Open Access Article
Open Access ArticleEngineering hyperthermostable rcSso7d as reporter molecule for in vitro diagnostic tests†
Ki-Joo
Sung
 a,
Eric A.
Miller
a and
Hadley D.
Sikes
a,
Eric A.
Miller
a and
Hadley D.
Sikes
 *ab
*ab
aDepartment of Chemical Engineering, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA. E-mail: sikes@mit.edu
bSingapore-MIT Alliance for Research and Technology Centre (SMART), 1 CREATE Way, INNOVATION Tower, 138602 Singapore
First published on 15th October 2018
Abstract
The hyperthermostable binding scaffold, rcSso7d, has recently been studied for use in in vitro diagnostic tests but has not yet been demonstrated as a soluble reporter protein. Here, we investigated several rcSso7d protein constructs and biotin conjugation methods to link a detectable signal to the presence of a target biomarker.
Design, System, ApplicationEngineered variants of the small thermostable protein, rcSso7d, show promise for use in low-cost diagnostic tests. Previous studies have demonstrated their use as surface-bound capture reagents, but in order to use rcSso7d as a reporter reagent, we must be able to conjugate a label to the protein while ensuring that these modifications do not interfere with function. To engineer rcSso7d as a soluble, monomeric reporter protein, we identified several design criteria beyond specific binding activity to enable successful integration into in vitro diagnostic tests. These include low-cost production and high expression yields in E. coli, a facile labeling procedure, function of the label, and strong retention of target-binding activity. To identify a protein variant that fulfills these criteria, we designed several genetic constructs, expressed the encoded proteins, and tested each protein in a paper-based assay format. We identified an rcSso7d construct that associates a detectable signal with the presence of target antigen, addressing a critical barrier for incorporation of this protein as both the capture and reporter species in a diagnostic assay. The use of rcSso7d-based reagents will enable the development of affordable and thermally stable assays to detect biomarkers in medical diagnostic applications. |
Introduction
In vitro diagnostic assays are necessary for healthcare, food safety, and environmental monitoring applications. Antibodies are commonly used as the capture and reporter reagents to detect specific proteins in samples via noncovalent binding interactions. Unfortunately, antibodies have inherent drawbacks when considering use in an in vitro diagnostic format, including high costs and development time, limitations in physical properties such as thermostability, and genetic complexity that constrains the attachment of anchoring groups, labels, or other modifications to chemical conjugation methods.1,2In recent years, many alternatives with improved qualities have been investigated, specifically those that can be engineered to bind to a target analyte. These include aptamers, DARPins, affibodies, anticalins, single-domain antibodies (sdAb), and Sso7d (comparison of properties in Table S1†).1–4 Many studies have proved antibody alternatives as promising entities for therapeutic applications;5 however, due to differing design criteria, additional studies must be conducted to establish their utility in in vitro diagnostics. Previous studies for diagnostic applications often used the alternate scaffold as the capture protein but used a reporter antibody6–9 or a phage-displayed variant of the scaffold with a reporter antibody9,10 to quantify target proteins in solution, or a biotinylated antigen to quantify capture efficiency and tolerance for thermal challenges.11–13 Soluble reporter proteins were investigated with sdAb by chemically conjugating biotins,6 genetically adding tags,7,14,15 or fusing the scaffold to a reporter enzyme.14–16 Although sdAb clones with high in vitro stability and excellent yields upon bacterial expression can be identified, these desirable properties are not universal, thus requiring additional screening during production of new clones. Therefore, the process for new binder development may be facilitated by using a scaffold with inherent thermostability and high expression levels in bacteria which could tolerate minor genetic mutations without significantly impacting these desirable properties.
The reduced-charged variant of Sso7d (rcSso7d)—a 7 kDa DNA-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus—fulfills our design criteria for an antibody replacement protein due to its intrinsic stability (wild-type melting temperature of 98 °C; engineered variants retained their thermostability with melting temperatures >60 °C and generally >90 °C); facile and high-yield bacterial expression; isolated binding face that has demonstrated high-affinity binding to various target molecules; and ease of genetic modification (Table S1†).4,9–13,17 Although studies have already demonstrated the use of rcSso7d as a soluble capture molecule,9,11–13 analogous studies have not yet been reported for the use of soluble rcSso7d as the reporter molecule. Further engineering is required to enable the protein to associate a detectable signal with the presence of a target antigen.
To this end, biotin is a versatile and popular choice for tagging the reporter protein. The wide availability of streptavidin-conjugates allows users to select from a variety of signal amplification methods, including fluorescence-based, chemiluminscence-based, colorimetric and electrochemical, using the biotin-labeled reporter protein as a universal reagent.18 We created several biotinylated rcSso7d constructs to compare signal intensities provided by each engineering approach, with fluorescent and colorimetric readouts as examples.
Here, we investigated two different methods of conjugating a biotin moiety to the rcSso7d: in vivo biotinylation and in vitro chemical biotinylation.19 After discovering issues of inaccessibility of biotin for binding by streptavidin with in vivo biotinylation and reduced activity with in vitro chemical conjugation, we studied the addition of a protein fusion partner, maltose-binding protein (MBP), as a remedy to these issues. Through this comparison of different protein constructs, we determined that an MBP-rcSso7d fusion protein conjugate yielded high signal output when used as the detection protein by increasing biotin accessibility while preserving target-binding activity and protein stability. We also demonstrated that the resulting soluble biotinylated rcSso7d proteins are effective in detecting immobilized antigen with different read-out methods, including fluorescence, enzymatic amplification, and polymerization-based amplification (PBA).
Results and discussion
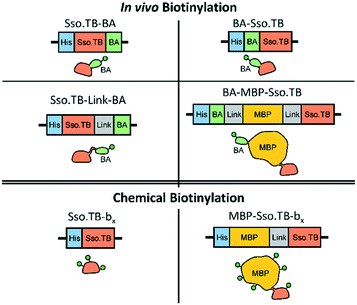
This study used rcSso7d.TB (termed Sso.TB), which is an engineered rcSso7d clone selected to bind against a urine-based active-tuberculosis (TB) biomarker Rv1656, following a similar selection process as reported previously (see ESI†).12,20 We constructed six variants of Sso.TB, as depicted in Fig. 1. Detail on the cloning procedure and genetic sequences can be found in the ESI.† We coupled biotin moieties to the proteins either via in vivo biotinylation through the action of biotin ligase on a biotin acceptor (BA) peptide or via chemical biotinylation with sulfo-N-hydroxysuccinimide (sulfo-NHS) ester crosslinking of biotin to primary amines (bx). Although chemical biotinylation adds an additional processing step, the resulting multiple biotins per protein may further improve detectable signal through multivalency effects. To develop the simplest in vivo biotinylated variants, we designed BA-Sso.TB (with N-terminal BA) and Sso.TB-BA (with C-terminal BA) to assess whether orientation affected accessibility of the biotin moiety for binding by streptavidin conjugates. For the simplest in vitro chemical biotinylated variant, we designed a construct with just the binding protein for chemical biotinylation (Sso.TB-bx). In addition to the simplest variants, we designed more complex constructs to improve the accessibility of the biotin label and prevent protein aggregation upon labelling. We introduced a flexible (G4S)2 linker sequence (Link) between the BA and Sso.TB sequences—Sso.TB-Link-BA—as a C-terminal modification to enhance biotin accessibility. We also introduced a 42.5 kDa maltose-binding protein (MBP) fusion partner to produce MBP-Sso.TB-bx and BA-MBP-Sso.TB. MBP was selected as the N-terminal fusion partner due to its documented positive effect on soluble expression yields.21,22 The larger protein mass also provides more primary amine sites for biotin conjugation (Table S4†). All gene constructs featured an N-terminal hexahistidine tag (6x-His), enabling purification via immobilized metal affinity chromatography (IMAC). Schematics of the developed gene constructs are included in Fig. 1.We expressed all of the Sso.TB constructs in BL21(DE3) E. coli and purified the proteins using IMAC columns as previously described and as detailed further in the ESI.†11,12 We quantified the protein concentrations using a bicinchoninic acid (BCA) assay and visualized purified proteins via SDS-PAGE to ensure product purity (Fig. 2).
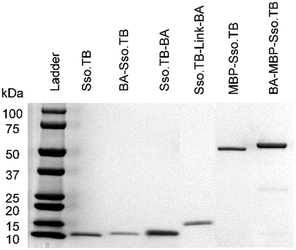 | ||
| Fig. 2 SDS-PAGE of all purified recombinant proteins. The theoretical MW are 9.3 kDa (Sso.TB), 11.4 kDa (BA-Sso.TB and Sso.TB-BA), 12.2 kDa (Sso.TB-Link-BA), 50.5 kDa (MBP-Sso.TB), and 53.7 kDa (BA-MBP-Sso.TB). Each lane was loaded with >1.5 μg of protein to ensure protein purity. The above image is a combination of three gel images for comprehension purposes (original images in ESI†). | ||
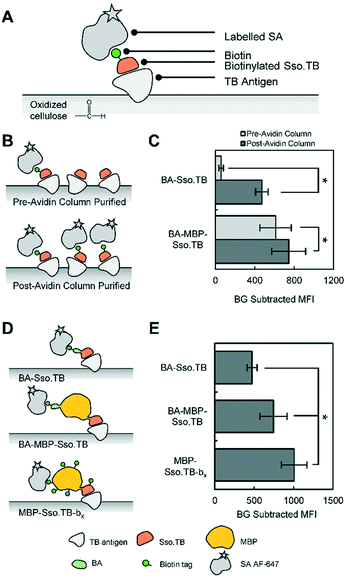
We first conducted preliminary tests of the simplest variants (BA-Sso.TB, Sso.TB-BA, and Sso.TB-bx) as the reporter protein to determine whether they would produce adequate signal without requiring further modifications. For these tests, we used TB antigen captured on oxidized cellulose strips and fluorescently labelled streptavidin (SA), via the procedure outlined in ESI† (schematic in Fig. S1). From these results, BA-Sso.TB and Sso.TB-BA were insufficient in generating detectable signals (Fig. S2†). Sso.TB-bx resulted in reduced signal (Fig. S3†), suggesting either inefficient chemical biotinylation or that chemical biotinylation detrimentally impacted the binding activity of rcSso7d. These results motivated the investigation into the other protein constructs to improve detectable signal. We explored the performance of each variant by 1) characterizing their yields and biotin functionality and then 2) assessing the dual functionality of Sso.TB binding to TB antigen and signal association with the antigen-Sso.TB complex.
Quantifying the biotinylation efficiency of all BA variants revealed biotinylation efficiencies of approximately 60% or below (Table 1). To eliminate this variable, we further purified aliquots of each BA protein using a monomeric avidin column to selectively capture and elute the biotinylated subpopulation. The purification yields are shown in Table 1. Almost all of the applied BA-Sso.TB and Sso.TB-BA proteins (∼95%) flowed through the column rather than binding to the avidin (Table 1). This low purification yield suggests that fusing BA directly to Sso.TB results in biotin moieties that are inaccessible by avidin—possibly due to incorporation of the hydrophobic biotins into the hydrophobic core of rcSso7d—which would lead to low detectable signals as shown in Fig. S2.† The additional linker sequence between BA and Sso.TB did not significantly improve biotin accessibility, as evidenced by similarly low avidin column purification yields. However, the majority of the BA-MBP-Sso.TB bound to the avidin column, resulting in a higher purification yield (approximately 50%). Therefore, adding a larger, structured protein mass in between BA and Sso.TB improved biotin accessibility.
| Protein | Approx. biotins per protein | Approx. avidin column purification yield | Approx. chemical conjugation product yield |
|---|---|---|---|
| a Could not quantify biotinylation extent of Sso.TB-bx due to low yields of recovered protein. b Did not purify chemically conjugated variants on the avidin column. c Proteins did not undergo chemical conjugation. | |||
| Sso.TB-bx | —a | —b | <0.2% |
| BA-Sso.TB | 0.35 | 5% | —c |
| Sso.TB-BA | 0.35 | 5% | —c |
| Sso.TB-Link-BA | 0.45 | 15% | —c |
| BA-MBP-Sso.TB | 0.60 | 50% | —c |
| MBP-Sso.TB-bx | 11 | —b | 50% |
Quantifying product yield after chemical biotinylation of Sso.TB-bx indicated significant losses from the process (total product yield <0.2% shown in Table 1, calculated by the total protein recovered over the total protein used for the conjugation reaction) by destabilizing the rcSso7d protein and causing precipitation, possibly due to the hydrophobic biotin moieties disrupting the protein solvation shell. The soluble product yield of Sso.TB-bx was too low to quantify biotinylation efficiency. In order to investigate whether the addition of MBP improves stability after chemical biotinylation, we chemically biotinylated MBP-Sso.TB to produce MBP-Sso.TB-bx. Product yield after chemical biotinylation was approximately 50% (Table 1), representing a >100-fold improvement on product yield compared to Sso.TB without MBP and confirming improved protein solubility. We then quantified the number of biotins per protein to verify MBP-Sso.TB-bx was biotinylated with an averaged 11 biotins per protein (Table 1).
To systematically compare how these different Sso.TB constructs perform as detection reagents, we captured TB antigens from solution using oxidized cellulose and assessed the viability of using the biotinylated Sso.TB variants and labelled SA to detect the antigens (Fig. 3A), following the same procedure as shown in Fig. S1.†
To assess the extent to which avidin column purification impacted performance of the protein variants, we tested both purification fractions of BA-Sso.TB and BA-MBP-Sso.TB (pre-avidin column purification with <100% biotinylation efficiency or post-avidin column purification with ∼100% biotinylation efficiency) in paper assays (Fig. 3B). Since only a portion of these pre-avidin column populations was biotinylated, the expected improvement from using 100% biotinylated protein population should reflect the relative increase in proportional biotinylation. For BA-Sso.TB, the performance after avidin column purification was much higher than the theoretical increase from 35% to 100% biotinylation (Fig. 3C). This significant increase—about an order of magnitude increase in background subtracted mean fluorescence intensity (MFI)—may be attributed to the inaccessibility of biotins on BA-Sso.TB. Through avidin column purification, only the proteins with biotins that were accessible to avidin were collected; therefore, the post-avidin column purified populations reflected the proteins with accessible biotins while the pre-avidin column purified population contained mainly proteins with inaccessible biotins. These inaccessible biotins still contributed to the biotin measurements (see ESI†), resulting in a higher value than the actual amount of accessible biotins. For BA-MBP-Sso.TB, the post-avidin column fraction demonstrated an increase in signal intensity as compared to the pre-avidin column population (Fig. 3C); this can be attributed to the increase in proportional biotinylation since this variant did not have biotin accessibility issues.
Since avidin column purification improved the performance of the BA-Sso.TB and BA-MBP-Sso.TB variants, we compared the avidin-purified proteins against the other variants to investigate their overall performance (Fig. 3D). Sso.TB-Link-BA did not improve the signal output compared to Sso.TB-BA (Fig. S4†), supporting the results from avidin column purification that the linker sequence did not improve biotin accessibility. N-terminal BA-Sso.TB showed moderately higher signal than C-terminal Sso.TB-BA after avidin column purification due to the orientation of the N-terminus away from the binding face (Fig. S5†). Compared to BA-Sso.TB, BA-MBP-Sso.TB demonstrated an increase in signal (Fig. 3E). This signal increase may be a result of the intrinsic improved accessibility of biotins on BA-MBP-Sso.TB and also potentially diminished steric hindrance effects, which may have caused the smaller Sso.TB to dissociate from the TB antigen as a result of streptavidin binding. Furthermore, MBP-Sso.TB-bx resulted in an additional increase in performance compared to BA-MBP-Sso.TB (Fig. 3E). This increase can be attributed to the increased biotin valency observed with the chemically biotinylated species, which allows more streptavidin-biotin pairs to form on a single complex. Additional controls were tested to verify specificity of binding to the TB antigen (Fig. S6 and S7†).
Through our binder engineering efforts, we were able to increase the detectable signal 17-fold between the simplest biotinylated variant (BA-Sso.TB) to the best-performing variant (MBP-Sso.TB-bx). This study also provides guidance for variants and processing techniques that may be used depending on the priorities of the system. For systems with antigen scarcity or low capture efficiency that require a greater degree of signal amplification, using the rcSso7d variant with the highest signal (MBP-Sso.TB-bx) may be optimal, even at the cost of additional processing steps and lower yields. For applications with adequate intrinsic sensitivity where minimization of processing steps is the priority, BA-MBP-Sso.TB may be better suited for ease of production.
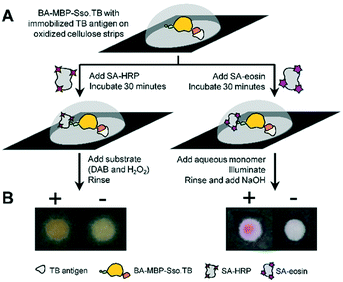
To extend our findings from fluorescence-based read-outs to colorimetric detection methods, we tested a variant with enzymatic amplification using HRP (horseradish peroxidase) and PBA.23 Similar to above, we captured TB antigen using oxidized cellulose strips and followed with BA-MBP-Sso.TB, which was chosen as the variant with minimal post-processing steps while still producing a high signal. In place of SA AF-647, either SA-HRP or SA-eosin were applied to the test surfaces and subsequent development steps were conducted, as shown in Fig. 4A. The resulting colorimetric responses were imaged (Fig. 4B). Both methods yielded visible color development compared to the negative samples lacking antigen, indicating that a naked-eye colorimetric response can be produced using Sso.TB as the detection reagent.
Conclusions
In summary, we demonstrated that the rcSso7d binding scaffold can be applied as a detection reagent in paper-based formats. We found that the addition of structured fusion partners allows in vivo biotinylation of these species while maintaining biotin accessibility. An additional purification step with an avidin column can isolate those proteins with accessible biotins to further increase signal output. Chemical modification of the small rcSso7d proteins impacts their stability and activity, which can be mitigated with fusion partners like MBP. The multivalency afforded by chemical biotinylation also yields higher signal than monovalent biotinylation from in vivo biotinylation methods. rcSso7d can be used as the reporter protein in conjunction with colorimetric amplification methods to distinguish between the presence or lack of the target antigen.Improving the detectable signal output with rcSso7d as the detection protein is another step towards using rcSso7d in place of antibodies to detect target antigens. This format could feasibly be used in an indirect ELISA format by capturing all relevant proteins from a solution onto a membrane and using a biotinylated rcSso7d variant to detect the presence of the target proteins. Future studies will involve the incorporation of rcSso7d into a clinically relevant indirect or sandwich ELISA format, using this scaffold as both the capture and detection reagent to demonstrate this species' applicability in a variety of diagnostic formats.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
KS acknowledges a National Science Foundation Graduate Research Fellowship (1122374). This work was further supported by the MIT Tata Center for Technology and Design, Singapore-MIT Alliance for Research and Technology (AMR IRG), MIT Deshpande Center for Technological Innovation, and the Cancer Center Support (Core) Grant P30CCA14051 from the National Cancer Institute. Authors would like to thank Emma Yee for valuable discussions.Notes and references
- H. K. Binz, P. Amstutz and A. Plückthun, Nat. Biotechnol., 2005, 23, 1257–1268 CrossRef CAS PubMed.
- S. Banta, K. Dooley and O. Shur, Annu. Rev. Biomed. Eng., 2013, 15, 93–113 CrossRef CAS PubMed.
- R. Vazquez-Lombardi, T. G. Phan, C. Zimmermann, D. Lowe, L. Jermutus and D. Christ, Drug Discovery Today, 2015, 20, 1271–1283 CrossRef CAS PubMed.
- N. Gera, M. Hussain, R. C. Wright and B. M. Rao, J. Mol. Biol., 2011, 409, 601–616 CrossRef CAS PubMed.
- R. Simeon and Z. Chen, Protein Cell, 2018, 9, 3–14 CrossRef CAS PubMed.
- D. Li, Y. Cui, C. Morisseau, S. J. Gee, C. S. Bever, X. Liu, J. Wu, B. D. Hammock and Y. Ying, Anal. Chem., 2017, 89, 6248–6256 CrossRef CAS PubMed.
- T. Wang, P. Li, Q. Zhang, W. Zhang, Z. Zhang, T. Wang and T. He, Sci. Rep., 2017, 7, 4348 CrossRef PubMed.
- C. Xie, C. Tiede, X. Zhang, C. Wang, Z. Li, X. Xu, M. J. McPherson, D. C. Tomlinson and W. Xu, Sci. Rep., 2017, 7, 9608 CrossRef PubMed.
- N. Zhao, J. Spencer, M. A. Schmitt and J. D. Fisk, Anal. Biochem., 2017, 521, 59–71 CrossRef CAS PubMed.
- N. Zhao, M. A. Schmitt and J. D. Fisk, FEBS J., 2016, 283, 1351–1367 CrossRef CAS PubMed.
- E. A. Miller, M. W. Traxlmayr, J. Shen and H. D. Sikes, Mol. Syst. Des. Eng., 2016, 1, 377–381 RSC.
- E. A. Miller, S. Baniya, D. Osorio, Y. J. Al Maalouf and H. D. Sikes, Biosens. Bioelectron., 2018, 102, 456–463 CrossRef CAS PubMed.
- E. A. Miller, Y. Jabbour Al Maalouf and H. D. Sikes, Anal. Chem., 2018, 90, 9472–9479 CrossRef CAS PubMed.
- M. Yamagata and J. R. Sanes, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 2126–2131 CrossRef PubMed.
- V. J. Bruce and B. R. McNaughton, Anal. Chem., 2017, 89, 3819–3823 CrossRef CAS PubMed.
- M. Mousli, M. Goyffon and P. Billiald, Biochim. Biophys. Acta, 1998, 1425, 348–360 CrossRef CAS.
- M. W. Traxlmayr, J. D. Kiefer, R. R. Srinivas, E. Lobner, A. W. Tisdale, N. K. Mehta, N. J. Yang, B. Tidor and K. D. Wittrup, J. Biol. Chem., 2016, 291, 22496–22508 CrossRef CAS PubMed.
- E. P. Diamandis and T. K. Christopoulos, Clin. Chem., 1991, 37, 625–636 CAS.
- B. K. Kay, S. Thai and V. V. Volgina, Methods Mol. Biol., 2009, 498, 185–196 CrossRef CAS PubMed.
- D. R. Napolitano, N. Pollock, S. S. Kashino, V. Rodrigues and A. Campos-Neto, Clin. Vaccine Immunol., 2008, 15, 638–643 CrossRef CAS PubMed.
- R. B. Kapust and D. S. Waugh, Protein Sci., 1999, 8, 1668–1674 CrossRef CAS PubMed.
- S. S. Ashraf, R. E. Benson, E. S. Payne, C. M. Halbleib and H. Grøn, Protein Expression Purif., 2004, 33, 238–245 CrossRef CAS PubMed.
- S. Lathwal and H. D. Sikes, Lab Chip, 2016, 16, 1374–1382 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8me00049b |
| This journal is © The Royal Society of Chemistry 2018 |