Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metallothioneins of the urochordate Oikopleura dioica have Cys-rich tandem repeats, large size and cadmium-binding preference†
Sara
Calatayud
 a,
Mario
Garcia-Risco
a,
Mario
Garcia-Risco
 b,
Natalia S.
Rojas
a,
Lizethe
Espinosa-Sánchez
a,
Sebastián
Artime
a,
Òscar
Palacios
b,
Natalia S.
Rojas
a,
Lizethe
Espinosa-Sánchez
a,
Sebastián
Artime
a,
Òscar
Palacios
 b,
Cristian
Cañestro
b,
Cristian
Cañestro
 a and
Ricard
Albalat
a and
Ricard
Albalat
 *a
*a
aDepartament de Genètica, Microbiologia i Estadística and Institut de Recerca de la Biodiversitat (IRBio), Facultat de Biologia, Universitat de Barcelona, Av. Diagonal 643, E-08028, Barcelona, Catalonia, Spain. E-mail: ralbalat@ub.edu
bDepartament de Química, Facultat de Ciències, Universitat Autònoma de Barcelona, E-08193 Cerdanyola del Vallès, Spain
First published on 28th September 2018
Abstract
The increasing levels of heavy metals derived from human activity are poisoning marine environments, threating zooplankton and ocean food webs. To protect themselves from the harmful effects of heavy metals, living beings have different physiological mechanisms, one of which is based on metallothioneins (MTs), a group of small cysteine-rich proteins that can bind heavy metals counteracting their toxicity. The MT system of urochordate appendicularians, an ecologically relevant component of the zooplankton, remained, however, unknown. In this work, we have characterized the MTs of the appendicularian species Oikopleura dioica, revealing that O. dioica has two MT genes, named OdMT1 and OdMT2, which encode for Cys-rich proteins, the former with 72 amino acids comparable with the small size MTs of other organisms, but the second with 399 amino acids representing the longest MT reported to date for any living being. Sequence analysis revealed that OdMT2 gene arose from a duplication of an ancestral OdMT1 gene followed by up to six tandem duplications of an ancestral repeat unit (RU) in the current OdMT2 gene. Interestingly, each RU contained, in turn, an internal repeat of a 7-Cys subunit (X3CX3CX2CX2CX3–6CX2CXCX), which is repeated up to 12 times in OdMT2. Finally, ICP-AES analyses of heterologously expressed OdMT proteins showed that both MTs were capable to form metal-complexes, with preference for cadmium ions. Collectively, our results provide the first characterization of the MT system in an appendicularian species as an initial step to understand the zooplankton response to metal toxicity and other environmental stress situations.
Significance to metallomicsThe analysis of the metallothioneins of Oikopleura dioica is important for understanding the evolution and function of the detoxification mechanisms that this zooplankton species might use to counteract the harmful effects of heavy metal exposure. This knowledge is fundamental to estimate the potential impact that an increase of heavy metal amounts may have on marine ecosystems. |
1. Introduction
Heavy metals such as zinc, iron or copper are essential for several biological processes, but toxic at high concentrations, while others such as cadmium, mercury or lead are highly poisonous even at low concentrations. Living beings have different physiological mechanisms to control the homeostasis of essential metals as well as to counteract the harmful effects of the non-essential ones. One of these mechanisms is based on metallothioneins (MTs), a group of metal-binding proteins originally discovered in 1957 in the horse kidney cortex,1 and classically considered a diverse family of Cys-rich (≈30%) and low molecular weight (<60 amino acids) proteins found in almost all organisms (reviewed in ref. 2 and 3). The amino acid sequence of different MTs is highly heterogeneous, particularly when MTs of distantly related taxa are compared. In such comparisons, sequence similarity appears restricted to an overall “Cys abundance” with distinctive cysteine motifs (i.e. CXC, CC and CCC), whereas the length of the protein, the number and distribution of cysteines and the intercalating residues are largely variable from one taxon to another.2 Vertebrate MT sequences, for instance, can be reliably aligned within the subphylum, but they are barely comparable with other MT sequences, even from their closest chordate relatives, amphioxus cephalochordates or ascidian urochordates.4,5From a structural perspective, MTs have been extensively analyzed showing that they are able to coordinate a number of heavy metal ions through the formation of metal–thiolate bonds.6,7 Depending on their metal-binding preferences, MTs can be classified in a stepwise gradation from extreme Zn/Cd-thioneins to extreme Cu-thioneins.8,9 The metal-binding preference of a given MT cannot be, however, directly predicted from its amino acid sequence, and metal-binding assays have to be performed to classify newly discovered forms.
From a physiological perspective, MTs are activated for controlling metal homeostasis and detoxification roles, but also for radical scavenging, oxidative stress protection or antiapoptotic defense (reviewed in ref. 6). The presence of Metal Responsive Elements (MRE) and different response elements for transcription factors (TF) involved in stress response (e.g. AREs, HSE, StRE) in the promoter regions has been associated to the transcriptional activation of MT genes in response to heavy metals and other stress situations (reviewed in ref. 10).
In the last century, what has been called the Anthropocene, human activities such as those related to the industries of mining, metal plating, fertilizer, paper, pesticides or batteries, are increasing the amount of heavy metals in aquatic environments,11,12 becoming a serious problem for coastal and marine ecosystems.13 In marine ecosystems, Appendicularians (a.k.a. Larvaceans; phylum Chordata, subphylum Urochordata) represent the second most abundant group of mesozooplankton grazers and an important component of food for fish and zooplankton species.14 Appendicularians are also ecologically relevant because they contribute to the vertical transport of carbon to deep ocean through the rapid sinking of fecal pellets and discarded houses.15–17 Among Appendicularians, Oikopleura dioica is the most studied species, and it is becoming a new laboratory model for comparative genetic and genomic analyses,18–22 developmental biology studies,23–30 as well as ecological and toxicological investigations.31–34
The ecological relevance of Appendicularians together with the potential of O. dioica as an experimental model35–38 prompted us to survey the MTs of O. dioica to characterize the MT system of an Appendicularian species. Here we report the identification of two MT genes in O. dioica, named OdMT1 and OdMT2, and the analysis of their gene structures and promoter regions. OdMT1 encode for a Cys-rich protein of 72 amino acids (theoretical molecular weight 7645 Da) comparable with the small size MTs of other organisms (human MT1 theoretical molecular weight 6120 Da), while OdMT2 encode for a Cys-rich protein of 399 amino acids (theoretical molecular weight 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 716 Da) representing the longest MT reported to date for any living being. Interestingly, sequence analysis reveals that OdMT2 gene arose from a duplication of an ancestral OdMT1 gene followed by up to six tandem duplications of an ancestral repeat unit (RU) in the current OdMT2 gene. OdMT promoters appear rich in stress-responsive elements, including binding sites for MREF, YAP1/CREB, RXR/PPAR and HSF transcription factors, envisaging a role of MT genes in adverse environmental conditions. Finally, heterologously expressed OdMT1 and OdMT2 proteins showed that they are capable to form metal complexes, with preference for cadmium ions. Overall, these results pave the way for a better understanding of the homeostasis of the physiological metals in O. dioica, as well as the genetic mechanisms that Appendicularians might use against the harmful effects of heavy metal exposure.
716 Da) representing the longest MT reported to date for any living being. Interestingly, sequence analysis reveals that OdMT2 gene arose from a duplication of an ancestral OdMT1 gene followed by up to six tandem duplications of an ancestral repeat unit (RU) in the current OdMT2 gene. OdMT promoters appear rich in stress-responsive elements, including binding sites for MREF, YAP1/CREB, RXR/PPAR and HSF transcription factors, envisaging a role of MT genes in adverse environmental conditions. Finally, heterologously expressed OdMT1 and OdMT2 proteins showed that they are capable to form metal complexes, with preference for cadmium ions. Overall, these results pave the way for a better understanding of the homeostasis of the physiological metals in O. dioica, as well as the genetic mechanisms that Appendicularians might use against the harmful effects of heavy metal exposure.
2. Material and methods
2.1. Genome survey of O. dioica MT genes
To identify O. dioica MT homologs we made low-restrictive tblastn searches39 in the genome database of O. dioica (Oikobase, http://oikoarrays.biology.uiowa.edu/Oiko) with a high value of expect threshold (1000) and no filtering for low complexity sequences, using the two only available MTs of Urochordate ascidians Ciona robusta and Herdmania curvata MTs (accession numbers ACN32211 and AY314949, respectively), as well as Cephalochordate and Vertebrate MTs as queries.4,5 Gene annotations were manually reviewed and corrected based on ESTs availability: EST FP794470.1 for OdMT1 and a consensus sequence derived from the assembly of 18 SRA sequences for OdMT2 (Fig. S1 and Table S1, ESI†).2.2. Promoter analysis and stress-responsive elements
The analysis of putative transcription factor binding sites for OdMT1 and OdMT2 genes was performed by means of the MatInspector 8.4.1 program with default parameters, using the MatBase 11.0 from the Genomatix software suite.40 The MatBase 11.0 includes 2029 weight matrices of 482 families, representing binding sites descriptions of more than 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 TF. We restricted our analysis to a subset of the 33 families that included TF involved in metal- or stress-response in animals, plants or fungi (Table S3, ESI†). A 1000-bp region upstream of predicted CDS of both OdMT genes was selected for the promoter analysis.
000 TF. We restricted our analysis to a subset of the 33 families that included TF involved in metal- or stress-response in animals, plants or fungi (Table S3, ESI†). A 1000-bp region upstream of predicted CDS of both OdMT genes was selected for the promoter analysis.
2.3. Cloning of OdMT1 and OdMT2 for recombinant expression
A full-length synthetic cDNA for predicted OdMT1 gene was synthesized by Integrated DNA Technologies Company (Coralville, IA, USA), while for technical limitations, the cDNA of OdMT2 was split in two fragments of 610 nt (5′-fragment 1) and 660 nt (3′-fragment 2), overlapping 52 nt among them. BamHI and XhoI restriction sites and 6–7 additional 5′-nucleotides were added to both OdMT cDNA ends to facilitate the cloning processes. The synthetic cDNA for OdMT1 was PCR amplified −94 °C 5 minutes (min); 25 cycles of 94 °C 30 seconds (s), 55 °C 30 s and 72 °C 30 s; and 72 °C 7 min – with specific primers (5′GGGGGATCCATGGATCCGGTTTGCTCTTTCCGCTG3′ and 5′GGGCTCGAGTTATTCCGCTGTGCTGGTCGGGCAG3′) using Expand High Fidelity PCR system® (Roche). The full-length synthetic cDNA for OdMT2 was reconstructed in two steps by adapting previous methods for assembling overlapping DNA fragments.41–44 In brief, in the first step, the two OdMT2 fragments were denaturalized at 98 °C for 3 min, annealed at 80 °C for 1 min, and extended at 94 °C for 30 s. Ten cycles of annealing and extension were repeated to generate a single full-length cDNA molecule from the assembly of the two overlapping fragments. In the second step, the resulting cDNA for OdMT2 was PCR-amplified – 15 cycles of 76 °C 30 s and 94 °C 30 s; and 76 °C 5 min – with specific end-terminal primers (5′GGGGGATCCATGGAAGTAAAACGACC3′ and 5′GGGCTCGAGTTAACAGCATTTTTTGG3′) using Phusion High Fidelity DNA polymerase (New England Biolabs, Thermo Scientific). OdMT1 and OdMT2 PCR products were BamHI/XhoI-digested and cloned into the E. coli pGEX-4T-1 expression vector (GE Healthcare) with the DNA Ligation kit 2.1 (Takara Bio Inc.), and transformed into E. coli Dh5α strain. Plasmid DNA was purified from bacteria using the GeneElute™ Plasmid Miniprep Kit (Sigma-Aldrich), screened for insert presence by digestion with ScaI enzyme, and sequenced at the Scientific and Technological Centers of the University of Barcelona, using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) in an automatic sequencer (ABIPRISM 310, Applied Biosystems). DNA from each recombinant OdMT-pGEX plasmid was used to transform E. coli BL21 strain, a protease deficient strain used for heterologous protein expression.2.4 Production of recombinant metal–OdMT1 and –OdMT2 complexes
For heterologous protein production 500 mL of LB medium with 100 μg mL−1 ampicillin were inoculated with E. coli BL21 cells transformed with the OdMT1 or OdMT2 recombinant plasmids. After overnight growth at 37 °C/250 rpm, the cultures were used to inoculate 5 L of fresh LB-100 μg mL−1 ampicillin medium. Gene expression was induced with 100 μM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3 hours (h). After the first 30 min of induction, cultures were supplemented with ZnCl2 (300 μM), CdCl2 (300 μM) or CuSO4 (500 μM) in order to generate metal–OdMT complexes. Cells were harvested by centrifugation for 5 min at 9100g (7700 rpm), and bacterial pellets were suspended in 125 mL of ice-cold PBS (1.4 M NaCl, 27 mM KCl, 101 mM Na2HPO4, 18 mM KH2PO4 and 0.5% v/v β-mercaptoethanol). Resuspended cells were sonicated (Sonifier Ultrasonic Cell Disruptor) 8 min at voltage 6 with pulses of 0.6 s, and then centrifuged for 40 min at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200g (12
200g (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) and 4 °C.
000 rpm) and 4 °C.
2.5. Purification of recombinant metal–OdMT complexes
Protein extracts containing GST–OdMT1 or GST–OdMT2 fusion proteins were incubated with glutathione sepharose beads (GE Healthcare) for 1 h at room temperature with gentle rotation. GST–OdMT fusion proteins bound to the sepharose beads were washed with 30 mL of cold 1× PBS bubbled with argon to prevent oxidation. After three washes, GST–MT fusion proteins were digested with thrombin (GE Healthcare, 25 U L−1 of culture) overnight at 17 °C, thus enabling separation of the metal–OdMT complexes from the GST that remained bound to the sepharose matrix. The eluted metal–OdMT complexes were concentrated with a 3 kDa Centripep Low Concentrator (Amicon, Merck), and fractionated on a Superdex-75 FPLC column (GE Healthcare) equilibrated with 20 mM Tris–HCl, pH 7.0 for OdMT1, and with fresh 50 mM amonium acetate, pH 7.0 for OdMT2, and run at 0.8 mL min−1. The protein-containing fractions, identified by their absorbance at 254 nm, were pooled and stored at −80 °C until use.2.6. Analysis of metal–OdMT complexes
Metal–OdMT complexes were analyzed by different techniques: inductively coupled plasma atomic emission spectroscopy (ICP-AES) for protein quantification and element composition (S, Zn, Cd and Cu) on a Polyscan 61E spectrometer (Thermo Jarrell Ash Corporation, Franklin, MA, USA) at appropriate wavelengths (S, 182.040 nm; Zn, 213.856 nm; Cd, 228.802 nm; Cu, 324.803 nm) under conventional (dilution with 2% HNO3 (v/v)) conditions.45 OdMT concentrations in the recombinant preparations were calculated from sulfur measurements, assuming the only contribution to their S content was that made by the OdMT peptides.Electrospray ionization mass spectrometry with a time-of-flight analyzer (ESI-TOF MS) was used to determine the molecular mass of the species formed, using a Micro Tof-Q Instrument (Bruker Daltonics Gmbh, Bremen, Germany) calibrated with ESI-L Low Concentration Tuning Mix (Agilent Technologies, Santa Clara, CA, USA), interfaced with a Series 1100 HPLC pump (Agilent Technologies) equipped with an autosampler, both controlled by the Compass Software. The experimental conditions for analyzing the proteins were as follows: 10–20 μL of the sample were injected at 30–50 μL min−1 at 3.5–5.0 kV capillary-counter electrode voltage, 90–110 °C desolvation temperature, and dry gas at 6 L min−1. Spectra were collected throughout an m/z range from 800 to 2000. The liquid carrier was a 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 mixture of 15 mM ammonium acetate and acetonitrile, pH 7.0. For the analysis at acidic pH the conditions used were the same as those used in the analysis of the divalent metals, except in the composition of the carrier liquid, which in this case was a 95
10 mixture of 15 mM ammonium acetate and acetonitrile, pH 7.0. For the analysis at acidic pH the conditions used were the same as those used in the analysis of the divalent metals, except in the composition of the carrier liquid, which in this case was a 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 mixture of formic acid and acetonitrile at pH 2.4. All samples were injected in duplicates to ensure reproducibility. In all cases, molecular masses were calculated according to the reported method.46
5 mixture of formic acid and acetonitrile at pH 2.4. All samples were injected in duplicates to ensure reproducibility. In all cases, molecular masses were calculated according to the reported method.46
3. Results and discussion
3.1. Identification, structural features and evolutionary origin of O. dioica MTs
We conducted an exhaustive survey of MT genes in the O. dioica genome.19,21 Because standard blast strategies39 were unsuccessful for identifying putative MT genes in O. dioica sequence databases, we used low-restrictive tblastn searches using C. robusta and H. curvata MTs as well as Cephalochordate and Vertebrate MTs as query sequences. We manually inspected all the retrieved sequences for Cys-rich ORFs with MT-distinctive Cys arrangements (i.e. CXC, CXXC, CC and CCC) as structural criteria for identification of new putative MT-coding genes. Two O. dioica genomic sequences satisfied the criteria, one in the scaffold 50 and another in the scaffold 16, which we named OdMT1 and OdMT2, respectively. OdMT1 gene spanned 317 nt and its coding region (CDS) was organized in 3 exons of 34 nt, 84 nt and 101 nt (Fig. 1A and Fig. S2, ESI†). OdMT2 gene extended over 1359 nt, and its CDS was organized in three small exons (exons 1, 3 and 4) of 67 nt, 84 nt and 107 nt, respectively, and one large exon (exon 2) of 942 nt (Fig. 1A and Fig. S2, ESI†).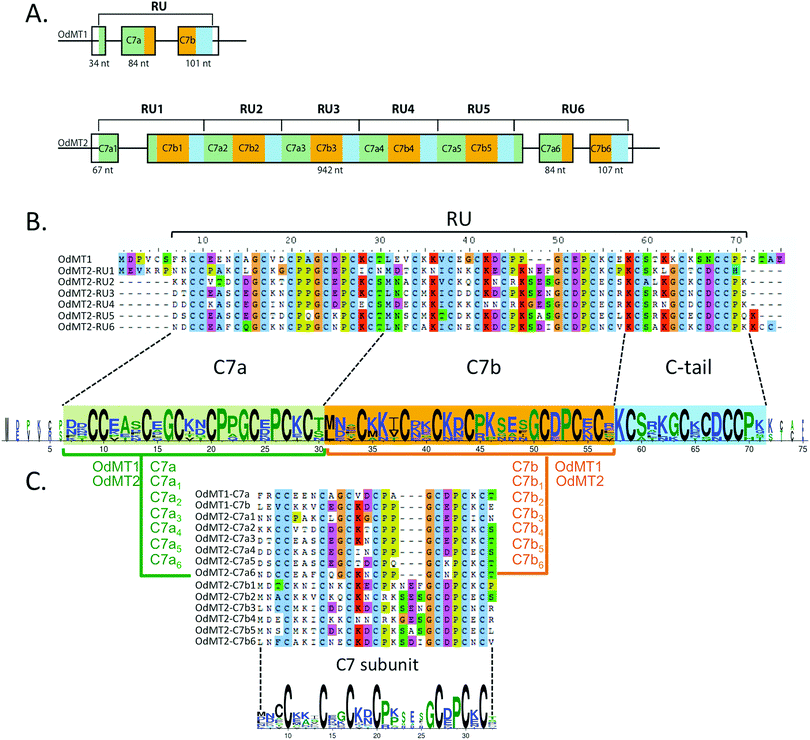 | ||
| Fig. 1 O. dioica metallothioneins. (A) Schematic representation of the exon/intron structure of OdMT1 and OdMT2 genes. OdMT1 is split in three exons and encodes for a protein made of a single repeat unit (RU) with an internal repeat of a C7 subunit – C7a (green box) and C7b (orange box) – followed by a C-terminal tail (blue box). OdMT2 is split in four exons and encodes for a protein made of 6 RU (RU1 to RU6) with the same C7a/C7b/C-tail structure. (B) Amino acid alignment of OdMT1 and RU1–RU6 of OdMT2, and graphical representation using WebLogo 378 highlighting the 20 conserved Cys in each RU. Each RU contains a repetition of a C7 subunit (C7a in green background, and C7b in orange background) followed by a C-terminal tail (in blue background). (C) Comparison of C7a and C7b defines the C7 subunit as a 24–27 amino acid sequence with 7 Cys (X3CX3CX2CX2CX3–6CX2CXCX). In the WebLogo, the overall height of the stack indicates the conservation degree at a given position, while the height of symbols within the stack indicates the relative frequency of each amino at that position. Color code represents amino acid hydrophobicity scale: hydrophobic amino acids (YVMCLFIW) in black, neutral (SGHTAP) in green, and hydrophilic (RKDENQ) in blue. | ||
OdMT1 encoded a 72 amino acid protein with 20 Cys (28%), and OdMT2, in contrast, encoded a much larger protein, with 399 amino acids containing 123 Cys (31%), being to our knowledge the longest MT so far described in any living being. A detail comparison of OdMT1 and OdMT2 sequences revealed that OdMT2 was made of 6 direct tandem repeat units (RU) of approximately 65 amino acids, each RU resembling to OdMT1 (Fig. 1A and B and Fig. S2, ESI†). Moreover, close inspection revealed that each RU was made of an internal repeat of 24–27 amino acids with 7 conserved Cys (C7a and C7b subunits: X2CX3CX2CX2CX3–6CX2CXCX; Fig. 1B and C) followed by a C-terminal tail of 14 amino acids with 5 additional Cys residues. The fact that OdMT1 corresponded to a single RU whereas OdMT2 had 6 RUs suggested that the latter might have been originated by duplication of an ancestral OdMT1 gene with one RU followed by successive tandem duplications of the primeval RU. These results indicated a modular and step-wise evolution of OdMTs, in which the unit of expansion was a RU made of two C7 subunits, likely related with functional and structural constraints that would link the number of RUs and C7 subunits to the metal-binding capacity of OdMTs. Interestingly, a modular structure based on different C7 subunits had been found in several long fungal MTs, that is, in Cryptococcus neoformans CnMT1 and CnMT247 and in Tremella mesenterica TmMT,48 although it had never been reported in multicellular eukaryotes, and never to the level reached by O. dioica MTs. Tandem amplifications of C7 subunits yielding MTs with high metal-binding capacity had been associated with biological adaptations, as virulence factors in the pathogenic fungus C. neoformans,47 or for Cu-providing to lignin-metabolizing enzymes in saprophytic fungus living in decaying woods as T. mesenterica.48 The selective advantage that MTs with high metal-binding capacity might provide to a marine animal as O. dioica is still unknown, but it has been suggested that the evolution of multi-domain MTs in some animal species (e.g. Littorina littorea and Pomatias elegans with 3-domain MTs49–51Crassostrea virginica with 4-domain MTs,52 and Alinda biplicata with 10-domain MTs53) might have improved their response to metal stress and adverse environmental conditions, offering a selective advantage to organisms that experience a greater exposure to metals due to their ecological niche.50,52
3.2. Promoter prediction analysis
Metals, stress and hormones regulate transcription of MT genes (reviewed in ref. 10). In animals, MT expression can be activated in response to metals by MRE-binding transcription factor 1 (MTF-1) that recognizes Metal Responsive Elements (MRE) present in MT promoters.54 MRE have been experimentally and bioinformatically identified in proximal MT promoters of most metazoans, from insects and mollusks to fish and mammals55–61 (reviewed in ref. 62). We surveyed for MRE in 1 kb upstream sequence from the predicted CDS of both OdMT1 and OdMT2 genes using MatInspector 8.4.1 program but, unexpectedly, we did not find any putative MRE sequence in their promoter regions. In agreement with the absence of MRE sequences, a genome survey for a O. dioica homolog of the MTF-1 gene was also unsuccessful, making thereby unlikely that these elements – MRE sequences and MTF-1 proteins – contributed to the transcriptional regulation of MT genes in this species. In contrast, bioinformatic analysis predicted the presence MREF-binding sequences (ACE1/AMT1) in the promoters of OdMT1 and OdMT2 genes (Fig. 2A and Fig. S2, ESI†), which are known to confer metal response to fungal metallothioneins CUP1 and CRS5 genes. Binding of metal regulatory elements factors (MREF) such as CUP2 (a.k.a ACE1 or AMT1 transcription factor) to ACE1/AMT1 sequences regulates CUP1 and CRS5 in response to copper levels,63,64 as well as they regulate fungal copper–zinc superoxide dismutase (Cu/ZnSOD1) genes.65 Although CUP2 orthologous have not been identified outside fungal species, ACE1/AMT1 sequences have been related to transcriptional regulation of Cu/ZnSOD1 genes in maize66 and Arabidopsis.67 Therefore, we also analyzed the promoters (1 kb upstream sequence from the CDS) of the five SOD1 genes predicted in O. dioica genome, and interestingly, we identified MREF (ACE1/AMT1) sequences in four of them (Fig. 2A), suggesting that as described in fungi, MT and SOD genes shared a common transcriptional response in O. dioica. | ||
| Fig. 2 Schematic representation of putative binding sites for transcription factors in the regulatory regions (1.0 kb upstream of CDS) of O. dioica genes. (A) MREF binding sites (orange boxes) were predicted in the OdMT1 and OdMT2 promoters as well as in the promoters of four out of five OdSOD1 genes. (B) In addition of the MREF elements, several stress-response elements were predicted in OdMT1 and OdMT2 promoters, including those for bZIP transcription factors such as YAP1 (violet boxes) and CREB (purple boxes), for RXR/PPAR (blue boxes) and for animals or yeast HSF (green and yellow boxes, respectively). A box above the lines indicates that the binding site was in the plus DNA strand, whereas a box below indicates that it was in the minus strand (see Fig. S2 and Table S3 for additional details, ESI†). | ||
Because MTs are activated in many stress circumstances, we extended our promoter analyses to 33 families of transcription factors with members involved in stress response in animals, plants and fungi (Table S2, ESI†). Interestingly, in addition to MRFE sequences, we discovered that OdMT1 and OdMT2 promoters shared binding sites for many stress response elements (Table S3, ESI†). First, OdMT promoters included binding sites for the family of fungal YAP1, most of them overlapping with sites for vertebrate (CREB) bZIP transcription factors (Fig. 2B, Fig. S2 and Table S3, ESI†). Fungal YAP1 transcription factor belongs to the bZIP (basic leucine zipper factors) family, and it is required for cadmium and oxidative stress tolerance in yeast.68 Vertebrate bZIP transcription factors of the CREB family (e.g. CREB, ATF-1, XBP1, E4BP4) might act also as activators of vertebrate MT genes,69 and CRE binding sites have been identified in the promoter of earthworm MT genes.70,71 Second, it has also been reported that transcription factors of the nuclear receptor family such as the steroid receptors, retinoid receptors and orphan receptors might be involved either directly (e.g. GR;72 PPAR56,57) or indirectly (e.g. RXR73) in the transcriptional regulation of MT expression. Our analysis of OdMT1 and OdMT2 genes revealed several binding sites for nuclear receptors, mainly for RXR/PPAR, in the promoter region (Fig. 2B, Fig. S2 and Table S3, ESI†). Third, it has been demonstrated that heat shock stress also induces MT expression through the binding of HSF to the heat shock elements (HSE),74,75 which appear remarkably abundant in some MT promoters.57 Our computational analysis revealed a preponderant abundance of putative HSF binding sites in the OdMT1 and OdMT2 promoters (Fig. 2B, Fig. S2 and Table S3, ESI†).
Overall, the promoter region of OdMT1 and OdMT2 genes did not appear to contain MRE sequences, which are a common feature of metazoans MT genes, but they had a MREF conserved motif, which is typical of fungal MT genes.63,64 The presence of a MREF in the OdMT promoters was shared by the promoters of O. dioica Cu/ZnSOD1 genes, and although the functionality of these sites needs to be experimentally demonstrated, they suggested a coordinated transcriptional activation of both (MT and SOD) stress-activated genes. OdMT promoters appeared, indeed, rich in stress-responsive elements, including binding sites for YAP1/CREB, RXR/PPAR and HSF transcription factors, envisaging a role of MT genes in adverse environmental conditions.
3.3. Metal-binding capacity of OdMT1 and OdMT2
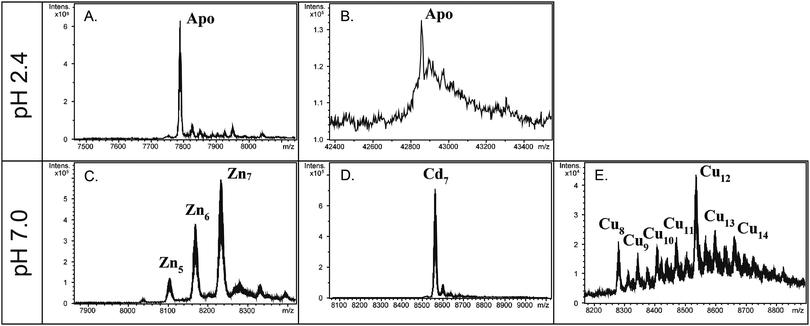
In order to verify the MT nature of the two O. dioica MTs, and to evaluate their metal-binding affinity and capacity, we studied the formation of metal–MT complexes of OdMT1 and OdMT2 proteins heterologously expressed in E. coli and grown in medium supplemented with cooper, cadmium or zinc salts. Metal–OdMT1 and –OdMT2 complexes were purified and analyzed by ICP-AES and ESI-MS. Acidification of the Zn–OdMT complexes yielded the corresponding apo-forms, with molecular masses of 7789 Da for OdMT1 and 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 Da for OdMT2 (Fig. 3A and B), fully concordant with the calculated average theoretical values for the synthesized products (7789.10 Da and 42860.48 Da, respectively; notice that recombinant proteins had two additional amino acids at N-terminus). This confirmed both the identity and purity of the recombinant proteins.
861 Da for OdMT2 (Fig. 3A and B), fully concordant with the calculated average theoretical values for the synthesized products (7789.10 Da and 42860.48 Da, respectively; notice that recombinant proteins had two additional amino acids at N-terminus). This confirmed both the identity and purity of the recombinant proteins.
The ICP-AES analyses showed that both recombinant OdMT1 and OdMT2 proteins could form metal complexes, principally with cadmium ions (9.7 and 55.0 Cd/protein ratios for OdMT1 and OdMT2, respectively; Table 1). ESI-MS analysis of the metal–OdMT1 species formed allowed us to observe the formation of a variety of species in the productions in Zn- and Cu-enriched media: from Zn4- to Zn7–OdMT1 (Fig. 3C), and from Cu8– to Cu14–OdMT1 (Fig. 3E), respectively. In contrast, the production in Cd-enriched media rendered a single Cd7–OdMT1 species, indicative of the formation of a highly favored species (Fig. 3D). Both ICP-AES and ESI-MS results indicated, therefore, that OdMT1 exhibits a clear preference for coordinating Cd2+. In agreement with the Cd preference, the yield of recombinant OdMT1 produced in Cd-media was higher than in Zn- or Cu-enriched media (Table 1) because metal binding might be contributing to stabilize recombinantly expressed protein. Regarding OdMT2, the high molecular weight of OdMT2 and its complex structure made of repeated modules may be responsible for the low concentration of the samples recovered from the metal–OdMT2 productions, even in the presence of Cd, impairing to obtain valid ESI-MS data of the metallated species. However, ICP data (Table 1), together with the fact that the yield of OdMT2 production was higher in Cd-enriched media than in Zn- and Cu-media (Table 1 and Fig. S3, ESI†) indicated a preference for Cd2+, likewise it has been demonstrated for OdMT1. In fact, OdMT2 seems to bind up to 6 more times higher amounts of Cd than OdMT1 (Table 1) accordingly with its high Cys content.
4. Conclusions
In this work, we have identified two MT genes in the chordate O. dioica revealing a particular modular structure of the encoded proteins based on repeat units of two C7 subunits. The structure of these chordate MTs together with data from fungal47,48 and protozoan MTs76,77 suggest that during metallothionein evolution, diverse living beings with different MT sequences have used a similar modular and step-wise mechanism for generating long proteins with high metal-binding capacities. In addition, sequence analyses have revealed the presence of metal- and stress-response elements in the regulatory regions of both genes, while ICP-AES analyses have shown the capacity of both O. dioica proteins to form metal–MT complexes, with preference for cadmium ions. These results support the MT nature of the O. dioica genes and proteins, and suggest that they might play a role in the physiological response of this animal against metal toxicity and other stress situations. Overall our data pave the way for better understanding the evolution and function of the detoxification mechanisms of zooplankton species, which might be crucial to counteract the increasing amounts of heavy metals in marine ecosystems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
In memory of Silvia Atrian, for so many years sharing ideas, projects and meetings in the Departament de Genètica of the Universitat de Barcelona. Her love for metallothioneins inspired this work and prompted us to look for new metallothionein in our favorite animal model. We miss her knowledge and advices, always accurate and opportune. The authors are grateful to Mercè Capdevila for her support and scientific advice, and for the opportunity to contribute to this special issue. We also thank all team members of the C. C. and R. A. laboratories for fruitful scientific discussions. We thank Josep Martí-Solans for the Oikopleura dioica picture. C. C. was supported by BFU2016-80601-P grant; O. P. was supported by BIO2015-67358-C2-2-P grant; and R. A. was supported by BIO2015-67358-C2-1-P grant from Ministerio de Economía y Competitividad (Spain). C. C. and R. A. were also supported by grant SGR2017-1665 from Generalitat de Catalunya. M. G.-R. acknowledges to the UAB the PIF grant. We thank the Centres Científics i Tecnològics (CCiT) de la Universitat de Barcelona (DNA sequencing) and the Servei d’Anàlisi Química (SAQ) de la Universitat Autònoma de Barcelona (ICP-AES, CD, UV-vis, ESI-MS) for allocating instrument time.References
- M. Margoshes and B. L. Vallee, A cadmium protein from equine kidney cortex, J. Am. Chem. Soc., 1957, 79, 4813–4814 CrossRef CAS.
- M. Capdevila and S. Atrian, Metallothionein protein evolution: a miniassay, JBIC, J. Biol. Inorg. Chem., 2011, 16, 977–989 CrossRef CAS PubMed.
- J. Calvo, H. Jung and G. Meloni, Copper metallothioneins, IUBMB Life, 2017, 69, 236–245 CrossRef CAS PubMed.
- N. Franchi, F. Boldrin, L. Ballarin and E. Piccinni, CiMT-1, an unusual chordate metallothionein gene in Ciona intestinalis genome: structure and expression studies, J. Exp. Zool., Part A, 2011, 315, 90–100 CrossRef CAS PubMed.
- M. Guirola, S. Perez-Rafael, M. Capdevila, O. Palacios and S. Atrian, Metal dealing at the origin of the Chordata phylum: the metallothionein system and metal overload response in amphioxus, PLoS One, 2012, 7, e43299 CrossRef CAS PubMed.
- M. Capdevila, R. Bofill, Ò. Palacios and S. Atrian, State-of-the-art of metallothioneins at the beginning of the 21st century, Coord. Chem. Rev., 2012, 256, 46–62 CrossRef CAS.
- J. S. Scheller, G. W. Irvine and M. J. Stillman, Unravelling the mechanistic details of metal binding to mammalian metallothioneins from stoichiometric, kinetic, and binding affinity data, Dalton Trans., 2018, 47, 3613–3637 RSC.
- R. Bofill, M. Capdevila and S. Atrian, Independent metal-binding features of recombinant metallothioneins convergently draw a step gradation between Zn- and Cu-thioneins, Metallomics, 2009, 1, 229–234 RSC.
- O. Palacios, S. Atrian and M. Capdevila, Zn- and Cu-thioneins: a functional classification for metallothioneins?, JBIC, J. Biol. Inorg. Chem., 2011, 16, 991–1009 CrossRef CAS PubMed.
- S. R. Davis and R. J. Cousins, Metallothionein expression in animals: a physiological perspective on function, J. Nutr., 2000, 130, 1085–1088 CrossRef CAS PubMed.
- J. O. Nriagu and J. M. Pacyna, Quantitative assessment of worldwide contamination of air, water and soils by trace metals, Nature, 1988, 333, 134–139 CrossRef CAS PubMed.
- S. Cheng, Heavy metal pollution in China: origin, pattern and control, Environ. Sci. Pollut. Res., 2003, 10, 192–198 CrossRef CAS PubMed.
- G. W. Bryan, The effects of heavy metals (other than mercury) on marine and estuarine organisms, Proc. R. Soc. London, Ser. B, 1971, 177, 389–410 CrossRef CAS.
- G. Gorsky and R. Fenaux, in The Biology of Pelagic Tunicates, ed. Q. Bone, Oxford University Press, Oxford, 1998 Search PubMed.
- P. Davoll and M. Youngbluth, Heterotrophic activity on appendicularian (Tunicata: Appendicularia) houses in mesopelagic regions and their potential contribution to particle flux, Deep-Sea Res., 1990, 37, 285–294 CrossRef.
- B. H. Robison, K. R. Reisenbichler and R. E. Sherlock, Giant larvacean houses: rapid carbon transport to the deep sea floor, Science, 2005, 308, 1609–1611 CrossRef CAS PubMed.
- K. Katija, R. E. Sherlock, A. D. Sherman and B. H. Robison, New technology reveals the role of giant larvaceans in oceanic carbon cycling, Sci. Adv., 2017, 3, e1602374 CrossRef PubMed.
- H.-C. Seo, M. Kube, R. B. Edvardsen, M. F. Jensen, A. Beck, E. Spriet, G. Gorsky, E. M. Thompson, H. Lehrach, R. Reinhardt and D. Chourrout, Miniature genome in the marine chordate Oikopleura dioica, Science, 2001, 294, 2506 CrossRef CAS PubMed.
- F. Denoeud, S. Henriet, S. Mungpakdee, J. M. Aury, C. Da Silva, H. Brinkmann, J. Mikhaleva, L. C. Olsen, C. Jubin, C. Cañestro, J. M. Bouquet, G. Danks, J. Poulain, C. Campsteijn, M. Adamski, I. Cross, F. Yadetie, M. Muffato, A. Louis, S. Butcher, G. Tsagkogeorga, A. Konrad, S. Singh, M. F. Jensen, E. H. Cong, H. Eikeseth-Otteraa, B. Noel, V. Anthouard, B. M. Porcel, R. Kachouri-Lafond, A. Nishino, M. Ugolini, P. Chourrout, H. Nishida, R. Aasland, S. Huzurbazar, E. Westhof, F. Delsuc, H. Lehrach, R. Reinhardt, J. Weissenbach, S. W. Roy, F. Artiguenave, J. H. Postlethwait, J. R. Manak, E. M. Thompson, O. Jaillon, L. Du Pasquier, P. Boudinot, D. A. Liberles, J. N. Volff, H. Philippe, B. Lenhard, H. Roest Crollius, P. Wincker and D. Chourrout, Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate, Science, 2010, 330, 1381–1385 CrossRef CAS PubMed.
- J. Marti-Solans, O. V. Belyaeva, N. P. Torres-Aguila, N. Y. Kedishvili, R. Albalat and C. Canestro, Coelimination and Survival in Gene Network Evolution: Dismantling the RA-Signaling in a Chordate, Mol. Biol. Evol., 2016, 33, 2401–2416 CrossRef CAS PubMed.
- G. Danks, C. Campsteijn, M. Parida, S. Butcher, H. Doddapaneni, B. Fu, R. Petrin, R. Metpally, B. Lenhard, P. Wincker, D. Chourrout, E. M. Thompson and J. R. Manak, OikoBase: a genomics and developmental transcriptomics resource for the urochordate Oikopleura dioica, Nucleic Acids Res., 2013, 41, D845–853 CrossRef CAS PubMed.
- K. Wang, T. Omotezako, K. Kishi, H. Nishida and T. A. Onuma, Maternal and zygotic transcriptomes in the appendicularian, Oikopleura dioica: novel protein-encoding genes, intra-species sequence variations, and trans-spliced RNA leader, Dev. Genes Evol., 2015, 225, 149–159 CrossRef CAS PubMed.
- S. Bassham, PhD, University of Oregon, 2002.
- H. C. Seo, R. B. Edvardsen, A. D. Maeland, M. Bjordal, M. F. Jensen, A. Hansen, M. Flaat, J. Weissenbach, H. Lehrach, P. Wincker, R. Reinhardt and D. Chourrout, Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica, Nature, 2004, 431, 67–71 CrossRef CAS PubMed.
- S. Bassham and J. H. Postlethwait, The evolutionary history of placodes: a molecular genetic investigation of the larvacean urochordate Oikopleura dioica, Development, 2005, 132, 4259–4272 CrossRef CAS PubMed.
- C. Cañestro, S. Bassham and J. H. Postlethwait, Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain, Dev. Biol., 2005, 285, 298–315 CrossRef PubMed.
- A. Nishino, Y. Satou, M. Morisawa and N. Satoh, Brachyury (T) gene expression and notochord development in Oikopleura longicauda (Appendicularia, Urochordata), Dev. Genes Evol., 2001, 211, 219–231 CrossRef CAS PubMed.
- R. B. Edvardsen, H. C. Seo, M. F. Jensen, A. Mialon, J. Mikhaleva, M. Bjordal, J. Cartry, R. Reinhardt, J. Weissenbach, P. Wincker and D. Chourrout, Remodelling of the homeobox gene complement in the tunicate Oikopleura dioica, Curr. Biol., 2005, 15, R12–R13 CrossRef CAS PubMed.
- S. Fujii, T. Nishio and H. Nishida, Cleavage pattern, gastrulation, and neurulation in the appendicularian, Oikopleura dioica, Dev. Genes Evol., 2008, 218, 69–79 CrossRef PubMed.
- C. Cañestro, S. Bassham and J. H. Postlethwait, Evolution of the thyroid: anterior–posterior regionalization of the Oikopleura endostyle revealed by Otx, Pax2/5/8, and Hox1 expression, Dev. Dyn., 2008, 237, 1490–1499 CrossRef PubMed.
- C. Troedsson, J.-M. Bouquet, C. Lobon, A. Novac, J. Nejstgaard, S. Dupont, S. Bosak, H. Jakobsen, N. Romanova, L. Pankoke, A. Isla, J. R. Dutz, A. Sazhin and E. Thompson, Effects of ocean acidification, temperature and nutrient regimes on the appendicularian Oikopleura dioica: a mesocosm study, Mar. Biol., 2013, 160, 2175–2187 Search PubMed.
- F. Yadetie, S. Butcher, H. E. Forde, C. Campsteijn, J. M. Bouquet, O. A. Karlsen, F. Denoeud, R. Metpally, E. M. Thompson, J. R. Manak, A. Goksoyr and D. Chourrout, Conservation and divergence of chemical defense system in the tunicate Oikopleura dioica revealed by genome wide response to two xenobiotics, BMC Genomics, 2012, 13, 55 CrossRef CAS PubMed.
- J. M. Bouquet, C. Troedsson, A. Novac, M. Reeve, A. K. Lechtenborger, W. Massart, K. S. Skaar, A. Aasjord, S. Dupont and E. M. Thompson, Increased fitness of a key appendicularian zooplankton species under warmer, acidified seawater conditions, PLoS One, 2018, 13, e0190625 CrossRef PubMed.
- N. P. Torres-Águila, J. Martí-Solans, A. Ferrández-Roldán, A. Almazán, V. Roncalli, S. D’Aniello, G. Romano, A. Palumbo, R. Albalat and C. Cañestro, Diatom bloom-derived biotoxins cause aberrant development and gene expression in the appendicularian chordate Oikopleura dioica, Commun. Biol., 2018, 1, 121 CrossRef PubMed.
- J. M. Bouquet, E. Spriet, C. Troedsson, H. Ottera, D. Chourrout and E. M. Thompson, Culture optimization for the emergent zooplanktonic model organism Oikopleura dioica, J. Plankton Res., 2009, 31, 359–370 CrossRef PubMed.
- H. Nishida, Development of the appendicularian Oikopleura dioica: culture, genome, and cell lineages, Dev., Growth Differ., 2008, 50(Suppl 1), S239–S256 CrossRef CAS PubMed.
- J. Marti-Solans, A. Ferrandez-Roldan, H. Godoy-Marin, J. Badia-Ramentol, N. P. Torres-Aguila, A. Rodriguez-Mari, J. M. Bouquet, D. Chourrout, E. M. Thompson, R. Albalat and C. Canestro, Oikopleura dioica culturing made easy: a low-cost facility for an emerging animal model in EvoDevo, Genesis, 2015, 53, 183–193 CrossRef PubMed.
- R. Albalat and C. Canestro, Evolution by gene loss, Nat. Rev. Genet., 2016, 17, 379–391 CrossRef CAS PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS PubMed.
- K. Cartharius, K. Frech, K. Grote, B. Klocke, M. Haltmeier, A. Klingenhoff, M. Frisch, M. Bayerlein and T. Werner, MatInspector and beyond: promoter analysis based on transcription factor binding sites, Bioinformatics, 2005, 21, 2933–2942 CrossRef CAS PubMed.
- D. G. Gibson, Enzymatic assembly of overlapping DNA fragments, Methods Enzymol., 2011, 498, 349–361 CAS.
- N. A. Shevchuk, A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland and S. Ladisch, Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously, Nucleic Acids Res., 2004, 32, e19 CrossRef PubMed.
- A. S. Xiong, Q. H. Yao, R. H. Peng, X. Li, H. Q. Fan, Z. M. Cheng and Y. Li, A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences, Nucleic Acids Res., 2004, 32, e98 CrossRef PubMed.
- A. S. Xiong, Q. H. Yao, R. H. Peng, H. Duan, X. Li, H. Q. Fan, Z. M. Cheng and Y. Li, PCR-based accurate synthesis of long DNA sequences, Nat. Protoc., 2006, 1, 791–797 CrossRef CAS PubMed.
- J. Bongers, C. D. Walton, D. E. Richardson and J. U. Bell, Micromolar protein concentrations and metalloprotein stoichiometries obtained by inductively coupled plasma atomic emission spectrometric determination of sulfur, Anal. Chem., 1988, 60, 2683–2686 CrossRef CAS PubMed.
- D. Fabris, J. Zaia, Y. Hathout and C. Fenselau, Retention of Thiol Protons in Two Classes of Protein Zinc Ion Coordination Centers, J. Am. Chem. Soc., 1996, 118, 12242–12243 CrossRef CAS.
- O. Palacios, A. Espart, J. Espin, C. Ding, D. J. Thiele, S. Atrian and M. Capdevila, Full characterization of the Cu-, Zn-, and Cd-binding properties of CnMT1 and CnMT2, two metallothioneins of the pathogenic fungus Cryptococcus neoformans acting as virulence factors, Metallomics, 2014, 6, 279–291 RSC.
- P. Iturbe-Espinoza, S. Gil-Moreno, W. Lin, S. Calatayud, O. Palacios, M. Capdevila and S. Atrian, The Fungus Tremella mesenterica Encodes the Longest Metallothionein Currently Known: Gene, Protein and Metal Binding Characterization, PLoS One, 2016, 11, e0148651 CrossRef PubMed.
- O. Palacios, E. Jimenez-Marti, M. Niederwanger, S. Gil-Moreno, O. Zerbe, S. Atrian, R. Dallinger and M. Capdevila, Analysis of Metal-Binding Features of the Wild Type and Two Domain-Truncated Mutant Variants of Littorina littorea Metallothionein Reveals Its Cd-Specific Character, Int. J. Mol. Sci., 2017, 18, 1452 CrossRef PubMed.
- C. Baumann, A. Beil, S. Jurt, M. Niederwanger, O. Palacios, M. Capdevila, S. Atrian, R. Dallinger and O. Zerbe, Structural Adaptation of a Protein to Increased Metal Stress: NMR Structure of a Marine Snail Metallothionein with an Additional Domain, Angew. Chem., Int. Ed., 2017, 56, 4617–4622 CrossRef CAS PubMed.
- L. Schmielau, M. Dvorak, M. Niederwanger, N. Dobieszewski, V. Pedrini-Martha, P. Ladurner, J. R. Pedregal, J. D. Marechal and R. Dallinger, Differential response to Cadmium exposure by expression of a two and a three-domain metallothionein isoform in the land winkle Pomatias elegans: valuating the marine heritage of a land snail, Sci. Total Environ., 2018, 648, 561–571 CrossRef PubMed.
- M. J. Jenny, S. L. Payton, D. A. Baltzegar and J. D. Lozier, Phylogenetic Analysis of Molluscan Metallothioneins: Evolutionary Insight from Crassostrea virginica, J. Mol. Evol., 2016, 83, 110–125 CrossRef CAS PubMed.
- R. Dallinger, Gastropod metallothioneins in evolution: new rules for an old protein family, Universität Innsbruck, 2018 Search PubMed.
- R. Heuchel, F. Radtke, O. Georgiev, G. Stark, M. Aguet and W. Schaffner, The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression, EMBO J., 1994, 13, 2870–2875 CrossRef CAS PubMed.
- P. Silar, L. Theodore, R. Mokdad, N. E. Erraiss, A. Cadic and M. Wegnez, Metallothionein Mto gene of Drosophila melanogaster: structure and regulation, J. Mol. Biol., 1990, 215, 217–224 CrossRef CAS PubMed.
- M. Egg, M. Hockner, A. Brandstatter, D. Schuler and R. Dallinger, Structural and bioinformatic analysis of the Roman snail Cd-Metallothionein gene uncovers molecular adaptation towards plasticity in coping with multifarious environmental stress, Mol. Ecol., 2009, 18, 2426–2443 CrossRef CAS PubMed.
- M. Niederwanger, M. Dvorak, R. Schnegg, V. Pedrini-Martha, K. Bacher, M. Bidoli and R. Dallinger, Challenging the Metallothionein (MT) Gene of Biomphalaria glabrata: Unexpected Response Patterns Due to Cadmium Exposure and Temperature Stress, Int. J. Mol. Sci., 2017, 18, 1747 CrossRef PubMed.
- M. Zafarullah, K. Bonham and L. Gedamu, Structure of the rainbow trout metallothionein B gene and characterization of its metal-responsive region, Mol. Cell. Biol., 1988, 8, 4469–4476 CrossRef CAS PubMed.
- A. Auf der Maur, T. Belser, G. Elgar, O. Georgiev and W. Schaffner, Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response, Biol. Chem., 1999, 380, 175–185 CAS.
- F. Radtke, M. Hug, O. Georgiev, K. Matsuo and W. Schaffner, Differential sensitivity of zinc finger transcription factors MTF-1, Sp1 and Krox-20 to CpG methylation of their binding sites, Biol. Chem. Hoppe-Seyler, 1996, 377, 47–56 CAS.
- G. Westin and W. Schaffner, A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene, EMBO J., 1988, 7, 3763–3770 CrossRef CAS PubMed.
- D. P. Giedroc, X. Chen and J. L. Apuy, Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation, Antioxid. Redox Signaling, 2001, 3, 577–596 CrossRef CAS PubMed.
- D. J. Thiele, ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene, Mol. Cell. Biol., 1988, 8, 2745–2752 CrossRef CAS PubMed.
- P. Zhou, M. S. Szczypka, T. Sosinowski and D. J. Thiele, Expression of a yeast metallothionein gene family is activated by a single metalloregulatory transcription factor, Mol. Cell. Biol., 1992, 12, 3766–3775 CrossRef CAS PubMed.
- E. B. Gralla, D. J. Thiele, P. Silar and J. S. Valentine, ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 8558–8562 CrossRef CAS.
- S. M. Ruzsa and J. G. Scandalios, Altered Cu metabolism and differential transcription of Cu/ZnSod genes in a Cu/ZnSOD-deficient mutant of maize: evidence for a Cu-responsive transcription factor, Biochemistry, 2003, 42, 1508–1516 CrossRef CAS PubMed.
- J. Xu, Y. S. Tian, R. H. Peng, A. S. Xiong, B. Zhu, X. F. Jin, J. J. Gao, X. L. Hou and Q. H. Yao, Yeast copper-dependent transcription factor ACE1 enhanced copper stress tolerance in Arabidopsis, BMB Rep., 2009, 42, 752–757 CrossRef CAS PubMed.
- A. Wu, J. A. Wemmie, N. P. Edgington, M. Goebl, J. L. Guevara and W. S. Moye-Rowley, Yeast bZip proteins mediate pleiotropic drug and metal resistance, J. Biol. Chem., 1993, 268, 18850–18858 CAS.
- R. K. Uddin and S. M. Singh, Ethanol-responsive genes: identification of transcription factors and their role in metabolomics, Pharmacogenomics J., 2007, 7, 38–47 CrossRef CAS PubMed.
- M. Hockner, R. Dallinger and S. R. Sturzenbaum, Metallothionein gene activation in the earthworm (Lumbricus rubellus), Biochem. Biophys. Res. Commun., 2015, 460, 537–542 CrossRef CAS PubMed.
- V. Drechsel, K. Schauer, M. Srut and M. Hockner, Regulatory Plasticity of Earthworm wMT-2 Gene Expression, Int. J. Mol. Sci., 2017, 18, 1113 CrossRef PubMed.
- M. Karin, A. Haslinger, H. Holtgreve, R. I. Richards, P. Krauter, H. M. Westphal and M. Beato, Characterization of DNA sequences through which cadmium and glucocorticoid hormones induce human metallothionein-IIA gene, Nature, 1984, 308, 513–519 CrossRef CAS PubMed.
- P. Martinez-Paz, M. Morales, P. Sanchez-Arguello, G. Morcillo and J. L. Martinez-Guitarte, Cadmium in vivo exposure alters stress response and endocrine-related genes in the freshwater snail Physa acuta. New biomarker genes in a new model organism, Environ. Pollut., 2017, 220, 1488–1497 CrossRef CAS PubMed.
- K. T. Tamai, X. Liu, P. Silar, T. Sosinowski and D. J. Thiele, Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways, Mol. Cell. Biol., 1994, 14, 8155–8165 CrossRef CAS PubMed.
- W. M. Yang, W. Gahl and D. Hamer, Role of heat shock transcription factor in yeast metallothionein gene expression, Mol. Cell. Biol., 1991, 11, 3676–3681 CrossRef CAS PubMed.
- S. Diaz, F. Amaro, D. Rico, V. Campos, L. Benitez, A. Martin-Gonzalez, E. P. Hamilton, E. Orias and J. C. Gutierrez, Tetrahymena metallothioneins fall into two discrete subfamilies, PLoS One, 2007, 2, e291 CrossRef PubMed.
- A. Espart, M. Marin, S. Gil-Moreno, O. Palacios, F. Amaro, A. Martin-Gonzalez, J. C. Gutierrez, M. Capdevila and S. Atrian, Hints for metal-preference protein sequence determinants: different metal binding features of the five tetrahymena thermophila metallothioneins, Int. J. Biol. Sci., 2015, 11, 456–471 CrossRef CAS PubMed.
- G. E. Crooks, G. Hon, J. M. Chandonia and S. E. Brenner, WebLogo: a sequence logo generator, Genome Res., 2004, 14, 1188–1190 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8mt00177d |
| This journal is © The Royal Society of Chemistry 2018 |