Open Access Article
Open Access ArticleStable and efficient generation of poly(β-amino ester)s for RNAi delivery†
P.
Dosta
 a,
V.
Ramos
*a and
S.
Borrós
a,
V.
Ramos
*a and
S.
Borrós
 *ab
*ab
aGrup d'Enginyera de Materials (GEMAT), Institut Químic de Sarrià, Universitat Ramon Llull, Via Augusta 390, 08017 Barcelona, Spain. E-mail: victor.ramos@iqs.url.edu; salvador.borros@iqs.url.edu
bCentro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Zaragoza, Spain
First published on 17th May 2018
Abstract
Cationic polymers are promising delivery systems for RNAi due to their ease of manipulation, scale-up conditions and transfection efficiency. However, some properties, such as stability and targeting, remain challenging to overcome. In this report, different modifications in poly(β-amino ester) (pBAE) structures have been explored to overcome these limitations. Recent studies have demonstrated that hydrophobicity plays a key role in controlling electrostatic interactions of plasma proteins with nanoparticles. Results show that a slight increase in the polymer hydrophobicity increases its siRNA packaging capacity, stability, and transfection efficiency. Consequently, polyplexes prepared with these hydrophobic structures are functional after incubation times longer than 48 hours in serum-containing medium. In addition, newly designed polymers were end-modified using different oligopeptide moieties in order to confer cell-specificity, as previously reported. Therefore, it can be concluded that these newly optimized pBAE polymers present great potential as delivery vectors to specifically drive therapeutic RNA-based nucleic acids in a cell-specific manner under physiological conditions.
Design, System, ApplicationThe development of efficient vectors in the field of RNAi is still challenging, especially due to the different harsh conditions and barriers that these vectors have to surpass and overcome when they are used for in vivo applications. In this paper, we have addressed one of the main limitations of synthetic vectors, which is their reduced stability in the presence of serum proteins. For that, we have synthesized hydrophobized versions of previously reported top performing poly(beta-amino ester) formulations using hexylamine, hexadecylamine, and cholesterol, making the resulting polyplexes stable against plasma proteins for more than 48 hours, which is really high for this class of transfection vectors. For RNAi complexation, the polymers were designed with terminated oligopeptides. Thus, the strategy chosen to attach the hydrophobic moieties was through the polymeric backbone, leaving the terminated oligopeptides free to complex with the RNAis. Moreover, the newly developed polymers present higher transfection efficiency than previously described pBAEs. Specifically, C6-50 and Cchol-50 polymers are promising delivery systems with improved stability, which may be useful for in vivo applications. Therefore, tailoring the hydrophilicity–hydrophobicity ratio of top performing polymer formulations described in this paper may result in significant advances with a high impact on the particle stability, packaging capacity and transfection efficiency of RNAi. |
Introduction
Small interfering RNA (siRNA) is a powerful tool to precisely downregulate expression of target proteins. This approach has enormous therapeutic potential to block the synthesis of disease-causing proteins. Despite its attractiveness, clinical trials have been limited due to the physiological barriers that siRNA molecules have to overcome in order to reach the intracellular site of target cells. Therefore, its translation into clinics is hampered due to the lack of appropriate delivery systems.1,2 Over the last few years, great efforts have been focused on developing stable RNAi-based drugs using different chemical modifications in their structure, increasing their lifetime under plasma conditions and reducing their off-target effects.3–5 However, the use of naked nucleic acids is still limited because they are rapidly eliminated by the kidneys6 and, if siRNA reaches the target cells, it lacks the mechanism to enter the cytoplasm.Different biomaterials have been used to protect RNAi-based drugs, increasing the RNAi functionality, such as lipid-based delivery systems, polymeric-based delivery systems, conjugated-based delivery systems, and cell-penetrating peptides.7–13 Although promising RNAi delivery systems have been developed, most of them present some drawbacks when they are applied in therapeutic applications. For instance, low endosomal escape has been observed when lipid-like nanoparticles enter by endocytosis, where 70% of the RNAi taken up by cells is not able to escape from the endosome, limiting their therapeutic utility.14 Moreover, cytotoxic effects have been observed after periodic administrations,15 which are essential to obtain an efficient RNAi therapy. Recently, conjugate-based RNAi delivery systems, linking delivery ligands to RNAi-based drugs, have been developed. In particular, the most clinically advanced formulations are Dynamic PolyConjugates (DPCs) and triantennary N-acetylgalactosamine (GalNAc), which are commonly used to target hepatocytes.16,17 However, targeting organs other than the liver using conjugate-based delivery systems is still challenging.
Recently, oligopeptide-modified pBAEs have shown great promise as delivery vectors in terms of transfection efficiency, biocompatibility, and cell specificity in vitro.18–20 However, pBAEs possess limited in vivo stability, which hampers their further development in clinical applications. Recently, it has been described that shielding the surface of pBAE–RNAi polyplexes using a hydrogel matrix is a promising way to stabilize the resulting formulation.21 Concretely, oligopeptide-modified pBAEs were combined with a hydrogel scaffold based on a polyamidoamine (PAMAM) dendrimer cross-linked with dextran aldehyde, which is able to protect them from degradation, obtaining a promising local delivery strategy to treat solid tumours.21 However, this technique cannot be used for systemic delivery, limiting its applicability in a wide range of therapeutic applications.
Alternatively, coating polyplexes with reactive hydrophilic polymers has also shown both lateral and steric stabilization, resulting in nanoparticles with greater circulation times in vivo.22,23 However, such strategies generate shielding around the nanoparticle that hampers the formation of protein corona, which has lately been shown to be beneficial for controlling uptake into specific cell types.24 In order to exploit the benefits of protein corona, it is required to generate protein-resistant pBAE–siRNA nanoparticles capable of maintaining their physicochemical properties in complex media. Then, such protein-resistant pBAE complexes are valuable candidates to study trafficking mechanisms of nanoparticles in vitro, as well as to broaden the use of pBAEs in in vivo applications. In addition, nanoparticles' surface composition is essential to describe their cell-entrance mechanism. It is described that a highly positive surface charge shows a different biodistribution than a slightly negative nanoparticle surface charge. Recently, different end-capped pBAEs using chemical compounds have been widely explored, obtaining polyplexes with varying behaviors,25 obtaining formulations more or less efficient depending on their final target.
In general, hydrophobization of polymers leads to increased stability of polyplexes, favoring the equilibrium between nanoparticles and plasma proteins.26 An increase of polymer hydrophobicity reduces protein absorption on the polyplex surface, making them more efficient to reach their final target or protect their RNAi-drug. Serum proteins inhibit the transfection efficiency of the resultant polyplexes or limit their stability.27 It has been described that amphiphilic polycations, such as Pluronics, were combined with other polymers, making them capable of forming stable complexes under plasma conditions.28 In addition, an increase in the polyplex hydrophobicity is able to enhance the packaging capacity of nucleic acids, reducing their nanometric size.27,29,30 Besides electrostatic forces, hydrophobicity plays a key role in complex formation between pBAEs and RNAi-drugs. Polymers with an optimized hydrophilic/hydrophobic ratio are able to condense higher RNA quantity, increasing the polyplex density. Furthermore, adding hydrophobic moieties to polymer formulations increases the polyplex-cell affinity to biological lipid membranes, improving their transfection efficiency in different cell lines.31 For example, a higher binding affinity to bone marrow stromal cells was observed when palmitic acid was conjugated to PEI.32 Also, cholesterol moieties are expected to enhance cellular transfection due to their high affinity with the lipid parts of the cell membrane.30
In this study, we explored different poly(β-amino ester) structures, tuning their hydrophobicity in order to enhance their stability, packaging capacity and transfection efficiency. Mainly, previously described pBAEs (C32 polymers)33 were further modified using different aliphatic amine chains, such as hexylamine, hexadecylamine, and cholesterol. As a result, we developed a wide range of stable natural-synthetic pBAEs capable of protecting RNAi-drugs from physiological media and maintaining their cell-specificity properties. Then, their composition differs from traditional cationic polymers due to their sufficient stability and functionality, increasing their potential for in vivo applications.
Experimental
Materials
Reagents and solvents used for polymer synthesis were purchased from Sigma-Aldrich and Panreac. Oligopeptide moieties used for polymer modification (H-Cys-Arg-Arg-Arg-NH2, H-Cys-Lys-Lys-Lys-NH2, H-Cys-His-His-His-NH2, H-Cys-Glu-Glu-Glu-NH2 and H-Cys-Asp-Asp-Asp-NH2) were obtained from GL Biochem Ltd (Shanghai) with a purity higher than 98%. For in vitro studies, silencer GFP siRNA (AM4626) was used for EGFP knockdown and ON-TARGETplus non-targeting control pool (D-001810-10) as scrambled siRNA control, both of them were obtained from Thermo GE Dharmacon. Labelled siRNA (AllStars Neg. siRNA AF 546) for uptake experiments was purchased from Qiagen. Polyplus Interferin reagent was purchased from VWR and used according to the manufacturer's instructions.Synthesis of C32, C6, and C16 polymers
Acrylate-terminated polymers were synthesized by an addition reaction of primary amines to 1,4-butanediol diacrylate (at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.1 molar ratio of amine
1.1 molar ratio of amine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) diacrylate). The C32 polymer was synthesized following the procedure described by Lynn et al. Briefly, 5-amino-1-pentanol (3.44 g, 33 mmol) and 1,4-butanediol diacrylate (7.93 g, 40 mmol) were polymerized under magnetic stirring at 90 °C for 24 h. The C32 polymer was characterized by 1H-NMR using DMSO-d6 as a solvent (ESI†).
diacrylate). The C32 polymer was synthesized following the procedure described by Lynn et al. Briefly, 5-amino-1-pentanol (3.44 g, 33 mmol) and 1,4-butanediol diacrylate (7.93 g, 40 mmol) were polymerized under magnetic stirring at 90 °C for 24 h. The C32 polymer was characterized by 1H-NMR using DMSO-d6 as a solvent (ESI†).
The C6 polymer was synthesized by conjugate addition of different ratios of hexylamine/5-amino-1-pentanol to 1,4-butanediol diacrylate using a slight excess of diacrylate (at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.1 molar ratio of amine
1.1 molar ratio of amine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) diacrylate). Briefly, C6-100 polymerization was performed using hexylamine (0.845 g, 8.3 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). C6-50 polymerization was performed using 5-amino-1-pentanol (0.426 g, 4.1 mmol), hexylamine (0.422 g, 4.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). C6-25 polymerization was performed using 5-amino-1-pentanol (0.639 g, 6.2 mmol), hexylamine (0.232 g, 2.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). All polymerizations were carried out under magnetic stirring at 90 °C for 24 h. Polymers were characterized by 1H-NMR using chloroform-d as a solvent (ESI†).
diacrylate). Briefly, C6-100 polymerization was performed using hexylamine (0.845 g, 8.3 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). C6-50 polymerization was performed using 5-amino-1-pentanol (0.426 g, 4.1 mmol), hexylamine (0.422 g, 4.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). C6-25 polymerization was performed using 5-amino-1-pentanol (0.639 g, 6.2 mmol), hexylamine (0.232 g, 2.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). All polymerizations were carried out under magnetic stirring at 90 °C for 24 h. Polymers were characterized by 1H-NMR using chloroform-d as a solvent (ESI†).
The C16 polymer was synthesized by conjugate addition of different ratios of hexadecylamine/5-amino-1-pentanol to 1,4-butanediol diacrylate using a slight excess of diacrylate (at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.1 molar ratio of amine
1.1 molar ratio of amine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) diacrylate). C16-50 polymerization was performed using 5-amino-1-pentanol (0.426 g, 4.1 mmol), hexadecylamine (1.1 g, 4.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). C16-25 polymerization was performed using 5-amino-1-pentanol (0.639 g, 6.2 mmol), hexadecylamine (0.550 g, 2.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). Polymerizations were carried out under magnetic stirring at 90 °C for 24 h and characterized by 1H-NMR using chloroform-d as a solvent (ESI†).
diacrylate). C16-50 polymerization was performed using 5-amino-1-pentanol (0.426 g, 4.1 mmol), hexadecylamine (1.1 g, 4.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). C16-25 polymerization was performed using 5-amino-1-pentanol (0.639 g, 6.2 mmol), hexadecylamine (0.550 g, 2.1 mmol) and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol). Polymerizations were carried out under magnetic stirring at 90 °C for 24 h and characterized by 1H-NMR using chloroform-d as a solvent (ESI†).
Synthesis of Cchol polymers
Cchol polymers were synthesized by esterification of hydroxyl groups from the C32 polymer with carboxylic acid-modified cholesterol (cholesterol-COOH). For instance, the Cchol-50 polymer was polymerized by esterification of 50% of hydroxyl groups mixing the C32 polymer (0.3 g, 0.14 mmol) and cholesterol-COOH (0.242 g, 0.49 mmol). The Cchol-25 polymer was polymerized by esterification of 25% of hydroxyl groups mixing C32 (0.3 g, 0.14 mmol) and cholesterol-COOH (0.121 g, 0.25 mmol). The Cchol-12.5 polymer was polymerized by esterification of 12.5% of hydroxyl groups mixing C32 (0.3 g, 0.14 mmol) and cholesterol-COOH (0.061 g, 0.12 mmol). All the polymerizations were carried out for 48 hours in tetrahydrofuran (THF). When the esterification was completed, dicyclohexylurea (DCU) salts were formed. Then, DCU salts were removed by filtration and THF was removed by rota-evaporation. Finally, the polymers were characterized by 1H-NMR using chloroform-d as a solvent.Synthesis of oligopeptide-modified pBAE polymers
In general, oligopeptide-modified pBAEs were obtained by end-modification of an acrylate-terminated polymer with thiol-terminated oligopeptides at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 molar ratio in dimethyl sulfoxide. The mixture was stirred overnight at room temperature, and the resulting polymer was obtained by precipitation in a mixture of diethyl ether and acetone (7
2.5 molar ratio in dimethyl sulfoxide. The mixture was stirred overnight at room temperature, and the resulting polymer was obtained by precipitation in a mixture of diethyl ether and acetone (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3; v/v).
3; v/v).
All the polymers were modified using arginine as an oligopeptide in order to study their stability/transfection efficiency and characterized by 1H-NMR (ESI†). After that, the top performing polymer formulations (C6-50 and Cchol-50) were further modified using arginine, lysine, histidine, aspartic acid and glutamic acid peptides. The different oligopeptide modifications were confirmed by 1H-NMR. C32 oligopeptide modifications have already described by Segovia et al.19
Biophysical characterization of oligopeptide end-modified pBAEs
Polyplexes were performed by mixing equal volumes of pBAEs and nucleic acids in acetate buffer at their appropriate concentration. For C32 polyplexes, 25 mM AcONa buffer was used, for C6 polyplexes 12.5 mM AcONa buffer was used, and for Cchol polyplexes 8.3 mM AcONa buffer was used. Briefly, pBAE stock solutions in DMSO (100 mg ml−1) were diluted at appropriate concentrations to obtain the desired polymer–RNAi weight/weight ratio. The pBAE was added to a solution of RNAi, mixed with pipetting for a few seconds and incubated at room temperature for 10 min. Then, the resulting nanoparticles were characterized by an agarose retardation assay and dynamic light scattering (DLS).To assess RNAi retardation, different RNA-to-polymer ratios (w/w) between 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 400
1 and 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were studied. pBAE–RNA complexes were freshly prepared and added to wells of agarose gel (2.5%, containing 1 μg ml−1 ethidium bromide). Samples were run at 80 V for 45 min (Apelex PS 305, France) and visualized by UV illumination.
1 were studied. pBAE–RNA complexes were freshly prepared and added to wells of agarose gel (2.5%, containing 1 μg ml−1 ethidium bromide). Samples were run at 80 V for 45 min (Apelex PS 305, France) and visualized by UV illumination.
The size and surface charge were determined by DLS (Malvern Instruments Ltd, United Kingdom, 4 mW laser). Polyplexes were synthesized as previously described. After 10 min of incubation at room temperature, 100 μl of nanoparticles were diluted with 900 μl of PBS 1× for further hydrodynamic size and Z-potential analysis.
Polymer stability study
Arginine-modified polyplexes using C32, C6-100, C6-50, C6-25, C16-50, C16-25, Cchol-50, Cchol-25, and Cchol-12.5 backbones were prepared as previously described and used for stability studies. 100 μl of polyplexes were precipitated in 900 μl of PBS 1×. Then, the size and polydispersity were determined using DLS at different time points. In contrast, the Z-potential was analyzed at the beginning and the end of the stability study. The C32-CR3 polymer was used as a control group.In vitro screening of newly developed hydrophilic/hydrophobic polymers using MDA MB-231
EGFP silencing screening using siRNA against GFP was performed using the new hydrophilic/hydrophobic polymer formulations (C32, C6-100, C6-50, C6-25, C16-50, C16-25, Cchol-50, Cchol-25, and Cchol-12.5) in the MDA MB 231 cell line. siRNA transfection was performed with polyplexes prepared as previously described, using AcONa buffer (25–8.3 mM, pH 5.0). Briefly, MDA MB 231 cells were seeded in 96 well plates at 10000 cells per well (DMEM, containing 10% fetal bovine serum, 100 units ml−1 penicillin, 100 μg ml−1 streptomycin, 0.1 mM MEM non-essential amino acids (NEAA), 2 mM glutamine) and incubated overnight to roughly 80% confluence prior to performing the transfection experiments. Firstly, screening of polyplexes were performed at different polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA w/w ratios using serum-free medium and the polyplexes were added to cells at a final RNAi concentration of 50 nM. After that, different siRNA concentrations, ranging from 50 nM to 12.5 nM, were studied using all the polymeric formulations at a 50
siRNA w/w ratios using serum-free medium and the polyplexes were added to cells at a final RNAi concentration of 50 nM. After that, different siRNA concentrations, ranging from 50 nM to 12.5 nM, were studied using all the polymeric formulations at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio. In both cases, cells were incubated with nanoparticles for 2 h at 37 °C in a 5% CO2 atmosphere. After that, polyplexes were removed and replaced with complete medium. GFP expression was analyzed at 48 hour post-transfection by flow cytometry (BD LSRFortessa cell analyzer). Polyplus Interferin was used as a transfection reagent control and untreated cells as a negative control.
siRNA ratio. In both cases, cells were incubated with nanoparticles for 2 h at 37 °C in a 5% CO2 atmosphere. After that, polyplexes were removed and replaced with complete medium. GFP expression was analyzed at 48 hour post-transfection by flow cytometry (BD LSRFortessa cell analyzer). Polyplus Interferin was used as a transfection reagent control and untreated cells as a negative control.
Cellular uptake assay
siRNA labelled with AF555 was used for the siRNA uptake experiment. MDA-MB 231 cells were seeded in 96 well plates at 10000 cells per well. After 24 hours, the uptake experiment was carried using different arginine-modified hydrophilic/hydrophobic polymer formulations. Briefly, polyplexes were synthesized as previously described using fluorescent siRNA. Then, cells were incubated with polyplexes at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and a final siRNA concentration of 50 nM for 2 h using serum-free medium. After that, the nanoparticles were removed, and the cells were washed twice with PBS 1× and collected according to standard protocols (fixed using 1% of paraformaldehyde) for flow cytometry analysis. Polyplus Interferin was employed following the manufacturer's instructions and used as a positive control.
1 ratio and a final siRNA concentration of 50 nM for 2 h using serum-free medium. After that, the nanoparticles were removed, and the cells were washed twice with PBS 1× and collected according to standard protocols (fixed using 1% of paraformaldehyde) for flow cytometry analysis. Polyplus Interferin was employed following the manufacturer's instructions and used as a positive control.
Cell viability assay
Immortalized MDA MB 231 cells were grown in 96 well plates at an initial seeding density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 200 μL growth medium. Cells were grown for 24 h, transfected with different polymer formulations at a final concentration of 50 nM (at a 50
000 cells per well in 200 μL growth medium. Cells were grown for 24 h, transfected with different polymer formulations at a final concentration of 50 nM (at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio) for 2 h using serum-free medium. Then, nanoparticles were removed, cells were washed once with PBS 1× and complete medium was added. 48 h post-transfection, the medium was removed, cells were washed with PBS, and complete medium supplemented with 20% MTS reagent (v/v) was added. Cells were incubated at 37 °C, and absorbance was measured at 490 nm using a microplate reader (Elx808 Biotek Instrument Ltd, USA). Cell viability was expressed as a relative percentage compared with untreated cells.
siRNA ratio) for 2 h using serum-free medium. Then, nanoparticles were removed, cells were washed once with PBS 1× and complete medium was added. 48 h post-transfection, the medium was removed, cells were washed with PBS, and complete medium supplemented with 20% MTS reagent (v/v) was added. Cells were incubated at 37 °C, and absorbance was measured at 490 nm using a microplate reader (Elx808 Biotek Instrument Ltd, USA). Cell viability was expressed as a relative percentage compared with untreated cells.
In vitro screening of oligopeptide-modified C6-50 and Cchol-50 polymers in the presence of serum
The transfection efficiency of our new oligopeptide end-modified C6-50 and Cchol-50 was studied in the presence of serum. Briefly, MDA MB 231 cells were grown in 96 well plates at an initial seeding density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in 200 μL of growth medium, obtaining 80% confluence prior to transfection. Then, cells were transfected using different oligopeptide end-modified pBAEs at a final siRNA concentration of 50 nM (at a 100
000 cells per well in 200 μL of growth medium, obtaining 80% confluence prior to transfection. Then, cells were transfected using different oligopeptide end-modified pBAEs at a final siRNA concentration of 50 nM (at a 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA w/w ratio). Polyplexes were incubated for 48 h. Then, the medium was removed, and the cells were washed with PBS 1×, fixed using 1% of paraformaldehyde and analyzed by flow cytometry. Polyplus Interferin was employed following the manufacturer's instructions and used as a positive control. Non-transfected cells and scrambled siRNA were used as negative controls.
siRNA w/w ratio). Polyplexes were incubated for 48 h. Then, the medium was removed, and the cells were washed with PBS 1×, fixed using 1% of paraformaldehyde and analyzed by flow cytometry. Polyplus Interferin was employed following the manufacturer's instructions and used as a positive control. Non-transfected cells and scrambled siRNA were used as negative controls.
Transfection efficiency of stable polymers at different time points
The transfection efficiency of C32-CR3, C6-50-CR3, and Cchol-50-CR3 at different time points (0, 24, 48, and 120 h) in the presence or absence of FBS was determined in MDA MB 231 cells. Briefly, 100 μl of nanoparticles condensing siGFP were added at different time points and incubated in complete or serum-free medium at 37 °C. After that, polyplexes were used to knockdown GFP expression using MDA MB 231 cells. Cells were seeded in 96 well plates at an initial seeding density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in order to obtain 80% confluence prior to performing the transfection. Then, cells were transfected using different polyplexes in complete medium and 48 h post-transfection GFP expression was analyzed by flow cytometry.
000 cells per well in order to obtain 80% confluence prior to performing the transfection. Then, cells were transfected using different polyplexes in complete medium and 48 h post-transfection GFP expression was analyzed by flow cytometry.
Statistical analysis
Statistical analyses were carried out with Graph-Pad Prism (GraphPad Software). All error bars reported are SD unless otherwise indicated. Pairwise comparisons were performed using one-way Student's t-tests. Differences between groups were considered significant at p values below 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001).Results and discussion
To obtain an efficient RNAi therapy, a well-designed delivery vector is needed. Vectors have to be able to protect RNAi-based nucleic acids from serum proteins, to reach the target cells, and to be therapeutically useful. Nowadays, there are a wide range of vectors, but none of them possess all these conditions. Therefore, the authors focused on enhancing our previously described poly(β-amino ester)s,18,19 whose cell-specificity can be modulated by changing their oligopeptide formulation and making them efficient and useful in physiological media.Synthesis and biophysical characterization of stable oligopeptide end-modified pBAEs
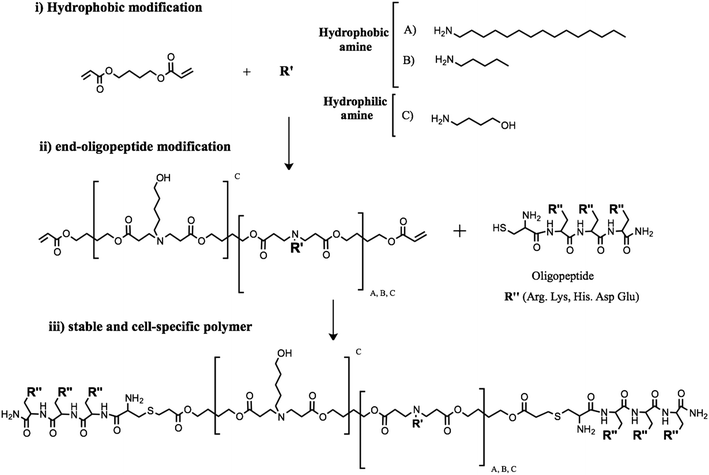
The polymer backbone structure plays a key role in controlling polyplexes' behavior in terms of stability and transfection efficiency.27 Then, poly(β-amino ester)s with different hydrophobicities were synthesized and biophysically characterized. Polymerization of new stable pBAEs was performed following the same methodology previously described in Materials and methods.33 In this study, pBAE polymers with different hydrophobicities were obtained combining three different monomers: 1,4-butanediol diacrylate, two different hydrophobic alkylamines, hexylamine or hexadecylamine, and 5-amino-1-pentanol, which was used as the hydrophilic amine. The hydrophobicity of the resulting polymers was controlled by the molar stoichiometry of the hydrophilic/hydrophobic amine, as shown in Fig. 1(i).An acrylate-terminated polymer intermediate was obtained by conjugate addition of hydrophilic/hydrophobic amine to 1,4-butanediol diacrylate using a slight excess of diacrylate. The C32 polymer was synthesized as previously described by Lynn et al.33 Briefly, the acrylate-terminated C32 intermediate polymer was obtained by addition of 5-amino-1-pentanol to 1,4-butanediol diacrylate. In contrast, a new family of hydrophobic polymers (C6-100, C6-50, C6-25, C16-50, and C16-25) was polymerized by combining different ratios of 5-amino-1-pentanol and hydrophobic amines, as previously explained in Materials and methods. For example, the C6-50 polymer was obtained using a stoichiometric proportion of 5-amino-1-pentanol/hexylamine and a slight excess of 1,4-butanediol. The resulting polymers were characterized in terms of their molecular structure by 1H-NMR.
The chemical structures of C6- or C16-containing polymers were confirmed by the decrease of –CH2-OH signals from the 5-amino-1-pentanol monomer and the presence of signals typically associated with the methyl group (–CH3) from hexylamine or hexadecylamine. At a higher percentage of hydrophobic amine, during pBAE polymerization, the methyl signal increased and the –CH2-OH signals decreased. Moreover, the molecular weight was determined using HPLC-SEC obtaining an average molecular weight of 2000–2500 g mol−1 (relative to polystyrene standards).
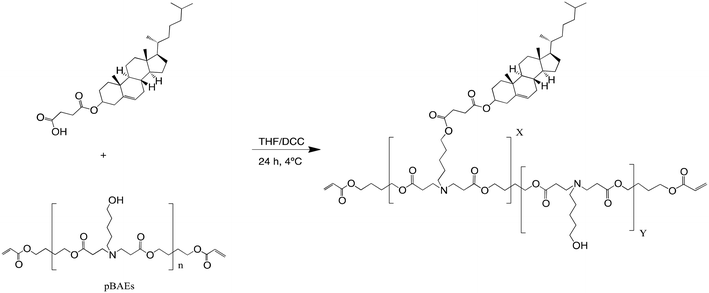
In addition, the hydrophobicity of the C32 polymer was further enhanced by conjugating carboxylic acid-modified cholesterol (Chol-COOH). Cchol polymers were obtained by esterification of the previously synthesized C32 polymer with carboxylic acid-modified cholesterol at different molar ratios, as shown Fig. 2. As reported previously, the resulting polymers were characterized in terms of their molecular structure by 1H-NMR. Then, the cholesterol percentage in the Cchol polymeric backbone was confirmed by the decrease of –CH2-OH signals from the 5-amino-1-pentanol monomer and the presence of signals typically associated with cholesterol, such as the terminal methyl from the aliphatic chain –CH-(CH3)2, C3 from Chol-COOH, and C6 from Chol-COOH. At a higher percentage of cholesterol in the pBAE backbone, –CH2-OH signals decreased and cholesterol signals increased. Moreover, the molecular weight was determined using HPLC-SEC obtaining an average molecular weight of 2000–3000 g mol−1 (relative to polystyrene standards).
Once the different backbone polymers were synthesized and characterized, the end-acrylate groups of the different polymers (C32, C6-100, C6-50, C6-25, Cchol-50, Cchol-25, Cchol-12.5, C16-50, and C16-25) were further modified using arginine oligopeptide moieties (CR3). Oligopeptide modification was carried out using a thiol reaction with cysteine-ended oligopeptides (Fig. 1-ii). Finally, polymer modifications were analyzed by 1H-NMR. The chemical structures of the new oligopeptide-modified pBAEs were confirmed by the disappearance of typical acrylate signals and the presence of signals associated with amino acid moieties. The 1H-NMR spectrum of the end-oligopeptide modified C32 polymer was in agreement with previously published data.19
Biophysical characterization of oligopeptide end-modified pBAEs
Once a new family of polymers has been synthesized, their ability to condense RNAi-drugs into discrete nanoparticles was studied. Polymer-nucleic acid binding capability was analyzed by a gel retardation assay (Fig. S1†). Results revealed that the newly synthesized hydrophobic pBAEs presented siRNA retardation at lower ratios than the previously synthesized C32-CR3 polymer. C6-50-CR3 and C16-50-CR3 polymers showed full siRNA retardation at ratios higher than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In addition, the Cchol-50-CR3 polymer presented full siRNA retardation at ratios higher than 25
1. In addition, the Cchol-50-CR3 polymer presented full siRNA retardation at ratios higher than 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. These results suggest that polymers modified with hydrophobic chains present a higher complexation capacity, making them good candidates for further studies.
1. These results suggest that polymers modified with hydrophobic chains present a higher complexation capacity, making them good candidates for further studies.
Once the polymer/siRNA ratios were determined, the polyplexes were further characterized using DLS. The hydrodynamic size and zeta potential of the resulting polyplexes were determined and summarized in Table 1.
| Polymer | Pendant group | Size (nm) | PDI | Z-pot (mV) | |
|---|---|---|---|---|---|
| Hydrophobic amine (%) | Hydrophilic amine (%) | ||||
| C32-CR3 | — | 5-Amino-1-pentanol (100%) | 222 ± 19 | 0.202 ± 0.038 | 18.5 ± 1.8 |
| C6-100-CR3 | Hexylamine (100%) | — | 184 ± 10 | 0.111 ± 0.012 | 18.2 ± 1.3 |
| C6-50-CR3 | Hexylamine (50%) | 5-Amino-1-pentanol (50%) | 111 ± 6 | 0.156 ± 0.017 | 17.2 ± 2.2 |
| C16-50-CR3 | Hexadecylamine (50%) | 267 ± 21 | 0.201 ± 0.076 | 17.9 ± 0.9 | |
| Cchol-50-CR3 | Chol-COOH (50%) | 112 ± 11 | 0.154 ± 0.023 | 15.7 ± 2.4 | |
| C6-25-CR3 | Hexylamine (25%) | 5-Amino-1-pentanol (75%) | 68 ± 7 | 0.266 ± 0.041 | 19.1 ± 0.5 |
| C16-25-CR3 | Hexadecylamine (25%) | 289 ± 37 | 0.232 ± 0.074 | 15.1 ± 1.8 | |
| Cchol-25-CR3 | Chol-COOH (25%) | 59 ± 3 | 0.166 ± 0.042 | 16.9 ± 1.6 | |
| Cchol-12.5-CR3 | Cchol-COOH (12.5%) | 5-Amino-1-pentanol (87.5%) | 113 ± 9 | 0.184 ± 0.008 | 17.1 ± 2.1 |
For this study, polyplexes were prepared at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio and, as before, an arginine moiety was used as the oligopeptide end-modifying moiety. Nevertheless, the C32 polymer was formulated at a 200
siRNA ratio and, as before, an arginine moiety was used as the oligopeptide end-modifying moiety. Nevertheless, the C32 polymer was formulated at a 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio, since this formulation required a higher polymer
siRNA ratio, since this formulation required a higher polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio to produce sufficiently stable particles for size analysis.18
siRNA ratio to produce sufficiently stable particles for size analysis.18
Polyplexes obtained using C32-CR3 as a polymer (5-amino-1-pentanol amine) showed a particle average size of 220 nm with a positive zeta potential, +18.5 mV. In general, dynamic light scattering analyses showed that some of the newly developed polymers were able to achieve smaller nanoparticles than the C32 polymer, while maintaining their positive surface charge. For example, the most promising formulations were able to form nanoparticles with hydrodynamic size ranging from 60 to 120 nm. In addition, low polydispersity indexes were observed in the resulting polyplexes. However, polyplexes obtained using hexadecylamine pBAE presented a considerably greater size, around 300 nm, maintaining a positive surface charge. Therefore, an increase of hydrophobicity due to the modification of the backbone polymer is able to reduce the size of the resultant nanoparticle, with the exception of hexadecylamine modification, maintaining the positive surface charge, which has been classically associated with a higher packaging capacity.27
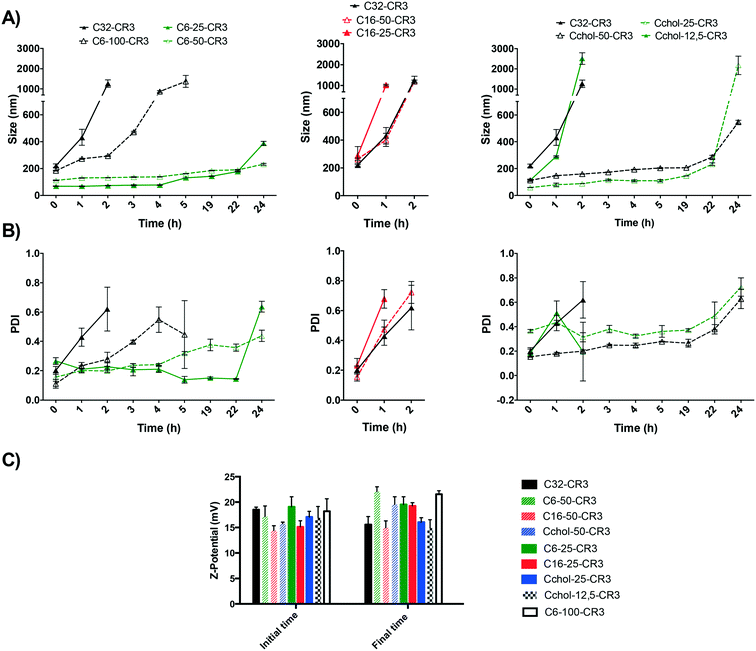
Stability effect of different hydrophobic modifications
Prior to transfection efficiency analysis, the influence of the modifying hydrophobic group on the stability of the resulting nanoparticles was evaluated in order to select the most stable polymer formulation. The stability of different hydrophobic modifications was determined using arginine as the oligopeptide. Briefly, nanoparticles were freshly prepared using hydrophobized pBAEs and siRNA and their size was evaluated as a function of time. The size of siRNA nanoparticles was determined using DLS over the course of 24 hours. Moreover, the initial and final nanoparticle surface charges were determined, as shown Fig. 3. Results showed that the basic poly(beta-amino ester), C32 polymer, described by Lynn et al. and further end-modified with arginine oligopeptides,18,19 exhibited low stability, reaching a size greater than 1000 nm, revealing a high degree of nanoparticle aggregation, i.e. average hydrodynamic diameter greater than 1000 nm, after a 2 hour incubation in PBS. However, an increase in particle diameter did not affect the nanoparticle surface charge. This result indicated that this polymeric formulation is able to completely condense the nucleic acids inside the polyplex, but it is not sufficiently stable to maintain its nanometric properties. In order to solve this limitation, an increase in polymer hydrophobicity was studied using hydrophobized polymers. Interestingly, results show that a slight increase in polymer hydrophobicity yields an important enhancement in nanoparticle stability. Results also showed that the C6 polymer modified with 25% hexylamine (C6-25) was able to maintain the size of the resulting nanoparticles below 250 nm up to 22 hours. Moreover, particles prepared with C6 polymer modified with 50% hexylamine (C6-50) were able to maintain a discrete nanometric size and a positive surface charge for more than 24 h in PBS. In contrast, nanoparticles obtained from 100% hexylamine-modified polymer (C6-100) presented greater stability than the C32 polymer, but lower than C6-25 and C6-60 polymers. C6-100 polyplexes were stable for approximately 2 hours, maintaining a particle size of 200 nm. Therefore, these results suggest that the hydrophobicity of the polymer backbone plays a major role in the stability of the resulting nanoparticle. Similar to the hexylamine modification, poly(beta-amino ester)s were modified using cholesterol (Cchol polymer). As previously discussed, the Cchol polymer was synthesized with different degrees of esterification using carboxylic acid-modified cholesterol, at 50% (Cchol-50), 25% (Cchol-25), 12.5% (Cchol-12.5), and 0% (C32). Results showed that 25% and 50% cholesterol modifications presented greater stability than 12.5%, maintaining the particle size below 300 nm for up to 22 h. In addition, nanoparticles containing 50% cholesterol remained smaller than polyplexes with 25% cholesterol after incubation in physiological medium for 22 hours. Moreover, formulations with a lower percentage of cholesterol than 25% did not present a significant improvement in their stability. Finally, modification of hexadecylamine at 50% and 25% was not able to further increase the stability of polyplexes in PBS. However, their surface charge remained positive due to their ability to efficiently condense siRNA.Taking into account the different biophysical proprieties, it can be observed that such nanoparticles that presented a smaller initial size are more stable than larger nanoparticles. Their ability to condense nucleic acids in smaller nanoparticles in a more efficient manner than the previously developed C32 polymer suggests that the new hydrophobic polymers may exhibit higher stability under physiological conditions. In addition, these results suggest that the nanoparticle composition plays a key role in nanoparticle–medium interactions. It is well described that hydrophilic polyplexes present a larger number of protein interactions than hydrophobic formulations. For instance, studies using n-iso-propylacrylamide/n-tert-butylacrylamide copolymer nanoparticles have shown a well described correlation between the nanoparticle core hydrophilicity and protein absorption, which describes their final protein corona.34 Therefore, it can be concluded that a similar correlation was observed here, showing that an increase in hydrophobicity is sufficient to dramatically improve the polyplexes' packaging capacity and stability in physiological medium.
In vitro study of slightly hydrophobic polyplexes in MDA MB 231 cells
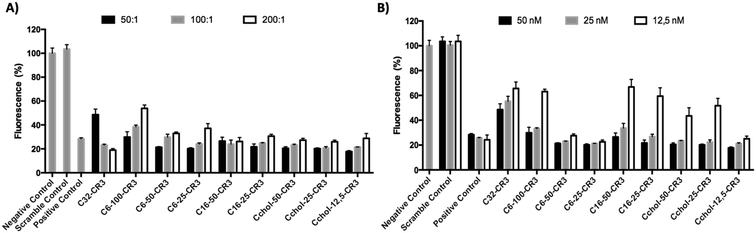
In order to study the hydrophobicity contribution in different pBAE formulations, EGFP knockdown was determined using anti-GFP siRNA in GFP-expressing MDA MB 231 cells. The silencing efficiency was evaluated by measuring the decrease in cell fluorescence by flow cytometry. The GFP knockdown of different hydrophobic/hydrophilic poly(beta-amino ester)s was compared to the previously described C32 polymer and commercial reagent, Polyplus Interferin, as shown in Fig. 4.As it has been already described, an increase in polyplex hydrophobicity results in an increase of packaging capacity and transfection efficiency.27 In this report, the transfection efficiency of different polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratios was tested, as shown in Fig. 4-A. The transfection efficiency using the C32-CR3 polymer increased with increasing polymer-to-siRNA ratio, indicating that the optimal C32 polymer
siRNA ratios was tested, as shown in Fig. 4-A. The transfection efficiency using the C32-CR3 polymer increased with increasing polymer-to-siRNA ratio, indicating that the optimal C32 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio is 200
siRNA ratio is 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In contrast, the opposite behavior was observed using hydrophobized polymers. Hexylamine-, hexadecylamine- or cholesterol-modified pBAEs achieved the highest reduction in cell fluorescence at low polymer
1. In contrast, the opposite behavior was observed using hydrophobized polymers. Hexylamine-, hexadecylamine- or cholesterol-modified pBAEs achieved the highest reduction in cell fluorescence at low polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratios, confirming their higher packaging capacity than the C32-CR3 formulation (Fig. 4-A). In addition, the incorporation of hydrophobic moieties into the pBAE structure may increase the nanoparticle interaction with the cell membrane and facilitate the subsequent entry into the cytoplasm.35 It is well known that amphiphilic or hydrophobic compounds show a high affinity for biological lipid membranes. In the case of polyplexes, their interaction should enhance polyplexes absorption or interaction to the cell membrane, which may ultimately favor cellular uptake. It has been described that the addition of palmitic acid, oleic acid, cholesterol, hexyl or dodecyl chains to polyplex formulations enhances endocytosis and, in consequence, increases the transfection efficiency.36,37
siRNA ratios, confirming their higher packaging capacity than the C32-CR3 formulation (Fig. 4-A). In addition, the incorporation of hydrophobic moieties into the pBAE structure may increase the nanoparticle interaction with the cell membrane and facilitate the subsequent entry into the cytoplasm.35 It is well known that amphiphilic or hydrophobic compounds show a high affinity for biological lipid membranes. In the case of polyplexes, their interaction should enhance polyplexes absorption or interaction to the cell membrane, which may ultimately favor cellular uptake. It has been described that the addition of palmitic acid, oleic acid, cholesterol, hexyl or dodecyl chains to polyplex formulations enhances endocytosis and, in consequence, increases the transfection efficiency.36,37
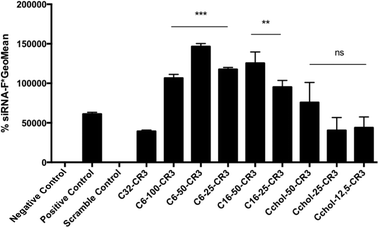
Once the most effective polymer/RNAi ratio was confirmed, a siRNA dose curve was obtained (Fig. 4-B) in order to determine the most efficient polymer formulation. The knockdown efficiency of the C32-CR3 polymer is limited at 50 nM. Then, when the siRNA concentration was decreased, C32-CR3 polyplexes were not able to silence GFP expression in MDA MB 231 cells. In contrast, C6-50-CR3 and C6-25-CR3 polymers are capable of maintaining reduced GFP expression at a siGFP concentration as low as 12.5 nM. Fully hexylamine-modified pBAEs present a limited transfection efficiency, showing a similar behavior to the C32-CR3 polymer. On the other hand, polyplexes prepared with hexadecylamine showed limited GFP knockdown when the siRNA concentration is reduced. Finally, slight modification of the pBAE backbone with cholesterol was able to silence more than 70% of the GFP expression at 12.5 nM. However, the silencing efficiency using highly modified pBAE-cholesterol (Cchol-50 and Cchol-25) is limited at 25 nM.
We can conclude that slightly hydrophobized polymers are capable of condensing siRNA more efficiently using lower polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) RNAi ratios and their hydrophobic component is able to efficiently interact with the cell membrane promoting their cellular entrance. Specifically, the stoichiometric mixture of 5-amino-1-pentanol/hexylamine (C6-50 polymer) is the most promising candidate in terms of transfection efficiency/stability.
RNAi ratios and their hydrophobic component is able to efficiently interact with the cell membrane promoting their cellular entrance. Specifically, the stoichiometric mixture of 5-amino-1-pentanol/hexylamine (C6-50 polymer) is the most promising candidate in terms of transfection efficiency/stability.
siRNA uptake using different synthesized hydrophobic/hydrophilic polymers
siRNA uptake was carried out in order to determine which polymer formulation is able to deliver nucleic acids into the cells more efficiently. For this study, different arginine-modified hydrophobic/hydrophilic polymers were tested. Briefly, polyplexes were obtained using different polymers and AF555-labeled siRNA at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. MDA MB 231 cells were incubated with nanoparticles for 2 hours and the resulting cellular fluorescence was determined by flow cytometry.
1 ratio. MDA MB 231 cells were incubated with nanoparticles for 2 hours and the resulting cellular fluorescence was determined by flow cytometry.
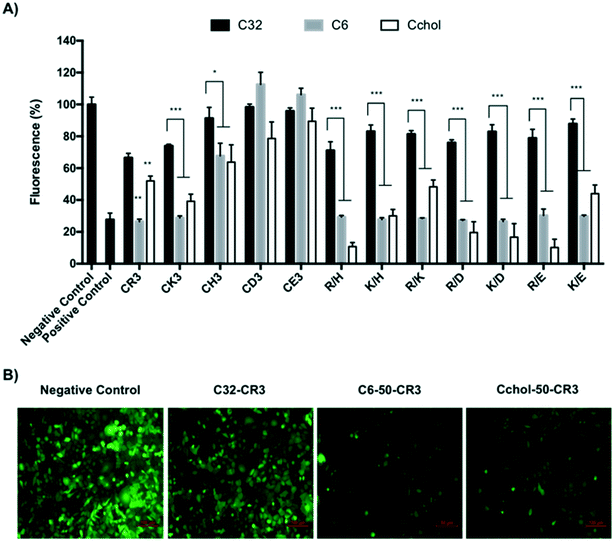
Uptake analysis of cells incubated with fluorescent polyplexes showed differential behaviors depending on the hydrophobic/hydrophilic amine used for poly(beta-amino ester) polymerization. Polyplexes prepared using the C32-CR3 polymer at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio showed lower siRNA uptake than the commercial reagent, which was in agreement with the observed knockdown. Polymer formulations containing hexylamine or hexadecylamine in their backbone showed the highest levels of cellular uptake, achieving a 2 to 3-fold higher fluorescence than the previously described C32 polymer and positive control. These results corroborated that hydrophobic groups enhance cellular entrance. In contrast, cholesterol-modified polymers showed lower siRNA uptake than hexylamine and hexadecylamine polymers, obtaining a similar cellular uptake to the C32-CR3 polymer and Polyplus Interferin. These results suggested that cholesterol-modified polyplexes are able to present higher GFP knockdown efficiency at lower cellular uptake, confirming that they are able to efficiently escape from the endosome after endocytosis. Consequently, cholesterol-containing polyplexes may interact with the endosome membrane and promote their endosomal escape (Fig. 5).
1 ratio showed lower siRNA uptake than the commercial reagent, which was in agreement with the observed knockdown. Polymer formulations containing hexylamine or hexadecylamine in their backbone showed the highest levels of cellular uptake, achieving a 2 to 3-fold higher fluorescence than the previously described C32 polymer and positive control. These results corroborated that hydrophobic groups enhance cellular entrance. In contrast, cholesterol-modified polymers showed lower siRNA uptake than hexylamine and hexadecylamine polymers, obtaining a similar cellular uptake to the C32-CR3 polymer and Polyplus Interferin. These results suggested that cholesterol-modified polyplexes are able to present higher GFP knockdown efficiency at lower cellular uptake, confirming that they are able to efficiently escape from the endosome after endocytosis. Consequently, cholesterol-containing polyplexes may interact with the endosome membrane and promote their endosomal escape (Fig. 5).
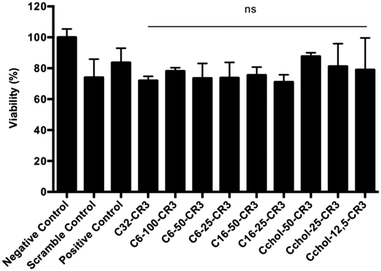
Cell cytotoxicity evaluation of hydrophobized polymers
Typically, highly efficient polymer formulations present some degree of cell toxicity due to their large capacity to cell-entry, which may limit their further use as RNAi delivery vehicles.38 Then, different polymer formulation factors have to be considered in order to avoid cytotoxicity problems. A polymeric formulation capable of delivering nucleic acids without altering the cellular mechanism is required. In this work, we evaluated the influence of hydrophobized pBAE formulations on cell viability. To determine the toxicity, a silencing assay using siRNA was carried out in MDA MB 231 cells and the viability was analyzed at 48 hour post-transfection by the MTS assay, as shown in Fig. 6. Polyplexes were freshly prepared at a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer
1 polymer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) siRNA ratio and cells were transfected at 50 nM siRNA concentration.
siRNA ratio and cells were transfected at 50 nM siRNA concentration.
Cell viability results did not show any significant differences between the hydrophobized pBAEs and the C32-CR3 polymer and commercial Polyplus Interferin polymer. Results showed that all the formulations were able to efficiently knockdown GFP expression with cell viabilities greater than 80%. Moreover, the scrambled control showed the same behavior as the hydrophobic modified pBAEs. These results suggest that hydrophobized PBAEs have low toxicity effects, maintaining a high transfection efficiency.
EGFP silencing in MDA MB231 cells using the most stable polymers under FBS conditions
Based on previous results, stoichiometric hexylamine/5-amino-1-pentanol amine (C6-50) and cholesterol/5-amino-1-pentanol (Cchol-50) polymers were chosen as the most promising polymers due to their high silencing efficiency, stability in physiological medium, and low cytotoxicity. Consequently, C6-50 and Cchol-50 were further modified with different oligopeptide moieties, such as arginine (R), lysine (K), histidine (H), glutamic acid (D) and aspartic acid (E). Tailoring the oligopeptide composition allows obtaining polyplexes with controlled surface properties (Fig. S2†), which have been shown to confer cell-specificity in different cells lines.18,19 In general, the hydrodynamic diameters of different oligopeptide-modified C6-50 and Ccho-50 nanoparticles were in the nanometric range, and were significantly smaller than those of polyplexes obtained from C32 polymers. These results suggest that hydrophobized polymers present a high capacity to condense RNAi-based drugs, making them promising delivery vectors for RNAi therapy, where the loading capacity is key for clinical translation to avoid systemic overexposure of vector materials.To assess conditions in complex media, transfection screening was performed in medium supplemented with FBS. In order to evaluate their behavior and efficiency, freshly prepared nanoparticles using different oligopeptide moiety combinations were compared with the previously characterized C32 polymer using MDA MB 231 as the cell line. The GFP fluorescence was determined at 48 hour post-transfection by flow cytometry, as shown in Fig. 7.
Different silencing behaviors of GFP knockdown using the different oligopeptide-modified C32, C6-50, and Cchol-50 polymers were observed. As previously noticed, the C32 transfection efficiency under serum conditions is limited, obtaining low levels of GFP knockdown due to its low stability under serum conditions. The C32-CR3 polymer showed the highest efficiency, obtaining 30% GFP silencing compared with the negative control.
However, hydrophobized polymers, such as C6-50, showed higher levels of GFP silencing than C32 polymer. 80% silencing was observed in all the oligopeptide formulations, with the exception of nanoparticles solely formulated with histidine-, aspartic acid- and glutamic acid-modified pBAEs. In addition, oligopeptide-modified C6-50 achieved a similar reduction in cell fluorescence to the commercial reagent. Furthermore, all siRNA complexes prepared with different oligopeptide end-modified Cchol-50 polymers showed high GFP silencing, with the exception of histidine-, aspartic acid- and glutamic acid oligopeptides. In addition, oligopeptide mixtures of arginine- and histidine or glutamic acid presented the highest GFP silencing in MDA MB213 cells, reaching 85–90% GFP knockdown. In addition, GFP knockdown was determined by confocal microscopy analysis using arginine-modified polymers, as shown Fig. 7-B. Results showed a direct correlation with previous data, showing the highest levels of GFP knockdown when cells are transfected with C6-50, C6-25 and Cchol-50 polymers. In contrast, the previously developed C32 polymer showed a lower silencing effect than hydrophobic formulations. Therefore, these results indicate that C6-50 and Cchol-50 polymers might be promising vectors to be used as nucleic acid carriers in the presence of plasma-proteins, making them interesting candidates for in vivo applications.
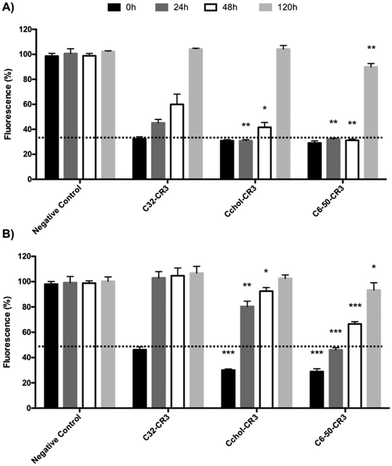
Stability vs. transfection efficiency study of C32, C6-50, and Cchol-50 in the presence of serum proteins
Finally, in order to further confirm the transfection efficiency in the presence of plasma proteins of formulations containing C6-50, Cchol-50 and C32 polymers, a dual experiment was performed where the stability and transfection efficiency were compared. Polyplexes were incubated in either serum-free or serum-containing medium for different time points. The resulting polyplexes were used to knockdown GFP expression in MDA MB 231 cells, as shown in Fig. 8.The serum-free stability experiment (Fig. 8-A) showed efficient EGFP silencing of polyplexes prepared with C32, Cchol-50 and C6-50 polymers, and at least a 70% knockdown effect was achieved, when polyplexes were freshly prepared and readily used for cell transfection (time point 0 h). In addition, polyplexes were incubated for 24 h, 48 h and 120 h in serum-free medium. Results showed that the knockdown effect of C32 nanoparticles was reduced, showing 50% silencing at 24 hours, 40% at 48 hours and no silencing after pre-incubation for 120 hours. In contrast, C6-50 polyplexes incubated in serum-free medium were able to maintain 70% GFP silencing for 48 hours. In addition, Cchol-50 polyplexes showed 70% GFP silencing for 24 hours, remaining at 60% silencing after pre-incubation for 48 hours. However, in all the cases, polyplexes' efficiency was dramatically decreased after 120 hours of incubation in serum-free medium, obtaining a moderate knockdown of 10% using the C6-50 polymer.
Different stability/transfection behaviors were observed when polyplexes were incubated in the presence of serum proteins. As previously demonstrated, hydrophobized pBAEs present higher transfection efficiency than the C32 polymer, reaching more than 70% GFP silencing compared to the 50% obtained with the C32 polymer. The silencing efficiency of C32 polyplexes is dramatically decreased when the polyplexes were incubated with medium containing serum proteins (Fig. 8-B). In contrast, hexylamine-modified polymers showed significant GFP knockdown when nanoparticles were incubated in complete medium, obtaining 50% silencing at 24 hours, 40% silencing at 48 hours, and 10% decrease at 120 hours. Lower stability/silencing was observed using cholesterol-modified pBAEs. The Cchol-50 polymer is able to efficiently deliver siRNA in the presence of serum, but when nanoparticles were incubated in serum proteins their efficiency was reduced, showing only a 20% GFP knockdown after 24 hours and 10% silencing at 48 hours. As previously discussed, intrinsic polyplex formulations could control protein corona absorption, making C6-50-CR3 a good candidate to protect and efficiently deliver siRNA in complex media.
Conclusions
The development of efficient vectors in the field of RNAi is still challenging, especially due to the different harsh conditions and barriers that these vectors have to surpass and overcome. In this report, the authors have addressed one of the main limitations of synthetic vectors, which is their reduced stability in the presence of serum proteins. Here, the authors have synthesized hydrophobized versions of previously reported top performing poly(beta-amino ester) formulations using hexylamine, hexadecylamine, and cholesterol, making the resulting polyplexes stable against plasma proteins for more than 24 hours. Moreover, the newly developed polymers present higher transfection efficiency than previously described pBAEs, e.g. C32. Specifically, C6-50 and Cchol-50 polymers are promising delivery systems with improved stability, which may be useful for in vitro and in vivo applications. Therefore, appropriate tailoring of the hydrophilicity–hydrophobicity ratio of top performing polymer formulations may result in significant advances with a high impact on particle stability, packaging capacity and transfection efficiency.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by Grup d'Enginyeria dels Materials (GEMAT). GEMAT would like to acknowledge Agència de Gestió d'Ajuts Universitaris i de Recerca, Generalitat de Catalunya (SGR 2014) no. 1170. Pere Dosta wishes to acknowledge the financial support received from AGAUR (Generalitat de Catalunya) 2017FI_B2 00141. Part of this work has been also supported by a grant from the Spanish Ministerio de Economia y Competitividad (MINECO) through the Grant: SAF2015-64927-C2-2-R. The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.References
- J. C. Burnett, J. J. Rossi and K. Tiemann, Current progress of siRNA/shRNA therapeutics in clinical trials, Biotechnol. J., 2011, 6(9), 1130–1146 CrossRef PubMed.
- J. E. Zuckerman and M. E. Davis, Clinical experiences with systemically administered siRNA-based therapeutics in cancer, Nat. Rev. Drug Discovery, 2015, 14(12), 843–856 CrossRef PubMed.
- G. R. Rettig and M. A. Behlke, Progress Toward In Vivo Use of siRNAs-II, Mol. Ther., 2012, 20(3), 483–512 CrossRef PubMed.
- Y. Chiu and T. M. Rana, siRNA function in RNAi: A chemical modification analysis, RNA, 2003, 9, 1034–1048 CrossRef PubMed.
- N. M. Snead, J. R. Escamilla-Powers, J. J. Rossi and A. P. McCaffrey, 5′ Unlocked Nucleic Acid Modification Improves siRNA Targeting, Mol. Ther.--Nucleic Acids, 2013, 2, e103 CrossRef PubMed.
- H. J. Kim, A. Kim, K. Miyata and K. Kataoka, Recent progress in development of siRNA delivery vehicles for cancer therapy, Adv. Drug Delivery Rev., 2016, 104, 61–77 CrossRef PubMed.
- W. Li and F. C. Szoka, Lipid-based Nanoparticles for Nucleic Acid Delivery, Pharm. Res., 2007, 24(3), 438–449 CrossRef PubMed.
- M. Thomas and A. M. Klibanov, Non-viral gene therapy: polycation-mediated DNA delivery, Appl. Microbiol. Biotechnol., 2003, 62(1), 27–34 CrossRef PubMed.
- M. E. Martin and K. G. Rice, Peptide-guided gene delivery, AAPS J., 2007, 9(1), E18–E29 CrossRef PubMed.
- D. E. Discher and F. Ahmed, POLYMERSOMES, Annu. Rev. Biomed. Eng., 2006, 8(1), 323–341 CrossRef PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Non-viral vectors for gene-based therapy, Nat. Rev. Genet., 2014, 15(8), 541–555 CrossRef PubMed.
- M. Foldvari, D. W. Chen, N. Nafissi, D. Calderon, L. Narsineni and A. Rafiee, Non-viral gene therapy: Gains and challenges of non-invasive administration methods, J. Controlled Release, 2016, 240, 165–190 CrossRef PubMed.
- L. Pärnaste, P. Arukuusk, K. Langel, T. Tenson and Ü. Langel, The Formation of Nanoparticles between Small Interfering RNA and Amphipathic Cell-Penetrating Peptides, Mol. Ther.--Nucleic Acids, 2017, 7, 1–10 CrossRef PubMed.
- G. Sahay, W. Querbes, C. Alabi, A. Eltoukhy, S. Sarkar and C. Zurenko, et al., Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling, Nat. Biotechnol., 2013, 31(7), 653–658 CrossRef PubMed.
- M. Breunig, U. Lungwitz, R. Liebl and A. Goepferich, Breaking up the correlation between efficacy and toxicity for nonviral gene delivery, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(36), 14454–14459 CrossRef PubMed.
- D. B. Rozema, D. L. Lewis, D. H. Wakefield, S. C. Wong, J. J. Klein and P. L. Roesch, et al., Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(32), 12982–12987 CrossRef PubMed.
- T. S. Zimmermann, V. Karsten, A. Chan, J. Chiesa, M. Boyce and B. R. Bettencourt, et al., Clinical Proof of Concept for a Novel Hepatocyte-Targeting GalNAc-siRNA Conjugate, Mol. Ther., 2017, 25(1), 71–78 CrossRef PubMed.
- P. Dosta, N. Segovia, A. Cascante, V. Ramos and S. Borrós, Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide-modified poly(beta-amino ester)s (PBAEs), Acta Biomater., 2015, 20, 82–93 CrossRef PubMed.
- N. Segovia, P. Dosta, A. Cascante, V. Ramos and S. Borrós, Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization, Acta Biomater., 2014, 10(5), 2147–2158 CrossRef PubMed.
- R. Núñez-Toldrà, P. Dosta, S. Montori, V. Ramos, M. Atari and S. Borrós, Improvement of osteogenesis in dental pulp pluripotent-like stem cells by oligopeptide-modified poly(β-amino ester)s, Acta Biomater., 2017, 53, 152–164 CrossRef PubMed.
- N. Segovia, M. Pont, N. Oliva, V. Ramos, S. Borrós and N. Artzi, Hydrogel Doped with Nanoparticles for Local Sustained Release of siRNA in Breast Cancer, Adv. Healthcare Mater., 2015, 4(2), 271–280 CrossRef PubMed.
- D. Oupicky, M. Ogris, K. A. Howard, P. R. Dash, K. Ulbrich and L. W. Seymour, Importance of Lateral and Steric Stabilization of Polyelectrolyte Gene Delivery Vectors for Extended Systemic Circulation, Mol. Ther., 2002, 5(4), 463–472 CrossRef PubMed.
- R. C. Carlisle, T. Etrych, S. S. Briggs, J. A. Preece, K. Ulbrich and L. W. Seymour, Polymer-coated polyethylenimine/DNA complexes designed for triggered activation by intracellular reduction, J. Gene Med., 2004, 6(3), 337–344 CrossRef PubMed.
- G. Caracciolo, L. Callipo, S. C. De Sanctis, C. Cavaliere, D. Pozzi and A. Laganà, Surface adsorption of protein corona controls the cell internalization mechanism of DC-Chol–DOPE/DNA lipoplexes in serum, Biochim. Biophys. Acta, Biomembr., 2010, 1798(3), 536–543 CrossRef PubMed.
- A. A. Eltoukhy, D. Chen, C. A. Alabi, R. Langer and D. G. Anderson, Degradable Terpolymers with Alkyl Side Chains Demonstrate Enhanced Gene Delivery Potency and Nanoparticle Stability, Adv. Mater., 2013, 25(10), 1487–1493 CrossRef PubMed.
- S. Y. Wong, J. M. Pelet and D. Putnam, Polymer systems for gene delivery—Past, present, and future, Prog. Polym. Sci., 2007, 32(8–9), 799–837 CrossRef.
- Z. Liu, Z. Zhang, C. Zhou and Y. Jiao, Hydrophobic modifications of cationic polymers for gene delivery, Prog. Polym. Sci., 2010, 35(9), 1144–1162 CrossRef.
- J. S. Kuo, Effect of Pluronic-block copolymers on the reduction of serum-mediated inhibition of gene transfer of polyethyleneimine – DNA complexes, Biotechnol. Appl. Biochem., 2003, 271, 267–271 CrossRef PubMed.
- A. Alshamsan, A. Haddadi, V. Incani, J. Samuel, A. Lavasanifar and H. Uludag, Formulation and Delivery of siRNA by Oleic, Acid, Mol. Pharmaceutics, 2008, 6(1), 188–200 Search PubMed.
- C.-J. Chen, J.-C. Wang, E.-Y. Zhao, L.-Y. Gao, Q. Feng and X.-Y. Liu, et al., Self-assembly cationic nanoparticles based on cholesterol-grafted bioreducible poly(amidoamine) for siRNA delivery, Biomaterials, 2013, 34(21), 5303–5316 CrossRef PubMed.
- D. Wang, A. S. Narang, M. Kotb, A. O. Gaber, D. D. Miller and S. W. Kim, et al., Novel Branched Poly(Ethylenimine)–Cholesterol Water-Soluble Lipopolymers for Gene Delivery, Biomacromolecules, 2002, 3(6), 1197–1207 CrossRef PubMed.
- P.-C. Chang, B.-Y. Liu, C.-M. Liu, H.-H. Chou, M.-H. Ho and H.-C. Liu, et al., Bone tissue engineering with novel rhBMP2-PLLA composite scaffolds, J. Biomed. Mater. Res., Part A, 2007, 81(4), 771–780 CrossRef PubMed.
- D. M. Lynn and R. Langer, Degradable Poly(β-amino esters): Synthesis, Characterization, and Self-Assembly with Plasmid DNA, J. Am. Chem. Soc., 2000, 122(44), 10761–10768 CrossRef.
- S. Lindman, I. Lynch, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Systematic investigation of the thermodynamics of HSA adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles. Effects of particle size and hydrophobicity, Nano Lett., 2007, 7(4), 914–920 CrossRef PubMed.
- V. Incani, A. Lavasanifar and H. Uludağ, Lipid and hydrophobic modification of cationic carriers on route to superior gene vectors, Soft Matter, 2010, 6(10), 2124 RSC.
- I. Khalil, S. Futaki, M. Niwa, Y. Baba, N. Kaji and H. Kamiya, et al., Mechanism of improved gene transfer by the N-terminal stearylation of octaarginine: enhanced cellular association by hydrophobic core formation, Gene Ther., 2004, 11(7), 636–644 CrossRef PubMed.
- D. Y. Takigawa and D. A. Tirrell, Phospholipid Packing by Branched Poly(ethy1enimine) Derivatives, Macromolecules, 1985, 18, 338–342 CrossRef.
- D. Fischer, Y. Li, B. Ahlemeyer, J. Krieglstein and T. Kissel, In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis, Biomaterials, 2003, 24(7), 1121–1131 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8me00006a |
| This journal is © The Royal Society of Chemistry 2018 |