Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quantitative temporal interrogation in 3D of bioengineered human cartilage using multimodal label-free imaging†
Catarina
Costa Moura
 ab,
Stuart A.
Lanham
ab,
Stuart A.
Lanham
 b,
Tual
Monfort
a,
Konstantinos N.
Bourdakos
a,
Rahul S.
Tare
b,
Tual
Monfort
a,
Konstantinos N.
Bourdakos
a,
Rahul S.
Tare
 bc,
Richard O. C.
Oreffo
bc,
Richard O. C.
Oreffo
 *b and
Sumeet
Mahajan
*b and
Sumeet
Mahajan
 *a
*a
aDepartment of Chemistry and Institute for Life Sciences, University of Southampton, Highfield Campus, Southampton SO17 1BJ, UK. E-mail: s.mahajan@soton.ac.uk
bBone and Joint Research Group, Centre for Human Development, Stem Cells and Regeneration, Institute of Developmental Sciences, Faculty of Medicine, University of Southampton, Southampton SO16 6YD, UK. E-mail: richard.oreffo@soton.ac.uk
cMechanical Engineering Department, Faculty of Engineering and the Environment, University of Southampton, Highfield Campus, Southampton, SO17 1BJ, UK
First published on 5th September 2018
Abstract
The unique properties of skeletal stem cells have attracted significant attention in the development of strategies for skeletal regeneration. However, there remains a crucial unmet need to develop quantitative tools to elucidate skeletal cell development and monitor the formation of regenerated tissues using non-destructive techniques in 3D. Label-free methods such as coherent anti-Stokes Raman scattering (CARS), second harmonic generation (SHG) and two-photon excited auto-fluorescence (TPEAF) microscopy are minimally invasive, non-destructive, and present new powerful alternatives to conventional imaging techniques. Here we report a combination of these techniques in a single multimodal system for the temporal assessment of cartilage formation by human skeletal cells. The evaluation of bioengineered cartilage, with a new parameter measuring the amount of collagen per cell, collagen fibre structure and chondrocyte distribution, was performed using the 3D non-destructive platform. Such 3D label-free temporal quantification paves the way for tracking skeletal cell development in real-time and offers a paradigm shift in tissue engineering and regenerative medicine applications.
Insight, innovation, integrationThis work reports the non-destructive temporal interrogation and quantitative assessment of bioengineered human cartilage using label-free multimodal imaging. Our imaging platform combines the chemically- and structurally-selective techniques of coherent anti-Stokes Raman scattering (CARS) and second harmonic generation (SHG) with two-photon excited auto-fluorescence (TPEAF) microscopy. Quantitative image analysis yields parameters that allow objective assessment of the engineered cartilage formed by human skeletal cells, and captures the interaction between collagen fibres and chondrocytes over time. This study represents an important step towards the provision of a novel tool to follow skeletal cell development and monitor the formation of newly engineered tissues in real-time, non-invasively and non-destructively, that will ultimately help realise the full potential of cell-based approaches in reparative and regenerative medicine. |
Introduction
Articular cartilage damage as a result of trauma, injury or degeneration can result in tissue loss and a deterioration in the specialised extracellular matrix.1 Skeletal stem cells (SSCs), present in bone marrow stroma, have the capacity to differentiate and give rise to the stromal lineages, namely bone, haematopoiesis-supportive stroma, fat and cartilage.2–4 The ability to enrich for skeletal stem cells from bone marrow has enabled the evaluation and application of this multipotent stem cell population in skeletal reparative strategies, including cartilage regeneration. Cartilage tissue engineering using, for example, three-dimensional (3D) cultures has been extensively applied for in vitro cartilage formation.2,5 However, these culture systems typically rely on two-dimensional (2D) imaging modalities to provide biological information.6,7 Furthermore, current available techniques for skeletal cell characterisation are: (i) invasive (involve labels or stains), (ii) require cell fixation or lysis, and/or (iii) are destructive. Thus, there remains a crucial unmet need to develop appropriate tools to follow skeletal stem cell development and to monitor the formation of regenerated tissues using non-destructive, non-invasive and label-free techniques that, importantly, enable temporal interrogation in 3D. The main objective of this work was to establish multimodal label-free imaging as a platform to quantitatively assess and evaluate the development of fetal skeletal cells towards a cartilage phenotype in 3D, non-destructively, and in the absence of any labelling with dyes or fluorophores.Techniques which rely on the measurement of vibrational information such as spontaneous Raman spectroscopy and coherent anti-Stokes Raman scattering (CARS) microscopy are inherently non-invasive, non-destructive, and chemically selective (reviewed in Moura et al.8). Although spontaneous Raman spectroscopy has been used to perform quantitative volumetric analysis of 3D stem cell cultures,6 the required long acquisition times preclude rapid (≈10 μs per pixel) or video-rate (>5 frames per second) imaging. In contrast, coherent Raman imaging techniques excite vibrational coherences in molecules which can enhance signals by more than 105 times.9
Research by Downes et al. and our group has demonstrated that CARS microscopy can be applied to image the differentiation of adipose-derived stem cells into osteoblasts,10 and skeletal stem cells into adipocytes,11 respectively. The application of coherent Raman techniques in assessing chondrogenesis, i.e. differentiation into cartilage, has however, not been investigated. Moreover, 3D imaging using CARS or its multimodal combination with other label-free techniques has not been applied to analyse chondrogenic cultures or bioengineered cartilage, obtained through differentiation of fetal skeletal cells into chondrocytes.
Crucially, technology has evolved to enable the use of higher harmonic generation techniques, such as second harmonic generation (SHG), in conjunction with CARS on a multimodal system.12 SHG is a second-order coherent process in which non-centrosymmetric structures, i.e. structures lacking a centre of symmetry, combine two lower energy photons from an excitation source to generate photons with exactly twice the incident frequency.13,14 Type I and Type II collagen have a hierarchy of non-centrosymmetric structures within each fibril and both form aligned striated fibres creating a super-structure (high degree of crystallinity) resulting in high SHG activity. Furthermore, polarisation resolved SHG microscopy can enhance molecular specificity in tissues15 and also allow Type I and Type II collagen to be distinguished.16 In contrast, Type IV collagen has no fibrillary structure and consequently the SHG signal generated is insufficient for imaging.17–19 Type II collagen is a principal component of cartilage tissue and represents more than 90% of the collagen in articular cartilage extracellular matrix.20–22 Since SHG has the established ability to directly (without labelling) visualise collagen fibres it is an ideal modality to provide comprehensive structural information on their arrangement within a 3D cell construct such as cartilage pellets.
In addition to CARS and SHG, two-photon excited autofluorescence (TPEAF) imaging offers another label-free modality to interrogate cellular composition and tissue development, providing complementary information.23–26 Two-photon excitation fluorescence has reduced phototoxicity/photodamage compared to single-photon fluorescence microscopy.24,25 Flavin adenine dinucleotide (FAD), a redox co-factor associated with mitochondria, is involved in redox reactions in living cells, as well as several other cellular pathways including biosynthetic processes. These co-enzymes contribute significantly to the cellular autofluorescence. Consequently TPEAF enables non-destructive imaging of living cells and tissues without the interference and, crucially, toxicity of exogenous dyes,24,27 and it has been applied to image cells undergoing chondrogenesis.28 TPEAF imaging of cells in cartilaginous pellets of fetal skeletal cells thus provides an invaluable method to elucidate the spatial organisation of chondrocytes within the extracellular matrix of cartilaginous pellets.
Given that CARS, SHG and TPEAF are all multiphoton microscopy techniques and therefore have inherent z-sectioning capability, this allows 3D imaging as well as the application of near infrared wavelengths enables deeper penetration into a 3D construct and thick tissue samples.29 The multimodal combination of complementary imaging techniques, namely CARS, SHG and TPEAF, is highly advantageous and could provide a holistic insight into the development and differentiation of skeletal cells. In this work, we have harnessed the power of multimodal, non-destructive imaging, namely CARS, SHG and TPEAF, to establish and detail, for the first time, a label-free platform for 3D quantitative assessment of human fetal skeletal cell development towards cartilage. The methodology developed here is superior to current prevalent methods to assess chondrogenic differentiation as there is no requirement for sample preparation, extensive and laborious staining procedures, is non-destructive and label-free, and allows the interrogation and, importantly, quantitative analysis of the bioengineered construct in 3D.
Materials and methods
Materials
Alpha minimum essential medium with deoxyribonucleotides, ribonucleotides and ultraglutamine (α-MEM), fetal calf serum (FCS), Dulbecco's phosphate buffered saline (PBS), and 0.025% trypsin–EDTA (w/v) solution with 0.05% glucose were purchased from Lonza (Switzerland). Bovine serum albumin (BSA) was obtained from GE Healthcare (United States of America). Collagenase B was purchased from Roche (Switzerland). TGF-β3 was purchased from PeproTech (United Kingdom). Anti-collagen Type I primary antibody, rabbit IgG, was a kind gift from Dr Larry Fisher (National Institutes of Health), and anti-collagen Type II primary antibody, rabbit IgG, was purchased from Calbiochem (Merck, Millipore, Germany). Alcian blue 8GX, ascorbate-2-phosphate, β-mercaptoethanol, biotinylated goat anti-rabbit secondary antibody, collagenase IV, dexamethasone, dimethyl sulfoxide (DMSO), DPX mountant for histology, ethanol, formaldehyde, hyaluronidase, hydrogen peroxide solution, ITS Liquid Media Supplement 100× (10 μg mL−1 recombinant human insulin, 5.5 μg mL−1 human transferrin and 5 ng mL−1 sodium selenite), molybdophosphoric acid, penicillin–streptomycin (100 U mL−1 penicillin and 100 mg mL−1 streptomycin), picric acid, sirius red F3B, Weigert's haematoxylin constituents, and oligonucleotide primers were supplied by Sigma-Aldrich. Histo-Clear and hydromount were purchased from National Diagnostics (United Kingdom).Cell culture
Gene expression analysis
| Transcript | Symbol | Primer sequence (5′–3′) |
|---|---|---|
| Aggrecan | ACAN | F: GACGGCTTCCACCAGTGT |
| R: GTCTCCATAGCAGCCTTCC | ||
| β-Actin | ACTB | F: GGCATCCTCACCCTGAAGTA |
| R: AGGTGTGGTGCCAGATTTTC | ||
| Alkaline phosphatase | ALPL | F: GGAACTCCTGACCCTTGACC |
| R: TCCTGTTCAGCTCGTACTGC | ||
| Alpha-1 Type I collagen | COL1A1 | F: GAGTGCTGTCCCGTCTGC |
| R: TTTCTTGGTCGGTGGGTG | ||
| Alpha-1 Type II collagen | COL2A1 | F: CCTGGTCCCCCTGGTCTTGG |
| R: CATCAAATCCTCCAGCCATC | ||
| Peroxisome proliferator activated receptor gamma | PPARG | F: GGGCGATCTTGACAGGAAAG |
| R: GGGGGGTGATGTGTTTGAACTTG | ||
| Fatty acid binding protein 4 | FABP4 | F: TAGATGGGGGTGTCCTGGTA |
| R: CGCATTCCACCACCAGTT | ||
| Transcription factor SOX9 | SOX9 | F: CCCTTCAACCTCCCACACTA |
| R: TGGTGGTCGGTGTAGTCGTA |
Histological analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in blocking buffer, and anti-collagen Type II, dilution 1
1000 in blocking buffer, and anti-collagen Type II, dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in blocking buffer) were added to the sections for an overnight incubation at 4 °C. Negative controls (omission of the primary antibody) were included in all immunohistochemistry procedures. Biotinylated secondary antibody (1
500 in blocking buffer) were added to the sections for an overnight incubation at 4 °C. Negative controls (omission of the primary antibody) were included in all immunohistochemistry procedures. Biotinylated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in blocking buffer) was applied for 1 hour before incubation with avidin-conjugated peroxidase. Sample sections were treated with 3-amino-9-ethylcarbazole for 3–4 minutes until visualisation of a red-brown reaction product. All sections were counter-stained with Alcian blue 8GX and mounted with hydromount mounting medium.
100 in blocking buffer) was applied for 1 hour before incubation with avidin-conjugated peroxidase. Sample sections were treated with 3-amino-9-ethylcarbazole for 3–4 minutes until visualisation of a red-brown reaction product. All sections were counter-stained with Alcian blue 8GX and mounted with hydromount mounting medium.
2D imaging
Sample sections at different time points (4, 7, 14 and 21 days) were imaged using SHG and TPEAF in a home-built laser scanning microscope (Fig. S1, ESI†). ScanImage 5.1 (Vidrio Technologies), an open source software, was used for acquiring images with the laser scanning microscope.32 A brief description of this system is as follows: a tuneable Ti:Sapphire oscillator (Spectra-Physics Mai Tai®, 700–1000 nm, 100 fs, 80 MHz) is coupled through a galvanometric scanner to an upright microscope configured for simultaneous epi-detection of SHG and TPEAF in different channels. A long pass dichroic beam splitter with cut-off at 775 nm is used to separate the laser excitation from the epi-collected emission. A further long pass dichroic beam splitter with cut-off at 458 nm (Semrock FF458-Di02) used in the collection path for separating the SHG and TPEAF signals which after going through narrow band pass filters centred at 400 nm and 500 nm respectively, are focused with short focal length lenses on the respective detectors (photomultiplier tubes; Hamamatsu: H10722-01 and H10722-20). The laser oscillator was tuned to 800 nm. Each sample was imaged using a 20×/0.5 NA water immersion objective, with 3× optical zoom using galvanometric scanning, and acquisition time of 16 ms per line for a 1024 × 1024 pixel image. The total incident power on the sample was approximately 70 mW.Raman spectroscopy
Fetal skeletal cell pellets cultured for 21 days were washed with PBS and fixed with a 4% (v/v) formaldehyde solution for 25 minutes at room temperature. Raman spectra were obtained using a Renishaw® inVia Raman microscope with a 633 nm laser and a 20×/0.5 NA water immersion objective, in combination with WiRE 3.4 software. For each spectrum 5 accumulations were collected using a 1200 lines per mm grating, 6 mW laser power at the sample, and an exposure time of 60 seconds. Cosmic ray artefacts were removed using WiRE 3.4. Final spectra were pre-processed with a 5th order polynomial baseline correction and wavelet de-noising using IRootLab,33 a MATLAB based toolbox for vibrational spectroscopy.3D imaging
Fetal skeletal cell pellets cultured for different time points (4, 7, 14 and 21 days) were washed with PBS and fixed with a 4% (v/v) formaldehyde solution for 25 minutes at room temperature. Cell pellets were washed with excess PBS, and images were captured using a home-built system (Fig. S2, ESI†) which allows for image acquisition with coherent anti-Stokes Raman scattering (CARS), SHG and TPEAF, simultaneously. This multimodal laser scanning microscope also used ScanImage 5.1 (Vidrio Technologies) for image acquisition.32 In more detail, for CARS imaging the fundamental of a fibre laser (1031 nm, 2 picosecond, 80 MHz, Emerald Engine, APE) was used as a Stokes beam, and the output of an optical parametric oscillator (OPO) (APE, Levante Emerald, 650–950 nm) which was synchronously pumped by the second harmonic (516 nm) of the fibre laser, was used as a pump beam. Our multimodal platform is unique in that it utilises 2 ps pulses. Non-linear interactions increase with shorter pulse-widths; however, with CARS since vibrational line-widths are of the order of 10 cm−1, therefore, 2 ps pulse-widths are near ideal for CARS in ensuring efficient excitation and reduction of non-resonant background9 and at the same time do not hugely compromise signal generation with other non-linear modalities such as SHG and TPEAF. The two beams were made collinear and then coupled through a galvanometric scanner to an inverted microscope (Nikon Ti-U) configured for epi-detection. Their temporal overlap was controlled with a delay line. For imaging lipids in the cell pellets, the C–H stretching mode at 2845 cm−1 was targeted, and for this reason the OPO was tuned to 797.8 nm. The total incident power on the sample was approximately 147 mW. The SHG (400 nm) and TPEAF signal (520 nm) was collected with the OPO beam at 800 nm. The detection was similar to the 2D imaging setup described above except that dichroic beamsplitters with cut-offs at 442 nm (Semrock Di02-R442) and 594 nm (Semrock Di02-R594). Each sample was imaged using a 20×/0.75 NA Nikon objective, with 6× optical zoom using galvanometric scanning, 29 ms per line period for a 512 × 512 pixel image.Image processing and analysis
Statistical analysis
All experiments were performed using three different human fetal samples. Graphs were prepared using GraphPad Prism 7 (San Diego, CA, USA) and statistical analysis was performed using IBM® SPSS® Statistics version 21.0 (IBM Corporation, Armonk, NY, USA). Data distributions were tested for normality using the Shapiro–Wilk test and statistical significance was tested using one-way analysis of variance (ANOVA) with Tukey's post hoc test for samples following a normal distribution or the Mann–Whitney U-test for samples not following a normal distribution. Differences were considered to be statistically significant at P ≤ 0.05.Results and discussion
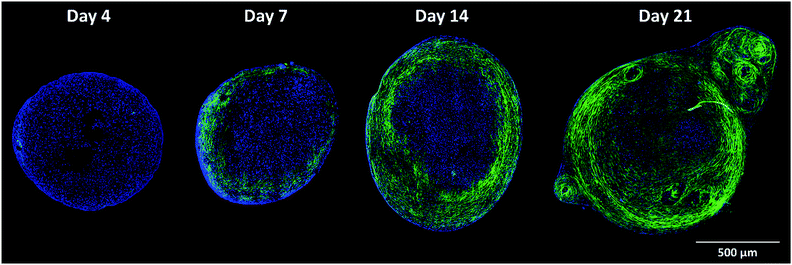
To investigate whether label-free, multimodal, non-linear techniques could be used to follow chondrogenic differentiation of skeletal cell populations, human fetal femur-derived skeletal stem/progenitor populations (fetal skeletal cells) were stimulated to differentiate into chondrocytes and generate cartilage tissue in an in vitro 3D pellet culture system over a period of 21 days in chondrogenic medium.37 Given that some scaffold materials may impair tissue formation and defect regeneration, a scaffold-free chondrogenic differentiation protocol was used.38,39In order to validate chondrogenic development, we initially imaged unstained histological sections of the pellets for collagen distribution and cellular autofluorescence (at 520 nm corresponding to FAD emission). Temporal analysis across the centre of the 3D cartilaginous pellet was performed. Fig. 1 shows multimodal images of the fetal skeletal cell pellet cultured in chondrogenic conditions for 4, 7, 14 and 21 days. Cellular autofluorescence was imaged using TPEAF (blue) and collagen fibres imaged using SHG (green) simultaneously. Pellets were formed by skeletal cell aggregation in chondrogenic media, which starts to form extracellular matrix to provide structural integrity to the bioengineered cartilaginous construct.40,41 Using this multimodal imaging platform, SHG enabled the temporal interrogation of chondrogenic differentiation of fetal skeletal cells in pellet culture. SHG allowed high resolution imaging of the concomitant extracellular deposition of fibrillar collagen that proceeds alongside the differentiation of progenitor cells into chondrocytes, as evidenced by the increase in SGH signals and gradual accumulation of collagen fibres over time. Interestingly, the images illustrate that collagen deposition commences from the periphery of the construct and progresses inwards, over time, as the cartilage pellet develops and matures. This pattern of collagen deposition can also be observed with conventional stains using histological analysis (Fig. 2). All sections were counter-stained with Alcian blue to visualise the dense proteoglycan matrix in the cartilage pellets. Sirius red staining showed formation of collagen fibres. Immunohistochemistry revealed expression of collagen Type I and strong expression of collagen Type II. However, histological stains can bind non-specifically while SHG is selective to non-centrosymmetric super-structures such as Type II collagen.42 The differences in collagen staining compared to Fig. 1 are also likely due to the fact that these were different planar sections from the same sample, highlighting a drawback of 2D analysis of a 3D sample. Furthermore, SHG provided a spatial resolution of <350 nm, considerably superior to resolutions observed using transmission/brightfield light microscopy in histology.
 | ||
| Fig. 2 Histological analysis of human fetal skeletal cells cultured in chondrogenic media for 4, 7, 14, and 21 days. Sirius red staining shows formation of collagen fibres. Immunohistochemistry reveals expression of collagen Type I and strong expression of collagen Type II. All sections were counter-stained with Alcian blue to visualise the dense proteoglycan matrix in the pellets. The staining was carried out on serial sections (at different planes) from the same sample as in Fig. 1. Scale bar corresponds to 500 μm. | ||
In addition to conventional histological analysis, chondrogenic differentiation of human fetal skeletal cells was also verified by analysis of gene expression using qPCR (Fig. 3). By day 21 of pellet culture, expression of SOX9, the key chondroinductive factor, COL2A1, encoding the α-chain of hyaline cartilage-specific Type II collagen, and ACAN, the major proteoglycan in cartilage, were all significantly up-regulated. Following 21 days of chondrogenic differentiation, SSCs showed a modest, but statistically non-significant, increase in expression of COL10A1, encoding the alpha chain of collagen Type X, a key constituent of fibrocartilage and bone matrix. As cartilage, bone, and marrow fat (adipocytes) are related to each other within the specific stromal developmental process and niche environment, and share a common progenitor (the skeletal stem cell),2 we also examined the expression profiles of characteristic adipogenic (PPARG and FABP4) and osteogenic genes (ALPL and COL1A1) at the corresponding time-points. No significant differences were observed in the temporal expression levels of the adipogenic genes, PPARG and FABP4. Furthermore, after 21 days of chondrogenic differentiation, no significant difference in ALPL expression, a key osteogenic marker, was observed. Although at day 21 the fetal skeletal cell populations demonstrated an increase in expression of COL1A1, a constituent of fibrocartilage and bone matrix, this was not statistically significant.
While 2D imaging of sections of bioengineered tissue can be used for assessment of tissue development by label-free imaging or conventional histological analysis, it is important to recognise that the actual tissue has a complex 3D architecture. Histological analysis necessitates a sample is sectioned using a microtome. In this work, cell pellets were sectioned every 5 μm for histology; thus, to analyse a 100 μm-thick sample, one would need 20 consecutive 5 μm-sections, assuming no loss of material during sample preparation. In addition, combination of multiple staining procedures simultaneously is challenging, while fixation, sectioning and staining can destroy functionality, potential preclinical evaluation and in vivo deployment. Moreover, 2D image analysis can lead to misinterpretation of the results, especially when analysing a 3D system with complex fibril structures such as collagen. Fig. 4 shows an example of potential misinterpretation following 2D image analysis. When the assessment of collagen content is based on a single image of the tissue-engineered construct, it is possible to infer the presence of either negligible (Fig. 4A) or extensive collagen fibres (Fig. 4B) in the same cartilaginous construct.
Elucidation of the three-dimensional architecture of the bioengineered tissue by non-perturbative 3D imaging is therefore crucial to determine whether the tissue structures are arranged appropriately and, critically, are functional for eventual therapeutic use. Moreover, in addition to imaging collagen as a marker for chondrocyte differentiation, it is vital to image cells in a non-invasive, non-destructive manner to obtain a holistic view of cartilage development.
Therefore, in a further development, we combined CARS with SHG and TPEAF in a multimodal platform to image chondrogenesis in 3D. Although CARS has, to date, predominantly been used to image lipid droplets, it can be potentially applied to image other biological components by targeting different vibrational frequencies.10 Here, we have applied CARS microscopy to image whole cells as well as lipid droplets in 3D in the developing cartilaginous pellet. We targeted the C–H stretch mode predominant in lipids at 2845 cm−1 (Raman spectrum is shown in Fig. S3, ESI†). This allowed visualisation of the distribution of lipid droplets during chondrogenesis. CARS is an ideal technique to image lipid droplets and we have shown in previous work that it is more sensitive than conventional Oil Red O staining for picking up minute changes on SSC differentiation into adipocytes.11 In an additional innovative step we have used the information provided by CARS imaging for visualisation of cells. While lipid droplet distribution is usually ignored during chondrogenesis (probably due to the lack of visualisation tools), lipids may have an important role informing metabolic activity. Imaging cells in a label-free manner is important to understand and characterise cell–matrix phenotypes, especially in cartilage tissue. Thus, the multimodal combination of CARS with SHG and TPEAF results in a powerful platform for label-free and non-destructive evaluation of chondrogenesis, informing matrix and lipid activity as well as resultant bioengineered cartilage architecture and composition.
Fig. 5 shows 3D multimodal images of fetal skeletal cells cultured in chondrogenic conditions for 4, 7, 14 and 21 days, with maximum intensity z-projections and 3D visualisation of the image stacks. Videos with 3D view are available in ESI,† Movies S1–S4. During the chondrogenic differentiation of skeletal cells and subsequent cartilage formation, the collagen fibres (green) were noted to change their shape and configuration. The cartilaginous pellet displayed rich fibrillar collagen from the early stages of differentiation, with chondrocytes (blue) distributed throughout the pellet within the network of collagen fibres, reminiscent of native articular cartilage.21 Additionally, the use of CARS provided information on the distribution of lipid droplets in the cartilaginous pellets (red). This is important, as while glucose is a key source of energy for chondrocytes, lipids within cartilage tissue are also essential for cartilage physiology.43 Chondrocytes have the ability to synthesise lipids and to harness lipids as an additional source of energy and as structural components and signalling molecules.43,44Fig. 5 demonstrates that chondrocytes synthesise lipids from day 4 of culture throughout the development of cartilage. The gene expression profile of adipogenic genes (PPARG and FAPB4) demonstrated negligible differences during chondrogenic differentiation (Fig. 3), indicating that the skeletal cells were not differentiating into adipocytes. However, as indicated above, temporal images of the 3D bioengineered cartilage demonstrate noticeable lipid production by skeletal cells during chondrocyte differentiation and chondrogenic maturation. 3D label-free imaging offers major advantages over standard imaging procedures, and with this detailed label-free approach, a 100 μm-thick sample can be imaged with no sample preparation, and without staining or application of dyes.
Critically, quantification of the changes during cartilage development using multimodal label-free techniques provides enhanced understanding of fetal skeletal cell chondrogenesis and cartilage growth. The SHG signal of human fetal skeletal cell pellet cultures was analysed using an established image analysis technique that enabled measurement of collagen fibre parameters (CT-FIRE35) to determine collagen fibre composition and distribution. Using image analysis, amounts as well as geometric parameters, namely length, width and straightness of each collagen fibre, were extracted from each z-stack image (Fig. 6A). Image analysis revealed that collagen fibre width did not change significantly during fetal skeletal cell differentiation along the chondrogenic lineage from day 4 to day 21 (Fig. 6B). The measured collagen fibre width was approximately between 2 and 2.5 μm. Published studies indicate that mature collagen fibres assemblies of such fibrils are typically wider than 2 μm;21,45–47 this is well within the resolution limit of our multimodal microscope (approximately 350 nm)48 and hence the changes we detail are not only reliable but consistent with literature reports. Quantitation of collagen fibre length over time demonstrated a significant increase in mean length from 22 μm at 4 days to 27 μm at 14 days of chondrogenic culture. There was a modest subsequent decrease at day 21 to give a mean collagen fibre length of 26 μm, indicating that collagen deposition occurs longitudinally (increase in length during cell culture time). Interestingly, there were no significant differences in fibre straightness in cartilaginous pellets of fetal skeletal cells between the different time-points of culture in chondrogenic medium. The trend in these results (n = 3) is for collagen fibres to become slightly straighter following culture to day 14. These results might indicate that initial deposition up to day 14 occurs by linear extension (length). By increasing the number of samples it would help to better understand any changes in straightness, possibly linked to the spirals of collagen fibres formed in discrete regions as chondrogenic maturation occurred (Fig. 5). It is noted that further work could also monitor collagen Type I and II through the differentiation process using techniques such polarisation resolved SHG microscopy and that will potentially provide added insight into cartilage formation.15,16
As pellet cultures provide an optimal 3D environment for robust cartilage development, quantitative cell analysis was extended to 3D imaging. Cell quantification is essential to understand cell–matrix interactions as, in combination with collagen quantification, it can yield an analytical parameter for evaluating the cartilage phenotype. The cells were counted using CARS images incorporating a modification of 3D particle analysis function in Fiji (Fig. 7A). Although not statistically significant (n = 3 patients) there is an increase in the number of cells per unit volume in fetal skeletal cell pellets cultured under chondrogenic conditions for 7 days before cell numbers decreased (Fig. 7B). Interestingly, the cell sizes in the corresponding volumes of interest showed a modest increase over time. Given that the overall size of the chondrocytic pellet increased, these observations indicate enhanced cell proliferation up to day 7 after which the cells grow in size. This individual increase in size of each cell accompanied by a decrease in cells numbers per unit volume indicates differentiation. The enhanced extra-cellular matrix deposition, due to collagen and proteoglycan formation typical of the chondrocyte phenotype, results in an overall increase in the size of the cartilage pellet.
In addition, here image analysis allowed us to calculate the amount of collagen fibres present in the same volume of interest to determine collagen production and cell number as a consequence of development of the cartilage tissue in the pellet (Fig. 8). The number of cells was observed to decline after day 7, possibly associated with the fact that in a confined volume, where the collagen fibre amount increases and a modest increase in cell size was observed (Fig. 7), there is limited space for additional cells in the same volume. Therefore, a new assessment parameter of collagen per cell (CpC) was defined, which captures the net balance between collagen deposition and proliferation of cells. While this needs further evaluation on a larger patient cohort, preliminarily studies using fetal skeletal cells from 3 subjects indicate a switch in chondrogenic differentiation from a proliferative state to a collagen-rich expansion state, suggesting that this parameter can be used to characterise a phenotypic change in chondrogenesis based on cell–matrix organisation. The use of this quantitative parameter could provide an important understanding in assessing the health/phenotype of bioengineered cartilage and measuring the effect of environmental and chemical modulators on engineered tissue.
The multimodal combination of CARS microscopy with SHG and TPEAF with integrated quantitative 3D imaging analysis offers unparalleled insight into the chondrogenic differentiation of human fetal skeletal progenitor and stem cell populations. Retrieval of 3D information using non-destructive approaches offers tissue engineers new platform technologies to follow the formation of new regenerated tissues, as well as new tools for stem cell biologists studying health and disease with 3D cartilage models in real-time.
Conclusions
In conclusion, a quantitative 3D label-free imaging system based on 2 ps lasers for optimal multimodal performance was devised, offering a non-invasive and non-destructive platform for the analysis of chondrogenic differentiation of human fetal skeletal cell populations, and enabling elucidation of temporal changes in cartilage development. Quantitative analysis of collagen and cells through multimodal microscopy (combining CARS, SHG and TPEAF) yielded parameters to objectively assess cartilage development. A new parameter based on quantitative analysis, collagen per cell or CpC, allows to capture the interplay between matrix deposition and cellular proliferation as the bioengineered tissue develops over time. Such non-perturbative label-free and quantitative analysis in 3D is essential for development of the field of human skeletal repair and regeneration research, with significant impact for tissue engineering and regenerative medicine applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Dr Emma Budd for helping with the collection and isolation of fetal tissues. This work was supported by the Institute for Life Sciences, University of Southampton, BBSRC (BB/L021072/1), ERC grant Nano-ChemBioVision 638258 and Wessex Medical Research. SM would also like to acknowledge the support provided by APE Gmbh for the laser system.Notes and references
- E. Budd, M. C. de Andres, T. Sanchez-Elsner and R. O. C. Oreffo, Sci. Rep., 2017, 7, 46704 CrossRef PubMed.
- P. Bianco and P. G. Robey, Development, 2015, 142, 1023–1027 CrossRef PubMed.
- A. Aarvold, J. O. Smith, E. R. Tayton, A. M. H. Jones, J. I. Dawson, S. Lanham, A. Briscoe, D. G. Dunlop and R. O. C. Oreffo, J. Tissue Eng. Regen. Med., 2014, 8, 779–786 CrossRef PubMed.
- J. I. Dawson, J. Kanczler, R. Tare, M. Kassem and R. O. C. Oreffo, Stem Cells, 2014, 32, 35–44 CrossRef PubMed.
- M. Serafini, B. Sacchetti, A. Pievani, D. Redaelli, C. Remoli, A. Biondi, M. Riminucci and P. Bianco, Stem Cell Res., 2014, 12, 659–672 CrossRef PubMed.
- C. Kallepitis, M. S. Bergholt, M. M. Mazo, V. Leonardo, S. C. Skaalure, S. A. Maynard and M. M. Stevens, Nat. Commun., 2017, 8, 14843 CrossRef PubMed.
- A. Khademhosseini and R. Langer, Nat. Protoc., 2016, 11, 1775–1781 CrossRef PubMed.
- C. C. Moura, R. S. Tare, R. O. C. Oreffo and S. Mahajan, J. R. Soc., Interface, 2016, 13 DOI:10.1098/rsif.2016.0182.
- J. X. Cheng and X. S. Xie, Coherent Raman Scattering Microscopy, Taylor & Francis, 2012 Search PubMed.
- A. Downes, R. Mouras, P. Bagnaninchi and A. Elfick, J. Raman Spectrosc., 2011, 42, 1864–1870 CrossRef PubMed.
- J. P. Smus, C. C. Moura, E. McMorrow, R. S. Tare, R. O. C. Oreffo and S. Mahajan, Chem. Sci., 2015, 6, 7089–7096 RSC.
- L. Mortati, C. Divieto and M. P. Sassi, J. Raman Spectrosc., 2012, 43, 675–680 CrossRef.
- P. Campagnola, Anal. Chem., 2011, 83, 3224–3231 CrossRef PubMed.
- G. Cox and E. Kable, in Cell Imaging Techniques: Methods and Protocols, ed. D. J. Taatjes and B. T. Mossman, Humana Press, Totowa, NJ, 2006, pp. 15–35 Search PubMed.
- R. Kumar, K. M. Grønhaug, E. I. Romijn, A. Finnøy, C. L. Davies, J. O. Drogset and M. B. Lilledahl, J. Biophotonics, 2015, 8, 730–739 CrossRef PubMed.
- P.-J. Su, W.-L. Chen, T.-H. Li, C.-K. Chou, T.-H. Chen, Y.-Y. Ho, C.-H. Huang, S.-J. Chang, Y.-Y. Huang, H.-S. Lee and C.-Y. Dong, Biomaterials, 2010, 31, 9415–9421 CrossRef PubMed.
- X. Chen, O. Nadiarynkh, S. Plotnikov and P. J. Campagnola, Nat. Protoc., 2012, 7, 654–669 CrossRef PubMed.
- J. K. Mouw, G. Ou and V. M. Weaver, Nat. Rev. Mol. Cell Biol., 2014, 15, 771–785 CrossRef PubMed.
- J.-Y. Exposito, U. Valcourt, C. Cluzel and C. Lethias, Int. J. Mol. Sci., 2010, 11, 407–426 CrossRef PubMed.
- J. A. Buckwalter, H. J. Mankin and A. J. Grodzinsky, Instructional course lectures, 2005, vol. 54, pp. 465–480 Search PubMed.
- A. H. Reddi, Articular Cartilage, CRC Press, 2013, pp. 1–50 Search PubMed.
- A. J. S. Fox, A. Bedi and S. A. Rodeo, Sports Health, 2009, 1, 461–468 CrossRef PubMed.
- A. Zoumi, A. Yeh and B. J. Tromberg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11014–11019 CrossRef PubMed.
- A. A. Heikal, Biomarkers Med., 2010, 4, 241–263 CrossRef PubMed.
- W. Denk, J. Strickler and W. Webb, Science, 1990, 248, 73–76 CrossRef PubMed.
- E. A. Shirshin, Y. I. Gurfinkel, A. V. Priezzhev, V. V. Fadeev, J. Lademann and M. E. Darvin, Sci. Rep., 2017, 7, 1171 CrossRef PubMed.
- A. Knight and N. Billington, Biophotonics International, 2001, 8, 42–50 Search PubMed.
- A. V. Meleshina, V. V. Dudenkova, A. S. Bystrova, D. S. Kuznetsova, M. V. Shirmanova and E. V. Zagaynova, Stem Cell Res. Ther., 2017, 8, 15 CrossRef PubMed.
- E. E. Hoover and J. A. Squier, Nat. Photonics, 2013, 7, 93–101 CrossRef PubMed.
- S. H. Mirmalek-Sani, R. S. Tare, S. M. Morgan, H. I. Roach, D. I. Wilson, N. A. Hanley and R. O. Oreffo, Stem Cells, 2006, 24, 1042–1053 CrossRef PubMed.
- R. S. Tare, D. Howard, J. C. Pound, H. I. Roach and R. O. Oreffo, Biochem. Biophys. Res. Commun., 2005, 333, 609–621 CrossRef PubMed.
- T. A. Pologruto, B. L. Sabatini and K. Svoboda, BioMedical Engineering Online, 2003, 2, 13 CrossRef PubMed.
- J. Trevisan, P. P. Angelov, A. D. Scott, P. L. Carmichael and F. L. Martin, Bioinformatics, 2013, 29, 1095–1097 CrossRef PubMed.
- S. Preibisch, S. Saalfeld and P. Tomancak, Bioinformatics, 2009, 25, 1463–1465 CrossRef PubMed.
- J. S. Bredfeldt, Y. Liu, C. A. Pehlke, M. W. Conklin, J. M. Szulczewski, D. R. Inman, P. J. Keely, R. D. Nowak, T. R. Mackie and K. W. Eliceiri, J. Biomed. Opt., 2014, 19, 16007 CrossRef PubMed.
- J. S. Bredfeldt, Y. Liu, M. W. Conklin, P. J. Keely, T. R. Mackie and K. W. Eliceiri, J. Pathol. Inform., 2014, 5, 28 CrossRef PubMed.
- S. Li, P. Glynne-Jones, O. G. Andriotis, K. Y. Ching, U. S. Jonnalagadda, R. O. Oreffo, M. Hill and R. S. Tare, Lab Chip, 2014, 14, 4475–4485 RSC.
- D. J. Huey, J. C. Hu and K. A. Athanasiou, Science, 2012, 338, 917–921 CrossRef PubMed.
- K. Shimomura, W. Ando, H. Fujie, D. A. Hart, H. Yoshikawa and N. Nakamura, J. Exp. Orthop., 2018, 5, 2 CrossRef PubMed.
- M. C. Lewis, B. D. MacArthur, R. S. Tare, R. O. C. Oreffo and C. P. Please, PLoS One, 2016, 11, e0147302 CrossRef PubMed.
- C. Gentili and R. Cancedda, Curr. Pharm. Des., 2009, 15, 1334–1348 CrossRef PubMed.
- J. D. Bancroft, C. Layton and S. K. Suvarna, Bancroft's theory and practice of histological techniques, 2013 Search PubMed.
- A. Villalvilla, R. Gomez, R. Largo and G. Herrero-Beaumont, Int. J. Mol. Sci., 2013, 14, 20793–20808 CrossRef PubMed.
- F. N. Ghadially, G. Meachim and D. H. Collins, Ann. Rheum. Dis., 1965, 24, 136–146 CrossRef PubMed.
- G. Cox, E. Kable, A. Jones, I. Fraser, F. Manconi and M. D. Gorrell, J. Struct. Biol., 2003, 141, 53–62 CrossRef PubMed.
- D. F. Holmes and K. E. Kadler, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17249–17254 CrossRef PubMed.
- C. K. Wen and M. C. Goh, Proteins, 2006, 64, 227–233 CrossRef PubMed.
- A. Diaspro, Confocal and Two-Photon Microscopy: Foundations, Applications and Advances, Wiley, 2001 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S3 and Videos S1–S4. See DOI: 10.1039/c8ib00050f |
| This journal is © The Royal Society of Chemistry 2018 |