Protein networks in the maturation of human iron–sulfur proteins
Simone
Ciofi-Baffoni
 *ab,
Veronica
Nasta
*ab,
Veronica
Nasta
 ab and
Lucia
Banci
ab and
Lucia
Banci
 *ab
*ab
aMagnetic Resonance Center-CERM, University of Florence, Via Luigi Sacconi 6, 50019, Sesto Fiorentino, Florence, Italy. E-mail: ciofi@cerm.unifi.it; banci@cerm.unifi.it; Fax: +39 055 4574923; Tel: +39 055 4574192 Tel: +39 055 4574273
bDepartment of Chemistry, University of Florence, Via della Lastruccia 3, 50019 Sesto Fiorentino, Florence, Italy
First published on 24th November 2017
Abstract
The biogenesis of iron–sulfur (Fe–S) proteins in humans is a multistage process occurring in different cellular compartments. The mitochondrial iron–sulfur cluster (ISC) assembly machinery composed of at least 17 proteins assembles mitochondrial Fe–S proteins. A cytosolic iron–sulfur assembly (CIA) machinery composed of at least 13 proteins has been more recently identified and shown to be responsible for the Fe–S cluster incorporation into cytosolic and nuclear Fe–S proteins. Cytosolic and nuclear Fe–S protein maturation requires not only the CIA machinery, but also the components of the mitochondrial ISC assembly machinery. An ISC export machinery, composed of a protein transporter located in the mitochondrial inner membrane, has been proposed to act in mediating the export process of a still unknown component that is required for the CIA machinery. Several functional and molecular aspects of the protein networks operative in the three machineries are still largely obscure. This Review focuses on the Fe–S protein maturation processes in humans with the specific aim of providing a molecular picture of the currently known protein–protein interaction networks. The human ISC and CIA machineries are presented, and the ISC export machinery is discussed with respect to possible molecules being the substrates of the mitochondrial protein transporter.
Introduction
Metal ions are essential for life, with more than one third of proteins encoded by the human genome depending on metals for their function.1–3 Nature has built several pathways responsible for metal handling and metalloprotein maturation in human cells. A large portion of metalloproteins themselves in the human proteome are involved in pathways dedicated to maintaining the delicate balance between metal uptake and usage, as essential nutrients, and the excretion of excess metals thus preventing any potential damage.4–6 Altered or lacking regulation of these cellular processes can lead to cell toxicity and death.7–9 Despite the extensive progress in the area of “metals in biology”, we still have only a partial understanding of: (i) how metalloproteins acquire the correct metals/metal cofactors, (ii) how cells establish and maintain the required metal levels, (iii) how the maturation of a metalloprotein occurs, and (iv) how these three aspects intersect each other and with other cellular processes. A molecular understanding of the cellular pathways related to all these processes needs to be thoroughly investigated through the identification and description of the individual components, of their modes of interaction to form networks, and of their interlinks among other cellular pathways. This knowledge provides an inescapable step to obtain a complete and realistic description of the cellular processes related to metals, and is particularly relevant because several human diseases, including a number of neurodegenerative diseases, cardiovascular disorders and certain types of cancer, have been associated with the malfunction of metal-based cellular mechanisms.10–14 A molecular description of the pathways responsible for obtaining the mature, active forms of metalloproteins thus represents an essential step in understanding the origin of the related human diseases and in developing effective treatments.Extensive studies in the past few decades have addressed, at a molecular level, the cellular pathways related to copper homeostasis and copper enzyme maturation in human cells.5,15–20 Those studies focused on the investigation of the structural and dynamic properties of proteins involved in copper transport, of their modes of interaction and of the factors driving copper transfer between protein partners along cellular routes.21,22 All these studies contributed to a reliable picture of the copper molecular systems biology in humans. Iron handling in human cells is by far much more complex than that of copper. The human iron proteome is, indeed, larger than that of copper, reflecting its extensive usage in human physiological processes,23–25 and, as a further degree of complexity with respect to copper, iron is physiologically present in various forms, i.e. either as individual metal ions or as bound to cofactors, mainly hemes and iron–sulfur (Fe–S) clusters. The latter are inorganic cofactors that bind to protein ligands, which, in the majority of the cases, are cysteines. Common Fe–S clusters include a rhomboid cluster composed of two iron ions and two inorganic sulfide ions ([2Fe–2S]),26 and a cubane composed of four iron and four sulfide ions ([4Fe–4S]).27 The [2Fe–2S] cluster can function as the building block to form [4Fe–4S] clusters. The capability of two rhombic [2Fe–2S] clusters to fuse to form a cubane [4Fe–4S] cluster has been, indeed, documented in vitro28 and in vivo.29 Much larger and more complex Fe–S clusters are found in enzymes such as hydrogenase and nitrogenase, and often also contain other metals, such as molybdenum.30 The biosynthesis of such complex Fe–S clusters requires specific maturation machinery for their proper synthesis and insertion into the enzyme.31
Fe–S clusters are present in all three kingdoms of life. The most common function of Fe–S clusters is based on their ability to accept or donate single electrons to carry out complex enzymatic reactions. This feature differentiates them from non-metal-containing prosthetic groups, such as NAD+, FMN or FAD, which are less versatile and are limited to accepting electron pairs once involved in a redox enzymatic reaction cycle. Moreover, the fact that electron transfer can occur without undergoing energetically costly protein reorganization32 favours Fe–S proteins to be responsible for fast electron transfer processes between different metal centres in large protein complexes. For example, in complex I of the mitochondrial respiratory chain, eight Fe–S clusters are positioned close to each other in a quasi-linear arrangement throughout the protein section that extends into the mitochondrial matrix.33 The increasing reduction potentials along the eight Fe–S clusters drive the electron transfer direction from NADH, a strong electron donor in the mitochondrial matrix, to ubiquinone, an electron acceptor and a carrier in the inner mitochondrial membrane.
The function of Fe–S clusters is not limited to electron transfer. Another well-characterized function of Fe–S clusters is enzymatic catalysis; a classic example being aconitase, in which a non-protein-coordinated Fe at one edge of a [4Fe–4S] cluster serves as a Lewis acid to assist H2O abstraction from citrate (the substrate), which is converted to isocitrate. Other well-known Fe–S enzymes are biotin synthase and lipoate synthase, which bind two Fe–S clusters each. One of these clusters is disassembled during the formation of the products biotin and lipoic acid, respectively, thus serving as a sulfur donor.34 Numerous other catalytic functions are known for eukaryotic Fe–S enzymes involved in metabolism.35 However, in many cases, the precise role of the Fe–S cluster is still unclear, and it is therefore possible that in some proteins the Fe–S cluster simply plays a structural role.
A third general role of Fe–S clusters is in sensing environmental or intracellular conditions to regulate gene expression.36 The best characterized example in mammals is provided by the cytosolic iron regulatory protein 1 (IRP1, also known as ACO1). Under iron-replete conditions, IRP1 holds a [4Fe–4S] cluster and functions as an aconitase. When the protein loses its labile cluster under iron deprivation, it can bind to stem–loop structures (termed iron-responsive elements (IREs)) in certain messenger RNAs of proteins involved in iron uptake, storage and distribution in the cell.37 Binding of apo-IRP1 to 5′-located IREs blocks translation by inhibiting ribosome scanning to the start AUG codon, whereas the association with 3′-located IREs protects the mRNAs from nucleolytic degradation, leading to increased translation.
A sequence-based bioinformatic approach38 has been applied to the human genome to identify the Fe–S proteome. This search mapped 70 unique genes (i.e. 0.35% of the human genes), with 61% of Fe–S proteins binding [4Fe–4S] clusters, 39% of them binding [2Fe–2S] clusters and only one (i.e. succinate dehydrogenase) binding a [3Fe–4S] cluster.39 Experimental verification of their ability to bind an iron–sulfur cofactor is available for most of these proteins either directly or by similarity to some close homologue. From this analysis it also emerges that the number of mitochondrial Fe–S proteins that bind a [2Fe–2S] cluster is very close to that of those binding the [4Fe–4S] cofactor.39 Instead, there is a higher number of [4Fe–4S] binding proteins in the cytoplasm, with the ratio of the [4Fe–4S]/[2Fe–2S] cluster-binding proteins being close to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Nuclear Fe–S proteins are almost exclusively of the [4Fe–4S] type, with the above ratio nearly 5
1. Nuclear Fe–S proteins are almost exclusively of the [4Fe–4S] type, with the above ratio nearly 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.39 Finally, the distribution of the identified human Fe–S proteins within cell compartments shows that the mitochondrial proteins are inherited from prokaryotic proteins of aerobes, whereas cytosolic and nuclear Fe–S proteins are inherited from anaerobic organisms.40
1.39 Finally, the distribution of the identified human Fe–S proteins within cell compartments shows that the mitochondrial proteins are inherited from prokaryotic proteins of aerobes, whereas cytosolic and nuclear Fe–S proteins are inherited from anaerobic organisms.40
Initially, researchers thought that the Fe–S clusters assemble spontaneously in vivo, given the fact that chemists are able to synthesize and to incorporate into proteins various Fe–S clusters in vitro using elemental iron and sulfur components as building blocks.28 However, the discovery of a bacterial operon (nitrogen fixation (nif)) that contains genes associated with the synthesis of the Fe–S clusters of nitrogenase in the bacterial plant symbiont Azotobacter vinelandii41 revolutionized the studies on cellular Fe–S cluster biogenesis. In bacterial DNA, the genes that function in a pathway are often grouped together in an operon that facilitates the co-expression of functionally related proteins. This is the case of the nif operon of A. vinelandii and of two related operons of Escherichia coli, the iron–sulfur cluster (isc) and the sulfur formation (suf) operons.26,42 In bacteria, the Fe–S protein biogenesis machineries were thus straightforwardly identified and showed how Fe–S clusters are synthesized and then provided to client proteins in a single cellular compartment, i.e. the cytoplasm. In mammalian cells, the synthesis and distribution of Fe–S clusters are more complex. Mitochondria are the major sites containing Fe–S clusters; the twelve Fe–S clusters of the respiratory chain complexes I–III and the Fe–S cluster of aconitase, a component of the citric acid cycle, are synthetized and incorporated into their respective mitochondrial proteins by the iron–sulfur cluster (ISC) assembly machinery.43,44 More recently, several cytosolic and nuclear proteins have been recognized as Fe–S proteins, including DNA primases, polymerases, and glycosylases, involved in base excision repair, and helicases, involved in the maintenance of genome stability.45,46 A cytosolic iron–sulfur assembly (CIA) machinery has been identified and shown to be responsible for the Fe–S cluster incorporation into cytosolic and nuclear Fe–S proteins.47 From the studies of yeast and human cells,48–51 it has been shown that their maturation, however, requires not only the CIA machinery, but also components of the mitochondrial ISC assembly machinery. From this finding, it was proposed that a sulfur-containing compound of still unknown identity (X–S) is exported from the mitochondria to the cytoplasm for use in the cytosolic and nuclear compartments of eukaryotic cells.52–54 An ISC export machinery has been proposed to act in mediating the export process of the X–S compound.51 The export mechanism is based on a protein transporter located in the mitochondrial inner membrane, which has been shown to have a key role for the maturation of the cytosolic and nuclear proteins.48 However, whether the mitochondrial and cytosolic Fe–S cluster assembly pathways are truly connected in mammals is still debated, and the paradigm that Fe–S clusters are assembled solely in the mitochondrial matrix does not necessarily extrapolate to mammalian cells.55,56 Indeed, a small amount of extra-mitochondrial isoforms of several components of the ISC assembly machinery responsible for assembling Fe–S clusters have been identified,57–61 and there is functional evidence that one of these components in its cytosolic isoform functions as a scaffold and a source for extra-mitochondrial Fe–S clusters in mammalian cells.59
This Review focuses on the Fe–S protein maturation processes in humans with the specific aim of providing a molecular picture of the currently known protein–protein interaction networks. The human ISC and CIA machineries are presented, and the ISC export machinery is discussed with respect to possible molecules being substrates of the mitochondrial protein transporter.
The mitochondrial iron–sulfur cluster (ISC) assembly machinery
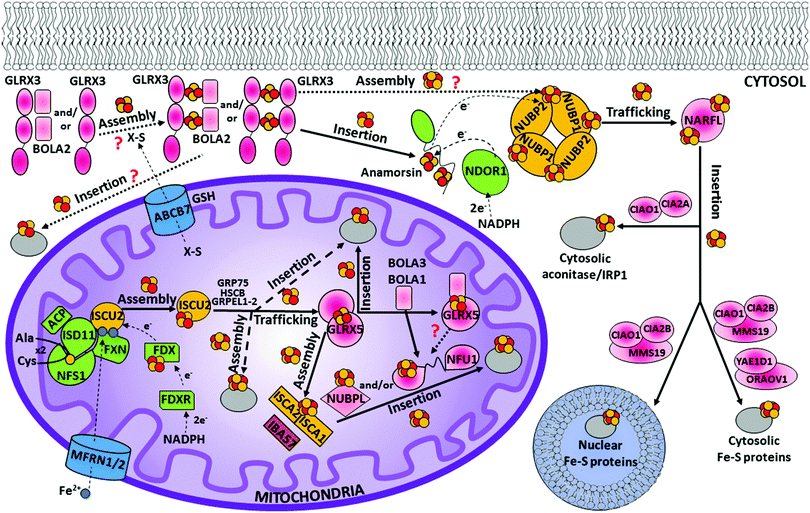
The mitochondrial ISC assembly machinery was inherited from endosymbiotic bacteria during evolution and is highly conserved in all eukaryotes.62,63 It is the crucial cellular component required for the maturation of mitochondrial and cytosolic/nuclear Fe–S proteins.51 In humans, this process encompasses 17 known proteins (Table 1), but the way in which they cooperate is still not well understood. The overall process can be dissected into three major functional steps: (i) the assembly of an initial [2Fe–2S] cluster on a scaffold protein, ISCU2; (ii) a chaperone-assisted [2Fe–2S] cluster transfer from ISCU2 to glutaredoxin-5 (GLRX5), which works as a [2Fe–2S] chaperone transferring the [2Fe–2S] clusters to several possible acceptors; and (iii) the assembly of a [4Fe–4S] cluster followed by its target-selective insertion into mitochondrial [4Fe–4S] target proteins.51,64,65| Short name | Full name | Functional cofactors | Proposed main function |
|---|---|---|---|
| ISC assembly factors involved in de novo Fe–S cluster synthesis | |||
| ISCU2 | Iron–sulfur cluster assembly enzyme 2 | [2Fe–2S] | Scaffold protein |
| NFS1 | Cysteine desulfurase | PLP | Sulfur donor |
| ISD11 | Iron–sulfur protein biogenesis desulfurase-interacting protein 11 | — | Stabilizer of NFS1 |
| ACP | Acyl carrier protein | S-dodecanoyl-4′-PPT | Metabolic sensor |
| FXN | Frataxin | Fe2+ | Iron donor, regulator of NFS1 |
| FDX | Ferredoxin | [2Fe–2S] | Electron transfer |
| FDXR | Ferredoxin reductase | FAD | Electron transfer |
| ISC factors involved in cluster transfer | |||
| GRP75 | 75 kDa glucose-regulated protein | ATP | Fe–S cluster transfer |
| HSCB | Iron–sulfur cluster co-chaperone protein | — | Fe–S cluster transfer |
| GRPEL1/2 | GrpE protein homolog 1/2 | — | Nucleotide exchange |
| GLRX5 | Glutaredoxin-5 | [2Fe–2S], GSH | Fe–S cluster transfer |
| Late-acting ISC factors involved in assembly and the insertion of Fe–S clusters into the target proteins | |||
| ISCA1 | Iron–sulfur cluster assembly 1 homolog | [2Fe–2S], [4Fe–4S] | [4Fe–4S] cluster assembly |
| ISCA2 | Iron–sulfur cluster assembly 2 homolog | [2Fe–2S], [4Fe–4S] | [4Fe–4S] cluster assembly |
| IBA57 | Iron–sulfur cluster assembly factor | — | [4Fe–4S] cluster assembly |
| NFU1 | Iron–sulfur cluster scaffold | [4Fe–4S] | Dedicated ISC targeting factor |
| BOLA3 | BolA-like protein 3 | — | Dedicated ISC targeting factor |
| NUBPL | Nucleotide-binding protein-like | [4Fe–4S] | Dedicated ISC targeting factor |
Assembly of a [2Fe–2S] cluster on the scaffold protein ISCU2
In humans, seven proteins act in the first step of the mitochondrial ISC assembly machinery (Table 1), five of them (ISCU2, NFS1, ISD11, ACP and FXN) forming a stable complex devoted to forming a [2Fe–2S]2+ cluster by assembling two Fe2+ ions and two sulfide ions (Fig. 1). Ferrous ions are imported into the mitochondrion by the intermembrane transporters mitoferrin-1 and mitoferrin-2 (MFRN1/2) to form a bioavailable iron pool in the mitochondrial matrix (Fig. 1), in which ferrous ions are believed to be bound to the still unidentified ligands.66 A recent approach based on liquid chromatography coupled with inductively coupled plasma mass spectrometry was used to analyse non-proteinaceous low-molecular-mass metal complexes in mitochondria.67 It was found that three iron complexes with masses of 580, 1100 and ∼1500 Da constitute the mitochondrial labile iron pool in mammals.68 This iron pool is used by the scaffold protein ISCU2 to assemble a [2Fe–2S] cluster (Fig. 1).69 It has been proposed that frataxin is responsible of supplying the iron to ISCU2, but concrete in vivo evidence of frataxin supplying iron ions is still lacking.70 It was also shown that the role of frataxin can go well beyond that of an iron transporter by working as a regulator of Fe–S cluster assembly.71 The origin of the two sulfide ions required to build a [2Fe–2S] cluster is instead clearer. They are indeed provided by the conversion of two cysteines into alanines. This process is performed by a sub-complex formed by the cysteine desulfurase NFS1, ISD11 and ACP proteins (Fig. 1) and requires four electrons. Assuming that two electrons are supplied by the oxidation of two ferrous ions,72 two additional electrons from an external source are required to make a [2Fe–2S]2+ cluster. They are provided by an electron donor chain formed by two other components acting in this first step, FDX and FDXR (Fig. 1).The sub-complex formed by NFS1, ISD11 and ACP proteins is a symmetric dimer in solution (Fig. 2),73 which is responsible for the conversion of a cysteine into alanine per each NFS1 subunit. The sulfur atom released from the cysteine is transiently bound as a persulfide group on a conserved cysteine residue of NFS1.74,75 NFS1 is formed by two domains, the larger one having a PLP binding site.73 NFS1 is inactive on its own and needs further proteins for its functioning, i.e. ISD11 and ACP.57,76,77 ISD11, also known as LYRM4, belongs to the LYR protein family characterized by a Leu-Tyr-Arg (LYR) motif near the N-terminus. It has been shown that the LYR motif and the N-terminus are essential for the role of ISD11 in the formation of a stable and active complex with NFS1.78 ISD11 is a small α-helical protein with three helix-forming regions (Fig. 2);73 and it has been demonstrated that it stabilizes NFS1 by preventing its self-aggregation through hydrophobic interactions.76 Recently, the mitochondrial acyl carrier protein (ACP) has been identified as part of this sub-complex, i.e. in the absence of ACP, the sub-complex is destabilized resulting in a profound depletion of Fe–S clusters throughout the cell.77,79,80 This role of ACP depends upon its covalently bound S-dodecanoyl-4′-phosphopantetheine (4′-PPT) to support maximal cysteine desulfurase activity.77 Crystallographic and EM structures of the co-expressed human NFS1-ISD11 and the E. coli ACP (ACPec 44% identical to the human mitochondrial ACP) dimeric complex, also called the SDAec complex, have been solved73 (Fig. 2). They are different from any previously determined prokaryotic cysteine desulfurase structures, identifying the bases for the differences in prokaryotic and eukaryotic cysteine desulfurase mechanisms. The SDAec complex shows an unprecedented architecture in which a pair of ISD11 subunits forms the dimeric core of the SDAec complex, explaining the critical role of ISD11 in eukaryotic assemblies. The structure also reveals that the 4′-PPT-conjugated acyl-group of ACP occupies a hydrophobic pocket of ISD11, explaining the basis of ACP stabilization (Fig. 2).73
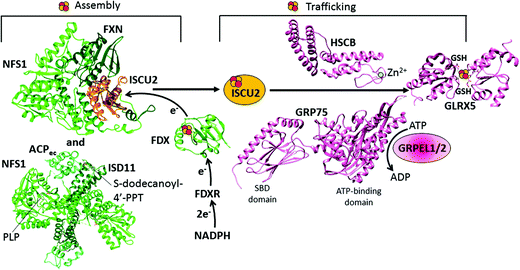 | ||
| Fig. 2 Structural overview of the early steps of the human mitochondrial iron–sulfur cluster (ISC) assembly machinery. The [2Fe–2S]2+ cluster biosynthesis is performed by a protein complex composed of NFS1, ISD11, ACP, FXN and ISCU2. The structure of this complex has been solved in pieces: the crystal structure of the [NFS1 ISD11 ACP] complex (PDB ID 5USR) and the cryo-EM structure of the [NFS1 FXN ISCU2] complex (PDB ID 5KZ5). For the electron transfer chain composed of FDX and FDXR, the X-ray structure of FDX is available (PDB ID 2Y5C). For the subsequent step consisting of the chaperone-mediated transfer of the assembled [2Fe–2S]2+ cluster from ISCU2 to GLRX5, structural information are available on some single players only: the crystal structures of the Zn-bound HSCB protein (PDB ID 3BVO), of the SBD (PDB ID 3N8E) and ATPase domains (PDB ID 4KBO) of GRP75, and of the homodimeric [2Fe–2S]2+ GLRX5 (PDB ID 2WUL). | ||
The [NFS1·ISD11·ACP] sub-complex can bind ISCU2 in the presence or absence of FXN.81,82 Binding of FXN to the sub-complex accelerates desulfurase activity81 and persulfide formation on NFS1, and is also thought to induce a conformational change in ISCU2 that enhances the transfer of sulfur from NFS1 to Cys 138 of ISCU2, the primary persulfide acceptor of ISCU2.83–85 The mechanism of sulfur transfer from the persulfide group on the conserved cysteine residue of NFS1 to the Fe–S cluster assembly site of ISCU2 involves the flexible loop of NFS1 containing the invariant cysteine residue.81,86,87 Ferrous ions further stimulate the cysteine desulfurase activity.81
The human ISCU protein has two different isoforms, ISCU1 and ISCU2, with different subcellular locations. It has been found by immunofluorescence microscopy that ISCU1 is localized to the cytosol and nucleus, whereas ISCU2 is localized to the mitochondria, being part of the ISC assembly machinery.88 Also human frataxin has two different isoforms, FXN42–210 and FXN81–210, with the 81–210 isoform being the most abundant species in normal individuals.89 Both are generated and localized within the mitochondria,89 and participate in the mitochondrial Fe–S cluster assembly using different modes of interaction with NFS1, ISD11 and ISCU2 as well as different iron binding capacities.90 These isoforms are generated through the sequential cleavage of the FXN protein precursor (FXN 1-210) by the mitochondrial processing peptidase upon import of FXN 1-210 to the mitochondrial matrix.91–93 As determined by size-exclusion chromatography of normal human cell extracts, the native state of FXN42–210 is a monomer–oligomer equilibrium, whereas the native state of FXN81–210 is monomeric.94 Complexes containing oligomeric FXN42–210, NFS1, ISD11 and ISCU2 could be isolated upon fractionation of normal human cell extracts by size-exclusion chromatography followed by co-immunoprecipitation,94–96 underscoring the stability of these complexes. In contrast, native FXN81–210 was only recovered in fractions that contained no detectable NFS1 and only traces of ISCU2. However, a [FXN81–210·NFS1·ISD11·ISCU2] complex has been isolated from bacterial cells upon the co-expression of all four components of the complex, and it could also be reconstituted in vitro from the purified [NFS1·ISD11·ISCU2] complex and the FXN81–210 monomer. However, in both cases, a large excess of monomeric FXN81–210 was required for complex formation,82,97 which may explain why this complex could not be detected under native conditions in human cells.94,95 On the basis of differentiated iron binding abilities of the two FXN isoforms under aerobic conditions,81,95,97 it has been proposed that, under physiological conditions, FXN81–210 may bind low levels of iron and support basal levels of Fe–S cluster assembly via transient interactions with the [NFS1·ISD11·ISCU2] complex. While the stable [FXN42–210·NFS1·ISD11·ISCU2] complex may provide a mechanism to increase the rate of Fe–S cluster assembly when the demand exceeds the low iron-binding capacity of FXN81–210.90
Recently, isolation, biochemical and structural characterization of the human Fe–S cluster assembly complex containing most of the proteins of the first step of the mitochondrial ISC assembly machinery (ISCU2, NFS1, ISD11 and FXN42–210) has been reported (Fig. 2).98 The structural data provided a first model of a coordinated mechanism for the transfer of iron and sulfur to ISCU2 for the synthesis of a [2Fe–2S]2+ cluster. It was shown that these four human proteins, once co-expressed in E. coli cell extracts, form a stable, active complex with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. This complex may also contribute to explain how human frataxin may serve as both the activator of NFS1 and the iron donor, an argument still largely debated in the literature.51,99 Indeed, in the context of the human [FXN81–210·NFS1·ISD11·ISCU2] complex having the shorter FXN isoform,97 one monomer of FXN81–210 is thought to bind in a pocket between NFS1 and ISCU2 through the conserved frataxin iron-binding surface,97,100 in such a way to block the ability of FXN81–210 to bind iron and thus contrasting with its role in iron delivery. In contrast, the location of ISCU2 relative to FXN42–210 and NFS1 in the human [FXN42–210·NFS1·ISD11·ISCU2] structure allows FXN42–210 and NFS1 to simultaneously bind to ISCU2 and to stimulate Fe–S cluster synthesis, consistent with the previous observation that ISCU2 and [NFS1·ISD11] can independently or simultaneously bind to oligomeric FXN42–210 in solution.101
1 stoichiometry. This complex may also contribute to explain how human frataxin may serve as both the activator of NFS1 and the iron donor, an argument still largely debated in the literature.51,99 Indeed, in the context of the human [FXN81–210·NFS1·ISD11·ISCU2] complex having the shorter FXN isoform,97 one monomer of FXN81–210 is thought to bind in a pocket between NFS1 and ISCU2 through the conserved frataxin iron-binding surface,97,100 in such a way to block the ability of FXN81–210 to bind iron and thus contrasting with its role in iron delivery. In contrast, the location of ISCU2 relative to FXN42–210 and NFS1 in the human [FXN42–210·NFS1·ISD11·ISCU2] structure allows FXN42–210 and NFS1 to simultaneously bind to ISCU2 and to stimulate Fe–S cluster synthesis, consistent with the previous observation that ISCU2 and [NFS1·ISD11] can independently or simultaneously bind to oligomeric FXN42–210 in solution.101
The electron transfer chain formed by ferredoxin (FDX) and ferredoxin reductase (FDXR), which receives an electron from NADH, accomplishes the task of providing the two electrons required to form a [2Fe–2S]2+ cluster on ISCU2 (Fig. 1).80 In human mitochondria, there are two different isoforms of ferredoxins, FDX1 and FDX2. Their X-ray structures are very similar even if the two ferredoxins share only 33% protein sequence identity (Fig. 2).80 By NMR spectroscopy, it has been demonstrated that both FDX1 and FDX2 interact directly with the [NFS1 ISD11 ACP] sub-complex and that their reduced forms, when added to a solution of the complex and L-cysteine, become oxidized, indicating that both ferredoxins are able to donate electrons leading to the reduction of the sulfur atom. Despite the fact that both FDX isoforms interact with the [NFS1 ISD11 ACP] sub-complex, FDX2 exhibits both a higher binding affinity for the complex and a much higher efficiency in assisting the Fe–S cluster assembly. Functional data indicated that only FDX2 is part of the electron transport chain required for de novo mitochondrial Fe–S protein biogenesis, while FDX1 is essentially devoted to the steroid metabolism in the adrenal gland.102 In contrast to this view, the RNA interference-mediated depletion of either FDX1 or FDX2 was reported to impair Fe–S protein maturation.103 An in vitro reconstitution system containing the yeast ISC members of the first step of the ISC assembly machinery showed that the yeast orthologue of FDX2, Yah1, in a reduced but not oxidized state tightly interacts with apo-Isu1 indicating a dynamic interaction between Yah1-apo-Isu1.104 Nuclear magnetic resonance structural studies identify the Yah1-apo-Isu1 interaction surface and suggest a pathway for the electron flow from reduced Yah1 to Isu1.104
A chaperone-assisted [2Fe–2S] cluster transfer from ISCU2 to glutaredoxin-5
In the second step, the de novo assembled [2Fe–2S]2+ cluster is released from the scaffold protein ISCU2 to the monothiol glutaredoxin GLRX5. This event is mediated by a dedicated chaperone system comprising the GRP75 chaperone (also named mortalin, HSPA9 or PBP74), which is a member of the ATP-dependent 70-kDa heat shock protein family Hsp70, its co-chaperone HSCB (also named HSC20), and the nucleotide exchange factors GRPEL1/2 (Table 1 and Fig. 1).105 The actual molecular model is based on the initial binding of the HSCB protein to [2Fe–2S] ISCU2, which then forms a complex with the chaperone GRP75.105–107 NMR data indicated that a conformational change of ISCU2 occurs upon HSCB binding, which specifically weakens the [2Fe–2S] ISCU2 interaction in the [FXN·ACP·NFS1·ISD11·ISCU2] complex, and strengthens its interaction with the co-chaperone HSCB, which targets the [2Fe–2S] [ISCU2 HSCB] complex to the chaperone GRP75.108 The C-terminal α-helical domain of the HSCB co-chaperone (Fig. 2) is directly responsible for binding ISCU2, with three highly conserved non-contiguous hydrophobic residues being of crucial importance for the HSCB–ISCU2 interaction.44,109–112 The N-terminal domain of HSCB (Fig. 2), which contains an invariant histidine, proline, and aspartate motif, is responsible for stimulating the ATPase activity of the GRP75 chaperone. Overall, the HSCB co-chaperone might have a dual function in stimulating the ATPase activity of the chaperone GRP75 and in promoting its recognition between ISCU2 and the chaperone GRP75. The N-terminus of the human HSCB co-chaperone shows distinctive features compared to the homologous co-chaperone proteins of bacteria and fungi (DnaJ type III), as it contains, downstream of the mitochondrial targeting sequence (residues 1–26), an additional domain, which harbors two CxxC stretches (C41/C44 and C58/C61) that were found to coordinate a zinc ion in vitro (Fig. 2).112 The physiological relevance of the unique N-terminal domain remains to be elucidated although it may facilitate protein dimerization.As for all Hsp70 family chaperones, GRP75 is composed of two domains, the crystal structures of them being determined: a ∼42 kDa N-terminal ATP-binding domain and a ∼25 kDa substrate-binding domain (SBD) involved in the ISCU2 recognition (Fig. 2).113 The two domains of GRP75 do not interact with each other but are tethered by a short interdomain linker. The sequence of the ATP-binding domain is highly conserved in all Hsp70 family members, while the SBD has several differences among family members. The ADP dissociation step on GRP75 is facilitated by the nucleotide exchange factors GRPEL1/2. Such dissociation enables GRP75 to undergo the next round of Fe–S cluster transfer. The ATP-binding domain of GRP75 is found to interact with the nucleotide exchange factors.114
On the basis of studies on the yeast Grx5 homologue,115–117 and considering that the Grx5 function is conserved throughout the eukaryotic kingdom,118–120 it has been proposed that GLRX5 mediates [2Fe–2S] cluster transfer from the [HSCB GRP75 [2Fe–2S] ISCU2] complex to [2Fe–2S] target proteins and to intermediate proteins responsible for the assembly of [4Fe–4S] clusters (Fig. 1). GLRX5 in solution binds a [2Fe–2S] cluster forming a [2Fe–2S]2+ cluster-bridged homodimer with two glutathione (GSH) molecules involved in cluster binding (Fig. 2).121 Gel filtration and 15N NMR relaxation data on the holo and apo states of GLRX5 showed that cluster binding induces a change in the quaternary structure from an apo-monomer to a holo dimer.121 The dimeric state of [2Fe–2S] GLRX5 prevents the cluster from being released in the presence of physiological concentrations of GSH, suggesting that GLRX5 can specifically transfer the cluster.
Recent studies have shown that the HSCB co-chaperone in the [HSCB GRP75 [2Fe–2S] ISCU2] complex can guide the selection of specific Fe–S recipient proteins for cluster delivery by binding to a conserved LYR motif present in specific proteins.122 In this regard, the best-characterized example is the succinate dehydrogenase subunit b (SDHB), which is the Fe–S cluster-containing subunit of the respiratory complex II.106 This protein contains three independent LYR binding sites, all close to the cysteines that coordinate the three Fe–S clusters of SDHB and in a position where the binding of the chaperone/co-chaperone [2Fe–2S] transfer apparatus can guide the release of the cluster from [2Fe–2S]–ISCU2 into the Fe–S binding sites of SDHB. The LYR motif family is present only in eukaryotes (conserved domains accession: cl05087),123 which lists other proteins related to the mitochondrial Fe–S protein biogenesis in humans, thus potentially being the HSCB-dependent recognition mechanism conserved in other Fe–S target insertion processes. One of the members of the LYR family is SDHAF1 (LYRM8), a succinate dehydrogenase complex assembly factor.124 SDHAF1 was shown to recruit the [HSCB GRP75 ISCU2] complex to the C-terminus of SDHB through direct binding of its N-terminal LYR motif to the HSCB co-chaperone.125 Another LYR protein is LYRM7, a complex III assembly factor,126 which interacts with the [HSCB GRP75 ISCU2] complex,106 suggesting that the LYRM7–HSCB interaction might guide the insertion of the [2Fe–2S] cluster into the Rieske Fe–S protein of complex III. LYR-containing subunits are also present in the mitochondrial complex I, and one of these has been suggested to be required for the proper incorporation of the N2 [4Fe–4S] center and for the function of complex I.123 A second consensus sequence, in addition to the LYR motif, is the KKx6–10KK motif, which has been found to interact with the HSCB co-chaperone.106 GLRX5 has a similar pattern of lysines at its C-terminus (K139K140x10K151K152) and has been found to interact with HSCB in vivo.106 All these findings suggest that the model in which GLRX5 is the central component for distributing [2Fe–2S] clusters to final [2Fe–2S] acceptors and to the proteins involved in the late steps of the mitochondrial ISC assembly machinery for maturing [4Fe–4S] target proteins might have to be revisited. Indeed, GLRX5 could be just one among the LYR and KKx6–10KK targets of the [HSCB GRP75 [2Fe–2S] ISCU2] complex, which, in contrast, could be the real distributor of [2Fe–2S] clusters (Fig. 1). However, there are several pieces of evidence that this new possible model is not able to answer. Among them, how [4Fe–4S] clusters are formed and inserted into the mitochondrial target proteins is fully undefined by this model. It has been shown, indeed, that the mammalian Fe–S assembly complex (i.e. the [FXN·ACP·NFS1·ISD11·ISCU2] complex) is able to synthesize exclusively [2Fe–2S] clusters.127 Therefore, the way that the [4Fe–4S] and [3Fe–4S] clusters of LYR-containing targets of the [HSC20 GRP75 [2Fe–2S] ISCU2] complex (i.e. mitochondrial respiratory complexes I–III) are formed, is still not addressed in this possible alternative model.
Reductive coupling of two [2Fe–2S]2+ clusters to form a [4Fe–4S]2+ cluster on ISCA proteins
Among the GLRX5 protein partners,121,128,129 two A-type Fe–S proteins named ISCA1 (iron–sulfur cluster assembly 1) and ISCA2 (iron–sulfur cluster assembly 2) are required for mitochondrial [4Fe–4S] protein maturation.130,131 ISCA1 and ISCA2 have been first proposed to function in concert with IBA57 to mature [4Fe–4S] proteins.130 However, the molecular relation between IBA57 and ISCA proteins has been recently debated. Indeed, while ISCA1 and ISCA2 are protein partners in vivo, IBA57 has been found to interact in vivo with ISCA2 but not with ISCA1, in contrast to what occurs for the yeast homologues, which both interact with Iba57.132 Surprisingly, it has been recently observed that only ISCA1 is required for the maturation of mitochondrial [4Fe–4S] proteins.128 A possible explanation for the differently observed ISCA2 phenotypes is that different mammalian cells and conditions were used in the two phenotyping experiments,128,130 and thus further in vivo data are required to resolve this matter. In vitro data133,134 support a model where a [ISCA1 ISCA2] complex converts [2Fe–2S]2+ clusters, received by GLRX5, into a [4Fe–4S]2+ cluster, with IBA57 not being required in this process (Fig. 1). In agreement with this model, it has been shown that GLRX5 can transfer a [2Fe–2S] cluster to a heterodimeric complex formed by human ISCA1 and ISCA2 through a cluster-mediated protein–protein interaction event,121,133 and that this complex acts as an assembler of a [4Fe–4S]2+ cluster through a reductive coupling process of two [2Fe–2S]2+–GLRX5 donated clusters (Fig. 3).133 In the latter reaction, dithiothreitol provides the two electrons required for the reductive coupling process. How these electrons are provided in vivo is still an open question. Finally, although ISCA2 alone can also bind a [4Fe–4S] cluster in a dimeric state, it has been observed that the formation of a heterodimeric [ISCA1 ISCA2] complex is thermodynamically favoured compared to the corresponding homodimeric forms, indicating that, at the cellular level, the heterodimeric complex should be the preferential species responsible of assembling [4Fe–4S]2+ clusters. This is in full agreement with the strong, specific interaction between ISCA1 and ISCA2 observed in vivo.128 Recently, a molecular picture of how the [4Fe–4S] cluster has been assembled on the heterodimeric [ISCA1 ISCA2] complex has been provided.134 In this model, the two fully conserved C-terminal cysteines, located in the unstructured and flexible C-terminal tail of the ISCA proteins,133 are required to extract the [2Fe–2S]2+ cluster from GLRX5, while the other fully conserved cysteine of ISCAs, Cys 79, is not essential for the cluster transfer step. This cysteine is required to promote the acquisition of the second [2Fe–2S]2+ cluster from another GLRX5 molecule via C-terminal cysteines of ISCAs, in such a way that the reductive coupling between the two [2Fe–2S]2+ clusters can occur on the [ISCA1 ISCA2] complex. This molecular model agrees with the in vivo data on yeast, which showed that the three conserved cysteines of yeast Isa1 and Isa2 are essential for the maturation of [4Fe–4S] proteins.135,136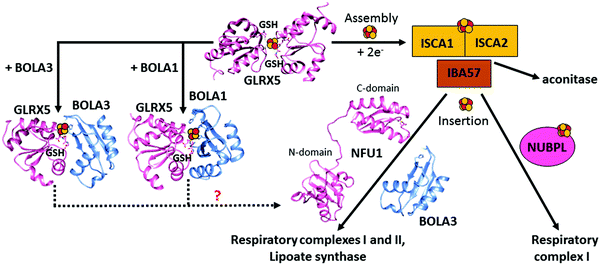 | ||
| Fig. 3 Structural overview of the late steps of the human mitochondrial iron–sulfur cluster (ISC) assembly machinery. In the late steps of the mitochondrial ISC assembly machinery, the [ISCA2 ISCA1] heterodimeric complex assembles a [4Fe–4S]2+ cluster by the reductive coupling of two [2Fe–2S]2+ clusters received by GLRX5.133 No evidence on the functional role of IBA57, which is part of this process, is available, nor is the physiological donor of the two electrons required in the [2Fe–2S] coupling process. NFU1 and NUBPL assist in the maturation of specific [4Fe–4S] target proteins, but how they work mechanistically in such a process is still undefined. [2Fe–2S] GLRX5 by interacting with BOLA1 and BOLA3 can be converted into [[2Fe–2S] BOLA1 GLRX5] and [[2Fe–2S] BOLA3 GLRX5] complexes, respectively, but their potential role in the maturation of specific [4Fe–4S] target proteins is not defined (dotted lines). The solution NMR structures of the C-domain (PDB ID 2M5O) and the N-domain (PDB ID 2LTM) of apo-NFU1, BOLA1 (PDB ID 5LCI) and BOLA3 (PDB ID 2NCL) have been solved. Experimentally-driven docking models of the two heterodimeric [BOLAs GLRX5] complexes are available in ref. 145. | ||
Target-selective insertion of the assembled [4Fe–4S] cluster into mitochondrial [4Fe–4S] target proteins
The [4Fe–4S] cluster assembled on the [ISCA1 ISCA2] complex is inserted into mitochondrial [4Fe–4S] target proteins with the possible assistance of dedicated ISC targeting factors, i.e. NFU1, BOLA3 and NUBPL (also named IND1) (Table 1). Specifically, some mitochondrial [4Fe–4S] target proteins could acquire their [4Fe–4S] cluster directly from the [ISCA1 ISCA2] complex, whereas others may derive their clusters after a prior transfer of the [4Fe–4S] cluster from the [ISCA1 ISCA2] complex to NFU1 and NUBPL. The latter can bind a [4Fe–4S] cluster, which facilitates the insertion of the cluster into specific mitochondrial [4Fe–4S] target proteins (Fig. 1).137–140 In particular, NFU1 acts as a late-acting factor that transfers the [4Fe–4S] clusters to the lipoic acid synthase and to the subunits of respiratory complexes I and II, facilitating their maturation,137,138 while NUBPL works as an assembly factor for the human respiratory complex I only.140 NFU1 was detected as an ISCA1 specific interacting partner only.128 However, in yeast, Nfu1 physically interacts with both Isa1 and Isa2 in the [Isa1 Isa2 Iba57] complex and with mitochondrial target proteins that need [4Fe–4S] clusters to function.141 The yeast data suggest that human NFU1 might specifically bind to the mitochondrial [4Fe–4S] target proteins independent of the [ISCA1 ISCA2] complex. Then, the binding of this complex with the [ISCA1 ISCA2] complex may serve to recruit the NFU1-associated [4Fe–4S] target proteins to the [ISCA1 ISCA2] complex, specifically assisting cluster transfer and insertion into these target proteins. As other possible model, NFU1 was proposed to be an alternative scaffold to ISCU2, thanks to its ability to assemble [4Fe–4S] clusters and transfer them to apo-proteins, thus operating upstream of the [ISCA1 ISCA2] complex.58 Human NFU1 comprises two domains: an N-terminal domain and a C-terminal domain, which contains the CXXC motif involved in [4Fe–4S] cluster binding (Fig. 3).58,142 While apo-NFU1 is largely monomeric in solution, [4Fe–4S] NFU1 consists of a trimer of dimers, with three [4Fe–4S] clusters bound by the two conserved cysteine residues in the CTDs of two NFU1 subunits forming the dimer.142 The functional relevance of the binding of three [4Fe–4S] clusters in a single aggregate is still unknown.The phenotypic similarity between the pathogenic mutations of NFU1 and of another mitochondrial protein named BOLA3 suggests that the two proteins function together in the late steps of the mitochondrial Fe–S protein maturation pathway (Fig. 1).137,138 Using yeast as a model system, it has been proposed that the yeast ortholog of human BOLA3 functions in the late step of transfer of [4Fe–4S] clusters to specific target proteins, together with yeast Nfu1.141 Despite the fact that yeast BolA3 and Nfu1 both act late in the mitochondrial Fe–S protein biogenesis, they appear to fulfil different tasks that cannot be taken over by the other protein.143 Proteomic studies did not detect any stable in vivo interaction between yeast Nfu1 and BolA3, suggesting a transient interaction in nature; in vitro this interaction has been found to have a μM affinity.143 Overall, these results in yeast might suggest that BOLA3 might function with NFU1 in the cluster insertion into specific mitochondrial [4Fe–4S] target proteins (Fig. 1), but this mechanism is still largely hypothetical. BOLA3 belongs to the highly conserved BOLA-like protein family, which also includes the human mitochondrial BOLA1.144 Recently, it has been shown in yeast that the yeast homolog BolA1 also facilitates the insertion of [4Fe–4S] clusters into a subset of mitochondrial target proteins, being unable to replace Nfu1, similarly to what occurs with BolA3 (Fig. 1).143 Even though BolA1 and BolA3 play some overlapping roles, their functions are not identical.143 The solution structures of BOLA3 and BOLA1 showed that they have a similar fold, but with local structural differences in the regions containing conserved sequence patterns comprising cluster ligands.143 It has been demonstrated that both BOLA1 and BOLA3 interact with apo-GLRX5 with Kd values of 3 μM and are able to form [2Fe–2S] cluster-bridged dimeric hetero-complexes with GLRX5.141,143 The [[2Fe–2S] BOLA1 GLRX5] complex is preferentially formed over the [[2Fe–2S] BOLA3 GLRX5] complex, because of a higher cluster-binding affinity.145 Experimentally driven structural models and spectroscopic data of these two heterodimeric holo complexes145 (Fig. 3) showed that: (i) cluster solvent accessibility is higher in the BOLA3 hetero-complex with respect to that in the BOLA1 heterocomplex; (ii) the [BOLA1 GLRX5] complex stabilizes a reduced, Rieske-type [2Fe–2S]1+ cluster, while an oxidized, ferredoxin-like [2Fe–2S]2+ cluster is present in the [BOLA3 GLRX5] complex; (iii) the [2Fe–2S] center in the [BOLA1 GLRX5] complex behaves like a stable redox active center able to perform electron transfer reactions, at variance with what occurs for the [[2Fe–2S] BOLA3 GLRX5] complex. This experimental evidence suggests that the [[2Fe–2S] BOLA1 GLRX5] complex might work in electron transfer reactions, while the [[2Fe–2S] BOLA3 GLRX5] complex might be involved in iron–sulfur cluster transfer processes versus target proteins along the Fe–S protein assembly pathway.145 In conclusion, the molecular scenarios of NFU1 and BOLA3 (alone or complexed with GLRX5) proteins acting late in the ISC pathway are still largely undefined, as well as the role of BOLA1 (alone or complexed with GLRX5) as the possible further ISC late-acting factor is still indefinite.
The other known targeting factor, NUBPL, is another [4Fe–4S] cluster binding protein, specifically required for the respiratory complex I assembly (Fig. 3).140 It is closely related in sequence to two cytosolic P-loop NTPases, NUBP1 and NUBP2, which act as a heteromeric scaffold complex in the synthesis of cytosolic [4Fe–4S] clusters (see later). It contains a highly conserved nucleotide binding domain and a Fe–S binding CPXC motif, which is required for the function of NUBPL and is proposed to provide the ligands for a transiently bound [4Fe–4S] cluster, probably inducing protein dimerization.140
The cytosolic iron–sulfur assembly (CIA) machinery
Up to now, thirteen CIA components participating in the maturation of human cytosolic and nuclear Fe–S proteins have been identified (Table 2).47,146,147 Most of these components are conserved in almost all eukaryotes.39,148 Six of these proteins act in the early stage of the CIA machinery, while all the others are late-acting CIA factors (Table 2).| Short name | Full name | Functional cofactors | Proposed main function |
|---|---|---|---|
| Early-acting CIA factors involved in Fe–S cluster assembly and transfer | |||
| NUBP1 | Nucleotide-binding protein 1 | [4Fe–4S] | Scaffold protein |
| NUBP2 | Nucleotide-binding protein 2 | [4Fe–4S] | Scaffold protein |
| NDOR1 | NADPH-dependent diflavin oxidoreductase 1 | FAD, FMN | Electron transfer |
| Anamorsin | Anamorsin | [2Fe–2S], [4Fe–4S] | Electron transfer |
| GLRX3 | Glutaredoxin-3 | [2Fe–2S], GSH | Fe–S cluster transfer, iron trafficking |
| BOLA2 | BolA-like protein 2 | — | Fe–S cluster transfer |
| Late-acting CIA factors involved in transfer and insertion of Fe–S clusters into the target proteins | |||
| NARFL | Fe–S cluster assembly factor | [4Fe–4S] | Mediates contact between the early and late parts of the CIA machinery |
| CIAO1 | Cytosolic iron–sulfur protein assembly | — | CIA targeting complex |
| CIA2B | Mitotic spindle-associated MMXD complex subunit MIP18 | — | CIA targeting complex |
| MMS19 | Nucleotide excision repair protein homolog | — | CIA targeting complex |
| CIA2A | MIP18 family protein FAM96A | — | Specific maturation factor of IRP1 |
| YAE1D1 | Yae1 domain-containing protein 1 | — | Specific maturation factor of the cytosolic ABCE1 protein |
| ORAOV1 | Oral cancer-overexpressed protein 1 | — | Specific maturation factor of the cytosolic ABCE1 protein |
The assembly of a [4Fe–4S] cluster in the early stage of the CIA machinery
The early stage of the CIA machinery involves two P-loop NTPase proteins, named NUBP1 and NUBP2 in humans (also known as NBP35 and CFD1, respectively). NUBP1 and NUBP2 are nuclear and cytosolic localized proteins containing several cysteine residues at their C- and N-termini.149,150 Sequence alignment of NUBP1 and NUBP2 with their respective eukaryotic homologues identified, in NUBP1, an invariant N-terminal Cx13Cx2Cx5C motif, which is absent in the NUBP2 family, and, in NUBP2, an invariant C-terminal Cx18Cx2Cx2C motif, which is partially conserved or totally absent in the NUBP1 family depending on the organism. In the yeast homologues, each of these two motifs binds a [4Fe–4S] cluster, such that the N-terminal domain is tightly bound, while that in the C-terminus is labile.151–153 No structural models of the cluster bound forms of NUBP1 and NUBP2 or of any eukaryotic homologues are available. This piece of information would be fundamental to address definitively how the two proteins bind the [4Fe–4S] cluster(s). Co-immunoprecipitation shows that the two proteins interact in the cellular context, suggesting that NUBP1 and NUBP2 proteins cooperate in the same molecular process of the CIA machinery.149In vitro and in vivo studies on the yeast homologous heterocomplex suggest the presence of bridging [4Fe–4S] clusters in a heterodimeric or heterotetrameric structural arrangement and that this complex performs a scaffold function by assembling a [4Fe–4S] cluster on the C-terminal motif in the early stage of the CIA machinery (Fig. 4).152,154 This function should be conserved in humans. Indeed, depletion of NUBP1 by RNA interference in HeLa cells resulted in a severe defect in the assembly of cytosolic and nuclear Fe–S proteins without an apparent effect on mitochondrial Fe–S protein maturation.149 However, how this heterocomplex works in the CIA machinery as a scaffold protein is unknown. The donors of Fe2+ and sulfide ions required to assemble a [4Fe–4S] cluster on the heterocomplex are not defined yet, nor are the cluster binding properties of the human heterocomplex well addressed. Since some early ISC components and the membrane transporter of the ISC export machinery are required for the functioning of the CIA machinery in yeast,48,50,155,156 it might be speculated that the sulfide ions for assembling the [4Fe–4S] cluster on the heterocomplex require the mitochondria-exported X–S compound.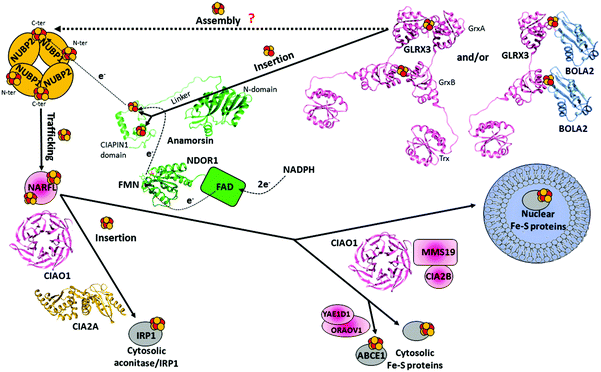 | ||
| Fig. 4 Structural overview of the human cytosolic iron–sulfur assembly (CIA) machinery. The tetrameric [NUBP1 NUBP2] complex might receive electrons from the NADPH-NDOR1-Anamorsin electron transfer chain to assemble a [4Fe–4S]2+ cluster on the C-terminus by reductively coupling two [2Fe–2S]2+ clusters. These might be received by the holo forms of the GLRX3 and/or [BOLA2 GLRX3] complex. The assembled [4Fe–4S]2+ cluster is transferred to NARFL, which then delivers it to cytosolic and nuclear target proteins with the contribution of several other CIA proteins. Three of them, CIAO1, MMS19 and CIA2B, form a complex (CIA targeting complex) assisting the transfer of the [4Fe–4S] cluster from NARFL to the majority of cytosolic and nuclear target proteins. The complex between CIAO1 and CIA2A specifically matures the cytosolic aconitase/IRP1, while a complex between the CIA targeting complex and the [ORAOV1 YAE1D1] complex specifically assists the maturation of the ABCE1 protein. The crystal structures of the single domains of apo-GLRX3 (the Trx domain PDB ID 2WZ9, the GrxA domain PDB ID 3ZYW and the GrxB domain PDB ID 2YAN), of the FMN-binding domain of NDOR1 (PDB ID 4H2D) and of CIAO1 (PDB ID 3FMNO) have been solved. Two different crystal structures of CIA2A are available (PDB ID 3UX2 and 3UX3); only one of them is shown in the figure. A homology model of BOLA2 was generated based on the available solution structure of the BOLA-like protein from Mus musculus (PDB ID 1V9J, sequence identity is 87%). A structural model of the [2Fe–2S] CIAPIN1 domain was calculated using diamagnetic and paramagnetic restraints,166 and the solution structure of the N-terminal domain of Anamorsin (PDB ID 2LD4) has been solved. | ||
The formation of the [4Fe–4S] cluster on the heterocomplex also requires electrons, and NADPH might provide these via the NDOR1 and Anamorsin CIA components (Fig. 1). Similar to the [NUBP1 NUBP2] complex, they operate indeed in the early stage of the CIA pathway by forming a stable complex.157,158 According to this proposal, a relatively weak, but specific interaction between NUBP1 and Anamorsin, compared to the much stronger interaction between NDOR1 and Anamorsin, has been detected from yeast-two-hybrid and surface plasmon resonance experiments in plants.159 Although plants lack the NUBP2 gene, thus not fully representing a fully reliable model for the molecular events in the human CIA machinery, the plant NUBP1 protein has a scaffold function similar to that of the human [NUBP1 NUBP2] complex.160
NDOR1 is a diflavin reductase consisting of two domains: the first binds FMN (FMN-binding domain, hereafter), and the second binds FAD and NADPH (FAD-binding domain, hereafter) (Fig. 4). On the basis of the well-known electron transfer mechanism occurring in diflavin reductase enzymes,161 the electrons are transferred from NADPH to FAD and then to FMN, which serves as a donor for one-electron terminal acceptors, i.e. the Fe–S cluster(s) of Anamorsin in this case. Anamorsin contains two domains: an N-terminal domain of 172 residues and a C-terminal domain of 90 residues, named cytokine-induced apoptosis inhibitor 1 (CIAPIN1 hereafter), containing two highly conserved, cysteine-rich motifs, CX8CX2CXC and CX2CX7CX2C (Fig. 4). An unstructured and flexible linker of 51 residues connects the two domains (Fig. 4). While the N-terminal domain is well-structured, the C-terminal CIAPIN1 domain is largely unstructured (Fig. 4).162 The CIAPIN1 domain of Anamorsin binds a [2Fe–2S] cluster in the first motif and a [2Fe–2S] or [4Fe–4S] cluster on the second motif depending on how the preparation of the protein sample has been conducted in vitro.163–165 To definitively define which is the physiologically relevant cluster bound to the second motif of Anamorsin, an in vivo approach would be required to define the cluster bound state of Anamorsin in the cellular context. The crystal structure of the FMN-binding domain of NDOR1 showed the classical fold of FMN-binding domains of diflavin reductases (Fig. 4).166 The crystal167 and solution162 structures of the N-terminal domain of Anamorsin (Fig. 4) showed an overall structure resembling a typical S-adenosylmethionine (SAM)-dependent methyltransferase fold but lacking the glycine-rich motif, which is typically responsible for the binding of the SAM cofactor in methyltransferases. As a result, Anamorsin does not show SAM-dependent methyltransferase activity.162 On this basis, it has been hypothesized that the N-terminal domain of Anamorsin acts as bait for protein–protein interactions. A structural model of the CIAPIN1 domain shows that the CX8CX2CXC and CX2CX7CX2C regions are connected by a stably formed α-helix and sample a restricted range of conformations that bring the two cluster-binding motifs close to each other (Fig. 4).166 Anamorsin and NDOR1 form a stable complex, thanks to the specific protein–protein recognition between the FMN-binding domain of NDOR1 and the unstructured linker region of Anamorsin separating the N-terminal domain from the C-terminal CIAPIN1 domain (specifically, residues 185–223).166 The N-terminal domain of Anamorsin is not involved in such a recognition process. The two protein partners interact permanently forming also a stable interaction in the cell,157 and no dissociation occurs during the electron transfer process.166 One electron transfer reaction was observed to occur from the hydroquinone state of the FMN moiety of NDOR1 to the oxidized [2Fe–2S]2+ cluster bound to the first motif of Anamorsin (N-terminal [2Fe–2S] cluster), while the Fe–S cluster bound to the second motif of Anamorsin does not receive electrons from FMN.163
The [Anamorsin NDOR1] complex might therefore supply electrons to the [NUBP1 NUBP2] complex, working similarly to the NADPH–FDX–FDXR electron transfer chain in the mitochondrial ISC assembly machinery.104 However, at variance with the ISC assembly machinery, the specific molecular targets of the electron transfer flow generated by the [Anamorsin NDOR1] complex are not identified yet. Depending on the molecular nature of the mitochondria-exported X–S compound, the complex might reduce the sulfur moiety of the X–S compound to sulfide, similar to the function of the NADPH–FDX–FDXR electron transfer chain in the mitochondrial ISC system,155,156 with the function of building a [2Fe–2S] cluster, and/or it could facilitate the fusion of two [2Fe–2S] clusters to a [4Fe–4S] cluster. The yeast homologues of NDOR1 and Anamorsin (named Tah18 and Dre2, respectively) have been shown not to be involved in the de novo formation of a [2Fe–2S] cluster.157 Assuming that the same holds in human cells, we could speculate that the electron transfer chain composed of NADPH–NDOR1–Anamorsin is involved in the fusion of two [2Fe–2S]2+ clusters to a [4Fe–4S]2+ cluster on the [NUBP1 NUBP2] complex. The X–S compound, which is needed for the CIA machinery function, should therefore operate upstream of the [NUBP1 NUBP2] complex, to build a [2Fe–2S] cluster, and therefore an alternative cytosolic pathway for the de novo synthesis of [2Fe–2S] clusters might be required. Some ISC components have been detected in small amounts in the cytosol and in the nucleus, being proposed to function in these compartments to assemble a [2Fe–2S] cluster,55,59 which is then used by the CIA machinery for the maturation of cytosolic and nuclear [4Fe–4S] proteins. However, evidence for a direct function of these extra-mitochondrial ISC proteins in Fe–S protein assembly is still under debate.50,151,168
Cytosolic [2Fe–2S] cluster trafficking by glutaredoxin-3 and BOLA2 protein
Another [2Fe–2S] cluster protein operating in the early stage of the CIA machinery is the cytosolic monothiol glutaredoxin GLRX3 (Fig. 1).169 GLRX3 consists of an N-terminal thioredoxin (Trx) domain that lacks a redox-active motif and two monothiol glutaredoxin (Grx) domains that both harbor a Cys-Gly-Phe-Ser active site.170 Crystal structures of the three single domains (Fig. 4) showed that their overall fold is very similar to that of other known monothiol glutaredoxins/thioredoxins. Upon [2Fe–2S] cluster binding, GLRX3 forms a dimeric complex bridging two [2Fe–2S] clusters between each of the two Grx domains ([2Fe–2S]2 GLRX32, Fig. 4).171,172 Each cluster is shared by the cysteine residue of the active site of each Grx domain and by a GSH molecule per each Grx domain (Fig. 4). Yeast has two homologous proteins of GLRX3, Grx3 and Grx4, which, similar to GLRX3, act in cytosolic/nuclear Fe–S protein maturation.173 Yeast Grx3 and Grx4 contain an N-terminal Trx domain, similar to GLRX3, but only one Grx domain, which binds one [2Fe–2S] cluster in a dimeric complex similarly to what occurs in GLRX3.174 In yeast, the binding/assembly of the [2Fe–2S] cluster in Grx3 requires the mitochondrial ISC assembly and export machineries, yet occurs independent of the two CIA proteins Nbp35 and Dre2.173,174 Conversely, Fe–S cluster binding on both Nar1 and Dre2 strictly depends on Grx3, placing Grx3 early in the CIA pathway. Applying this yeast model to humans, GLRX3 might be the primary target of the X–S compound after its exit from the mitochondria (Fig. 1). This compound might determine the formation of the [2Fe–2S] cluster bound state of GLRX3, which might then act as a cytoplasmic [2Fe–2S] cluster trafficking protein in the early phase of the CIA pathway (Fig. 1), in analogy to the proposed function of mitochondrial GLRX5. Now, the arising question is how GLRX3 is reconstituted with [2Fe–2S] clusters. The crucial point to provide the answer is the identification of the chemical nature of the X–S compound.The first evidence showing that GLRX3 can work as a [2Fe–2S] cluster trafficking protein came from an in vitro study.175 Indeed, it was shown that GLRX3 matures Anamorsin by transferring two [2Fe–2S] clusters to the cysteine-rich motif of the CIAPIN1 domain (Fig. 1). Anamorsin and GLRX3 are protein partners in the cellular context as identified by the yeast-two-hybrid assay.176 It was shown that the transfer mechanism was dependent on the formation of a specific protein–protein complex between the N-terminal domains of GLRX3 and of Anamorsin, with part of the linker of Anamorsin having a further, specific role in stabilizing the interaction.175 Their interaction is the fundamental requisite to observe the transfer of both [2Fe–2S] clusters from Grx domains of GLRX3 to the two cluster-binding motifs of the CIAPIN1 domain of Anamorsin. In conclusion, GLRX3 plays a key role in maturing Anamorsin, specifically working as an intracellular [2Fe–2S] cluster-trafficking protein. This indirectly links GLRX3 to all Anamorsin-dependent cellular processes. In vivo data showed that silencing of human GLRX3 expression in HeLa cells decreased the activities of the cytosolic Fe–S proteins IRP1 and GPAT.169 This behavior can be interpreted as an impairment of the GLRX3-dependent Anamorsin maturation process, making the CIA machinery unable to function, since it lacks the electron transfer chain required to form [4Fe–4S] clusters on the [NUBP1 NUBP2] complex. Similarly, in vivo data in yeast173 showed a decrease of Fe–S cluster insertion levels into cytosolic [4Fe–4S] target proteins upon Grx4 depletion. Deficiency in yeast Grx3/4 also leads to the impairment of the cytosolic ribonucleotide reductase (RNR) enzyme despite cytosolic iron overload.173 Similar to the CIA machinery, the maturation of the di-iron cofactor of RNR depends on the Dre2-Tah18 electron transfer chain,177,178 but iron loading into RNR does not require Dre2, while it requires Grx3/Grx4 proteins.179 On this basis, an iron-trafficking function of cytosolic monothiol glutaredoxins can also be proposed. According to this view, the depletion of GLRX3 in humans and of Grx3 or Grx4 in yeast specifically also impaired the synthesis of heme.173 However, direct evidence showing that monothiol Grx proteins are able to transfer iron ions from the bound [2Fe–2S] clusters to target proteins is still missing, and such data would be required to validate the proposed iron-trafficking function of cytosolic Grx proteins.
GLRX3 forms a complex in vitro with the cytosolic protein BOLA2.171,180 Specifically, the in vitro studies showed that an apo-complex is formed, with each Grx domain of apo-GLRX3 binding a BOLA2 molecule to form a heterotrimeric complex (Fig. 1).180 This apo-complex is able to bind two [2Fe–2S]2+ clusters by chemical reconstitution, with each cluster being bridged between BOLA2 and a Grx domain of GLRX3 ([[2Fe–2S]2 GLRX3 BOLA22], Fig. 4).171 The same cluster binding state (i.e. two [2Fe–2S] clusters per trimer) is observed once the heterotrimeric complex was isolated by cells co-expressing the two BOLA2 and GLRX3 proteins.171,180 The [GLRX3 BOLA2] complex was also observed in mammalian cells.181 This complex is, however, not as abundant as apo-form in mammalian cells, likely due to the low protein–protein affinity (Kd = 25 μM), but it accumulates upon [2Fe–2S] cluster binding, which stabilizes its interaction.181In vitro studies180 showed that the [[2Fe–2S]2 GLRX3 BOLA22] complex transfers both its [2Fe–2S]2+ clusters to apo-Anamorsin producing its mature holo state (Fig. 1 and 4). The mechanism of cluster transfer relies on the interaction between the N-terminal domain of Anamorsin with that of GLRX3, similarly to what was observed in the cluster transfer from [2Fe–2S]2 GLRX32 to apo-Anamorsin. In vivo studies confirmed that this direct [2Fe–2S] cluster transfer process occurs in mammalian cells upon complex formation between [[2Fe–2S] GLRX3 BOLA2] and [apo-Anamorsin NDOR1] complexes.181 Collectively, the experimental data suggested that the heterotrimeric complex can work as a [2Fe–2S] cluster transfer component in the CIA machinery, similarly to what the homodimeric [2Fe–2S]2 GLRX32 does (Fig. 1). Now, the arising question is whether and how the cells handle homodimeric [2Fe–2S]2 GLRX32 and heterotrimeric [[2Fe–2S]2 GLRX3 BOLA22] complexes, i.e. which is the predominant species acting as a [2Fe–2S] cluster chaperone at the cellular level. In vivo data181 support a model in which the heterotrimeric [[2Fe–2S]2 GLRX3 BOLA22] complex is the species transferring the [2Fe–2S] clusters under physiological conditions. However, we cannot exclude that, in human cells, the [2Fe–2S]2 GLRX32 homodimers transfer their own clusters to a number of target proteins larger than the [[2Fe–2S]2 GLRX3 BOLA22] hetero-complex and, in doing so, they do not accumulate to a detectable degree in cells. Consistent with this hypothesis, the bridged clusters in the GLRX3 homodimer were labile in vitro, much more than in the [[2Fe–2S]2 GLRX3 BOLA22] complex,171,182 which, conversely, accumulates in human cells.181 In the hypothesis that the [[2Fe–2S]2 GLRX3 BOLA22] complex works as the major species in the cytoplasm of human cells transferring the clusters to several target proteins, we would expect similar effects on the in vivo Fe–S cluster loading into several target proteins by deleting BOLA2 or GLRX3, besides Anamorsin. We believe that such in vivo studies would help in further elucidating the role of the [[2Fe–2S]2 GLRX3 BOLA22] complex as the cytoplasmic distributor of [2Fe–2S] clusters. In such studies, alternative sources of Fe–S cluster loading into the target proteins might be operative and need to be taken into account. This is indeed what occurs for Anamorsin, where significant amounts of residual iron are retained in Anamorsin complexes in the cells lacking GLRX3 or BOLA2.181 The mitoNEET/miner1 family of [2Fe–2S] proteins has been proposed to be an alternative source of Anamorsin for acquiring [2Fe–2S] clusters, since mitoNEET/miner1 proteins can transfer their [2Fe–2S] clusters to apo-Anamorsin in vitro,183 and human mitoNEET can transfer [2Fe–2S] clusters to apo-proteins in the cells exposed to oxidative stress.184–186 This alternative source of Fe–S clusters could also account for the relatively milder phenotypes associated with the depletion of GLRX3 in mammalian cells versus yeast, which does not have mitoNEET homologues. However, the role of the mitoNEET/miner1 family in the CIA machinery is still controversial, since still there is no clear evidence from functional and interactomic data supporting the role of this protein family in cluster transfer under physiological conditions. In conclusion, the [2Fe–2S]2+ GLRX32 homodimer and/or the [[2Fe–2S]2 GLRX3 BOLA22] heterotrimer might work as cytoplasmic [2Fe–2S] chaperones inserting [2Fe–2S]2+ clusters into cytosolic [2Fe–2S] target proteins, such as Anamorsin, or into the [NUBP1 NUBP2] heterotetrameric complex to assemble a [4Fe–4S]2+ cluster (Fig. 1).
Target-selective insertion of the assembled [4Fe–4S] cluster into cytosolic/nuclear [4Fe–4S] target proteins
The late steps of the CIA machinery are quite complex and involve the majority of the CIA proteins. Several studies in the past few years investigated how these proteins work, all showing that the principal component of this process is a stable complex composed of three proteins, [CIAO1 MMS19 CIA2B], commonly named the ternary CIA targeting complex (Fig. 1).187,188 MMS19 contains several HEAT repeats, which are commonly involved in protein–protein interactions. CIA2B (also named FAM96B or MIP18) and CIAO1 (also named CIA1) interact with the C-terminal HEAT repeats of MMS19 to form a docking site for the majority of cytosolic and nuclear [4Fe–4S] target proteins.189 CIA2B directly interacts with the C-terminal HEAT repeats of MMS19, while CIAO1 requires the presence of CIA2B for a stable interaction with MMS19. The direct interaction of MMS19 with the C-terminus of CIA2B protects CIA2B from proteosomal degradation.189 A structural analysis of the CIA targeting complex will be of fundamental relevance for understanding whether the interaction between MMS19 and CIAO1 is solely mediated by CIA2B or whether CIAO1 displays a weak affinity for MMS19 that is greatly enhanced by CIA2B. Such structural investigation will also be able to clarify which domains/proteins are in direct physical contact with cytoplasmic and nuclear [4Fe–4S] target proteins. The most straightforward model is that MMS19, CIA2B and CIAO1, jointly, form a docking site for the [4Fe–4S] target proteins, but we cannot exclude that the formation of the ternary complex induces a conformational change in one of the proteins that exposes an interaction site for the [4Fe–4S] target proteins, otherwise not accessible. The majority of the [4Fe–4S] target proteins bind the CIA targeting complex only in the presence of all three proteins of the complex, i.e. MMS19, CIA2B and CIAO1.189 The only exception is the transcription factor XPD, which binds to the N-terminal HEAT repeats of MMS19 independent of CIA2B and CIAO1.189 However, it cannot be excluded that XPD features two interaction sites with the CIA targeting complex: one with the N-terminus of MMS19 and one with the “canonical” [4Fe–4S] docking site at the C-terminus of MMS19 that depends on CIA2B and CIAO1. The only structure available among the proteins of the CIA targeting complex is the crystal structure of CIAO1, solved at a resolution of 1.7 Å, showing a β-propeller fold with seven pseudo-symmetrically orientated blades around a central axis (Fig. 4).190 This doughnut-shaped geometry is highly conserved among the large family of WD40 proteins, which serve as docking sites in a multitude of cellular networks and mediate the molecular recognition of partner proteins mainly through the top and side surfaces.191In addition to the cytosolic and nuclear Fe–S target proteins, the CIA targeting complex interacts with another CIA component, the NARFL protein (also named IOP1) (Fig. 1).192,193 NARFL has been proposed to also interact with the [NUBP1 NUBP2] complex because of the specific interaction between NUBP2 and NARFL.194 NARFL can thus function at the interface between the [NUBP1 NUBP2] complex and the CIA targeting complex in the CIA pathway, but its precise molecular role remains to be elucidated (Fig. 1). The protein NARFL exhibits a sequence homology to bacterial [FeFe] hydrogenases,195,196 yet the absence of hydrogenase activity in yeast suggests that, instead of a [4Fe–4S] cluster linked to the Fe2(CO)3(CN)2 active-site cluster, NARFL only retains the [4Fe–4S] cluster.197 Nevertheless, NARFL coordinates two [4Fe–4S] cofactors that are similar to those in hydrogenases.197–199 Studies on the yeast homolog Nar1 showed that these clusters are bound to the N- and C-terminal motifs with four conserved cysteine residues each.197,198 Homology modelling and stability studies suggested that the C-terminal cluster is buried in the protein and that the more labile N-terminal cluster is surface-exposed, thus potentially being transferred to protein partners.197,198 Both motifs are crucial for the function of NARFL as CIA component.198 In yeast, the depletion of all four early-acting CIA factors (Cdf1, Nbp35, Tah18, and Dre2) impairs the Fe–S cluster incorporation into Nar1 in vivo.152,157,197 Conversely, depletion of Nar1 has no influence on the Fe–S cluster assembly on Cfd1, Nbp35, or Dre2, but Nar1 is required for the maturation of cytosolic and nuclear [4Fe–4S] target proteins. In contrast, none of the late-acting CIA factors are required for the [4Fe–4S] cluster assembly on Nar1.188,200 Protein interaction studies indicated that Nar1 interacts with the yeast homologues of CIAO1 and CIA2B (Cia1 and Cia2, respectively),200,201i.e. with the CIA targeting complex, as observed for the human homologues. Assuming that most of the in vivo findings on yeast hold in human cells, the most reliable model is that the CIA targeting complex binds NARFL, and that the full complex (the CIA targeting complex + NARFL) may catalyse the late CIA reactions transferring [4Fe–4S] clusters, received by the [NUBP1 NUBP2] complex, to the majority of cytosolic and nuclear [4Fe–4S] target proteins (Fig. 1 and 4). As a result, it might be possible that the observed NARFL–NUBP2 interaction,194 discussed above, originates both from the maturation of NARFL processed by the early-acting CIA components, and from the [4Fe–4S] cluster transfer from the [NUBP1 NUBP2] complex to an already matured (i.e. containing the C-terminal cluster only) NARFL protein bound to the CIA targeting complex.
NARFL has also been found to be a protein partner of another late-acting CIA component, i.e. the CIA2A protein (also named FAM96A), which, in turn, forms a complex with CIAO1 (Fig. 1 and 4).202 The [CIAO1 CIA2A] complex has been proposed to be implicated in the [4Fe–4S] cluster maturation of exclusively IRP1, although no direct interaction with IRP1 was detected, possibly because of a rather labile interaction between apo-IRP1 and the [CIAO1 CIA2A] complex.202 An NMR solution structure of a monomeric form of human CIA2A203 and two different crystal structures of dimeric CIA2A forms204 have been reported (Fig. 4). So far, the available structural information has not provided any decisive insights into the molecular function of CIA2A. A cysteine residue appears to perform a crucial role in the CIA2A function for maturing Fe–S target proteins.205 Elucidation of the molecular role of this reactive cysteine residue might clarify not only the role of CIA2A but also that of the entire CIA targeting complex in Fe–S protein maturation.
In conclusion, the proposed model assumes NARFL as the CIA component distributing [4Fe–4S] clusters assembled on the [NUBP1 NUBP2] complex to [CIAO1 CIA2A] and CIA targeting complexes. However, since NARFL cannot replace the cellular function of Nar1 in yeast,197 more robust data need to be provided to prove the existence of a physical interaction between NARFL and the [NUBP1 NUBP2] complex.
Recently, two newly identified CIA factors, termed ORAOV1 and YAE1D1, were shown to function as specific adaptors connecting the CIA targeting complex with a specific [4Fe–4S] target protein, a cytosolic ABCE1 protein (Fig. 4). YAE1D1 and ORAOV1 form a complex that, in turn, interacts with the CIA targeting complex.193 Both proteins contain an evolutionary conserved deca-GX3 motif of 40 residues that is not found in any other eukaryotic protein.193 ORAOV1 additionally carries a conserved C-terminal tryptophan (phenylalanine in some organisms).193 A chain of binding events facilitates efficient ABCE1 maturation: (i) the CIA targeting complex interacts with ORAOV1 in the [YAE1D1 ORAOV1] complex via its C-terminal tryptophan; (ii) the two deca-GX3 motifs are crucial for [YAE1D1 ORAOV1] complex formation; (iii) YAE1D1 in the [YAE1D1 ORAOV1] complex mediates contact with the ABCE1 protein.193,206 Interestingly, a characteristically different but related maturation strategy is followed by the radical-SAM Fe–S enzyme, viperin, an interferon-induced antiviral defense component.207 This protein also uses its conserved C-terminal tryptophan to directly associate with the CIA targeting complex. The removal of this residue abolishes complex formation with the CIA targeting complex, the assembly of viperin's Fe–S cluster, and, consequently, the antiviral function.207 Although it has been reported that CIAO1 forms mutually exclusive complexes with either CIA2A or CIA2B,202 in a very recent work investigating the maturation pathway of the [4Fe–4S] cluster protein viperin, it has been found that both CIA targeting complex and CIA2A interact with viperin.208 Specifically, the CIA targeting complex interacts with the C-terminus of viperin, while CIA2A with the N-terminus. Despite this multitude of viperin interactors, only CIAO1 is required for the Fe–S cluster incorporation into viperin. The role of all the other interacting components is still unclear, but it might be possible that a concerted mechanism of cluster insertion is operative, in which CIAO1 is the essential component as it drives the association of all the other interactors.
The mitochondrial iron–sulfur cluster (ISC) export machinery
The main component of the mitochondrial ISC export machinery is an inner membrane ABCB transporter belonging to the ABCB7 group (Fig. 1).48,209 This group includes the human ABCB7, yeast Atm1 and Arabidopsis thaliana ATM3 transporters and can be found in virtually all eukaryotic species and in some bacterial organisms. In vivo data indicate that the members of this subfamily have the same function.209–211 In a variety of organisms, this transporter specifically supports the maturation of cytosolic and nuclear Fe–S proteins.48,209,211,212 Its depletion is also associated with defects in cellular iron homeostasis resulting in cytosolic iron deficiency and a concomitant iron accumulation in the mitochondria, that is, a similar phenotype observed for the proteins involved in the early stage of ISC protein deficiencies.48,213The other well-established component of the ISC export machinery is glutathione, which has been found to be essential for the maturation of extra-mitochondrial Fe–S proteins and for iron regulation.52,214 It was observed that GSH molecules can form an in vitro tetra-GSH-coordinated [2Fe–2S] cluster (defined as (GS)4–[2Fe–2S]).215–217 The (GS)4–[2Fe–2S] complex is shown to significantly stimulate the ATPase activity of an ABCB7-type transporter in both solution and proteoliposome-bound forms. On the basis of this in vitro evidence, it has been suggested that the (GS)4–[2Fe–2S] complex is the molecule transiting from the mitochondria to the cytosol being the physiological substrate of the membrane transporter.218,219 On the other hand, the ATPase activity of the purified, liposome-integrated membrane transporter can be stimulated by many thiol-containing compounds including GSH and GSSG (the oxidized form of GSH),53 questioning the physiological relevance of such an in vitro approach. In particular, the stimulatory effect of (GS)4–[2Fe–2S] was 30-fold weaker than that observed with Cys-containing small peptides. Moreover, the affinity of (GS)4–[2Fe–2S] binding to Atm1 was in the same range as that of GSH alone, questioning again the specificity of the (GS)4–[2Fe–2S] interaction with Atm1 in vitro.218–220 Furthermore, it still needs to be documented that the (GS)4–[2Fe–2S] complex remains intact under physiological transport conditions, and to verify the existence of a (GS)4–[2Fe–2S] complex in a living cell. In conclusion, it is possible and reasonable that the (GS)4–[2Fe–2S] complex is the substrate of the membrane transporter, but further experimental data are required to definitively confirm this conclusion.
The crystal structures of free and glutathione-bound Atm1 were solved and they are virtually identical.219 The main outcome of the structure is that a V-shaped molecule enclosing a 6900 Å3 positively charged cavity is present. The cavity is close to the hydrophilic phase of the matrix side of the inner membrane, and likely serves as the substrate-binding site. The fact that Atm1 and a related bacterial ABC transporter219 bind GSH or its derivatives suggests that these transporters may exhibit a similar substrate specificity. Both proteins bind GSH or its derivatives in the putative substrate-binding pocket comprised by the large, positively charged cavity close to the internal membrane surface and near to the putative exit channel formed by the twelve trans-membrane helices of Atm1. The cavity features much more space than that occupied by GSH alone, and therefore it is reasonable to suggest that GSH is a part of a larger Atm1 substrate and is required for substrate binding. In agreement with GSH being directly involved in the substrate export process, the Atm1-deficient mitochondria show an increased content of GSH,221 and the ATPase activity of Atm1 is stimulated by GSH.53 In particular, the largest stimulation of ATP hydrolysis by Atm1 from Novosphingobium aromaticivorans was observed for the Ag and Hg complexes of GSH.222 In the crystal structure of Atm1 from Novosphingobium aromaticivorans, an oxidized glutathione molecule, GSSG, is bound into the transmembrane region in a location similar to GSH, suggesting that this interaction is specific and that this region of Atm1 family members may allow the binding of a GSH-containing substrate. Another study has suggested the molecule GSSSG, i.e. formed by the oxidized form of GSSH (glutathione persulfide) and GSH, as the Atm1 substrate and a potential sulfur carrier.223 In conclusion, the putative Atm1-like substrate is currently under debate. The identification on the nature of the molecule exported from the mitochondria to the cytosol by the ABCB transporter will be crucial to define whether a de novo synthesis of [2Fe–2S] clusters is required in the cytosol.
In yeast, another proposed component of the ISC export machinery is the sulfhydryl oxidase Erv1 located in the mitochondrial intermembrane space. This proposal is based on a similar phenotype to that observed for Atm1 deficiency.224 However, it has been recently shown that the observed phenotype was a consequence of strongly decreased glutathione levels, ruling out significant roles for Erv1 in cytosolic Fe–S protein biogenesis and iron regulation.225
Concluding remarks
Recent research has greatly advanced our understanding of the assembly and insertion of Fe–S clusters into mitochondrial, cytosolic and nuclear target proteins in human cells. The majority of the proteins involved in such processes have been identified, but a complete picture of how they interact and work at a molecular level is still ill-defined. The former steps of the mitochondrial ISC assembly machinery devoted to the assembly of a [2Fe–2S] cluster are best characterized at a molecular level. Nevertheless, the high molecular complexity of the process still does not provide a conclusive model on how [2Fe–2S] clusters are assembled and on the specific role of all components that are part of this complex mechanism. Even more “molecular chaos” exists in the late stages of the ISC assembly machinery. In particular, the role of IBA57 is still not defined, and how the [4Fe–4S] cluster assembled in mitochondria is inserted into the final target proteins has to be understood. The most relevant CIA machinery components in humans have been identified, but a molecular picture of their function is still largely missing. The available functional data do not provide a clear view on how the CIA machinery components cooperate at the molecular level. Even the numerous interactomic data provided over the past few years are not conclusive in this respect. The investigation of the interactions of the CIA components at a molecular level and the associated structural data are required to make a step forward on defining how the CIA machinery works. We believe that this information will be fundamental to understand the degree of dependence of the CIA machinery on the ISC assembly and export machineries in humans. Finally, the identification of the nature of the molecule exported from mitochondria to the cytosol is a fundamental step to be attained in order to define the role of the ISC isoforms that are present in the cytosol in a de novo cytosolic synthesis of [2Fe–2S] clusters. In conclusion, only a combination of structural, biochemical and cell biological approaches can move forward the understanding of the molecular mechanisms responsible for the assembly and insertion of Fe–S clusters into target proteins in humans.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support from the iNEXT Project 653706 “Infrastructure for NMR, EM and X-ray Crystallography for Translational Research”. The authors acknowledge the support and the use of resources of Instruct-ERIC, a Landmark ESFRI project, and specifically CERM/CIRMMP Italy Centre. This article is also based upon work from COST Action CA15133, supported by COST (European Cooperation in Science and Technology).References
- K. J. Waldron, J. C. Rutherford, D. Ford and N. J. Robinson, Metalloproteins and metal sensing, Nature, 2009, 460, 823–830 CrossRef CAS PubMed.
- J. J. R. Frausto da Silva and R. J. P. Williams, The biological chemistry of the elements: the inorganic chemistry of life, Oxford University Press, New York, 2001 Search PubMed.
- I. Bertini, H. B. Gray, E. I. Stiefel and J. S. Valentine, Biological Inorganic Chemistry, University Science Books, Sausalito, California, 2006 Search PubMed.
- L. A. Finney and T. V. O'Halloran, Transition Metal Speciation in the Cell: Insights from the Chemistry of Metal Ion Receptors, Science, 2003, 300, 931–936 CrossRef CAS PubMed.
- B. E. Kim, T. Nevitt and D. J. Thiele, Mechanisms for copper acquisition, distribution and regulation, Nat. Chem. Biol., 2008, 4, 176–185 CrossRef CAS PubMed.
- A. Atkinson and D. R. Winge, Metal acquisition and availability in the mitochondria, Chem. Rev., 2009, 109, 4708–4721 CrossRef CAS PubMed.
- G. J. Brewer, Copper in medicine, Curr. Opin. Chem. Biol., 2003, 7, 207–212 CrossRef CAS PubMed.
- M. Valko, H. Morris and M. T. Cronin, Metals, toxicity and oxidative stress, Curr. Med. Chem., 2005, 12, 1161–1208 CrossRef CAS PubMed.
- R. Lill and U. Muhlenhoff, Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases, Annu. Rev. Biochem., 2008, 77, 669–700 CrossRef CAS PubMed.
- T. A. Rouault, Iron metabolism in the CNS: implications for neurodegenerative diseases, Nat. Rev. Neurosci., 2013, 14, 551–564 CrossRef CAS PubMed.
- M. R. Bleackley and R. T. Macgillivray, Transition metal homeostasis: from yeast to human disease, Biometals, 2011, 24, 785–809 CrossRef CAS PubMed.
- A. Sheftel, O. Stehling and R. Lill, Iron-sulfur proteins in health and disease, Trends Endocrinol. Metab., 2010, 21, 302–314 CrossRef CAS PubMed.
- E. Madsen and J. D. Gitlin, Copper and iron disorders of the brain, Annu. Rev. Neurosci., 2007, 30, 317–337 CrossRef CAS PubMed.
- K. J. Barnham and A. I. Bush, Metals in Alzheimer's and Parkinson's diseases, Curr. Opin. Chem. Biol., 2008, 12, 222–228 CrossRef CAS PubMed.
- A. V. Davis and T. V. O'Halloran, A place for thioether chemistry in cellular copper ion recognition and trafficking, Nat. Chem. Biol., 2008, 4, 148–151 CrossRef CAS PubMed.
- D. L. Huffman and T. V. O'Halloran, Function, structure, and mechanism of intracellular copper trafficking proteins, Annu. Rev. Biochem., 2001, 70, 677–701 CrossRef CAS PubMed.
- A. K. Boal and A. C. Rosenzweig, Structural biology of copper trafficking, Chem. Rev., 2009, 109, 4760–4779 CrossRef CAS PubMed.
- J. M. Leitch, P. J. Yick and V. C. Culotta, The right to choose: multiple pathways for activating copper, zinc superoxide dismutase, J. Biol. Chem., 2009, 284, 24679–24683 CrossRef CAS PubMed.
- L. Banci, I. Bertini, F. Cantini and S. Ciofi-Baffoni, Cellular copper distribution: a mechanistic systems biology approach, Cell. Mol. Life Sci., 2010, 67, 2563–2589 CrossRef CAS PubMed.
- F. S. La, R. Burke and D. P. Giedroc, Mammalian copper biology: hitting the pause button in celebration of three pioneers and four decades of discovery, Metallomics, 2016, 8, 810–812 RSC.
- L. Banci and A. Rosato, Structural genomics of proteins involved in copper homeostasis, Acc. Chem. Res., 2003, 36, 215–221 CrossRef CAS PubMed.
- L. Banci, I. Bertini, S. Ciofi-Baffoni, T. Kozyreva, K. Zovo and P. Palumaa, Affinity gradients drive copper to cellular destinations, Nature, 2010, 465, 645–648 CrossRef CAS PubMed.
- C. Andreini, L. Banci, I. Bertini, S. Elmi and A. Rosato, Non-heme iron through the three domains of life, Proteins: Struct., Funct., Bioinf., 2007, 67, 317–324 CrossRef CAS PubMed.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, Occurence of copper through the three domains of life: a bioinformatic approach, J. Proteome Res., 2008, 1, 209–216 CrossRef PubMed.
- A. C. Dlouhy and C. E. Outten, The iron metallome in eukaryotic organisms, Met. Ions Life Sci., 2013, 12, 241–278 Search PubMed.
- D. C. Johnson, D. R. Dean, A. D. Smith and M. K. Johnson, Structure, function, and formation of biological iron-sulfur clusters, Annu. Rev. Biochem., 2005, 74, 247–281 CrossRef CAS PubMed.
- H. Beinert, R. H. Holm and E. Munck, Iron-sulfur clusters: nature's modular, multipurpose structures, Science, 1997, 277, 653–659 CrossRef CAS PubMed.
- P. Venkateswara Rao and R. H. Holm, Synthetic Analogues of the Active Sites of Iron–Sulfur Proteins, Chem. Rev., 2004, 104, 527–559 CrossRef CAS PubMed.
- P. J. Kiley and H. Beinert, The role of Fe–S proteins in sensing and regulation in bacteria, Curr. Opin. Microbiol., 2003, 6, 181–185 CrossRef CAS PubMed.
- Y. Hu and M. W. Ribbe, A journey into the active center of nitrogenase, J. Biol. Inorg. Chem., 2014, 19, 731–736 CrossRef CAS PubMed.
- E. M. Shepard, E. S. Boyd, J. B. Broderick and J. W. Peters, Biosynthesis of complex iron–sulfur enzymes, Curr. Opin. Chem. Biol., 2011, 15, 319–327 CrossRef CAS PubMed.
- R. A. Marcus and N. Sutin, Electron transfer in chemistry and biology, Biochim. Biophys. Acta, 1985, 811, 265–275 CrossRef CAS.
- J. Hirst, Mitochondrial complex I, Annu. Rev. Biochem., 2013, 82, 551–575 CrossRef CAS PubMed.
- S. J. Booker, R. M. Cicchillo and T. L. Grove, Self-sacrifice in radical S-adenosylmethionine proteins, Curr. Opin. Chem. Biol., 2007, 11, 543–552 CrossRef CAS PubMed.
- J. Meyer, Iron-sulfur protein folds, iron-sulfur chemistry, and evolution, J. Biol. Inorg. Chem., 2008, 13, 157–170 CrossRef CAS PubMed.
- E. L. Mettert and P. J. Kiley, Fe–S proteins that regulate gene expression, Biochim. Biophys. Acta, 2015, 1853, 1284–1293 CrossRef CAS PubMed.
- T. A. Rouault, The role of iron regulatory proteins in mammalian iron homeostasis and disease, Nat. Chem. Biol., 2006, 2, 406–414 CrossRef CAS PubMed.
- C. Andreini, I. Bertini and A. Rosato, Metalloproteomes: a bioinformatic approach, Acc. Chem. Res., 2009, 42, 1471–1479 CrossRef CAS PubMed.
- C. Andreini, L. Banci and A. Rosato, Exploiting bacterial operons to illuminate human iron-sulfur proteins, J. Proteome Res., 2016, 15, 1308–1322 CrossRef CAS PubMed.
- C. Andreini, A. Rosato and L. Banci, The Relationship between Environmental Dioxygen and Iron-Sulfur Proteins Explored at the Genome Level, PLoS One, 2017, 12, e0171279 Search PubMed.
- L. Zheng, V. L. Cash, D. H. Flint and D. R. Dean, Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii, J. Biol. Chem., 1998, 273, 13264–13272 CrossRef CAS PubMed.
- B. Roche, L. Aussel, B. Ezraty, P. Mandin, B. Py and F. Barras, Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity, Biochim. Biophys. Acta, 2013, 1827, 455–469 CrossRef CAS PubMed.
- O. Stehling, C. Wilbrecht and R. Lill, Mitochondrial iron-sulfur protein biogenesis and human disease, Biochimie, 2014, 100, 61–77 CrossRef CAS PubMed.
- N. Maio and T. A. Rouault, Iron-sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery, Biochim. Biophys. Acta, 2015, 1853, 1493–1512 CrossRef CAS PubMed.
- J. O. Fuss, C. L. Tsai, J. P. Ishida and J. A. Tainer, Emerging critical roles of Fe–S clusters in DNA replication and repair, Biochim. Biophys. Acta, 2015, 1853, 1253–1271 CrossRef CAS PubMed.
- O. A. Lukianova and S. S. David, A role for iron-sulfur clusters in DNA repair, Curr. Opin. Chem. Biol., 2005, 9, 145–151 CrossRef CAS PubMed.
- D. J. Netz, J. Mascarenhas, O. Stehling, A. J. Pierik and R. Lill, Maturation of cytosolic and nuclear iron-sulfur proteins, Trends Cell Biol., 2014, 24, 303–312 CrossRef CAS PubMed.
- G. Kispal, P. Csere, C. Prohl and R. Lill, The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins, EMBO J., 1999, 18, 3981–3989 CrossRef CAS PubMed.
- A. Biederbick, O. Stehling, R. Rosser, B. Niggemeyer, Y. Nakai, H. P. Elsasser and R. Lill, Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation, Mol. Cell. Biol., 2006, 26, 5675–5687 CrossRef CAS PubMed.
- J. Gerber, K. Neumann, C. Prohl, U. Muhlenhoff and R. Lill, The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins, Mol. Cell. Biol., 2004, 24, 4848–4857 CrossRef CAS PubMed.
- R. Lill, R. Dutkiewicz, S. A. Freibert, T. Heidenreich, J. Mascarenhas, D. J. Netz, V. D. Paul, A. J. Pierik, N. Richter, M. Stumpfig, V. Srinivasan, O. Stehling and U. Muhlenhoff, The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins, Eur. J. Cell Biol., 2015, 94, 280–291 CrossRef CAS PubMed.
- K. Sipos, H. Lange, Z. Fekete, P. Ullmann, R. Lill and G. Kispal, Maturation of cytosolic iron-sulfur proteins requires glutathione, J. Biol. Chem., 2002, 277, 26944–26949 CrossRef CAS PubMed.
- G. Kuhnke, K. Neumann, U. Muhlenhoff and R. Lill, Stimulation of the ATPase activity of the yeast mitochondrial ABC transporter Atm1p by thiol compounds, Mol. Membr. Biol., 2006, 23, 173–184 CrossRef CAS PubMed.
- C. A. Chen and J. A. Cowan, Characterization of Saccharomyces cerevisiae Atm1p: functional studies of an ABC7 type transporter, Biochim. Biophys. Acta, 2006, 1760, 1857–1865 CrossRef CAS PubMed.
- T. A. Rouault, Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease, Dis. Models & Mech., 2012, 5, 155–164 CAS.
- N. Maio and T. A. Rouault, Iron-sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery, Biochim. Biophys. Acta, 2015, 1853, 1493–1512 CrossRef CAS PubMed.
- Y. Shi, M. C. Ghosh, W. H. Tong and T. A. Rouault, Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis, Hum. Mol. Genet., 2009, 18, 3014–3025 CrossRef CAS PubMed.
- W. H. Tong, G. N. Jameson, B. H. Huynh and T. A. Rouault, Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe–4S] cluster, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9762–9767 CrossRef CAS PubMed.
- W. H. Tong and T. A. Rouault, Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis, Cell Metab., 2006, 3, 199–210 CrossRef CAS PubMed.
- F. Acquaviva, I. De Biase, L. Nezi, G. Ruggiero, F. Tatangelo, C. Pisano, A. Monticelli, C. Garbi, A. M. Acquaviva and S. Cocozza, Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2, J. Cell Sci., 2005, 118, 3917–3924 CrossRef CAS PubMed.
- T. Land and T. A. Rouault, Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization, Mol. Cell, 1998, 2, 807–815 CrossRef CAS PubMed.
- M. W. Gray, G. Burger and B. F. Lang, Mitochondrial evolution, Science, 1999, 283, 1476–1481 CrossRef CAS PubMed.
- C. G. Kurland and S. G. Andersson, Origin and evolution of the mitochondrial proteome, Microbiol. Mol. Biol. Rev., 2000, 64, 786–820 CrossRef CAS PubMed.
- T. A. Rouault and W. H. Tong, Iron-sulfur cluster biogenesis and human disease, Trends Genet., 2008, 24, 398–407 CrossRef CAS PubMed.
- X. M. Xu and S. G. Moller, Iron-sulfur clusters: biogenesis, molecular mechanisms, and their functional significance, Antioxid. Redox Signaling, 2011, 15, 271–307 CrossRef CAS PubMed.
- F. Pierrel, P. A. Cobine and D. R. Winge, Metal Ion availability in mitochondria, Biometals, 2007, 20, 675–682 CrossRef CAS PubMed.
- P. A. Lindahl and M. J. Moore, Labile Low-Molecular-Mass Metal Complexes in Mitochondria: Trials and Tribulations of a Burgeoning Field, Biochemistry, 2016, 55, 4140–4153 CrossRef CAS PubMed.
- S. P. McCormick, M. J. Moore and P. A. Lindahl, Detection of Labile Low-Molecular-Mass Transition Metal Complexes in Mitochondria, Biochemistry, 2015, 54, 3442–3453 CrossRef CAS PubMed.
- J. Gerber, U. Muhlenhoff and R. Lill, An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1, EMBO Rep., 2003, 4, 906–911 CrossRef CAS PubMed.
- D. J. Lane, A. M. Merlot, M. L. Huang, D. H. Bae, P. J. Jansson, S. Sahni, D. S. Kalinowski and D. R. Richardson, Cellular iron uptake, trafficking and metabolism: key molecules and mechanisms and their roles in disease, Biochim. Biophys. Acta, 2015, 1853, 1130–1144 CrossRef CAS PubMed.
- A. Pastore and H. Puccio, Frataxin: a protein in search for a function, J. Neurochem., 2013, 126(suppl 1), 43–52 CrossRef CAS PubMed.
- R. Lill, B. Hoffmann, S. Molik, A. J. Pierik, N. Rietzschel, O. Stehling, M. A. Uzarska, H. Webert, C. Wilbrecht and U. Muhlenhoff, The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism, Biochim. Biophys. Acta, 2012, 1823, 1491–1508 CrossRef CAS PubMed.
- S. A. Cory, J. G. Van Vranken, E. J. Brignole, S. Patra, D. R. Winge, C. L. Drennan, J. Rutter and D. P. Barondeau, Structure of human Fe–S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5325–E5334 CrossRef CAS PubMed.
- L. Zheng, R. H. White, V. L. Cash and D. R. Dean, Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product, Biochemistry, 1994, 33, 4714–4720 CrossRef CAS PubMed.
- J. T. Kaiser, T. Clausen, G. P. Bourenkow, H. D. Bartunik, S. Steinbacher and R. Huber, Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly, J. Mol. Biol., 2000, 297, 451–464 CrossRef CAS PubMed.
- P. P. Saha, S. Srivastava, S. K. P. Kumar, D. Sinha and P. D'Silva, Mapping Key Residues of ISD11 Critical for NFS1-ISD11 Subcomplex Stability: Implications in the Development of Mitochondrial Disorder, COXPD19, J. Biol. Chem., 2015, 290, 25876–25890 CrossRef CAS PubMed.
- J. G. Van Vranken, M. Y. Jeong, P. Wei, Y. C. Chen, S. P. Gygi, D. R. Winge and J. Rutter, The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis, Elife, 2016, 5, e17828 Search PubMed.
- M. Friemel, Z. Marelja, K. Li and S. Leimkuhler, The N-Terminus of Iron-Sulfur Cluster Assembly Factor ISD11 Is Crucial for Subcellular Targeting and Interaction with L-Cysteine Desulfurase NFS1, Biochemistry, 2017, 56, 1797–1808 CrossRef CAS PubMed.
- K. Cai, R. O. Frederick, M. Tonelli and J. L. Markley, Mitochondrial Cysteine Desulfurase and ISD11 Coexpressed in Escherichia coli Yield Complex Containing Acyl Carrier Protein, ACS Chem. Biol., 2017, 12, 918–921 CrossRef CAS PubMed.
- K. Cai, M. Tonelli, R. O. Frederick and J. L. Markley, Human Mitochondrial Ferredoxin 1 (FDX1) and Ferredoxin 2 (FDX2) Both Bind Cysteine Desulfurase and Donate Electrons for Iron-Sulfur Cluster Biosynthesis, Biochemistry, 2017, 56, 487–499 CrossRef CAS PubMed.
- C. L. Tsai and D. P. Barondeau, Human frataxin is an allosteric switch that activates the Fe–S cluster biosynthetic complex, Biochemistry, 2010, 49, 9132–9139 CrossRef CAS PubMed.
- S. Schmucker, A. Martelli, F. Colin, A. Page, M. Wattenhofer-Donze, L. Reutenauer and H. Puccio, Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex, PLoS One, 2011, 6, e16199 CAS.
- J. Bridwell-Rabb, N. G. Fox, C. L. Tsai, A. M. Winn and D. P. Barondeau, Human frataxin activates Fe–S cluster biosynthesis by facilitating sulfur transfer chemistry, Biochemistry, 2014, 53, 4904–4913 CrossRef CAS PubMed.
- N. G. Fox, D. Das, M. Chakrabarti, P. A. Lindahl and D. P. Barondeau, Frataxin Accelerates [2Fe–2S] Cluster Formation on the Human Fe–S Assembly Complex, Biochemistry, 2015, 54, 3880–3889 CrossRef CAS PubMed.
- A. Parent, X. Elduque, D. Cornu, L. Belot, J. P. Le Caer, A. Grandas, M. B. Toledano and B. d'Autreaux, Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols, Nat. Commun., 2015, 6, 5686 CrossRef CAS PubMed.
- R. Shi, A. Proteau, M. Villarroya, I. Moukadiri, L. Zhang, J. F. Trempe, A. Matte, M. E. Armengod and M. Cygler, Structural basis for Fe–S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions, PLoS Biol., 2010, 8, e1000354 Search PubMed.
- E. N. Marinoni, J. S. de Oliveira, Y. Nicolet, E. C. Raulfs, P. Amara, D. R. Dean and J. C. Fontecilla-Camps, (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer, Angew. Chem., Int. Ed., 2012, 51, 5439–5442 CrossRef CAS PubMed.
- W. H. Tong and T. Rouault, Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells, EMBO J., 2000, 19, 5692–5700 CrossRef CAS PubMed.
- S. Schmucker, M. Argentini, N. Carelle-Calmels, A. Martelli and H. Puccio, The in vivo mitochondrial two-step maturation of human frataxin, Hum. Mol. Genet., 2008, 17, 3521–3531 CrossRef CAS PubMed.
- R. A. Vaubel and G. Isaya, Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia, Mol. Cell. Neurosci., 2013, 55, 50–61 CrossRef CAS PubMed.
- P. Cavadini, J. Adamec, F. Taroni, O. Gakh and G. Isaya, Two-step processing of human frataxin by mitochondrial processing peptidase. Precursor and intermediate forms are cleaved at different rates, J. Biol. Chem., 2000, 275, 41469–41475 CrossRef CAS PubMed.
- S. Schmucker, M. Argentini, N. Carelle-Calmels, A. Martelli and H. Puccio, The in vivo mitochondrial two-step maturation of human frataxin, Hum. Mol. Genet., 2008, 17, 3521–3531 CrossRef CAS PubMed.
- R. K. Jobling, M. Assoum, O. Gakh, S. Blaser, J. A. Raiman, C. Mignot, E. Roze, A. Durr, A. Brice, N. Levy, C. Prasad, T. Paton, A. D. Paterson, N. M. Roslin, C. R. Marshall, J. P. Desvignes, N. Roeckel-Trevisiol, S. W. Scherer, G. A. Rouleau, A. Megarbane, G. Isaya, V. Delague and G. Yoon, PMPCA mutations cause abnormal mitochondrial protein processing in patients with non-progressive cerebellar ataxia, Brain, 2015, 138, 1505–1517 CrossRef PubMed.
- H. Li, O. Gakh, D. Y. Smith, W. K. Ranatunga and G. Isaya, Missense mutations linked to friedreich ataxia have different but synergistic effects on mitochondrial frataxin isoforms, J. Biol. Chem., 2013, 288, 4116–4127 CrossRef CAS PubMed.
- A. J. Andrew, J. Y. Song, B. Schilke and E. A. Craig, Posttranslational regulation of the scaffold for Fe–S cluster biogenesis, Isu, Mol. Biol. Cell, 2008, 19, 5259–5266 CrossRef CAS PubMed.
- O. Gakh, T. Bedekovics, S. F. Duncan, D. Y. Smith, D. S. Berkholz and G. Isaya, Normal and Friedreich ataxia cells express different isoforms of frataxin with complementary roles in iron-sulfur cluster assembly, J. Biol. Chem., 2010, 285, 38486–38501 CrossRef CAS PubMed.
- F. Colin, A. Martelli, M. Clemancey, J. M. Latour, S. Gambarelli, L. Zeppieri, C. Birck, A. Page, H. Puccio and C. S. Ollagnier de, Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly, J. Am. Chem. Soc., 2013, 135, 733–740 CrossRef CAS PubMed.
- O. Gakh, W. Ranatunga, D. Y. Smith, E. C. Ahlgren, S. Al-Karadaghi, J. R. Thompson and G. Isaya, Architecture of the Human Mitochondrial Iron-Sulfur Cluster Assembly Machinery, J. Biol. Chem., 2016, 291, 21296–21321 CrossRef CAS PubMed.
- T. L. Stemmler, E. Lesuisse, D. Pain and A. Dancis, Frataxin and mitochondrial FeS cluster biogenesis, J. Biol. Chem., 2010, 285, 26737–26743 CrossRef CAS PubMed.
- J. D. Cook, K. C. Kondapalli, S. Rawat, W. C. Childs, Y. Murugesan, A. Dancis and T. L. Stemmler, Molecular details of the yeast frataxin-Isu1 interaction during mitochondrial Fe–S cluster assembly, Biochemistry, 2010, 49, 8756–8765 CrossRef CAS PubMed.
- H. Li, O. Gakh, D. Y. Smith and G. Isaya, Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis, J. Biol. Chem., 2009, 284, 21971–21980 CrossRef CAS PubMed.
- A. D. Sheftel, O. Stehling, A. J. Pierik, H. P. Elsasser, U. Muhlenhoff, H. Webert, A. Hobler, F. Hannemann, R. Bernhardt and R. Lill, Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 11775–11780 CrossRef CAS PubMed.
- Y. Shi, M. Ghosh, G. Kovtunovych, D. R. Crooks and T. A. Rouault, Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis, Biochim. Biophys. Acta, 2012, 1823, 484–492 CrossRef CAS PubMed.
- H. Webert, S. A. Freibert, A. Gallo, T. Heidenreich, U. Linne, S. Amlacher, E. Hurt, U. Muhlenhoff, L. Banci and R. Lill, Functional reconstitution of de novo mitochondrial Fe/S cluster synthesis reveals an essential role of ferredoxin, Nat. Commun., 2014, 5, 5013–5025 CrossRef CAS PubMed.
- H. Uhrigshardt, A. Singh, G. Kovtunovych, M. Ghosh and T. A. Rouault, Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis, Hum. Mol. Genet., 2010, 19, 3816–3834 CrossRef CAS PubMed.
- N. Maio, A. Singh, H. Uhrigshardt, N. Saxena, W. H. Tong and T. A. Rouault, Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery, Cell Metab., 2014, 19, 445–457 CrossRef CAS PubMed.
- Y. Shan and G. Cortopassi, HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis, Hum. Mol. Genet., 2012, 21, 1457–1469 CrossRef CAS PubMed.
- K. Cai, R. O. Frederick, J. H. Kim, N. M. Reinen, M. Tonelli and J. L. Markley, Human Mitochondrial Chaperone (mtHSP70) and Cysteine Desulfurase (NFS1) Bind Preferentially to the Disordered Conformation, Whereas Co-chaperone (HSC20) Binds to the Structured Conformation of the Iron-Sulfur Cluster Scaffold Protein (ISCU), J. Biol. Chem., 2013, 288, 28755–28770 CrossRef CAS PubMed.
- A. K. Fuzery, J. J. Oh, D. T. Ta, L. E. Vickery and J. L. Markley, Three hydrophobic amino acids in Escherichia coli HscB make the greatest contribution to the stability of the HscB-IscU complex, BMC Biochem., 2011, 12, 3 CrossRef CAS PubMed.
- S. J. Ciesielski, B. A. Schilke, J. Osipiuk, L. Bigelow, R. Mulligan, J. Majewska, A. Joachimiak, J. Marszalek, E. A. Craig and R. Dutkiewicz, Interaction of J-protein co-chaperone Jac1 with Fe–S scaffold Isu is indispensable in vivo and conserved in evolution, J. Mol. Biol., 2012, 417, 1–12 CrossRef CAS PubMed.
- J. R. Cupp-Vickery and L. E. Vickery, Crystal structure of Hsc20, a J-type Co-chaperone from Escherichia coli, J. Mol. Biol., 2000, 304, 835–845 CrossRef CAS PubMed.
- E. Bitto, C. A. Bingman, L. Bittova, D. A. Kondrashov, R. M. Bannen, B. G. Fox, J. L. Markley and G. N. Phillips, Jr., Structure of human J-type co-chaperone HscB reveals a tetracysteine metal-binding domain, J. Biol. Chem., 2008, 283, 30184–30192 CrossRef CAS PubMed.
- J. Amick, S. E. Schlanger, C. Wachnowsky, M. A. Moseng, C. C. Emerson, M. Dare, W. I. Luo, S. S. Ithychanda, J. C. Nix, J. A. Cowan, R. C. Page and S. Misra, Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone, Protein Sci., 2014, 23, 833–842 CrossRef CAS PubMed.
- H. Schroder, T. Langer, F. U. Hartl and B. Bukau, DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage, EMBO J., 1993, 12, 4137–4144 CAS.
- U. Muhlenhoff, J. Gerber, N. Richhardt and R. Lill, Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p, EMBO J., 2003, 22, 4815–4825 CrossRef PubMed.
- M. A. Uzarska, R. Dutkiewicz, S. A. Freibert, R. Lill and U. Muhlenhoff, The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation, Mol. Biol. Cell, 2013, 24, 1830–1841 CrossRef CAS PubMed.
- P. Shakamuri, B. Zhang and M. K. Johnson, Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins, J. Am. Chem. Soc., 2012, 134, 15213–15216 CrossRef CAS PubMed.
- M. M. Molina-Navarro, C. Casas, L. Piedrafita, G. Belli and E. Herrero, Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria, FEBS Lett., 2006, 580, 2273–2280 CrossRef CAS PubMed.
- R. A. Wingert, J. L. Galloway, B. Barut, H. Foott, P. Fraenkel, J. L. Axe, G. J. Weber, K. Dooley, A. J. Davidson, B. Schmid, B. H. Paw, G. C. Shaw, P. Kingsley, J. Palis, H. Schubert, O. Chen, J. Kaplan and L. I. Zon, Deficiency of glutaredoxin 5 reveals Fe–S clusters are required for vertebrate haem synthesis, Nature, 2005, 436, 1035–1039 CrossRef CAS PubMed.
- M. T. Rodriguez-Manzaneque, J. Tamarit, G. Belli, J. Ros and E. Herrero, Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes, Mol. Biol. Cell, 2002, 13, 1109–1121 CrossRef CAS PubMed.
- L. Banci, D. Brancaccio, S. Ciofi-Baffoni, R. Del Conte, R. Gadepalli, M. Mikolajczyk, S. Neri, M. Piccioli and J. Winkelmann, [2Fe–2S] cluster transfer in iron-sulfur protein biogenesis, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6203–6208 CrossRef CAS PubMed.
- N. Maio and T. A. Rouault, Mammalian Fe–S proteins: definition of a consensus motif recognized by the co-chaperone HSC20, Metallomics, 2016, 8, 1032–1046 RSC.
- H. Angerer, Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes, Biology, 2015, 4, 133–150 CrossRef PubMed.
- D. Ghezzi, P. Goffrini, G. Uziel, R. Horvath, T. Klopstock, H. Lochmuller, P. D'Adamo, P. Gasparini, T. M. Strom, H. Prokisch, F. Invernizzi, I. Ferrero and M. Zeviani, SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy, Nat. Genet., 2009, 41, 654–656 CrossRef CAS PubMed.
- N. Maio, D. Ghezzi, D. Verrigni, T. Rizza, E. Bertini, D. Martinelli, M. Zeviani, A. Singh, R. Carrozzo and T. A. Rouault, Disease-Causing SDHAF1 Mutations Impair Transfer of Fe–S Clusters to SDHB, Cell Metab., 2016, 23, 292–302 CrossRef CAS PubMed.
- J. L. Faria, M. Berberan-Santos and M. J. Prieto, A comment on the localization of cyanine dye binding to brush-border membranes by the fluorescence quenching of n-(9-anthroyloxy) fatty acid probes, Biochim. Biophys. Acta, 1990, 1026, 133–134 CrossRef CAS.
- N. G. Fox, M. Chakrabarti, S. P. McCormick, P. A. Lindahl and D. P. Barondeau, The Human Iron-Sulfur Assembly Complex Catalyzes the Synthesis of [2Fe–2S] Clusters on ISCU2 That Can Be Transferred to Acceptor Molecules, Biochemistry, 2015, 54, 3871–3879 CrossRef CAS PubMed.
- L. K. Beilschmidt, C. S. Ollagnier de, M. Fournier, I. Sanakis, M. A. Hograindleur, M. Clemancey, G. Blondin, S. Schmucker, A. Eisenmann, A. Weiss, P. Koebel, N. Messaddeq, H. Puccio and A. Martelli, ISCA1 is essential for mitochondrial Fe4S4 biogenesis in vivo, Nat. Commun., 2017, 8, 15124 CrossRef PubMed.
- K. D. Kim, W. H. Chung, H. J. Kim, K. C. Lee and J. H. Roe, Monothiol glutaredoxin Grx5 interacts with Fe–S scaffold proteins Isa1 and Isa2 and supports Fe–S assembly and DNA integrity in mitochondria of fission yeast, Biochem. Biophys. Res. Commun., 2010, 392, 467–472 CrossRef CAS PubMed.
- A. D. Sheftel, C. Wilbrecht, O. Stehling, B. Niggemeyer, H. P. Elsasser, U. Muhlenhoff and R. Lill, The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe–4S] protein maturation, Mol. Biol. Cell, 2012, 23, 1157–1166 CrossRef CAS PubMed.
- I. Cozar-Castellano, M. M. del Valle, E. Trujillo, M. F. Arteaga, T. Gonzalez, P. Martin-Vasallo and J. Avila, hIscA: a protein implicated in the biogenesis of iron-sulfur clusters, Biochim. Biophys. Acta, 2004, 1700, 179–188 CrossRef CAS PubMed.
- U. Muhlenhoff, N. Richter, O. Pines, A. J. Pierik and R. Lill, Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe–4S] proteins, J. Biol. Chem., 2011, 286, 41205–41216 CrossRef PubMed.
- D. Brancaccio, A. Gallo, M. Mikolajczyk, K. Zovo, P. Palumaa, E. Novellino, M. Piccioli, S. Ciofi-Baffoni and L. Banci, Formation of [4Fe–4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery, J. Am. Chem. Soc., 2014, 136, 16240–16250 CrossRef CAS PubMed.
- D. Brancaccio, A. Gallo, M. Piccioli, E. Novellino, S. Ciofi-Baffoni and L. Banci, [4Fe–4S] Cluster Assembly in Mitochondria and Its Impairment by Copper, J. Am. Chem. Soc., 2017, 139, 719–730 CrossRef CAS PubMed.
- L. T. Jensen and V. C. Culotta, Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis, Mol. Cell. Biol., 2000, 20, 3918–3927 CrossRef CAS PubMed.
- A. Kaut, H. Lange, K. Diekert, G. Kispal and R. Lill, Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function, J. Biol. Chem., 2000, 275, 15955–15961 CrossRef CAS PubMed.
- A. Navarro-Sastre, F. Tort, O. Stehling, M. A. Uzarska, J. A. Arranz, T. M. Del, M. T. Labayru, J. Landa, A. Font, J. Garcia-Villoria, B. Merinero, M. Ugarte, L. G. Gutierrez-Solana, J. Campistol, A. Garcia-Cazorla, J. Vaquerizo, E. Riudor, P. Briones, O. Elpeleg, A. Ribes and R. Lill, A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe–S proteins, Am. J. Hum. Genet., 2011, 89, 656–667 CrossRef CAS PubMed.
- J. M. Cameron, A. Janer, V. Levandovskiy, N. MacKay, T. A. Rouault, W. H. Tong, I. Ogilvie, E. A. Shoubridge and B. H. Robinson, Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes, Am. J. Hum. Genet., 2011, 89, 486–495 CrossRef CAS PubMed.
- K. Bych, S. Kerscher, D. J. Netz, A. J. Pierik, K. Zwicker, M. A. Huynen, R. Lill, U. Brandt and J. Balk, The iron-sulphur protein Ind1 is required for effective complex I assembly, EMBO J., 2008, 27, 1736–1746 CrossRef CAS PubMed.
- A. D. Sheftel, O. Stehling, A. J. Pierik, D. J. Netz, S. Kerscher, H. P. Elsasser, I. Wittig, J. Balk, U. Brandt and R. Lill, Humanind1, an iron-sulfur cluster assembly factor for respiratory complex I, Mol. Cell. Biol., 2009, 29, 6059–6073 CrossRef CAS PubMed.
- A. Melber, U. Na, A. Vashisht, B. D. Weiler, R. Lill, J. A. Wohlschlegel and D. R. Winge, Role of Nfu1 and Bol3 in iron-sulfur cluster transfer to mitochondrial clients, Elife, 2016, 5, e15991 Search PubMed.
- K. Cai, G. Liu, R. O. Frederick, R. Xiao, G. T. Montelione and J. L. Markley, Structural/Functional Properties of Human NFU1, an Intermediate [4Fe–4S] Carrier in Human Mitochondrial Iron-Sulfur Cluster Biogenesis, Structure, 2016, 24, 2080–2091 CrossRef CAS PubMed.
- M. A. Uzarska, V. Nasta, B. D. Weiler, F. Spantgar, S. Ciofi-Baffoni, M. R. Saviello, L. Gonnelli, U. Muhlenhoff, L. Banci and R. Lill, Mitochondrial Bol1 and Bol3 function as assembly factors for specific iron-sulfur proteins, Elife, 2016, 5, e16673 Search PubMed.
- H. Li and C. E. Outten, Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe–2S] binding partners in iron homeostasis, Biochemistry, 2012, 51, 4377–4389 CrossRef CAS PubMed.
- V. Nasta, A. Giachetti, S. Ciofi-Baffoni and L. Banci, Structural insights into the molecular function of human [2Fe–2S] BOLA1-GRX5 and [2Fe–2S] BOLA3-GRX5 complexes, Biochim. Biophys. Acta, 2017, 1861, 2119–2131 CrossRef CAS PubMed.
- V. D. Paul and R. Lill, Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability, Biochim. Biophys. Acta, 2015, 1853, 1528–1539 CrossRef CAS PubMed.
- A. K. Sharma, L. J. Pallesen, R. J. Spang and W. E. Walden, Cytosolic iron-sulfur cluster assembly (CIA) system: factors, mechanism, and relevance to cellular iron regulation, J. Biol. Chem., 2010, 285, 26745–26751 CrossRef CAS PubMed.
- A. D. Tsaousis, E. Gentekaki, L. Eme, D. Gaston and A. J. Roger, Evolution of the cytosolic iron-sulfur cluster assembly machinery in Blastocystis species and other microbial eukaryotes, Eukaryotic Cell, 2014, 13, 143–153 CrossRef CAS PubMed.
- O. Stehling, D. J. Netz, B. Niggemeyer, R. Rosser, R. S. Eisenstein, H. Puccio, A. J. Pierik and R. Lill, Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis, Mol. Cell. Biol., 2008, 28, 5517–5528 CrossRef CAS PubMed.
- T. Okuno, H. Yamabayashi and K. Kogure, Comparison of intracellular localization of Nubp1 and Nubp2 using GFP fusion proteins, Mol. Biol. Rep., 2010, 37, 1165–1168 CrossRef CAS PubMed.
- A. Hausmann, D. J. Aguilar Netz, J. Balk, A. J. Pierik, U. Muhlenhoff and R. Lill, The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 3266–3271 CrossRef CAS PubMed.
- D. J. Netz, A. J. Pierik, M. Stumpfig, U. Muhlenhoff and R. Lill, The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol, Nat. Chem. Biol., 2007, 3, 278–286 CrossRef CAS PubMed.
- L. J. Pallesen, N. Solodovnikova, A. K. Sharma and W. E. Walden, Interaction with Cfd1 increases the kinetic lability of FeS on the Nbp35 scaffold, J. Biol. Chem., 2013, 288, 23358–23367 CrossRef CAS PubMed.
- E. J. Camire, J. D. Grossman, G. J. Thole, N. M. Fleischman and D. L. Perlstein, The Yeast Nbp35-Cfd1 Cytosolic Iron-Sulfur Cluster Scaffold Is an ATPase, J. Biol. Chem., 2015, 290, 23793–23802 CrossRef CAS PubMed.
- H. Lange, A. Kaut, G. Kispal and R. Lill, A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1050–1055 CrossRef CAS.
- J. Li, S. Saxena, D. Pain and A. Dancis, Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis, J. Biol. Chem., 2001, 276, 1503–1509 CrossRef CAS PubMed.
- D. J. Netz, M. Stumpfig, C. Dore, U. Muhlenhoff, A. J. Pierik and R. Lill, Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis, Nat. Chem. Biol., 2010, 6, 758–765 CrossRef CAS PubMed.
- L. Vernis, C. Facca, E. Delagoutte, N. Soler, R. Chanet, B. Guiard, G. Faye and G. Baldacci, A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast, PLoS One, 2009, 4, e4376 Search PubMed.
- E. L. Bastow, K. Bych, J. C. Crack, N. E. Le Brun and J. Balk, NBP35 interacts with DRE2 in the maturation of cytosolic iron-sulphur proteins in Arabidopsis thaliana, Plant J., 2017, 89, 590–600 CrossRef CAS PubMed.
- K. Bych, D. J. Netz, G. Vigani, E. Bill, R. Lill, A. J. Pierik and J. Balk, The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage, J. Biol. Chem., 2008, 283, 35797–35804 CrossRef CAS PubMed.
- M. B. Murataliev, R. Feyereisen and F. A. Walker, Electron transfer by diflavin reductases, Biochim. Biophys. Acta, 2004, 1698, 1–26 CrossRef CAS PubMed.
- L. Banci, I. Bertini, S. Ciofi-Baffoni, F. Boscaro, A. Chatzi, M. Mikolajczyk, K. Tokatlidis and J. Winkelmann, Anamorsin is a 2Fe2S cluster-containing substrate of the Mia40-dependent mitochondrial protein trapping machinery, Chem. Biol., 2011, 18, 794–804 CrossRef CAS PubMed.
- L. Banci, S. Ciofi-Baffoni, M. Mikolajczyk, J. Winkelmann, E. Bill and M. E. Pandelia, Human anamorsin binds [2Fe–2S] clusters with unique electronic properties, J. Biol. Inorg. Chem., 2013, 18, 883–893 CrossRef CAS PubMed.
- Y. Zhang, C. Yang, A. Dancis and E. Nakamaru-Ogiso, EPR studies of wild type and mutant Dre2 identify essential [2Fe–2S] and [4Fe–4S] clusters and their cysteine ligands, J. Biochem., 2017, 161, 67–78 CrossRef PubMed.
- D. J. Netz, H. M. Genau, B. D. Weiler, E. Bill, A. J. Pierik and R. Lill, The conserved protein Dre2 uses essential [2Fe–2S] and [4Fe–4S] clusters for its function in cytosolic iron-sulfur protein assembly, Biochem. J., 2016, 473, 2073–2085 CrossRef CAS PubMed.
- L. Banci, I. Bertini, V. Calderone, S. Ciofi-Baffoni, A. Giachetti, D. Jaiswal, M. Mikolajczyk, M. Piccioli and J. Winkelmann, Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7136–7141 CrossRef CAS PubMed.
- G. Song, C. Cheng, Y. Li, N. Shaw, Z. C. Xiao and Z. J. Liu, Crystal structure of the N-terminal methyltransferase-like domain of anamorsin, Proteins, 2014, 82, 1066–1071 CrossRef CAS PubMed.
- U. Muhlenhoff, J. Balk, N. Richhardt, J. T. Kaiser, K. Sipos, G. Kispal and R. Lill, Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae, J. Biol. Chem., 2004, 279, 36906–36915 CrossRef PubMed.
- P. Haunhorst, E. M. Hanschmann, L. Brautigam, O. Stehling, B. Hoffmann, U. Muhlenhoff, R. Lill, C. Berndt and C. H. Lillig, Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation, Mol. Biol. Cell, 2013, 24, 1895–1903 CrossRef CAS PubMed.
- E. Herrero and M. A. de la Torre-Ruiz, Monothiol glutaredoxins: a common domain for multiple functions, Cell. Mol. Life Sci., 2007, 64, 1518–1530 CrossRef CAS PubMed.
- H. Li, D. T. Mapolelo, S. Randeniya, M. K. Johnson and C. E. Outten, Human glutaredoxin 3 forms [2Fe–2S]-bridged complexes with human BolA2, Biochemistry, 2012, 51, 1687–1696 CrossRef CAS PubMed.
- P. Haunhorst, C. Berndt, S. Eitner, J. R. Godoy and C. H. Lillig, Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein, Biochem. Biophys. Res. Commun., 2010, 394, 372–376 CrossRef CAS PubMed.
- U. Muhlenhoff, S. Molik, J. R. Godoy, M. A. Uzarska, N. Richter, A. Seubert, Y. Zhang, J. Stubbe, F. Pierrel, E. Herrero, C. H. Lillig and R. Lill, Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster, Cell Metab., 2010, 12, 373–385 CrossRef PubMed.
- H. Li, D. T. Mapolelo, N. N. Dingra, S. G. Naik, N. S. Lees, B. M. Hoffman, P. J. Riggs-Gelasco, B. H. Huynh, M. K. Johnson and C. E. Outten, The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe–2S] cluster with cysteinyl and histidyl ligation, Biochemistry, 2009, 48, 9569–9581 CrossRef CAS PubMed.
- L. Banci, S. Ciofi-Baffoni, K. Gajda, R. Muzzioli, R. Peruzzini and J. Winkelmann, N-terminal domains mediate [2Fe–2S] cluster transfer from glutaredoxin-3 to anamorsin, Nat. Chem. Biol., 2015, 11, 772–778 CrossRef CAS PubMed.
- Y. Saito, H. Shibayama, H. Tanaka, A. Tanimura, I. Matsumura and Y. Kanakura, PICOT is a molecule which binds to anamorsin, Biochem. Biophys. Res. Commun., 2011, 408, 329–333 CrossRef CAS PubMed.
- Y. Zhang, H. Li, C. Zhang, X. An, L. Liu, J. Stubbe and M. Huang, Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1695–E1704 CrossRef CAS PubMed.
- Y. Zhang, L. Liu, X. Wu, X. An, J. Stubbe and M. Huang, Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins, J. Biol. Chem., 2011, 286, 41499–41509 CrossRef CAS PubMed.
- H. Li, M. Stumpfig, C. Zhang, X. An, J. Stubbe, R. Lill and M. Huang, The diferric-tyrosyl radical cluster of ribonucleotide reductase and cytosolic iron-sulfur clusters have distinct and similar biogenesis requirements, J. Biol. Chem., 2017, 292, 11445–11451 CrossRef CAS PubMed.
- L. Banci, F. Camponeschi, S. Ciofi-Baffoni and R. Muzzioli, Elucidating the molecular function of human BOLA2 in GRX3-Dependent anamorsin maturation pathway, J. Am. Chem. Soc., 2015, 137, 16133–16134 CrossRef CAS PubMed.
- A. G. Frey, D. J. Palenchar, J. D. Wildemann and C. C. Philpott, A Glutaredoxin-BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery, J. Biol. Chem., 2016, 291, 22344–22356 CrossRef CAS PubMed.
- X. Nuttle, G. Giannuzzi, M. H. Duyzend, J. G. Schraiber, I. Narvaiza, F. Camponeschi, S. Ciofi-Baffoni, P. H. Sudmant, O. Penn, G. Chiatante, M. Malig, J. Huddleston, C. Benner, H. A. F. Stessman, M. C. N. Marchetto, L. Denman, L. Harshman, C. Baker, A. Raja, K. Penewit, W. J. Tang, M. Ventura, F. Antonacci, J. M. Akey, C. T. Amemiya, L. Banci, F. H. Gage, A. Reymond and E. E. Eichler, Emergence of a Homo sapiens-specific gene family and the evolution of autism risk at chromosome 16p11.2, Nature, 2016, 536, 205–209 CrossRef CAS PubMed.
- C. H. Lipper, M. L. Paddock, J. N. Onuchic, R. Mittler, R. Nechushtai and P. A. Jennings, Cancer-Related NEET Proteins Transfer 2Fe–2S Clusters to Anamorsin, a Protein Required for Cytosolic Iron-Sulfur Cluster Biogenesis, PLoS One, 2015, 10, e0139699 Search PubMed.
- I. Ferecatu, S. Goncalves, M. P. Golinelli-Cohen, M. Clemancey, A. Martelli, S. Riquier, E. Guittet, J. M. Latour, H. Puccio, J. C. Drapier, E. Lescop and C. Bouton, The diabetes drug target MitoNEET governs a novel trafficking pathway to rebuild an Fe–S cluster into cytosolic aconitase/iron regulatory protein 1, J. Biol. Chem., 2014, 289, 28070–28086 CrossRef CAS PubMed.
- M. P. Golinelli-Cohen, E. Lescop, C. Mons, S. Goncalves, M. Clemancey, J. Santolini, E. Guittet, G. Blondin, J. M. Latour and C. Bouton, Redox Control of the Human Iron-Sulfur Repair Protein MitoNEET Activity via Its Iron-Sulfur Cluster, J. Biol. Chem., 2016, 291, 7583–7593 CrossRef CAS PubMed.
- F. Camponeschi, S. Ciofi-Baffoni and L. Banci, Anamorsin/Ndor1 Complex Reduces [2Fe–2S]-MitoNEET via a Transient Protein-Protein Interaction, J. Am. Chem. Soc., 2017, 139, 9479–9482 CrossRef CAS PubMed.
- K. Gari, A. M. Leon Ortiz, V. Borel, H. Flynn, J. M. Skehel and S. J. Boulton, MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism, Science, 2012, 337, 243–245 CrossRef CAS PubMed.
- O. Stehling, A. A. Vashisht, J. Mascarenhas, Z. O. Jonsson, T. Sharma, D. J. Netz, A. J. Pierik, J. A. Wohlschlegel and R. Lill, MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity, Science, 2012, 337, 195–199 CrossRef CAS PubMed.
- D. C. Odermatt and K. Gari, The CIA Targeting Complex Is Highly Regulated and Provides Two Distinct Binding Sites for Client Iron-Sulfur Proteins, Cell Rep., 2017, 18, 1434–1443 CrossRef CAS PubMed.
- C. Xu and J. Min, Structure and function of WD40 domain proteins, Protein Cell, 2011, 2, 202–214 CrossRef CAS PubMed.
- V. Srinivasan, D. J. Netz, H. Webert, J. Mascarenhas, A. J. Pierik, H. Michel and R. Lill, Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis, Structure, 2007, 15, 1246–1257 CrossRef CAS PubMed.
- M. Seki, Y. Takeda, K. Iwai and K. Tanaka, IOP1 protein is an external component of the human cytosolic iron-sulfur cluster assembly (CIA) machinery and functions in the MMS19 protein-dependent CIA pathway, J. Biol. Chem., 2013, 288, 16680–16689 CrossRef CAS PubMed.
- V. D. Paul, U. Muhlenhoff, M. Stumpfig, J. Seebacher, K. G. Kugler, C. Renicke, C. Taxis, A. C. Gavin, A. J. Pierik and R. Lill, The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion, Elife, 2015, 4, e08231 Search PubMed.
- E. L. Huttlin, L. Ting, R. J. Bruckner, F. Gebreab, M. P. Gygi, J. Szpyt, S. Tam, G. Zarraga, G. Colby, K. Baltier, R. Dong, V. Guarani, L. P. Vaites, A. Ordureau, R. Rad, B. K. Erickson, M. Wuhr, J. Chick, B. Zhai, D. Kolippakkam, J. Mintseris, R. A. Obar, T. Harris, S. Artavanis-Tsakonas, M. E. Sowa, C. P. De, J. A. Paulo, J. W. Harper and S. P. Gygi, The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell, 2015, 162, 425–440 CrossRef CAS PubMed.
- Y. Nicolet and J. C. Fontecilla-Camps, Structure-function relationships in [FeFe]-hydrogenase active site maturation, J. Biol. Chem., 2012, 287, 13532–13540 CrossRef CAS PubMed.
- D. W. Mulder, E. M. Shepard, J. E. Meuser, N. Joshi, P. W. King, M. C. Posewitz, J. B. Broderick and J. W. Peters, Insights into [FeFe]-hydrogenase structure, mechanism, and maturation, Structure, 2011, 19, 1038–1052 CrossRef CAS PubMed.
- J. Balk, A. J. Pierik, D. J. Netz, U. Muhlenhoff and R. Lill, The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins, EMBO J., 2004, 23, 2105–2115 CrossRef CAS PubMed.
- E. Urzica, A. J. Pierik, U. Muhlenhoff and R. Lill, Crucial role of conserved cysteine residues in the assembly of two iron-sulfur clusters on the CIA protein Nar1, Biochemistry, 2009, 48, 4946–4958 CrossRef CAS PubMed.
- C. Cavazza, L. Martin, S. Mondy, J. Gaillard, P. Ratet and J. C. Fontecilla-Camps, The possible role of an [FeFe]-hydrogenase-like protein in the plant responses to changing atmospheric oxygen levels, J. Inorg. Biochem., 2008, 102, 1359–1365 CrossRef CAS PubMed.
- J. Balk, D. J. Aguilar Netz, K. Tepper, A. J. Pierik and R. Lill, The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly, Mol. Cell. Biol., 2005, 25, 10833–10841 CrossRef CAS PubMed.
- M. Costanzo, B. VanderSluis, E. N. Koch, A. Baryshnikova, C. Pons, G. Tan, W. Wang, M. Usaj, J. Hanchard, S. D. Lee, V. Pelechano, E. B. Styles, M. Billmann, L. J. van Leeuwen, D. N. van Dyk, Z. Y. Lin, E. Kuzmin, J. Nelson, J. S. Piotrowski, T. Srikumar, S. Bahr, Y. Chen, R. Deshpande, C. F. Kurat, S. C. Li, Z. Li, M. M. Usaj, H. Okada, N. Pascoe, B. J. San Luis, S. Sharifpoor, E. Shuteriqi, S. W. Simpkins, J. Snider, H. G. Suresh, Y. Tan, H. Zhu, N. Malod-Dognin, V. Janjic, N. Przulj, O. G. Troyanskaya, I. Stagljar, T. Xia, Y. Ohya, A. C. Gingras, B. Raught, M. Boutros, L. M. Steinmetz, C. L. Moore, A. P. Rosebrock, A. A. Caudy, C. L. Myers, B. Andrews and C. Boone, A global genetic interaction network maps a wiring diagram of cellular function, Science, 2016, 353 Search PubMed.
- O. Stehling, J. Mascarenhas, A. A. Vashisht, A. D. Sheftel, B. Niggemeyer, R. Rosser, A. J. Pierik, J. A. Wohlschlegel and R. Lill, Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins, Cell Metab., 2013, 18, 187–198 CrossRef CAS PubMed.
- B. OuYang, L. Wang, S. Wan, Y. Luo, L. Wang, J. Lin and B. Xia, Solution structure of monomeric human FAM96A, J. Biomol. NMR, 2013, 56, 387–392 CrossRef CAS PubMed.
- K. E. Chen, A. A. Richards, J. K. Ariffin, I. L. Ross, M. J. Sweet, S. Kellie, B. Kobe and J. L. Martin, The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 637–648 CAS.
- E. Weerapana, C. Wang, G. M. Simon, F. Richter, S. Khare, M. B. Dillon, D. A. Bachovchin, K. Mowen, D. Baker and B. F. Cravatt, Quantitative reactivity profiling predicts functional cysteines in proteomes, Nature, 2010, 468, 790–795 CrossRef CAS PubMed.
- C. Zhai, Y. Li, C. Mascarenhas, Q. Lin, K. Li, I. Vyrides, C. M. Grant and B. Panaretou, The function of ORAOV1/LTO1, a gene that is overexpressed frequently in cancer: essential roles in the function and biogenesis of the ribosome, Oncogene, 2014, 33, 484–494 CrossRef CAS PubMed.
- A. S. Upadhyay, K. Vonderstein, A. Pichlmair, O. Stehling, K. L. Bennett, G. Dobler, J. T. Guo, G. Superti-Furga, R. Lill, A. K. Overby and F. Weber, Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity, Cell. Microbiol., 2014, 16, 834–848 CrossRef CAS PubMed.
- A. S. Upadhyay, O. Stehling, C. Panayiotou, R. Rosser, R. Lill and A. K. Overby, Cellular requirements for iron-sulfur cluster insertion into the antiviral radical SAM protein viperin, J. Biol. Chem., 2017, 292, 13879–13889 CrossRef CAS PubMed.
- P. Csere, R. Lill and G. Kispal, Identification of a human mitochondrial ABC transporter, the functional orthologue of yeast Atm1p, FEBS Lett., 1998, 441, 266–270 CrossRef CAS PubMed.
- S. Kushnir, E. Babiychuk, S. Storozhenko, M. W. Davey, J. Papenbrock, R. R. De, G. Engler, U. W. Stephan, H. Lange, G. Kispal, R. Lill and M. Van Montagu, A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik, Plant Cell, 2001, 13, 89–100 CrossRef CAS.
- S. Chen, R. Sanchez-Fernandez, E. R. Lyver, A. Dancis and P. A. Rea, Functional characterization of AtATM1, AtATM2, and AtATM3, a subfamily of Arabidopsis half-molecule ATP-binding cassette transporters implicated in iron homeostasis, J. Biol. Chem., 2007, 282, 21561–21571 CrossRef CAS PubMed.
- M. Martinez-Garcia, J. Campos-Salinas, M. Cabello-Donayre, E. Pineda-Molina, F. J. Galvez, L. M. Orrego, M. P. Sanchez-Canete, S. Malagarie-Cazenave, D. M. Koeller and J. M. Perez-Victoria, LmABCB3, an atypical mitochondrial ABC transporter essential for Leishmania major virulence, acts in heme and cytosolic iron/sulfur clusters biogenesis, Parasites Vectors, 2016, 9, 7 CrossRef PubMed.
- N. Mitsuhashi, T. Miki, H. Senbongi, N. Yokoi, H. Yano, M. Miyazaki, N. Nakajima, T. Iwanaga, Y. Yokoyama, T. Shibata and S. Seino, MTABC3, a novel mitochondrial ATP-binding cassette protein involved in iron homeostasis, J. Biol. Chem., 2000, 275, 17536–17540 CrossRef CAS PubMed.
- C. Kumar, A. Igbaria, B. d'Autreaux, A. G. Planson, C. Junot, E. Godat, A. K. Bachhawat, A. Delaunay-Moisan and M. B. Toledano, Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control, EMBO J., 2011, 30, 2044–2056 CrossRef CAS PubMed.
- W. Qi, J. Li, C. Y. Chain, G. A. Pasquevich, A. F. Pasquevich and J. A. Cowan, Glutathione complexed Fe–S centers, J. Am. Chem. Soc., 2012, 134, 10745–10748 CrossRef CAS PubMed.
- J. Li, S. A. Pearson, K. D. Fenk and J. A. Cowan, Glutathione-coordinated [2Fe–2S] cluster is stabilized by intramolecular salt bridges, J. Biol. Inorg. Chem., 2015, 20, 1221–1227 CrossRef CAS PubMed.
- W. Qi, J. Li, C. Y. Chain, G. A. Pasquevich, A. F. Pasquevich and J. A. Cowan, Glutathione-complexed iron-sulfur clusters. Reaction intermediates and evidence for a template effect promoting assembly and stability, Chem. Commun., 2013, 49, 6313–6315 RSC.
- J. Li and J. A. Cowan, Glutathione-coordinated [2Fe–2S] cluster: a viable physiological substrate for mitochondrial ABCB7 transport, Chem. Commun., 2015, 51, 2253–2255 RSC.
- W. Qi, J. Li and J. A. Cowan, A structural model for glutathione-complexed iron-sulfur cluster as a substrate for ABCB7-type transporters, Chem. Commun., 2014, 50, 3795–3798 RSC.
- V. Srinivasan, A. J. Pierik and R. Lill, Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1, Science, 2014, 343, 1137–1140 CrossRef CAS PubMed.
- G. Kispal, P. Csere, B. Guiard and R. Lill, The ABC transporter Atm1p is required for mitochondrial iron homeostasis, FEBS Lett., 1997, 418, 346–350 CrossRef CAS PubMed.
- J. Y. Lee, J. G. Yang, D. Zhitnitsky, O. Lewinson and D. C. Rees, Structural basis for heavy metal detoxification by an Atm1-type ABC exporter, Science, 2014, 343, 1133–1136 CrossRef CAS PubMed.
- T. A. Schaedler, J. D. Thornton, I. Kruse, M. Schwarzlander, A. J. Meyer, H. W. van Veen and J. Balk, A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly, J. Biol. Chem., 2014, 289, 23264–23274 CrossRef CAS PubMed.
- H. Lange, T. Lisowsky, J. Gerber, U. Muhlenhoff, G. Kispal and R. Lill, An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins, EMBO Rep., 2001, 2, 715–720 CrossRef CAS PubMed.
- H. K. Ozer, A. C. Dlouhy, J. D. Thornton, J. Hu, Y. Liu, J. J. Barycki, J. Balk and C. E. Outten, Cytosolic Fe–S Cluster Protein Maturation and Iron Regulation Are Independent of the Mitochondrial Erv1/Mia40 Import System, J. Biol. Chem., 2015, 290, 27829–27840 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2018 |