Fig. 6).(i) Vibrioferrin: both AC strains produced vibrioferrin A as the major product associated with the vibrioferrin gene cluster (Fig. 4). The structure of vibrioferrin A was confirmed by comparison of m/z, retention time, and MS/MS fragmentation to an isolated standard (Fig. S3, ESI†).
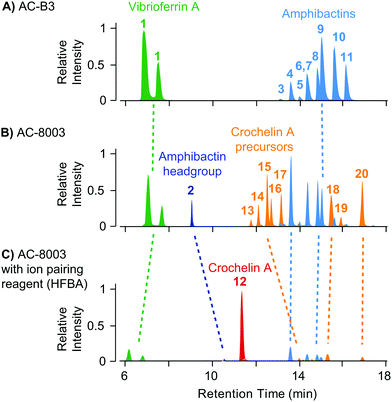 | ||
| Fig. 4 Comparison of extracted ion chromatograms of the major identified siderophores detected in spent media of AC-B3 (panel A) and AC-8003 (panels B and C) during late exponential growth in Fe limited Burk's medium (5 μM Fe, 100 μM EDTA). Both strains produce vibrioferrin and a suite of soluble hydrophilic amphibactins (main products with fatty acid tail lengths C10, C12, C14, see Fig. 5). Strain AC-8003 makes an additional amphibactin headgroup. The major product of AC-8003, the positively charged, hydrophilic crochelin A, is only observed after adding a strong ion-pairing reagent to the mobile phase buffer (heptafluorobutyric acid, HFBA) as shown in panels B and C. A number of previously described lipopetidic crochelin A precursors were also observed in the medium (fatty acid chain length C10 and C12, see Fig. 5 for structures). Tables with detailed LC-MS results for all detected siderophores can be found in the ESI† (Tables S8–S10). | ||
(ii) Amphibactins: both AC strains produced several analogs of soluble amphibactins detected in culture supernatants (Fig. 4, 5 and Table S8, ESI†). Cell pellet extracts also contained minor amounts of more hydrophobic amphibactin analogs (Table S9, ESI†). One of the amphibactin analogs produced by AC-B3 corresponds in mass, retention time, and MS/MS fragmentation to an amphibactin S standard (fatty acid tail C14:1), a siderophore previously reported from marine Vibrio species (Fig. S4, ESI†). Another structure corresponds to amphibactin T (saturated C12:0 tail).36 Apart from amphibactins S and T, we isolated several new hydrophilic amphibactins and quantified their concentrations by 1H-NMR spectroscopy (Fig. S5, ESI†). Two major new analogs were identified with 3-hydroxydecanoyl (4, amphibactin ACA) and decanoyl (6, amphibactin ACB) fatty-acyl groups (Fig. 5). Additional analogs were detected with C10–C14 chain lengths of the fatty-acyl groups. The fatty acid tails were fully saturated, contained one double bond or one hydroxyl group. Strain AC-8003 secreted the amphibactin headgroup without a fatty acid tail (Fig. 4 and 5), which originated presumably from the activity of a fatty acid hydrolase enzyme associated with the crochelin cluster (CroM, see also section crochelins below). Minor additional peaks corresponding to more hydrophobic analogs were detected in cell-pellet extracts, including compounds matching to the previously described amphibactin D (C14:0 fatty-acyl group) and amphibactin E (C16:1).36,38,44 For a detailed list of all detected amphibactins and a comparison between relative abundances for AC-8003 and AC-B3, see Tables S8–S10 (ESI†).
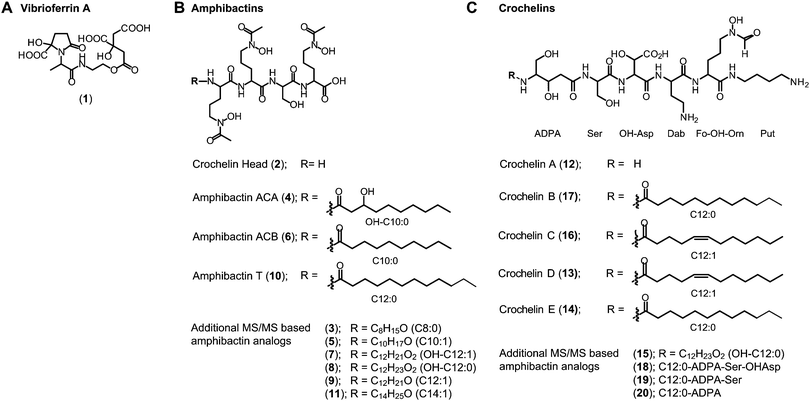 | ||
| Fig. 5 Siderophore structures from AC-8003 and AC-B3. Vibrioferrin, amphibactins, and crochelins were identified by comparison of retention time and high-resolution MS and MS/MS spectra to standards. Both strains produced a suite of new hydrophilic amphibactins with short-chain fatty acid tails (C10, C12 for AC-8003 and C10, C12, C14 for AC-B3). Two of the new amphibactins were isolated for NMR quantification and characterization (Amphibactin ACA and ACB). Other new hydrophilic amphibactin analogs had the same head-group but different fatty acid tails as shown by high-resolution MS/MS analyses. Detailed LC-MS results can be found in the ESI† (Tables S8–S10). | ||
(iii) Crochelins: in addition to vibrioferrin and amphibactins, strain AC-8003 also made crochelins – siderophores synthesized by the cro cluster.16 We previously characterized crochelin A from AC-8003, a siderophore with mixed chelating groups that include a new Fe chelating moiety amino-dihydroxy-pentanoic acid (ADPA).16 The biosynthesis of crochelin A involves the production of lipopeptide precursors with a fatty acyl tail bound to the terminal free amine in ADPA (Fig. 5). During maturation of crochelin A to the active siderophore the fatty acyl group is cleaved by a hydrolase (CroM). We found crochelin A to be the major product in the spent medium with relatively minor contributions of several crochelin A precursors (<10% of the crochelin A concentration, Fig. 4C). The lipopeptidic precursors related to crochelin A had fatty acyl chains identical to those in amphibactins (main analogs with carbon chain length C10 and C12 with full saturation, single unsaturation, or hydroxylation, Fig. 5).
As mentioned above, the amphibactin headgroup (without the fatty acyl tail) was only observed in cultures of AC-8003. This organism also encodes the CroM fatty acid acylase as part of the crochelin biosynthesis cluster, suggesting that the production of the amphibactin headgroup may result from the hydrolysis of fatty acid tails from amphibactins by CroM due to substrate promiscuity.
Function of siderophores: production, Fe availability, and possible synergies
To better understand the function of vibrioferrin, amphibactins, and crochelin A, we measured growth and siderophore production of AC-8003 and AC-B3 with different levels of Fe availability: (A) precipitated amorphous Fe oxides (no added EDTA) for high Fe availability, (B) 5 μM Fe with 100 μM EDTA for intermediate Fe availability, and (C) 0.1 μM Fe with 100 μM EDTA for low Fe availability (Fig. 6). These conditions were applied in a previous study of siderophore production by A. vinelandii.14 Consistent with increasing severity of Fe limitation from condition A to condition C, we observed slower growth rates and yields for AC-8003 (Fig. S6, ESI†). Correspondingly, measurements of dissolved Fe concentrations in AC-8003 spent media showed that condition A was associated with dissolution of Fe during exponential growth, reaching ∼12 μM Fe (without EDTA) at higher optical densities. In contrast, condition B showed a slow reduction in dissolved Fe concentrations (initially added as 5 μM Fe and 100 μM EDTA) indicating slow Fe uptake in the presence of EDTA. Condition C (initially added as 0.1 μM Fe and 100 μM EDTA) had Fe concentrations consistently below the detection limit (<0.1 μM) (Fig. S7, ESI†). Siderophore production at low cell density (OD620nm of ∼0.1) increased from condition A to C (condition A, total siderophore concentration = 0.6 μM at OD620nm = 0.13; B, total siderophore concentration = 4.4 μM at OD620nm = 0.08; C, total siderophore concentration = 25.2 μM at OD = 0.08). AC-B3 and A. vinelandii14 showed similar exponential growth under conditions A and B and slow linear growth under condition C. The combined observations from AC-8003, AC-B3, and A. vinelandii14 reflect decreasing Fe bioavailability from condition A to C.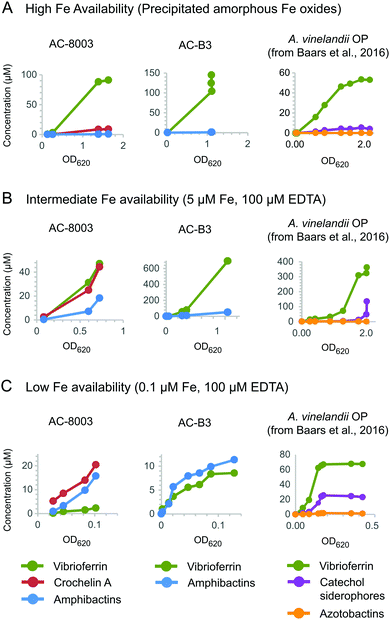 | ||
| Fig. 6 Concentrations of vibrioferrin, amphibactins, and crochelin A secreted by AC-8003 and AC-B3 at different Fe availabilities. (A) High Fe availability, indicated by fast growth and high optical densities in medium with amorphous Fe oxides (no added EDTA). (B) Intermediate Fe availability, indicated by slower growth in medium with 5 μM Fe with an excess of EDTA (100 μM). (C) Low Fe availability (severe Fe limitation), indicated by slow non-exponential growth and low optical densities in medium with 0.1 μM Fe and 100 μM EDTA. Siderophore production by A. vinelandii OP14 under the same Fe treatments is shown for comparison. Corresponding optical densities and total dissolved iron concentrations are shown in Fig. S6 and S7 (ESI†). | ||
Both AC strains produced high vibrioferrin A concentrations when Fe was provided as amorphous Fe oxides ([vibrioferrin A] ≤ 91 μM for AC-8003 and 145 μM for AC-B3) or as 5 μM Fe complexed to EDTA ([vibrioferrin A] = 45 μM for AC-8003 and 695 μM for AC-B3), the two conditions representing high and intermediate Fe bioavailability14 (Fig. 6). In the case of AC-8003, vibrioferrin production became suppressed with stronger Fe limitation under conditions B and C (Fig. 6). In contrast to the low affinity siderophore vibrioferin, the total concentration of high affinity siderophores was low under condition A (<10 μM for AC-8003, <1 μM for AC-B3), then increased under conditions B and C, when Fe was less available (36 μM for AC-8003, a ∼4 fold increase, and 11.3 μM for AC-B3, a ∼12 fold increase, at 10× lower optical densities in condition C compared to condition A) (Fig. 6).
Because the low affinity siderophore vibrioferrin was produced even under condition A (the highest Fe availability), we wanted to confirm its production in response to Fe limitation. Therefore, we cultured AC strains in medium with 5 μM, 0.5 μM, or no added Fe under non-nitrogen fixing conditions (2 mM NH4+) with the weaker chelator NTA (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 15.945) instead of EDTA (log
K = 15.945) instead of EDTA (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 25.145) and a lower concentration of carbon source (1 g L−1 for glucose and mannitol instead of 10 g L−1). Such conditions might result in decreased vibrioferrin production due a decreased cellular need for Fe and increased Fe bioavailability (NTA instead of EDTA). Indeed, vibrioferrin production was low or undetectable with 5 μM added Fe in the NTA chelated medium (<0.5 μM) (Fig. S8, ESI†). When Fe concentration was reduced in the same medium to 0.5 μM, vibrioferrin production was significant with both AC strains (4–6 μM). When no Fe was added, vibrioferrin production decreased in AC-8003 but not in AC-B3 (AC-8003, 1.6 μM; AC-B3, 10 μM), in agreement with the results from conditions A to C described above. For the high-affinity siderophores produced in the NTA-chelated medium (Fig. S8, ESI†), we observed trends similar to those of conditions A to C – increasing concentrations of high-affinity siderophores in medium with less Fe (5 μM, 0.5 μM, no added total Fe). With AC-8003, amphibactins and crochelins were not detected at 5 μM added Fe and increased ∼1.5 and ∼3-fold between the 0.5 μM Fe and no added Fe conditions. With AC-B3, amphibactins were only detected in the no added Fe condition (3.7 μM) (Fig. S8, ESI†).
K = 25.145) and a lower concentration of carbon source (1 g L−1 for glucose and mannitol instead of 10 g L−1). Such conditions might result in decreased vibrioferrin production due a decreased cellular need for Fe and increased Fe bioavailability (NTA instead of EDTA). Indeed, vibrioferrin production was low or undetectable with 5 μM added Fe in the NTA chelated medium (<0.5 μM) (Fig. S8, ESI†). When Fe concentration was reduced in the same medium to 0.5 μM, vibrioferrin production was significant with both AC strains (4–6 μM). When no Fe was added, vibrioferrin production decreased in AC-8003 but not in AC-B3 (AC-8003, 1.6 μM; AC-B3, 10 μM), in agreement with the results from conditions A to C described above. For the high-affinity siderophores produced in the NTA-chelated medium (Fig. S8, ESI†), we observed trends similar to those of conditions A to C – increasing concentrations of high-affinity siderophores in medium with less Fe (5 μM, 0.5 μM, no added total Fe). With AC-8003, amphibactins and crochelins were not detected at 5 μM added Fe and increased ∼1.5 and ∼3-fold between the 0.5 μM Fe and no added Fe conditions. With AC-B3, amphibactins were only detected in the no added Fe condition (3.7 μM) (Fig. S8, ESI†).
Taken together, these results show a consistent pattern across all Azotobacter species (A. chroococcum str. B-3, A. chroococcum str. NCIMB 8003, and A. vinelandii OP): vibrioferrin was produced at mild Fe limitation whereas the high-affinity siderophores were employed only under more severe Fe limitation. The co-production of both low- and high-affinity siderophore structures by AC strains under moderate to severe Fe limitation (Fig. 6B and C) was previously observed with A. vinelandii. This may reflect a “bucket-brigade mechanism” whereby low- and high-affinity siderophores interact synergistically to enable bacterial access to distant sources of Fe.36 The amphiphilic nature of strong AC siderophores, amphibactins and crochelins, may aid synergistic Fe acquisition: production of hydrophilic amphiphilic siderophores can accelerate Fe exchange from EDTA, making Fe available to membrane-bound variants of the amphiphilic siderophores (Table S10, ESI†). Regardless of the specific mechanisms by which siderophores interact, the ability to make functionally distinct siderophores has clear benefits for bacteria living in heterogeneous environments like soils where Fe bioavailability may vary based on mineralogy, organic matter content and composition, hydrology, oxygen content, and microbiology. Indeed, the use of multiple siderophore systems in the distantly related soil bacterium, Pseudomonas aeruginosa, was shown to be critical to its survival in environments with changing Fe availability.46 Given evidence that functionally redundant siderophores are lost from microbial genomes,47 the preservation of multiple siderophores of varying Fe binding strength in AC and other species attests to the variable Fe availability of Azotobacter's natural environment.
Conclusion
This study of A. chroococcum, a widespread N2 fixer in soils and a common agricultural inoculant, shows the existence of “core” and lineage-specific siderophores in the Azotobacter genus that have different functions in Fe acquisition. Production of the low-affinity siderophore vibrioferrin, a vertically inherited, conserved feature of Azotobacter physiology, is favored under mild cellular Fe limitation, whereas lineage-specific, high-affinity siderophores (e.g. amphibactins, crochelins), which originate from genetic recombination, are produced once Fe limitation becomes more pronounced. Together with studies of A. vinelandii,14,23–27 these results suggest that a central characteristic of Azotobacter physiology may be the ability to produce a combination of low- and high-affinity Fe-binding structures to optimize growth over a range of Fe availability.Experimental methods
Bacterial cultures
Wild type Azotobacter chroococcum strain B3 (ATCC strain 7486) and strain NCIMB 8003 (ATCC strain 4412) were grown aerobically in a modified Burk's medium under diazotrophic conditions by shaking at room temperature as previously described.14,48 For HR-LC-MS analysis of siderophores, the availability of Fe was limited by addition of 100 μM EDTA to 0.1 μM FeCl3 for AC-B3, whereas a concentration of 5 μM FeCl3 was added to achieve sufficient growth with AC-8003. To study the effect of Fe sources, Fe was added as (1) 100 μM EDTA and 0.1 μM FeCl3; (2) 100 μM EDTA and 5 μM FeCl3; and (3) precipitated amorphous Fe oxides. Bacterial growth was monitored at an optical density (OD) of 620 nm (OD620nm).HR-LC-MS siderophore analysis
Cell pellets, collected from a 30 mL culture for AC-B3 and from 11 × 80 mL cultures for AC-8003, were extracted overnight in 20 mL methanol on a shaker plate at 4 °C. The extracts were centrifuged and the methanolic extract was filtered (0.2 μm). The filtered extract was then dried in a SpeedVac and reconstituted with 25% methanol for extraction on a reversed phase SPE column (Oasis HLB Plus, Waters). The SPE cartridge was washed with 20 mL water and eluted with 100% methanol. The extract was dried in a SpeedVac and re-constituted with 25% methanol added to aqueous buffer prior to LC-MS.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (at m/z = 400). MS/MS spectra were simultaneously acquired using CID in the Orbitrap using a parent ion intensity threshold >10
000 (at m/z = 400). MS/MS spectra were simultaneously acquired using CID in the Orbitrap using a parent ion intensity threshold >10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and targeting either the most abundant species in the full-scan spectrum or selectively the three most abundant species on a parent ion list.
000 and targeting either the most abundant species in the full-scan spectrum or selectively the three most abundant species on a parent ion list.
Siderophore quantification
To follow concentrations of siderophores in AC-B3, 1 mL sample aliquots were taken at different times throughout the growth, filtered through 0.2 μm syringe filters, acidified (0.1% acetic acid and 0.1% formic acid) and analyzed by direct injection on a single quadrupole LC-MS system (Agilent 6120). The analysis of vibrioferrin and amphibactins was performed with a C18 column (Agilent Eclipse Plus C18 3.5 μm, 4.6 × 100 mm) and a gradient of solution A (water + 0.1% FA + 0.1% acetic acid) and B (acetonitrile + 0.1% FA + 0.1% acetic acid; gradient from 0–100% A over 30 min; flow rate of 0.8 mL min−1). To achieve sufficient retention on the same C18 column, the analysis of crochelin A required the use of the ion-pairing reagent heptafluorobutyric acid (HFBA) added to solutions A (water + 0.05% HFBA) and B (acetonitrile + 0.05% HFBA) with a gradient from 0–100% A over 30 min; flow rate of 0.8 mL min−1. The column outflow was diverted to waste for the first 5.25 min ensuring that the sample was completely desalted before introduction into the mass spectrometer. Siderophores were quantified by single ion monitoring (SIM) using a list of target m/z values of the identified siderophores from the HR-LC-MS analysis (Table S8, ESI†). For quantification, LC-MS peak areas were determined and converted to concentrations by calibration with standards of vibrioferrin A,16 amphibactins ACA, S, and T (isolated as described below, see also Fig. S5, ESI†), and crochelin A.16 Seven technical replicates of a spent medium siderophore mix “standard” showed relative standard deviations of <10% when present at concentrations above 0.5 μM. The detection limit of vibrioferrin A, amphibactins, and crochelin A in the supernatant sample was in the range of 0.02 to 0.10 μM.Purification of amphibactins
Amphibactins were purified for NMR spectroscopic analysis and to use as purification standard by growing batch cultures (11 × 80 mL) on a shaker table under nitrogen fixing conditions with 100 μM EDTA and 5 μM FeCl3. This condition enabled exponential growth which became strongly Fe limited before reaching stationary phase, resulting in a strong production of amphibactins. The incubations were combined once they reached stationary phase, centrifuged and filtered (0.2 μm). The supernatants were passed over reversed phase resins (Oasis HLB Plus), activated with methanol (30 mL) and conditioned with Milli-Q water (30 mL). The cartridges were washed with Milli-Q (30 mL) and crude amphiphilic crochelins were eluted with 75% methanol. The extracts were dried down (SpeedVac) and reconstituted in 2 mL of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol/water. The extracts were further purified by HPLC with a water acetonitrile gradient with added 0.1% formic acid and acetic acid. The elution of amphibactins was monitored at 220 nm. Combined fractions were analyzed by HPLC-MS and HPLC-UV/vis before they were dried down (SpeedVac, ThermoFisher) and reconstituted with 2% d4-methanol for 1H-NMR spectroscopic quantification using an external standard (PULCON method).
1 methanol/water. The extracts were further purified by HPLC with a water acetonitrile gradient with added 0.1% formic acid and acetic acid. The elution of amphibactins was monitored at 220 nm. Combined fractions were analyzed by HPLC-MS and HPLC-UV/vis before they were dried down (SpeedVac, ThermoFisher) and reconstituted with 2% d4-methanol for 1H-NMR spectroscopic quantification using an external standard (PULCON method).
Dissolved Fe concentrations
Iron (56Fe) concentrations in the supernatants were measured by inductively coupled plasma-mass spectrometry (Thermo iCAP-Q in KED mode) after acidification (10% HNO3) and dilution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). The detection limit for iron (56Fe) was approximately 0.10 μM.
5). The detection limit for iron (56Fe) was approximately 0.10 μM.
Genome sequencing of A. chroococcum strain B3
AC-B3 genomic DNA was isolated using a Qiagen Genomic Tip DNA extraction kit. Samples of a 10 kb insert library were prepared at the Molecular Biology Core Facility at Princeton University Library prior to genome sequencing at the UC Irvine Genomics High Throughput Sequencing Facility. Sequencing reads, generated from 5 single-molecule real-time (SMRT) cells of a Pacific Biosciences RS2 sequencer system (Pacific Biosciences, Menlo Park, CA), yielded greater than 20× average genome coverage. Read sequences were assembled de novo using continuous long reads (CLR) following the Hierarchical Genome Assembly Process (HGAP) workflow (PacBio DevNet; Pacific Biosciences) as available in SMRT Analysis v2.0. The PacBio workflow followed by manual assembly of the largest fragment yielded a draft genome of AC-B3 consisting of single closed circular chromosome (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 575
575![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 910 bp) and 3 linear plasmids (pacX50fB3, 306
910 bp) and 3 linear plasmids (pacX50fB3, 306![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 103 bp; pacX50dB3a, 74
103 bp; pacX50dB3a, 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 783 bp; pacX50dB3b, 66
783 bp; pacX50dB3b, 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259 bp). These sequences were annotated with the NCBI (National Center for Biotechnology Information) Prokaryotic Genomes Automatic Annotation Pipeline. Genome sequences have been deposited as draft whole-genome shotgun projects in GenBank under the accession numbers CP011835–CP011838. The versions described in this paper are the first versions.
259 bp). These sequences were annotated with the NCBI (National Center for Biotechnology Information) Prokaryotic Genomes Automatic Annotation Pipeline. Genome sequences have been deposited as draft whole-genome shotgun projects in GenBank under the accession numbers CP011835–CP011838. The versions described in this paper are the first versions.
Genome analysis
Putative siderophore genes were identified in Azotobacter genome sequences (see Table S3, ESI†) using the secondary metabolite genome mining software AntiSmash,49 the gene annotation software RAST,50 and targeted BLAST homology analyses. Siderophore genes were compared to those of A. vinelandii strain CA (GenBank accession number CP005094), previously described by ref. 14, 51 and 52. Phylogenies were constructed using the MUSCLE aligner53 and PHYML options within the Geneious software environment54 (version 11.0.4) and the SILVA rRNA database.55Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank Wei Wang and Lance Parsons of the Lewis-Sigler Institute for Integrative Genomics (Princeton University) for help with genomic sequencing, Mohammed Seyedsayamdost for useful discussions (Princeton University), the National Science Foundation (grant EAR-1631814 to X. Z. and OCE 1315200 to F. M. M. M.,) and the Princeton Environmental Institute Innovative Research Award (to O. B.) for support of this work. O. B. was supported by the National Institute of Food and Agriculture Hatch project 201367 during analyses and writing stages of this work.Notes and references
- J. P. Thompson and V. B. D. Skerman, Azotobacteraceae: the Taxonomy and Ecology of the Aerobic Nitrogen-Fixing Bacteria, Academic Press, London, UK, 1979 Search PubMed.
- R. L. Robson, R. Jones, R. M. Robson, A. Schwartz and T. H. Richardson, Azotobacter genomes: The genome of Azotobacter chroococcum NCIMB 8003 (ATCC 4412), PLoS One, 2015, 10, e0127997 CrossRef.
- J. H. Becking, in The Prokaryotes, ed. M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer and E. Stackebrandt, Springer, New York, NY, USA, 3rd edn, 2006, vol. 6, pp. 759–783 Search PubMed.
- S. A. Wani, S. Chand, M. A. Wani, M. Ramzan and K. R. Hakeem, in Soil Science: Agricultural and Environmental Prospectives, ed. K. R. Hakeem, J. Akhtar and M. Sabir, Springer International Publishing, Cham, 2016, pp. 333–348, DOI:10.1007/978-3-319-34451-5_15.
- K. V. B. R. Tilak, K. K. Pal and R. Dey, Microbes for Sustainable Agriculture, I.K. International Publishing House, New Delhi, India, 2010 Search PubMed.
- M. V. Martinez Toledo, J. Gonzalez-Lopez, T. de la Rubia, J. Moreno and A. Ramos-Cormenzana, Effect of inoculation with Azotobacter chroococcum on nitrogenase activity of Zea mays roots grown in agricultural soils under aseptic and non-sterile conditions, Biol. Fertil. Soils, 1988, 6, 170–173 CrossRef.
- M. E. Brown and S. K. Burlingham, Production of plant growth substances by Azotobacter chroococcum, J. Gen. Microbiol., 1968, 53, 135–144 CrossRef CAS.
- M. Kelly, Some properties of purified nitrogenase of Azotobacter chroococcum, Biochim. Biophys. Acta, 1969, 171, 9–22 CrossRef CAS.
- R. L. Robson, R. R. Eady, T. H. Richardson, R. W. Miller, M. Hawkins and J. R. Postgate, The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme, Nature, 1986, 322, 388–390 CrossRef CAS.
- R. L. Robson, P. R. Woodley, R. N. Pau and R. R. Eady, Structural genes for the vanadium nitrogenase from Azotobacter chroococcum, EMBO J., 1989, 8, 1217–1224 CrossRef CAS.
- C. C. Walker and M. G. Yates, The hydrogen cycle in nitrogen-fixing Azotobacter chroococcum, Biochimie, 1978, 60, 225–231 CrossRef CAS.
- L. Du, K. Tibelius, E. Souza, R. Garg and M. Yates, Sequences, organization and analysis of the hupZMNOQRTV genes from the Azotobacter chroococcum hydrogenase gene cluster, J. Mol. Biol., 1994, 243, 549–557 CrossRef CAS.
- C. Ford, N. Garg, R. Garg, K. Tibelius, M. Yates, D. Arp and L. Seefeldt, The identification, characterization, sequencing and mutagenesis of the genes (hupSL) encoding the small and large subunits of the H2-uptake hydrogenase of Azotobacter chroococcum, Mol. Microbiol., 1990, 4, 999–1008 CrossRef CAS.
- O. Baars, X. Zhang, F. M. M. Morel and M. Seyedsayamdost, The siderophore metabolome of Azotobacter vinelandii, Appl. Environ. Microbiol., 2016, 82, 27–39 CrossRef CAS.
- D. L. McRose, O. Baars, F. M. M. Morel and A. M. L. Kraepiel, Siderophore production in Azotobacter vinelandii in response to Fe-, Mo- and V-limitation, Environ. Microbiol., 2017, 19, 3595–3605 CrossRef CAS.
- O. Baars, X. Zhang, M. I. Gibson, A. T. Stone, F. M. M. Morel and M. R. Seyedsayamdost, Crochelins: siderophores with an unprecedented iron-chelating moiety from the nitrogen-fixing bacterium Azotobacter chroococcum, Angew. Chem., 2018, 130, 545–550 CrossRef.
- S. A. Amin, D. H. Green, F. C. Küpper and C. J. Carrano, Vibrioferrin, an unusual marine siderophore: Iron binding, photochemistry, and biological Implications, Inorg. Chem., 2009, 48, 11451–11458 CrossRef CAS.
- E. F. Wawrousek and J. V. McArdle, Spectroelectrochemistry of ferrioxamine B, ferrichrome, and ferrichrome A, J. Inorg. Biochem., 1982, 17, 169–183 CrossRef CAS.
- I. Spasojević, S. K. Armstrong, T. J. Brickman and A. L. Crumbliss, Electrochemical behavior of the Fe(III) Complexes of the cyclic hydroxamate siderophores alcaligin and desferrioxamine E, Inorg. Chem., 1999, 38, 449–454 CrossRef.
- S. Suneja, N. Narula, R. C. Anand and K. Lakshminarayana, Relationship of Azotobacter chroococcum siderophores with nitrogen fixation, Folia Microbiol., 1996, 41, 154–158 CrossRef CAS.
- W. J. Page, Iron-dependent production of hydroxamate by sodium-dependent Azotobacter chroococcum, Appl. Environ. Microbiol., 1987, 53, 1418–1424 CAS.
- F. A. Fekete, R. A. Lanzi, J. B. Beaulieu, D. C. Longcope, A. W. Sulya, R. N. Hayes and G. A. Mabbott, Isolation and preliminary characterization of hydroxamic acids formed by nitrogen-fixing Azotobacter chroococcum B-8, Appl. Environ. Microbiol., 1989, 55, 298–305 CAS.
- A. Cornish and W. Page, Production of the triacetecholate siderophore protochelin by Azotobacter vinelandii, Biometals, 1995, 8, 332–338 CrossRef CAS.
- W. A. Bulen and J. R. LeComte, Isolation and properties of a yellow-green fluorescent peptide from Azotobacter medium, Biochem. Biophys. Res. Commun., 1962, 9, 523–528 CrossRef CAS.
- W. J. Page and M. v. Tigerstrom, Aminochelin, a catecholamine siderophore produced by Azotobacter vinelandii, J. Gen. Microbiol., 1988, 134, 453–460 CAS.
- J. L. Corbin and W. A. Bulen, Isolation and identification of 2,3-dihydroxybenzoic acid and N2,N6-di(2,3-dihydroxybenzoyl)-L-lysine formed by iron-deficient Azotobacter vinelandii, Biochemistry, 1969, 8, 757–762 CrossRef CAS.
- W. Page, S. K. Collinson, P. Demange, A. Dell and M. Abdallah, Azotobacter vinelandii strains of disparate origin produce azotobactin siderophores with identical structures, Biol. Met., 1991, 4, 217–222 CrossRef CAS.
- A. S. Cornish and W. J. Page, Role of molybdate and other transition metals in the accumulation of protochelin by Azotobacter vinelandii, Appl. Environ. Microbiol., 2000, 66, 1580–1586 CrossRef CAS.
- J. M. Harrington, J. R. Bargar, A. A. Jarzecki, J. G. Roberts, L. A. Sombers and O. W. Duckworth, Trace metal complexation by the triscatecholate siderophore protochelin: structure and stability, Biometals, 2012, 25, 393–412 CrossRef CAS.
- A. L. Crumbliss and J. M. Harrington, Iron sequestration by small molecules: thermodynamic and kinetic studies of natural siderophores and synthetic model compounds, Adv. Inorg. Chem., 2009, 61, 179–250 CrossRef CAS.
- O. Baars, F. M. M. Morel and D. H. Perlman, ChelomEx: Isotope-assisted discovery of metal chelates in complex media using high-resolution LC-MS, Anal. Chem., 2014, 86, 11298–11305 CrossRef CAS PubMed.
- K. Blin, H. U. Kim, M. H. Medema and T. Weber, Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters, Brief. Bioinform., 2017, bbx146, DOI:10.1093/bib/bbx146.
- G. L. Challis, A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases, ChemBioChem, 2005, 6, 601–611 CrossRef CAS.
- A. J. van Tonder, S. Mistry, J. E. Bray, D. M. C. Hill, A. J. Cody, C. L. Farmer, K. P. Klugman, A. von Gottberg, S. D. Bentley, J. Parkhill, K. A. Jolley, M. C. J. Maiden and A. B. Brueggemann, Defining the estimated core genome of bacterial populations using a Bayesian decision model, PLoS Comput. Biol., 2014, 10, e1003788 CrossRef.
- S. K. Thode, E. Rojek, M. Kozlowski, R. Ahmad and P. Haugen, Distribution of siderophore gene systems on a Vibrionaceae phylogeny: Database searches, phylogenetic analyses and evolutionary perspectives, PLoS One, 2018, 13, e0191860 CrossRef.
- J. S. Martinez, J. N. Carter-Franklin, E. L. Mann, J. D. Martin, M. G. Haygood and A. Butler, Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 3754–3759 CrossRef CAS.
- J. Vraspir, P. Holt and A. Butler, Identification of new members within suites of amphiphilic marine siderophores, Biometals, 2011, 24, 85–92 CrossRef CAS.
- M. P. Kem, H. K. Zane, S. D. Springer, J. M. Gauglitz and A. Butler, Amphiphilic siderophore production by oil-associating microbes, Metallomics, 2014, 6, 1150–1155 RSC.
- R. C. MacLean and A. San Millan, Microbial evolution: Towards resolving the plasmid paradox, Curr. Biol., 2015, 25, R764–R767 CrossRef CAS.
- U. Antonenka, C. Nölting, J. Heesemann and A. Rakin, Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid, Mol. Microbiol., 2005, 57, 727–734 CrossRef CAS.
- C. R. Osorio, A. J. Rivas, M. Balado, J. C. Fuentes-Monteverde, J. Rodríguez, C. Jiménez, M. L. Lemos and M. K. Waldor, A transmissible plasmid-borne pathogenicity island confers piscibactin biosynthesis in the fish pathogen Photobacterium damselae subsp. piscicida, Appl. Environ. Microbiol., 2015, 81, 5867 CrossRef CAS.
- H. Naka, M. Liu, L. A. Actis and J. H. Crosa, Plasmid- and chromosome-encoded siderophore anguibactin systems found in marine vibrios: Biosynthesis, transport and evolution, Biometals, 2013, 26, 537–547 CrossRef CAS.
- M. R. Seyedsayamdost, S. Cleto, G. Carr, H. Vlamakis, M. João Vieira, R. Kolter and J. Clardy, Mixing and matching siderophore clusters: Structure and biosynthesis of serratiochelins from Serratia sp. V4, J. Am. Chem. Soc., 2012, 134, 13550–13553 CrossRef CAS.
- J. Gauglitz and A. Butler, Amino acid variability in the peptide composition of a suite of amphiphilic peptide siderophores from an open ocean Vibrio species, JBIC, J. Biol. Inorg. Chem., 2013, 18, 489–497 CrossRef CAS.
- A. E. Martell, R. M. Smith and R. J. Motekaitis, NIST Critically Selected Stability Constants of Metal Complexes Database, Database 46 Version 7.0 for Windows, Gaithersburg, MD, USA, 2003 Search PubMed.
- Z. Dumas, A. Ross-Gillespie and R. Kümmerli, Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments, Proc. R. Soc. B, 2013, 280, 20131055 CrossRef PubMed.
- H. Bruns, M. Crüsemann, A.-C. Letzel, M. Alanjary, J. O. McInerney, P. R. Jensen, S. Schulz, B. S. Moore and N. Ziemert, Function-related replacement of bacterial siderophore pathways, ISME J., 2017, 12, 320 CrossRef.
- C. K. Horner, D. Burk, G. E. Allison and M. S. Sherman, Nitrogen fixation by Azotobacter as influenced by molybdenum and vanadium, J. Agric. Res., 1942, 65, 173–193 Search PubMed.
- T. Weber, K. Blin, S. Duddela, D. Krug, H. U. Kim, R. Bruccoleri, S. Y. Lee, M. A. Fischbach, R. Müller, W. Wohlleben, R. Breitling, E. Takano and M. H. Medema, antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters, Nucleic Acids Res., 2015, 43, W237–W243 CrossRef CAS.
- R. Overbeek, R. Olson, G. D. Pusch, G. J. Olsen, J. J. Davis, T. Disz, R. A. Edwards, S. Gerdes, B. Parrello, M. Shukla, V. Vonstein, A. R. Wattam, F. Xia and R. Stevens, The SEED and the rapid annotation of microbial genomes using Subsystems Technology (RAST), Nucleic Acids Res., 2014, 42, D206–D214 CrossRef CAS.
- A. E. Tindale, M. Mehrotra, D. Ottem and W. J. Page, Dual regulation of catecholate siderophore biosynthesis in Azotobacter vinelandii by iron and oxidative stress, Microbiology, 2000, 146, 1617–1626 CrossRef CAS PubMed.
- F. Yoneyama, M. Yamamoto, W. Hashimoto and K. Murata, Azotobacter vinelandii gene clusters for two types of peptidic and catechol siderophores produced in response to molybdenum, J. Appl. Microbiol., 2011, 111, 932–938 CrossRef CAS.
- R. C. Edgar, MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res., 2004, 32, 1792–1797 CrossRef CAS.
- M. Kearse, R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, A. Cooper, S. Markowitz, C. Duran, T. Thierer, B. Ashton, P. Meintjes and A. Drummond, Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data, Bioinformatics, 2012, 28, 1647–1649 CrossRef.
- E. Pruesse, C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies and F. O. Glockner, SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB, Nucleic Acids Res., 2007, 35, 7188–7196 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8mt00236c |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |
