Identification and analysis of the cleavage site in a signal peptide using SMOTE, dagging, and feature selection methods†
ShaoPeng
Wang‡
 a,
Deling
Wang‡
b,
JiaRui
Li
*a,
Tao
Huang
a,
Deling
Wang‡
b,
JiaRui
Li
*a,
Tao
Huang
 *c and
Yu-Dong
Cai
*c and
Yu-Dong
Cai
 *a
*a
aSchool of Life Sciences, Shanghai University, Shanghai 200444, People's Republic of China. E-mail: wsptfb@163.com; jerryakii@qq.com; caiyudong@staff.shu.edu.cn; Fax: +86-21-66136109; Tel: +86-21-66135166 Tel: +86-21-66136132
bDepartment of Medical Imaging, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, 651 Dong feng Road East, Guangzhou, Guangdong 510060, People's Republic of China. E-mail: wangdl@sysucc.org.cn
cInstitute of Health Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, People's Republic of China. E-mail: huangtao@sibs.ac.cn; Tel: +86-21-54923269
First published on 20th December 2017
Abstract
The cleavage site of a signal peptide located in the C-region can be recognized by the signal peptidase in eukaryotic and prokaryotic cells, and the signal peptides are typically cleaved off during or after the translocation of the target protein. The identification of cleavage sites remains challenging because of the diverse lengths of signal peptides and the weak conservation of the motif recognized by the signal peptidase. In this study, we applied a fast and accurate computational method to identify cleavage sites in signal peptides based on protein sequences. We collected 2683 protein sequences with experimentally validated N-terminus signal peptides from the newly released UniProt database. A 20 amino acid-length peptide segment flanking the cleavage site was extracted from each protein, and four types of features were used to encode the peptide segment. We applied the synthetic minority oversampling technique, maximum relevance minimum redundancy, and incremental feature selection, together with dagging and random forest algorithms, to identify the optimal features that can lead to the optimal identification of the cleavage sites. The optimal dagging and random forest classifiers constructed on the optimal features yielded Youden's indexes of 0.871 and 0.736, respectively. The sensitivity, specificity, and accuracy yielded by the optimal dagging classifier all exceeded 0.9, which demonstrated the high prediction ability of the optimal dagging classifier. These optimal features that resulted from the dagging algorithm, predominantly the position-specific scoring matrix and the amino acid factor, played crucial roles in identifying the cleavage sites by a literature review. The prediction method proposed in this study was confirmed to be a powerful tool for recognizing cleavage sites from protein sequences.
1. Introduction
Protein targeting and translocation are essential for the living of prokaryotic and eukaryotic cells. As initially demonstrated in 1975, the N-terminal signal peptide was confirmed to guide proteins to the endoplasmic reticulum (ER) after their synthesis from ribosomes and through a channel in the membrane.1,2 The lengths of signal peptides are diverse, ranging from 15 to 40 amino acids, with certain exceptions of much longer lengths.3 In the process of targeting to the ER, besides the signal peptide, another three molecules are also critical: the cytosolic signal recognition particle,4,5 the ER membrane-bound SRP receptor,6 and the ER membrane-spanning hetero-trimetric Sec61 complex.7 In normal circumstances, the SRP recognizes the hydrophobic domain of a signal peptide and binds to it in a newly translated precursor protein in eukaryotes, which arrests the process of translation and directs the ribosome with the precursor protein to the SRP receptor located in the ER membrane.4,5 Then, the Sec61 complex forms a channel in the ER membrane and the precursor protein can enter the ER lumen by passing through it.8 After this step, the signal peptide is cleaved off by the type I signal peptidase (SP-I)9 in the activated cleavage site, thus starting the protein translocation in the ER. The cleaved signal peptide is then released into the membrane of the ER and digested by the signal peptide peptidase.10 However, in several exceptional examples, the signal peptide is not cleaved and can execute diverse post-targeting functions.11,12Traditional methods based on the weight matrix have limited power in identifying cleavage sites in signal peptides due to the diversity of the composition and the length of the sequences flanking the cleavage sites.13 With the development of data mining, several machine learning algorithms14–18 have been designed to speed up the identification procedures for different biological problems. Thus, the tendency for identifying cleavage sites has shifted to machine learning methods to obtain a better prediction performance.19–21 Among them, the most well-known and accurate method was demonstrated to be SignalP (including ANN and HMM versions).22 Simultaneously, several other methods including TargetP, LipoP, and PredSi were proposed to predict signal peptides and cleavage sites in eukaryotes and prokaryotes.23–25 In addition, Hiss and Schneider also developed a prediction method to recognize long signal peptides, which considered both the overall length and secondary structure of long signal peptides.26
However, the convenience in updating proposed methods usually cannot catch up to the database that contained newly released protein sequences with cleavage sites in signal peptides. For example, the newly updated SignalP method with version 4.0 that was used to distinguish signal peptides from transmembrane regions was published in 2011,27 and the most recent method involved in predicting signal peptide cleavage sites was reported in 2014.28 Thus, several newly released signal peptides with cleavage sites were not used in these methods. Another factor that causes the difficulty in identifying the cleavage sites is data imbalance, which means the positive sample size is much larger than the negative one.
In this study, we proposed a machine learning based prediction framework to identify the cleavage sites of signal peptides based on the protein sequence. A total of 2683 protein sequences with experimentally validated signal peptides and cleavage sites were retrieved from the newly released UniProt database. Four types of widely used features, including amino acids (AAs), the position-specific scoring matrix (PSSM), the AA factor, and the disorder status, were used to encode peptide segments surrounding the cleavage sites in our constructed dataset. To solve the data imbalance, the synthetic minority oversampling technique (SMOTE)29 was used to generate synthesized positive samples, and several classifiers obtained in later steps were built on the new balanced datasets. Features ranked by the maximum-relevance minimum-redundancy (mRMR)30 were then selected through the incremental feature selection (IFS) method and used to build a series of dagging17 classifiers, which were evaluated using the 10-fold cross-validation test. We identified the optimal dagging classifier that produced a Youden's index31 of 0.871, which suggests its considerable power in recognizing the cleavage sites. As a comparison, we also used the random forest (RF)18 algorithm to predict cleavage sites using the same dataset, which produced the optimal RF classifier with a relatively lower Youden's index of 0.736. These features selected for building the optimal dagging classifier were further analyzed through a literature review, which confirmed their relevance in locating the cleavage sites.
2. Materials and methods
2.1 Datasets
To collect protein sequences with signal peptides and cleavage sites on their N-terminus, the UniProt database32 (http://www.uniprot.org/, released: 2016_07) was retrieved and the “signal peptide” was used as the keyword. As a result, 45![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 534 reviewed items were obtained and those annotated with “By similarity” or “Sequence analysis” were discarded. Furthermore, proteins shorter than 50aa or proteins with signal peptides shorter than 15aa were discarded. In addition, 371 protein sequences containing hydrophobic transmembrane domains (TDs) were also screened out. Finally, we obtained 2683 protein sequences annotated by their UniProt IDs and organisms, which are provided in the ESI,† S1. Because these protein sequences were derived from 722 heterogeneous organisms, we also provided the number of sequences in each individual organism, whose information is listed in the ESI,† S2.
534 reviewed items were obtained and those annotated with “By similarity” or “Sequence analysis” were discarded. Furthermore, proteins shorter than 50aa or proteins with signal peptides shorter than 15aa were discarded. In addition, 371 protein sequences containing hydrophobic transmembrane domains (TDs) were also screened out. Finally, we obtained 2683 protein sequences annotated by their UniProt IDs and organisms, which are provided in the ESI,† S1. Because these protein sequences were derived from 722 heterogeneous organisms, we also provided the number of sequences in each individual organism, whose information is listed in the ESI,† S2.
For each sequence, we investigated the neighboring sequences of the cleavage sites similar to previous studies.33 The last 15 residues in the target peptide and five residues closely proximal to it downstream were combined as a 20-amino acid length peptide segment, which was indicated as a positive sample. The peptide segment can be represented as A−14A−13…A0…A4A5, which consisted of the upstream 15 residues of the signal peptides and the five downstream residues of the mature proteins after cleavage at the site between A0 and A1.
Meanwhile, a sliding window strategy toward the C-terminus with a walking distance of one residue was used to extract every 20-amino acid length peptide segment in the next 49 residues of each sequence. Besides the positive peptide segment, 29 peptide segments can also be extracted from each sequence and were regarded as negative samples. From all of the 2683 sequences, 2683 positive samples and 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833 negative samples were accordingly used in our study (listed in the ESI,† S3).
833 negative samples were accordingly used in our study (listed in the ESI,† S3).
2.2 Feature vectors
Four types of features, namely, AA features, PSSM conservation scores, the AA factor, and the disorder status, were used to encode the samples in our dataset as described in previous studies to capture key features of the extracted twenty-amino acid length peptide segments and construct an effective prediction classifier.34–41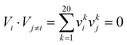 | (1) |
Accordingly, we extracted 20 × 20 = 400 features for this type of feature.
In total, 920 (400 + 400 + 100 + 20) features can be extracted from each peptide segment. The distribution of the 920 features is listed in Table 1.
| Feature type | Description | Number of features |
|---|---|---|
| AA | Composition of residues | 400 |
| PSSM | Conservation status of a residue | 400 |
| AA factor | 5 factors represent properties | 100 |
| Disorder | Probability of a residue in a disorder region | 20 |
| Total | — | 920 |
2.3 SMOTE
As mentioned above, the number of the negative samples is much larger than that of the positive ones (approximately 29 times). The classification model based on an imbalanced dataset could hardly be trained. To tackle an imbalanced dataset, some advanced techniques have been proposed, such as ensemble classification,48–50 and choosing high-quality samples from the majority class.51 In this study, we adopted SMOTE29 to challenge this problem. SMOTE is an oversampling method, which can generate a predefined number of synthetic samples from the samples in the minority class. The oversampling synthesis procedure can be described as follows. For each sample x in the minority class, its Euclidean distance to all other samples in this class was calculated. Then, k (k is a parameter) nearest neighbors of this sample were selected. One neighbor from these neighbors, such as y, was randomly selected to generate a new sample z by the following equation:| z = x + a·(y − x) | (2) |
Weka52 is a suite of software that collects several machine learning algorithms and data processing tools. The tool named “SMOTE” implements the above mentioned SMOTE method. In our work, we applied “SMOTE” to generate newly synthesized positive samples that were 28 times as many as original positive samples and thus balance the positive and negative samples.
2.4 Feature selection
In the original feature vector, features did not contribute equally to identifying signal peptides and some of them played key roles in depicting their essential attributes. Some feature selection techniques should be employed to analyze them and extract the important ones. To date, several feature selection methods have been proposed in the field of data mining, such as maximum relevance maximum distance (MRMD),53,54 Monte-Carlo Feature Selection (MCFS),55 and ReliefF.56 Here, we adopted two popular feature selection methods, namely, mRMR and IFS, which were used sequentially in this study to evaluate the importance of the 920 features in the feature vector and select the optimal feature subset.As a mutual information-based method, the relevance and redundancy of one feature between target variables and other features were all defined as their mutual information, which can be formulated in the following equation:
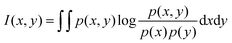 | (3) |
For a classification or regression problem involving N features, the mRMR method executed a loop procedure with N rounds. Let Ω be a set with N features, Ωs be a set containing the already selected features (it is an empty set before the method executed), and Ωt be a set including the to-be selected features, i.e., Ωt = Ω − Ωs. For each feature f in Ωt, calculate the following two values:
| D = I(f, c) | (4) |
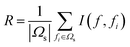 | (5) |
| F = [f1,f2,…,fN] | (6) |
2.5 Classification algorithm
Given a dataset, it is quite difficult to select a proper classification algorithm because lots of classification algorithms, like a Support Vector Machine (SVM)14,15 and LibD3C,16 have been proposed and each of them has its own advantages and disadvantages. In this study, we tried two classic machine learning algorithms: dagging17 and random forest18 and selected the best one to construct the classifier.Dagging17 is one type of algorithm used to construct meta-classifiers. For a given training dataset with N samples, M (M is a free parameter) datasets can be built and derived from the original training dataset. Each dataset has a number of n (n < N) samples and no two datasets have common samples. The sampling technique used in dagging is the so-called sampling without replacement. With the increased number of sample datasets, any sample in the original training dataset can be selected as a component of a given dataset. Thus, a basic classifier was constructed on each of the sample datasets, and finally many classifiers will be obtained after the training process. For a query sample in the testing dataset, its target variable is determined by the most votes of predicted outputs of all the basic classifiers.
The RF algorithm18 is also widely known as the notion of random decision forests,64 which, as an ensemble learning algorithm, can be applied to classification, regression and other tasks. In this algorithm, two innovative techniques, called the bootstrap (bagging method)65 and random selection of sub-features,17 were combined to train classifiers for promoting their predicted accuracy. In the bootstrap procedure, a training dataset with n samples was repeatedly sampled B times (as a free parameter). Each time, randomly selected n samples from the dataset were replaced as the new training dataset. Then, a subset of features was randomly selected by splitting the original feature space. As the authors suggested in the classification tasks with p features, 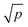 (rounded down) were selected and used in each split. At last, B decision trees were constructed to form random forests. In a query sample, its class is decided by the majority votes from all decision trees.
(rounded down) were selected and used in each split. At last, B decision trees were constructed to form random forests. In a query sample, its class is decided by the majority votes from all decision trees.
In this study, tools named “Dagging” and “RandomForest” in Weka66 software were used to build classifiers using features in feature subsets derived from the IFS method. For convenience, these two tools were executed with their default parameters.
2.6 Measurements
Based on one classification algorithm, one classifier can be constructed using features in each feature subset. The prediction abilities of these classifiers were evaluated using the widely adopted 10-fold cross-validation test, which obtained an approximated result and consumed lower computation cost compared to the jackknife cross-validation test.67,68 During the 10-fold cross-validation test, the SMOTE method was used after each splitting of the original dataset and applied only on the training sub-dataset.To quantitatively measure the prediction performance of classifiers, four measurements, namely, sensitivity (SN), specificity (SP), accuracy (ACC), and Youden's index (J),31 were used in our study, which are defined as follows:
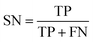 | (7) |
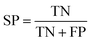 | (8) |
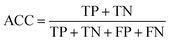 | (9) |
| J = SN + SP − 1 | (10) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833 negative samples and 2683 positive samples. The negative samples were about 29 times as many as the positive ones. It is a highly imbalanced dataset. In this case, as suggested in some previous studies,69–74 Youden's index was recommended as a better evaluation measurement than the Mathews correlation coefficient (MCC),75,76 a well-known balanced measurement for an imbalanced dataset, for a highly imbalanced dataset because it directly integrates the prediction accuracies of positive and negative samples. Thus, we adopted it as a major one to evaluate the prediction abilities of the constructed classifiers in this study.
833 negative samples and 2683 positive samples. The negative samples were about 29 times as many as the positive ones. It is a highly imbalanced dataset. In this case, as suggested in some previous studies,69–74 Youden's index was recommended as a better evaluation measurement than the Mathews correlation coefficient (MCC),75,76 a well-known balanced measurement for an imbalanced dataset, for a highly imbalanced dataset because it directly integrates the prediction accuracies of positive and negative samples. Thus, we adopted it as a major one to evaluate the prediction abilities of the constructed classifiers in this study.
3. Results
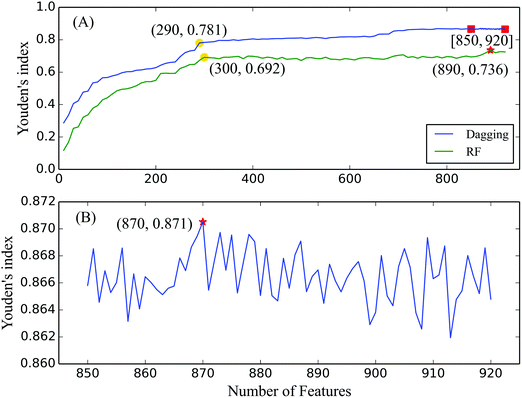
As introduced in Sections 2.2 and 2.4, each sample in our dataset was represented as 920 features and the mRMR method was adopted to rank them, which produced MaxRel and mRMR feature lists (ESI,† S4). Based on the mRMR feature list, a two-step strategy of classification and evaluation by the IFS method using dagging and RF algorithms was employed to identify the optimal feature subsets.3.1 Result obtained from the dagging algorithm
In the first step, classifiers were built using the dagging algorithm by incrementally adding ten features (see Fig. 1(A)). The prediction performance of each classifier was evaluated using the 10-fold cross-validation test, which produced four measurements (provided in the ESI,† S5) to represent their prediction abilities in identifying the cleavage sites in peptide segments. As shown in the IFS curve in Fig. 1(A), high Youden's indexes are obtained when feature numbers ranged from 850 to 920, which suggested that the optimal number of features was located in this interval. In addition, by carefully checking the IFS curve of the dagging algorithm, we found that the turning point is located at x = 290, which yielded a Youden's index of 0.781. After that point, the increase of the IFS curve of the dagging algorithm becomes slow.In the second step, 71 classifiers were built on the feature subsets with increased feature numbers from 850 to 920, which were also evaluated using the 10-fold cross-validation test (ESI,† S5). Based on the IFS curve in Fig. 1(B), when the first 870 features were used, the corresponding classifier with a Youden's index of 0.871 was regarded as the optimal classifier. Simultaneously, the first 870 features were regarded as the optimal features. Besides the Youden's index, the SN, SP, and ACC yielded by the optimal classifier are listed in Table 2. All three measurements exceeded 0.9, indicating the good prediction ability of the optimal classifier.
| Classification algorithm | Number of features | SN | SP | ACC | Youden's index |
|---|---|---|---|---|---|
| Dagging | 870 | 0.917 | 0.953 | 0.952 | 0.871 |
| Random forest | 890 | 0.750 | 0.986 | 0.978 | 0.736 |
3.2 Result obtained from the RF algorithm
As a comparison, we also applied the RF algorithm to build classifiers and identify cleavage sites on the first series of feature subsets. Similar to that of the dagging algorithm, each classifier was tested using the 10-fold cross-validation test and the obtained four measurements are listed in the ESI,† S5. To make a clear comparison, we also plotted its IFS curve in Fig. 1(A). The optimal Youden's index of the RF algorithm is 0.736 when the top 890 features were used, which is about 0.15 less than that of the optimal dagging classifier, suggesting that the optimal dagging classifier yielded a better performance than the optimal RF classifier.4. Discussion
As introduced in Section 3.1, the first 870 features in the mRMR feature list were regarded as the optimal features. To examine the relevance of these optimal features to the cleavage sites, we analyzed the top 10% features in the MaxRel feature list, which are regarded as the most important features due to their high ranks. We also selected the top 100 features in the mRMR feature list for a literature review to further confirm their roles in identifying cleavage sites. A detailed analysis is provided below.4.1 Analysis of the top 10% features in the MaxRel feature list
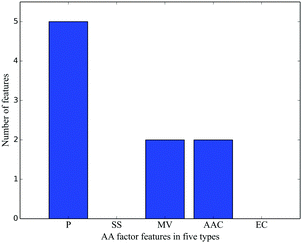
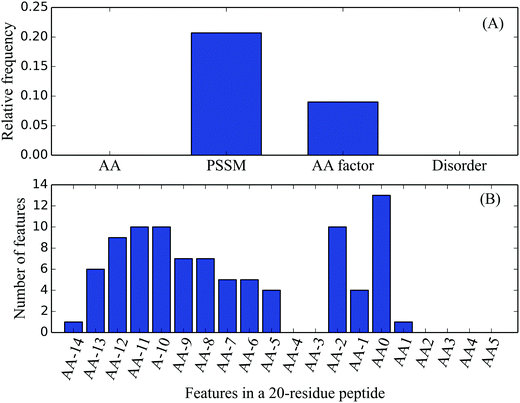
Among the top 10% MaxRel features, 83 belonged to PSSM features, and nine were AA factor features. According to the number of features in each feature type, their relative frequencies can be calculated. For example, there were a total of 400 PSSM features, its relative frequency for the top 10% MaxRel features was 0.208 (83/400). The relative frequencies of the four feature types are listed in Table 3 and Fig. 2(A). As shown in Fig. 2(A), the PSSM and AA factor have much larger relative frequencies than others, indicating the strong relevance of these two types of features with the cleavage sites. In contrast, Fig. 2(B) shows the distribution of the top 10% MaxRel features in each site of the 20-residue length peptide segment. These features are enriched in two groups of sites: the first group ranges from sites −14 to −5 and the second one contains sites −2 to 1. Site 0 had the most number of features compared to the other sites, which reveals that the residues within the signal peptides but not the mature proteins are particularly important in identifying the cleavage sites.| Feature type | Number of features | Relative frequency |
|---|---|---|
| AA | 0 | 0 |
| PSSM | 83 | 0.208 |
| AA factor | 9 | 0.090 |
| Disorder | 0 | 0 |
 | ||
| Fig. 2 Distributions of the top 10% features in the MaxRel feature list on four feature types and a 20-residue length peptide segment. | ||
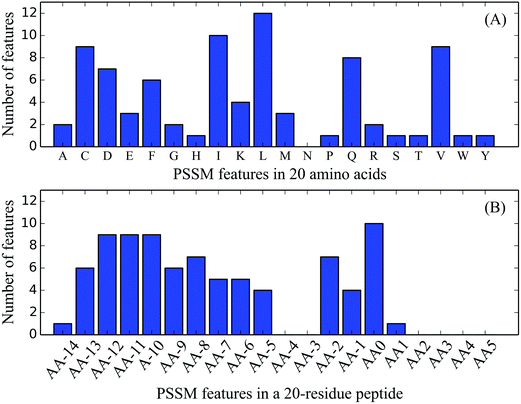 | ||
| Fig. 3 Distributions of the 83 optimal PSSM features on 20 native AAs and a 20-residue length peptide segment. | ||
Although the sequence similarity is limited, the signal peptides share three general regions: (1) a slightly positively charged N-region whose length is rather diverse; (2) a hydrophobic core H-region; and (3) a polar and uncharged C-region with the cleavage sites.77,78 The net positive charge introduced by the AA residues in the N-domain has been investigated to enhance the processing and translocation rates of a precursor protein.79 However, some of the precursor proteins with neutral or negatively charged N-domains can also be processed, leading to reduced rates.80 Therefore, the above-mentioned findings can be used as one of the possible reasons to elucidate the occurrence of K, D, and E AAs in Fig. 3(A).
The hydrophobic H-region located in the middle of the signal peptide with a length ranging from seven to fifteen amino acids is an essential element to the function of the signal peptide.80 Its critical role is partially proven by the observation that mutations in other sites of the signal peptide can be compensated for by the increased hydrophobicity in the H-region.81,82 Our result showed that most of the PSSM features representing hydrophobic residues were located in the H-region, including F, I, L, and V, which suggested that these residues in the H-region might play a role in determining the cleavage sites.
In the C-region, the most attractive characteristic is the peptidase cleavage site. The type I peptidase is responsible for ordinary pre-proteins with conserved residues located in sites −3 to −1 (sites −2 and 0 in our study) in the C-domain,83 which forms a B1–X–B2 motif, and B1 and B2 are preferred to be small and uncharged residues, such as glycine.84 Our results showed that two features were exactly located in −2 and 0 sites (Site-2_PSSM_A and Site0_PSSM_A), which were the top two PSSM features. In addition, the 22nd feature was a glycine AA located in site 0. All the results are consistent with previous knowledge and emphasize again the critical roles of these two sites in determining the cleavage sites.
In Fig. 3(B), the distribution of PSSM features on 20 sites is similar to that of all features shown in Fig. 2(B), which further indicates the biological importance of these conserved sites. Therefore, according to the positions of the three regions, most of the sites in the first group can be in the H-region and the majority of (except one feature in site 1) the sites in the second group belong to the C-region. Accordingly, the combination of site- and residue-preferences in PSSM features would be beneficial to identify cleavage sites by considering the aforementioned analyses.
4.2 Analysis of the non-redundant features in the mRMR feature list
In the first 100 mRMR features, most were included in the top 10% MaxRel features, except 13 features (Table 4). Among these features, nine were PSSM and four were derived from AA factor features. Among these nine PSSM features, eight were located in sites −4 to 0 (except one feature in site 2), which belongs to the C-domain, suggesting these nine features to be relevant to cleavage sites as discussed in Section 4.1.1. For the four AA factor features, three of them belonged to bipolar, and the analysis on this property is provided in Section 4.1.2.| No. | Feature name | Score |
|---|---|---|
| 62 | Site2_PSSM_E | 0.059 |
| 70 | Site-3_PSSM_M | 0.056 |
| 74 | Site2_AA_factor_4 | 0.055 |
| 78 | Site-5_AA_factor_1 | 0.054 |
| 81 | Site-1_PSSM_D | 0.052 |
| 85 | Site-4_PSSM_K | 0.052 |
| 89 | Site-8_AA_factor_1 | 0.051 |
| 90 | Site0_PSSM_F | 0.051 |
| 93 | Site-1_PSSM_W | 0.050 |
| 94 | Site-4_PSSM_T | 0.050 |
| 95 | Site-12_AA_factor_1 | 0.050 |
| 97 | Site-3_PSSM_T | 0.050 |
| 99 | Site0_PSSM_N | 0.049 |
4.3 Residue-preferences on the peptide segments with cleavage sites
To further illustrate the occurrence frequencies of residues on each site of the peptide segments, the WebLogo program85 was used to plot their distributions. As shown in Fig. 5, residues that can have higher occurrence in each site are placed in the upper positions of the site and different colors represent their side-chain properties. At sites −14 to −5, the most preferred residues are L, A, V, and F/I. Residue distributions on sites −2 to 0 satisfied the aforementioned motif B1–X–B2 with A as a more preferred residue.83 For residues in the mature protein at sites 1 to 5, residues A and P are more prevalent. Except that, no strongly obvious residue tendency was observed and many different types of residues can occur on these sites. According to the analyses in Sections 4.1 and 4.2, the distributions of the selected 92 features were basically consistent with the residue- and site-preferences in peptide segments with cleavage sites, which indicates the effectiveness of these features and our method. | ||
| Fig. 5 Residue preferences on a peptide segment with a cleavage site. The increased position of one residue favors its occurrence on the site. | ||
5. Conclusion
In this study, we built an effective framework to search for sequence properties that can identify cleavage sites from protein sequences via the SMOTE, mRMR, and IFS methods. A detailed analysis on the top 10% features in the MaxRel list and the first 100 mRMR features showed their crucial roles in locating the cleavage sites. Our results may elucidate the molecular mechanisms of the recognition of cleavage sites by the signal peptidases and provide a new perspective in the identification of cleavage sites in a signal peptide.Conflicts of interest
No potential conflict of interest was reported by the authors.Funding
This work was supported by the National Natural Science Foundation of China [grant number 31371335].References
- G. Blobel and B. Dobberstein, J. Cell Biol., 1975, 67, 835–851 CrossRef CAS PubMed.
- G. Blobel and B. Dobberstein, J. Cell Biol., 1975, 67, 852–862 CrossRef CAS PubMed.
- K. H. Choo, T. W. Tan and S. Ranganathan, BMC Bioinf., 2005, 6, 249 CrossRef PubMed.
- V. Siegel and P. Walter, Cell, 1988, 52, 39–49 CrossRef CAS PubMed.
- S. L. Wolin and P. Walter, J. Cell Biol., 1993, 121, 1211–1219 CrossRef CAS PubMed.
- R. Gilmore, P. Walter and G. Blobel, J. Cell Biol., 1982, 95, 470–477 CrossRef CAS PubMed.
- R. J. Deshaies, S. L. Sanders, D. A. Feldheim and R. Schekman, Nature, 1991, 349, 806–808 CrossRef CAS PubMed.
- T. A. Rapoport, FEBS J., 2008, 275, 4471–4478 CrossRef CAS PubMed.
- E. A. Evans, R. Gilmore and G. Blobel, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 581–585 CrossRef CAS.
- A. Weihofen, K. Binns, M. K. Lemberg, K. Ashman and B. Martoglio, Science, 2002, 296, 2215–2218 CrossRef CAS PubMed.
- R. S. Hegde, Mol. Cell, 2002, 10, 697–698 CrossRef CAS PubMed.
- M. K. Lemberg and B. Martoglio, Mol. Cell, 2002, 10, 735–744 CrossRef CAS PubMed.
- G. von Heijne, Nucleic Acids Res., 1986, 14, 4683–4690 CrossRef CAS PubMed.
- D. Meyer, F. Leisch and K. Hornik, Neurocomputing, 2003, 55, 169–186 CrossRef.
- V. V. Corinna Cortes, Mach. Learn., 1995, 20, 273–297 Search PubMed.
- C. Lin, W. Chen, C. Qiu, Y. Wu, S. Krishnan and Q. Zou, Neurocomputing, 2014, 123, 424–435 CrossRef.
- K. M. Ting and I. H. Witten, presented in part at the Fourteenth International Conference on Machine Learning, San Francisco, CA., 1997.
- L. Breiman, Mach. Learn., 2001, 45, 5–32 CrossRef.
- G. Schneider and U. Fechner, Proteomics, 2004, 4, 1571–1580 CrossRef CAS PubMed.
- H. Nielsen, J. Engelbrecht, S. Brunak and G. von Heijne, Protein Eng., 1997, 10, 1–6 CrossRef CAS PubMed.
- H. Nielsen and A. Krogh, Int. Conf. Intell. Syst. Mol. Biol., 1998, 6, 122–130 CAS.
- J. D. Bendtsen, H. Nielsen, G. von Heijne and S. Brunak, J. Mol. Biol., 2004, 340, 783–795 CrossRef PubMed.
- K. Hiller, A. Grote, M. Scheer, R. Munch and D. Jahn, Nucleic Acids Res., 2004, 32, W375–379 CrossRef CAS PubMed.
- A. S. Juncker, H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen and A. Krogh, Protein Sci., 2003, 12, 1652–1662 CrossRef CAS PubMed.
- O. Emanuelsson, H. Nielsen, S. Brunak and G. von Heijne, J. Mol. Biol., 2000, 300, 1005–1016 CrossRef CAS PubMed.
- J. A. Hiss and G. Schneider, Briefings Bioinf., 2009, 10, 569–578 CrossRef CAS PubMed.
- T. N. Petersen, S. Brunak, G. von Heijne and H. Nielsen, Nat. Methods, 2011, 8, 785–786 CrossRef CAS PubMed.
- S. W. Zhang, T. H. Zhang, J. N. Zhang and Y. Huang, Mol. Inf., 2014, 33, 230–239 CrossRef CAS PubMed.
- N. V. Chawla, K. W. Bowyer, L. O. Hall and W. P. Kegelmeyer, J. Artif. Intell. Res., 2002, 16, 321–357 Search PubMed.
- H. Peng, F. Long and C. Ding, IEEE Trans. Pattern Anal. Mach. Intell., 2005, 27, 1226–1238 CrossRef PubMed.
- Y. W. Youden, Cancer, 1950, 3, 32–35 CrossRef PubMed.
- A. Bairoch, U. Consortium, L. Bougueleret, S. Altairac, V. Amendolia, A. Auchincloss, G. Argoud-Puy, K. Axelsen, D. Baratin, M. C. Blatter, B. Boeckmann, J. Bolleman, L. Bollondi, E. Boutet, S. B. Quintaje, L. Breuza, A. Bridge, E. Decastro, L. Ciapina, D. Coral, E. Coudert, I. Cusin, G. Delbard, D. Dornevil, P. D. Roggli, S. Duvaud, A. Estreicher, L. Famiglietti, M. Feuermann, S. Gehant, N. Farriol-Mathis, S. Ferro, E. Gasteiger, A. Gateau, V. Gerritsen, A. Gos, N. Gruaz-Gumowski, U. Hinz, C. Hulo, N. Hulo, J. James, S. Jimenez, F. Jungo, V. Junker, T. Kappler, G. Keller, C. Lachaize, L. Lane-Guermonprez, P. Langendijk-Genevaux, V. Lara, P. Lemercier, V. Le Saux, D. Lieberherr, T. D. Lima, V. Mangold, X. Martin, P. Masson, K. Michoud, M. Moinat, A. Morgat, A. Mottaz, S. Paesano, I. Pedruzzi, I. Phan, S. Pilbout, V. Pillet, S. Poux, M. Pozzato, N. Redaschi, S. Reynaud, C. Rivoire, B. Roechert, M. Schneider, C. Sigrist, K. Sonesson, S. Staehli, A. Stutz, S. Sundaram, M. Tognolli, L. Verbregue, A. L. Veuthey, L. Yip, L. Zuletta, R. Apweiler, Y. Alam-Faruque, R. Antunes, D. Barrell, D. Binns, L. Bower, P. Browne, W. M. Chan, E. Dimmer, R. Eberhardt, A. Fedotov, R. Foulger, J. Garavelli, R. Golin, A. Horne, R. Huntley, J. Jacobsen, M. Kleen, P. Kersey, K. Laiho, R. Leinonen, D. Legge, Q. Lin, M. Magrane, M. J. Martin, C. O’Donovan, S. Orchard, J. O’Rourke, S. Patient, M. Pruess, A. Sitnov, E. Stanley, M. Corbett, G. di Martino, M. Donnelly, J. Luo, P. van Rensburg, C. Wu, C. Arighi, L. Arminski, W. Barker, Y. X. Chen, Z. Z. Hu, H. K. Hua, H. Z. Huang, R. Mazumder, P. McGarvey, D. A. Natale, A. Nikolskaya, N. Petrova, B. E. Suzek, S. Vasudevan, C. R. Vinayaka, L. S. Yeh and J. Zhang, Nucleic Acids Res., 2009, 37, D169–D174 CrossRef PubMed.
- Y. D. Cai, S. L. Lin and K. C. Chou, Peptides, 2003, 24, 159–161 CrossRef CAS.
- Y. Cai, T. Huang, L. Hu, X. Shi, L. Xie and Y. Li, Amino Acids, 2012, 42, 1387–1395 CrossRef CAS PubMed.
- L. L. Hu, S. B. Wan, S. Niu, X. H. Shi, H. P. Li, Y. D. Cai and K. C. Chou, Biochimie, 2011, 93, 489–496 CrossRef CAS PubMed.
- L. L. Hu, Z. Li, K. Wang, S. Niu, X. H. Shi, Y. D. Cai and H. P. Li, Biopolymers, 2011, 95, 763–771 CAS.
- Y. Zhou, N. Zhang, B. Q. Li, T. Huang, Y. D. Cai and X. Y. Kong, J. Biomol. Struct. Dyn., 2015, 33, 2479–2490 CAS.
- S. Niu, L. L. Hu, L. L. Zheng, T. Huang, K. Y. Feng, Y. D. Cai, H. P. Li, Y. X. Li and K. C. Chou, J. Biomol. Struct. Dyn., 2012, 29, 650–658 Search PubMed.
- Y. Cai, J. He and L. Lu, J. Biomol. Struct. Dyn., 2011, 28, 797–804 CAS.
- S. Niu, T. Huang, K. Feng, Y. Cai and Y. Li, J. Proteome Res., 2010, 9, 6490–6497 CrossRef CAS PubMed.
- X. Xu, D. Yu, W. Fang, Y. Cheng, Z. Qian, W. Lu, Y. Cai and K. Feng, J. Proteome Res., 2008, 7, 4521–4524 CrossRef CAS PubMed.
- S. F. Altschul, T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller and D. J. Lipman, Nucleic Acids Res., 1997, 25, 3389–3402 CrossRef CAS PubMed.
- S. Kawashima and M. Kanehisa, Nucleic Acids Res., 2000, 28, 374 CrossRef CAS PubMed.
- W. R. Atchley, J. P. Zhao, A. D. Fernandes and T. Druke, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 6395–6400 CrossRef CAS PubMed.
- F. Ferron, S. Longhi, B. Canard and D. Karlin, Proteins: Struct., Funct., Bioinf., 2006, 65, 1–14 CrossRef CAS PubMed.
- O. Noivirt-Brik, J. Prilusky and J. L. Sussman, Proteins: Struct., Funct., Bioinf., 2009, 77, 210–216 CrossRef CAS PubMed.
- K. Peng, P. Radivojac, S. Vucetic, A. K. Dunker and Z. Obradovic, BMC Bioinf., 2006, 7, 208 CrossRef PubMed.
- S. Wan, Y. Duan and Q. Zou, Proteomics, 2017, 17 DOI:10.1002/pmic.201700262.
- L. Chen, Z. L. Qian, K. Y. Fen and Y. D. Cai, J. Comput. Chem., 2010, 31, 1766–1776 CAS.
- L. Chen, C. Chu, Y.-H. Zhang, M.-Y. Zheng, L. Zhu, X. Kong and T. Huang, Curr. Bioinf., 2017 DOI:10.2174/1574893611666160618094219.
- L. Wei, M. Liao, Y. Gao, R. Ji, Z. He and Q. Zou, IEEE/ACM Trans. Comput. Biol. Bioinf., 2014, 11, 192–201 CrossRef PubMed.
- I. H. Witten and E. Frank, Data Mining: Practical Machine Learning Tools and Techniques, Morgan, Kaufmann, San Francisco, 2005 Search PubMed.
- Q. Zou, J. Zeng, L. Cao and R. Ji, Neurocomputing, 2016, 173, 346–354 CrossRef.
- Q. Zou, S. Wan, Y. Ju, J. Tang and X. Zeng, BMC Syst. Biol., 2016, 10, 114 CrossRef PubMed.
- M. Draminski, A. Rada-Iglesias, S. Enroth, C. Wadelius, J. Koronacki and J. Komorowski, Bioinformatics, 2008, 24, 110–117 CrossRef CAS PubMed.
- I. Kononenko, E. Simec and M. RobnikSikonja, Appl. Intell., 1997, 7, 39–55 CrossRef.
- T. Huang, L. Chen, Y. Cai and C. Chou, PLoS One, 2011, 6, e25297 CAS.
- L. Liu, L. Chen, Y. H. Zhang, L. Wei, S. Cheng, X. Kong, M. Zheng, T. Huang and Y. D. Cai, J. Biomol. Struct. Dyn., 2017, 35, 312–329 CAS.
- L. Chen, Y. H. Zhang, G. Lu, T. Huang and Y. D. Cai, Artif. Intell. Med., 2017, 76, 27–36 CrossRef PubMed.
- M. Radovic, M. Ghalwash, N. Filipovic and Z. Obradovic, BMC Bioinf., 2017, 18, 9 CrossRef PubMed.
- L. Chen, S. Wang, Y.-H. Zhang, J. Li, Z.-H. Xing, J. Yang, T. Huang and Y.-D. Cai, IEEE Access, 2017 DOI:10.1109/ACCESS.2017.2775703.
- L. Chen, C. Chu and K. Feng, Comb. Chem. High Throughput Screening, 2016, 19, 136–143 CrossRef CAS PubMed.
- L. Chen, Y.-H. Zhang, G. Huang, X. Pan, S. Wang, T. Huang and Y.-D. Cai, Mol. Genet. Genomics, 2017 DOI:10.1007/s00438-017-1372-7.
- T. K. Ho, Random Decision Forests, Montreal, QC, 1995 Search PubMed.
- T. K. Ho, IEEE Trans. Pattern Anal. Mach. Intell., 1998, 20, 832–844 CrossRef.
- M. Hall, E. Frank, G. Holmes, B. Pfahringer, P. Reutemann and I. H. Witten, SIGKDD Explor., 2009, 10–18 CrossRef.
- K. C. Chou and H. B. Shen, Nat. Protoc., 2008, 3, 153–162 CrossRef CAS PubMed.
- L. Chen, W.-M. Zeng, Y.-D. Cai, K.-Y. Feng and K.-C. Chou, PLoS One, 2012, 7, e35254 CAS.
- L. Zhang, C. Zhang, R. Gao, R. Yang and Q. Song, BMC Bioinf., 2016, 17, 225 CrossRef PubMed.
- M. Martinot-Peignoux, M. Lapalus, C. Laouenan, O. Lada, A. C. Netto-Cardoso, N. Boyer, M. P. Ripault, R. Carvalho-Filho, T. Asselah and P. Marcellin, J. Clin. Virol., 2013, 58, 401–407 CrossRef CAS PubMed.
- I. Naseem, S. Khan, R. Togneri and M. Bennamoun, IEEE/ACM Trans. Comput. Biol. Bioinf., 2016 DOI:10.1109/TCBB.2016.2617337.
- Y. H. Lee, H. Choi, S. Park, B. Lee and G. S. Yi, BMC Bioinf., 2017, 18, 226 CrossRef PubMed.
- S. H. Wu, R. S. Schwartz, D. J. Winter, D. F. Conrad and R. A. Cartwright, Bioinformatics, 2017, 33, 2322–2329 CrossRef PubMed.
- W. H. Yu, H. Hovik and T. Chen, Bioinformatics, 2010, 26, 1423–1430 CrossRef CAS PubMed.
- B. W. Matthews, Biochim. Biophys. Acta, 1975, 405, 442–451 CrossRef CAS.
- L. Chen, K. Y. Feng, Y. D. Cai, K. C. Chou and H. P. Li, BMC Bioinf., 2010, 11, 293 CrossRef PubMed.
- G. von Heijne, Nature, 1998, 396(111), 113 Search PubMed.
- G. von Heijne, J. Mol. Biol., 1985, 184, 99–105 CrossRef CAS PubMed.
- J. Gennity, J. Goldstein and M. Inouye, J. Bioenerg. Biomembr., 1990, 22, 233–269 CrossRef CAS PubMed.
- P. Fekkes and A. J. Driessen, Microbiol. Mol. Biol. Rev., 1999, 63, 161–173 CAS.
- C. Hikita and S. Mizushima, J. Biol. Chem., 1992, 267, 12375–12379 CAS.
- J. Macfarlane and M. Muller, Eur. J. Biochem., 1995, 233, 766–771 CAS.
- G. von Heijne, J. Mol. Biol., 1984, 173, 243–251 CrossRef CAS PubMed.
- A. P. Pugsley, Microbiol. Rev., 1993, 57, 50–108 CAS.
- G. E. Crooks, G. Hon, J. M. Chandonia and S. E. Brenner, Genome Res., 2004, 14, 1188–1190 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7mo00030h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |