Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Streptomyces cell-free systems for natural product discovery and engineering
Simon J.
Moore
 *ab,
Hung-En
Lai
*ab,
Hung-En
Lai
 c,
Jian
Li
c,
Jian
Li
 d and
Paul S.
Freemont
*e
d and
Paul S.
Freemont
*e
aSchool of Biosciences, University of Kent, UK
bSchool of Biological and Behavioural Sciences, Queen Mary University of London, UK. E-mail: simon.moore@qmul.ac.uk
cSchool of Biological Sciences, Victoria University of Wellington, New Zealand
dSchool of Physical Science and Technology, ShanghaiTech University, China
eDepartment of Medicine, Imperial College London, UK. E-mail: p.freemont@imperial.ac.uk
First published on 7th November 2022
Abstract
Streptomyces bacteria are a major microbial source of natural products, which are encoded within so-called biosynthetic gene clusters (BGCs). This highlight discusses the emergence of native Streptomyces cell-free systems as a new tool to accelerate the study of the fundamental chemistry and biology of natural product biosynthesis from these bacteria. Cell-free systems provide a prototyping platform to study plug-and-play reactions in microscale reactions. So far, Streptomyces cell-free systems have been used to rapidly characterise gene expression regulation, access secondary metabolite biosynthetic enzymes, and catalyse cell-free transcription, translation, and biosynthesis of example natural products. With further progress, we anticipate the development of more complex systems to complement existing experimental tools for the discovery and engineering of natural product biosynthesis from Streptomyces and related high G + C (%) bacteria.
Introduction
Streptomyces is a genus of Gram-positive bacteria, which are a major source of natural products. These environmental bacteria have led to the discovery of hundreds of drugs and commercialised chemicals, including about two-thirds of derived antibiotic scaffolds.1 With the rise of genome sequencing,2 there is renewed vigour for studying the fundamental chemistry and biology of Streptomyces and the natural products that they produce.3,4,5 However, it is challenging to study and engineer Streptomyces to access new natural products. This review will discuss the emergence of specialised Streptomyces cell-free lysate systems and how these platforms help complement existing experimental and computational approaches in natural product discovery. The motivation for developing Streptomyces cell-free lysate systems is the potential to study gene expression regulation, enzymes and biosynthetic pathways encoded within so-called “silent” or “cryptic” biosynthetic gene clusters (BGCs) native to Streptomyces genomes.6 We will first summarise existing approaches within the natural products field, before discussing current and potential future applications of Streptomyces cell-free lysate systems.Natural product discovery from Streptomyces
Streptomyces are found in terrestrial and aquatic environments, whose genomes have a high guanine and cytosine (G + C) content ranging between 67 to 72%.5 Historically, laboratory fermentation of environmental microbes, including Streptomyces, inspired a golden era of antibiotic discovery during the 1940–1960s.6 Shortly after, rediscovery of antibiotics and the rise of antimicrobial resistance initially suggested that this discovery pipeline was exhausted.7 However, in the early 2000s, genome sequencing of the model Streptomyces coelicolor A3(2) strain revealed clusters of DNA encoding proteins with similarity to known biosynthetic enzymes.8 These co-localised regions were recognised as BGCs linked to natural product biosynthesis. Of the 20 BGCs identified, five produced known natural products.8 The remaining BGCs are silent, in terms of gene expression, under standard laboratory conditions.6,8 With the rapid rise of next-generation DNA sequencing, there is now a rich availability of genomic and metagenomic sequences from diverse habitats.3,4 Specifically, Streptomyces genomes (∼6 to 11 Mb) can encode up to 100 BGCs that vary greatly in size from approximately 5 to 100 kb of DNA. Together with multi-omics data,4,9,10 new natural product classes,11 and improved computational BGC prediction and annotation tools,3,12 these combined advances have enabled the prediction of natural product scaffolds based on input DNA sequence – an approach called genome mining.13 This is especially true for well-defined natural product classes such as polyketides, non-ribosomal and ribosomal peptides. Although, there is always scope to discover novel natural products from understudied biosynthetic pathways not accurately predicted by current algorithms.14 Because of the extensive diversity of BGCs available to study, the prioritisation of novel and/or potentially bioactive BGCs is also an important focus.4 However, there is a disparity between the number of predicted BGCs and experimentally characterised natural products. From bacteria alone, the number of chemical classes documented in the Natural Product Atlas10 (currently ∼12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) is approximately 3% of the total predicted potential classes.4 Therefore, there is a continuous need to develop higher throughput and better experimental tools to drive new discoveries in natural products.
500) is approximately 3% of the total predicted potential classes.4 Therefore, there is a continuous need to develop higher throughput and better experimental tools to drive new discoveries in natural products.
Current microbial cell approaches to study natural products encoded by BGCs
Most BGCs are either transcriptionally or translationally silent in terms of gene expression when studied in the laboratory environment.6,8,15 Therefore, there are two general approaches to study natural products encoded by BGCs – native or heterologous expression. The traditional approach is the “one strain many compounds” (OSMAC) method to study BGCs that are naturally active in native hosts under laboratory conditions. This involves culturing environmental bacterial or fungal isolates under a wide range of conditions to induce BGC gene expression and natural product biosynthesis. This is because conditions that imitate environmental factors can serendipitously activate “silent” BGCs through unknown gene expression regulators.16 Other OSMAC approaches included small molecule elicitor library screening17 and co-culturing,18 which can simulate complex microbiome interactions in native microenvironments to trigger BGC gene expression and natural product biosynthesis.18 However, the OSMAC approach is best applied to novel strains, since key limitations include rediscoveries due to screening bias towards common targets (i.e., antimicrobials), while many BGCs will remain switched-off in terms of gene expression.19,20 Most often, the environmental triggers that activate BGC gene expression are absent under laboratory conditions, requiring extensive screening experiments to mimic these triggers fortuitously.Alternatively, heterologous expression represents a major approach on activating silent BGCs.19 There are multiple genetic engineering strategies used to isolate or engineer target BGCs from source (meta)genomes – for more detail on this subject, we recommend the following review.20 Although successful in expressing a wide range of natural products and complex BGCs that are otherwise not produced in native hosts, heterologous expression remains a painstaking, trial-and-error process, as it is difficult to predict the optimum expression host and culture condition for each specific BGC.21 In addition, many BGCs expressed in heterologous hosts, remain silent in terms of gene expression.22 This means further genetic manipulation and metabolic engineering is often required to activate silent BGCs in heterologous hosts.
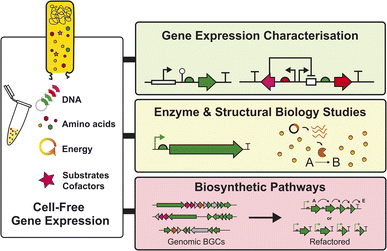
Some of the problems discussed with studying BGC expression – either native or heterologous – are often associated with the inherent challenges of working with complex biological systems. This includes unknown genetic regulation, poor genetic tools, variable product titres and complex metabolite profiles. Herein, this highlight aims to discuss an emerging cell-free synthetic biology tool that bypasses some of the limitations discussed previously (Fig. 1). We will first discuss what a cell-free system is, before focusing on the development of specialised Streptomyces cell-free gene expression (CFE) systems as a tool to study the biology and chemistry of natural product biosynthesis from Streptomyces, and related high G + C (%) bacteria.
Cell-free synthetic biology
Cell-free systems are an emerging platform technology within synthetic biology – a sub-field referred to as cell-free synthetic biology. Rapid advances within the past two decades have demonstrated potential for cell-free systems to compete or replace traditional cell-based approaches.23,24 Specifically, cell-free systems allow the study of enzyme biosynthesis within a test tube, an approach that draws parallels to total synthesis from organic chemistry. In comparison, cell-free systems start from known substrates and proceed to completion through exhaustion of substrates or the accumulation of inhibitory products in batch reactions. Since the reactions are biological, reactions do not require elevated temperatures, organic solvents, or heavy metal catalysts, except for some metalloenzymes. To extend reaction time, scaled-up continuous or fed-batch methods can increase the duration, efficiency and yields of reactions.24 At this point, we highlight that there are two distinct cell-free approaches. First, enzyme pathways can be assembled in a single “one-pot” biosynthetic reaction, using purified enzymes or crude cell extracts, combined with cofactors and substrates. For this, the key advantage of cell-free “one-pot” systems provide is the ability to make complex natural products, or toxic intermediates, that are challenging to make within a cell, or to probe specific chemical mechanism questions. Although not covered in detail within this highlight, we refer the reader to some recent literature reviews.25,26 We will instead focus on discussing the emerging crude extract-based Streptomyces CFE systems – also called cell-free transcription-translation or cell-free protein synthesis – for studying and engineering natural product biosynthesis. We will discuss specific advantages and the potential of these systems compared to traditional cell-based expression.CFE systems require a cell extract, DNA, energy, and amino acids to catalyse coupled messenger RNA (mRNA) and protein synthesis in a one-pot reaction.27,28 Early CFE studies used cell extract and a synthetic energy regeneration system (e.g., creatine phosphate), along with purified mRNA or a T7 bacteriophage RNA polymerase system, to make recombinant proteins. Importantly, CFE systems are devoid of cell wall, cell membrane, and genomic DNA, which provides specific advantages in comparison to cell-based expression, such as resistance to toxic intermediates and products.24 In addition, the crude cell extract processing steps deplete certain metabolites and mRNA carried over from cell growth. This creates a quasi-chemical reactor system, where individual chemical or physical parameters can be fine-tuned to make mRNA, protein, or chemicals as required. Cell extracts are also more complex than originally thought. This is because a bacterial cell extract contains not only the essential components to perform coupled transcription–translation, but also for other metabolic pathways. For protein synthesis alone, over 100 distinct proteins/RNA components are required.24,27 This includes RNA polymerase (α2ββ′ω), sigma factors, translation factors, 20 aminoacyl-transfer RNA (tRNA) synthetases, 30S/50S ribosomal subunits and tRNA (i.e., 83 in E. coli). Although poorly characterised and variable between different cell extract types, a few hundred proteins essential to cell growth are also present. This includes primary metabolic enzymes involved in energy and amino acid biogenesis, as well as vital supporting processes such as RNA degradation, transcription factors and protein folding chaperones. To activate the cell-free systems to make RNA and protein, the reactions require both a primary and secondary energy source.27 The primary energy source is nucleotide triphosphates, which drive initial mRNA and protein synthesis. The secondary energy source is required to regenerate the primary energy source from a high-energy substrate, such as glucose, L-glutamate or certain glycolysis or Krebs cycle intermediates.27–29 This is important since many of the native primary metabolic pathways are active in cell extracts. For example, catabolism drives adenosine triphosphate (ATP) regeneration from a secondary energy source (e.g., glucose, 3-phosphoglycerate) to leverage maximal protein synthesis.30 Importantly, many metabolic enzymes present in the cell extract have phosphorylase activity, which contributes to overall energy loss in the cell-free systems. In a recent study, approximately 98% of total energy (adenosine triphosphate) was shown to be lost to waste.29 This is a key limitation within CFE systems; therefore, energy regeneration is a major focal point for CFE optimisation. Finally, primary metabolites, and the free energy generated by catabolic metabolism, provide the basic building blocks of secondary metabolism, which we refer to as natural products. Therefore, there is potentially an untapped capacity of cell-free systems to fuel secondary metabolite biosynthesis. For example, background metabolic pathways could also provide electrons for redox cofactors (e.g., nicotinamide adenine nucleotide, flavins), amino acids for peptide biosynthesis, as well as acetyl-CoA, malonyl-CoA, and related analogs for specific polyketide biosynthetic pathways. We will now review the recent advances in the development of specialised Streptomyces CFE systems, as a new tool to study the chemistry and biology of Streptomyces.
Streptomyces cell-free gene expression (CFE) systems
Recently, there has been a rising interest in E. coli CFE systems to study enzymes and biosynthetic pathways from secondary metabolism.31 This is because E. coli is the most well-characterized CFE system and capable of making high recombinant protein yields up to 4 mg mL−1 in batch reactions.27,30 Alternatively, a range of Streptomyces CFE systems have emerged, compatible with multiple strains,32 for high-yield protein production.32–37 Currently, Streptomyces venezuelae and Streptomyces lividans CFE systems have been shown to reach up to 0.45 mg mL−135,37 and 0.52 mg mL−1,33,38 respectively. These protein yields are close to commercial E. coli cell-free systems, which were developed over several decades of research and include many genetic changes to increase recombinant protein yields. While still in development, we suggest the Streptomyces CFE systems present several opportunities over the better characterised E. coli CFE systems. This includes the provision of a native protein folding environment, high G + C (%) tRNA pool and background metabolism, which may contain enzymes, cofactors or metabolites required for specific natural product biosynthetic pathways. For example, polyketide and non-ribosomal peptide biosynthesis is catalysed by large modular enzymes, which contain an acyl or peptidyl carrier protein (ACP/PCP) domain, respectively. The ACP/PCP domain is post-translationally modified by specialised phosphopantetheinyl transferase (PPTase) enzymes, found only in microbes that make these natural products. While this causes an issue for heterologous expression systems, such as E. coli, the Streptomyces cell-free extracts may contain PPTase enzymes to activate ACP/PCP domains, although further work is required. On this point, there are also wider opportunities to work with other Streptomyces and related actinobacteria strains, especially those with poor genetic tractability. This is because several other Streptomyces strains have also been used to establish CFE systems using the same preparation procedure.32 These recent studies have provided user-friendly protocols,32–39 which will likely lead to the development of more productive Streptomyces CFE systems in the future for natural product research. We will now discuss the current advances for these specialised CFE systems, before summarising the next steps for their continued and wider use.Regulatory factors controlling BGC gene expression
It is important to study the fundamental mechanisms of gene expression regulation in order to discover new natural products encoded by BGCs. Focusing on Streptomyces, the genetic determinants that control BGC gene expression occur at the level of transcription40 and post-transcription regulation, which are highly complex in comparison to better characterised models (Fig. 2). This includes both non-coding and coding DNA sequences that regulate these levels of gene expression. As an example of this complexity, one recent study in Streptomyces coelicolor A3(2) showed that RNA and protein levels are not necessarily correlated for secondary metabolism genes, due to efficient translation of even low levels of mRNA.41 Other regulatory control points also include alternative start codons,37 pleiotropic regulatory factors,42 and other regulatory elements, which are summarised in Fig. 2. In addition, unlike better characterised models, the fundamental biological numbers that control protein synthesis, are largely uncharacterised in Streptomyces, bacteria. We envision that CFE provides an emerging opportunity to help address some of these questions, not only in Streptomyces, but potentially other related actinobacteria and microbial sources of natural products. This is possible because CFE systems are emerging in a range of microbial strains and enable to study of native gene expression regulatory elements from these hosts. A key point of interest in natural product discovery has been refactoring BGCs with optimised regions of non-coding DNA. One example is using strong constitutive promoters to drive gene expression, such as the recent study showing the activation of a “cryptic” streptophenazine BGC in a marine actinobacteria.44 This is important within the context of synthetic biology since a repository of characterised DNA parts is a prerequisite for engineering cells. Here, CFE can rapidly characterise regulatory DNA parts in hours, which with the assistance of computational modelling, has been shown to provide strong correlation to cell-based data.29 In contrast to slow-growing or genetically intractable strains of Streptomyces, the equivalent experimental timeframe is weeks to months (Fig. 2). As discussed, the Streptomyces cell extracts contain all the core proteins to catalyse transcription and translation. Another complexity here is that each strain of Streptomyces contains between 50–100 Sigma factors, compared to E. coli that has seven. The main E. coli Sigma factor is σ70, which is active during exponential growth to regulate a range of essential genes. The equivalent homolog in Streptomyces is HrdB, which is also uniquely post-translationally modified to activate its function.40 Interestingly, the core HrdB recognition sequence is homologous to E. coli σ70 promoters. Originally, a strong Streptomyces HrdB-dependent promoter, called kasOp*, was developed with 20-fold more active than the widely used ermEp* promoter.40 From this DNA sequence, a synthetic promoter (SP1-SP44) library was created and tested in S. venezuelae cells, along with a range of synthetic ribosome-binding site (RBS) parts. This created a diverse combinatorial library with activity up to 1000-fold dynamic range.45 The strongest promoter, SP44, was two-fold more active than kasOp*. Two recent CFE studies characterised a selection of this promoter and RBS library in S. venezuelae ATCC 1071237 and S. lividans.33 Findings in these two distinct studies were remarkably similar: the SP44 promoter was the strongest in both studies, proving even stronger than the popular T7 RNA polymerase bacteriophage system used widely in biotechnology.33 Furthermore, we can also modularly control translation of proteins using RBSs with different strengths.33,37 Importantly, CFE plasmids can be constructed with a series of promoters and RBSs to control gene expression levels. Therefore, co-expression of enzymes from a natural product gene cluster can be fine-tuned using different regulatory parts to balance the metabolic flux for pathway optimization. Another advantage of CFE systems is compatibility with high-throughput experiments, such as next-generation sequencing (NGS). Park et al. showed a CFE NGS approach, to decode a library of non-coding 5′UTR regulatory sequences associated with predicted BGCs.43 A library pool was rapidly characterised both in CFE and in vivo, which showed strong positive correlation for these regulatory features. This experiment revealed a snapshot of potential BGCs that are likely transcriptionally active in a specific cell extract. A potential extension of this approach is a diverse selection of Streptomyces cell extracts that might provide a screening platform to identify a suitable host cell for heterologous expression.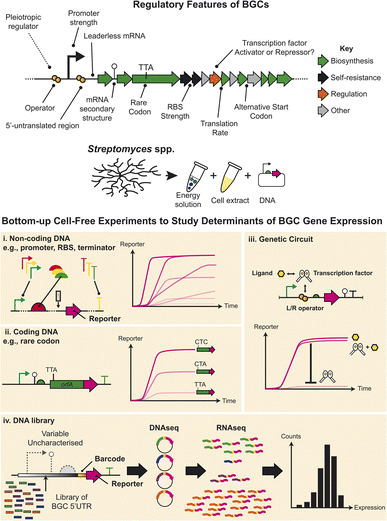 | ||
| Fig. 2 A hypothetical BGC and the regulatory factors that can control gene expression. Streptomyces CFE systems offer an opportunity to study individual native regulatory components native from the bottom-up. Potential cell-free experiments include the study of regulatory factors in (i) non-coding and (ii) coding regions of DNA,33,37 and (iii) transcription factors.34 (iv) CFE also offers the potential to study regulatory components at high-throughput scale, such as the 5′-untranslated region (5′-UTR) of BGCs, which was studied using next-generated sequencing.43 | ||
In addition, CFE also provides the ability to study coding regulatory features (Fig. 2). For example, there is an abundance of alternative start codons in BGCs, which although context dependent (i.e., different between genes/non-coding sequences), is likely to play a key control point for translational efficiency along with the 5′-untranslated region (5′UTR) and initial coding sequence of BGC genes. The S. venezuelae CFE system showed that GTG is a weak alternative start codon, while ATG and TTG were equivalent in activity.37 Last, there is limited literature concerning how protein synthesis is controlled or can be optimised for Streptomyces. For this specific question, Streptomyces CFE can be used to study individual rate limiting factors during protein synthesis.38
Natural product biosynthetic enzymes and pathways
While there has been a rising interest in using E. coli CFE systems for studying natural product biosynthetic pathways,46 the Streptomyces CFE systems potentially provide new advantages. Recent examples of Streptomyces CFE applied to natural product biosynthesis include the synthesis of a range of oxytetracycline biosynthetic enzymes,34,37 single module non-ribosomal peptide synthetase (NRPS) domains37,38 and combined transcription, translation and enzyme biosynthesis of some model enzyme pathways from haem and melanin biosynthesis.37 A potential advantage for Streptomyces CFE is the production of correctly folded and soluble biosynthetic enzymes for biochemical characterisation and structural studies. For example, a large proportion of predicted BGCs encode multidomain type I polyketide synthases (PKS) and NRPS enzymes. These so-called megasynthases can reach over a megadalton in size. Here, open questions remain on how bacterial ribosomes can even make such colossal proteins. Therefore, CFE potentially provides a tool to help fold and solubilise either single or multidomain NRPS/PKS enzymes. Currently, structural biology studies of NRPS/PKS enzymes rely on truncated modules using conventional E. coli recombinant protein production, while computational modelling also helps predict how large NRPS/PKS assembly lines are likely folded. So far, Streptomyces CFE systems have successfully produced significant yields (microgram level) of NRPS enzymes from valinomycin and tambromycin pathways,46 and an uncharacterised single domain NRPS (NH08_RS0107360) from Streptomyces rimosus.37 Interestingly, Streptomyces CFE systems can improve the solubility of proteins originated from Streptomyces when compared to E. coli-based CFE.34,39,47 This could be attributed to the presence of some yet unknown accessory proteins (e.g., molecular chaperones) or metabolite factors in Streptomyces cell extracts. Another reason could be that a strain-specific physicochemical environment (e.g., metabolites) that mimics the native Streptomyces cytoplasm may play a role in protein folding and solubility.In terms of Streptomyces biosynthetic pathways, three pigmented proof-of-concept examples of combined cell-free transcription, translation, and biosynthesis within a single reaction have been shown.37 A simple example is the activity of the β-glucuronidase enzyme from E. coli codon-adapted for expression in S. venezuelae CFE system. Moving a step further, a two-gene S. venezuelae operon encoding tyrosinase and its metallochaperone “caddy” protein were activated, after the addition of copper, post-translation. Here, copper-dependent tyrosinase activity leads to the accumulation of 5,6-dihydroxyphenyalanine that tautomerises to the reactive dopaquinone, which then spontaneously reacts to form a range of melanin pigments (Fig. 3). Last, an operon encoding the early stages of haem biosynthesis to uroporphyrinogen III, the precursor of all tetrapyrroles, was active in the S. venezuelae CFE system (Fig. 3). Interestingly, these reactions could be improved by scale-up in a semi-continuous dialysis reaction to afford the uroporphyrin III product in the metabolite feed.37 However, both haem and melanin are actively produced during S. venezuelae cell growth; haem is an essential molecule for cell growth, while melanin synthesis occurs under specific growth conditions (i.e., mannitol soya flour). Therefore, gene expression regulation is not a limitation for these biosynthetic pathways. In contrast, the study of genes from “silent” BGCs may require further promoter/RBS refactoring and codon optimisation. With further development, native Streptomyces CFE systems could provide a rapid prototyping environment to optimise gene expression and biosynthetic pathways.24
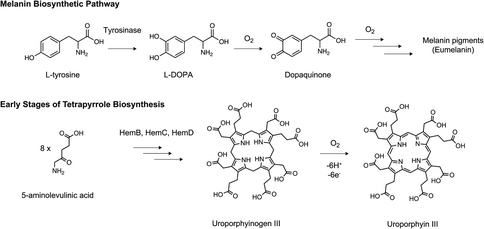 | ||
| Fig. 3 Example biosynthetic pathways produced using Streptomyces CFE including melanin and haem biosynthetic intermediates.37 Reproduced with permission from ref. 37. Copyright 2021 American Chemical Society. | ||
Future opportunities
With further developments, both Streptomyces and E. coli CFE systems provide an alternative and new tool to study uncharacterised BGCs encoding enzymes and biosynthetic pathways from natural product biosynthesis. Specifically, CFE systems offer potential to couple primary metabolism – through cell-lysate energy and cofactor regeneration – with natural product biosynthetic pathways. It is also easier to control metabolites and feed precursors or analogs (e.g., non-canonical amino acids) in a CFE system, whereas cell-based approaches are both time- and resource-consuming to engineer, as well as challenging to achieve consistent production titres. Furthermore, it is desirable to make natural products in a cleaner metabolite background since the chromatography fractions of organic extracts from cultivated Streptomyces and other environmental microbes are complex and often challenging to deconvolute. Perhaps the most striking natural product class with immediate potential for CFE systems is the ribosomally-synthesised and post-translationally modified peptides (RiPPs). RiPPs are suitable because the BGCs are relatively small yet encode remarkable chemical and structural diversity through a plethora of post-translational modifications. In addition, RiPP precursor peptides are notoriously challenging to solubilise, wherein CFE systems may provide an advantage. So far, E. coli CFE has been used to make diverse RiPP libraries, including combinatorial peptide libraries. This includes improved antibiotic variants of nisin48 and millions of sequence-diverse lasso peptides.49 There is an abundance of RiPPs in Streptomyces and related genomes, and therefore, a well-suited target for the Streptomyces CFE systems described. Here, other technologies, such as automation, microfluidics, and machine-learning models, can complement CFE approaches.A key priority for Streptomyces CFE systems is the need to increase protein yields, potentially to levels equivalent or exceeding E. coli CFE systems. This will enable further progress and the ability to study more challenging targets (i.e., NRPS/PKS enzymes). For this to occur, it is essential to characterise the precise proteins and metabolites present in the cell extracts, using different analytical methods. While this will differ substantially between extracts obtained from different growth stages, conditions, and strains, it can be used to inform our overall understanding of how we can improve key bottlenecks in CFE systems. These bottlenecks may include regeneration of energy or the supply of biosynthetic precursors, which will be important for optimising natural product titres in CFE systems. Moreover, if specific limiting factors can be identified in the gene expression machinery,38 then overall productivity may also be improved through both upstream and downstream engineering strategies. For instance, strain engineering (e.g., modifying nuclease and protease genes), bio-process engineering (e.g., high cell density cultivation in large-scale bioreactors), and reaction engineering (e.g., scale-up, semi-continuous, and continuous reaction modes) are achievable with CFE systems.24 Another important focus for Streptomyces CFE will include implementing the systems to study the genetic basis of “silent” or “cryptic” BGCs6 and the scalability of CFE systems, to make specific proteins for enzymology or structural biology studies37,39 and natural products37 to complement existing tools in studying natural product biosynthesis (Fig. 4).
Conclusions
In summary, the key strengths of Streptomyces CFE systems are the increased throughput and dexterity to explore a range of biosynthetic or gene expression regulatory questions specific to natural product biosynthetic pathways associated with these specialist environmental bacteria (Fig. 4). Understanding gene expression in these microbes is a fundamental question, and one that if addressed, is likely to lead to new developments in natural product discovery. So far, Streptomyces CFE systems have specifically allowed the study of native DNA regulatory features associated with BGCs. This concept can be expanded to other Streptomyces, related actinobacteria,32 or wider microbial producers of natural products (e.g., Myxococcus, Burkholderia). Alternatively, the expression of single genes or pathways from metagenomic targets is another attractive target for CFE. This is beneficial, since CFE does not require any genetic manipulation and native CFE systems may provide additional metabolites and cofactors that are not provided in hosts such as E. coli. In addition, CFE systems provide a reduced metabolite background,37 which with further progress, could be advantageous for the study and isolation of more complex natural product biosynthetic pathways. Currently, it is unrealistic to use CFE systems to express large (>20–100 kb) uncharacterised BGCs, particularly those that encode large modular megasynthases that are still difficult to activate in cell-based heterologous expression. Here, there is a limit to energy exhaustion and substrate depletion in the CFE systems during the entire reaction, due to the increased need for nucleotides, energy, and amino acids for larger BGCs/NRPS/PKS pathway targets. Possible solutions might include developing fed-batch CFE reactions,37 which have proved remarkably efficient and scalable for large-scale recombinant protein production.24 Finally, it is currently challenging to use CFE to study completely novel BGCs that may encode uncharacterised enzymes or require bespoke cofactors or substrates, although this is also a challenge for cell-based expression. One example is the diverse radical S′-adenosyl-L-methionine (SAM) enzyme family that are challenging to study and require anaerobic conditions. Instead, CFE is potentially better suited for the production and study of single NRPS/PKS enzymes, RiPPs and some well-defined biosynthetic pathways. Also, promiscuous enzymes or biosynthetic pathways that allow the combinatorial feeding of substrate analogs to make new-to-nature natural products, is also an advantage for CFE, where rapid screening is possible.43,49 This is advantageous to CFE systems compared to cell-based systems, as CFE systems lack a cell membrane, which may hinder substrate transport. In addition, CFE systems are resistant to substrate/product toxicity issues and can be studied in microscale conditions, which is advantageous for increased throughput and reduced cost. In conclusion, Streptomyces CFE systems are an emerging development within synthetic biology that complements and advances existing cell-based expression approaches for natural product discovery.Author contributions
All authors contributed equally to the preparation and writing of the review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. J. M. acknowledges funding from the Royal Society (RGS/R1/191186), Wellcome Trust (217528/Z/19/Z) and Leverhulme Trust (RPG-2021-018). P. S. F. acknowledges funding from EPSRC (EP/K038648/1). J. L. acknowledges funding from National Natural Science Foundation of China (31971348).References
- E. A. Barka, et al., Taxonomy, physiology, and natural products of Actinobacteria, Microbiol. Mol. Biol. Rev., 2016, 80, 1–43 CrossRef PubMed.
- S. Lautru, R. J. Deeth, L. M. Bailey and G. L. Challis, Discovery of a new peptide natural product by Streptomyces coelicolor genome mining, Nat. Chem. Biol., 2005, 1, 265–269 CrossRef CAS PubMed.
- M. H. Medema, et al., antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences, Nucleic Acids Res., 2011, 39, W339–W346 CrossRef CAS.
- A. Gavriilidou, et al., Compendium of specialized metabolite biosynthetic diversity encoded in bacterial genomes, Nat. Microbiol., 2022, 7, 726–735 CrossRef CAS PubMed.
- T. Kieser, M. J. Bibb and M. J. Buttner, Practical Streptomyces Genetics, John Innes Foundation, 2000, 613 Search PubMed.
- P. A. Hoskisson and R. F. Seipke, Cryptic or Silent? The Known Unknowns, Unknown Knowns, and Unknown Unknowns of Secondary Metabolism, mBio, 2020, 11(5), e02642-20 CrossRef.
- M. Miethke, et al., Towards the sustainable discovery and development of new antibiotics, Nat. Rev. Chem., 2021, 5, 726–749 CrossRef CAS.
- S. D. Bentley, et al., Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2), Nature, 2002, 417, 141–147 CrossRef.
- J. A. van Santen, S. A. Kautsar, M. H. Medema and R. G. Linington, Microbial natural product databases: moving forward in the multi-omics era, Nat. Prod. Rep., 2021, 38, 264–278 RSC.
- J. A. van Santen, et al., The Natural Products Atlas: An open access knowledge base for microbial natural products discovery, ACS Cent. Sci., 2019, 5, 1824–1833 CrossRef CAS PubMed.
- P. G. Arnison, et al., Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- M. Alanjary, et al., The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery, Nucleic Acids Res., 2017, 45, W42–W48 CrossRef CAS.
- M. Zerikly and G. L. Challis, Strategies for the discovery of new natural products by genome mining, Chembiochem, 2009, 10, 625–633 CrossRef CAS.
- D. J. McClung, et al., Harnessing rare actinomycete interactions and intrinsic antimicrobial resistance enables discovery of an unusual metabolic inhibitor, mBio, 2022, e0039322 CrossRef.
- S. Hwang, et al., Primary transcriptome and translatome analysis determines transcriptional and translational regulatory elements encoded in the Streptomyces clavuligerus genome, Nucleic Acids Res., 2019, 47, 6114–6129 CrossRef CAS PubMed.
- J. Schwarz, G. Hubmann, K. Rosenthal and S. Lütz, Triaging of culture conditions for enhanced secondary metabolite diversity from different bacteria, Biomolecules, 2021, 11, 193 CrossRef CAS.
- F. Xu, B. Nazari, K. Moon, L. B. Bushin and M. R. Seyedsayamdost, Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens, J. Am. Chem. Soc., 2017, 139, 9203–9212 CrossRef CAS PubMed.
- R. Wang, S. Zhao, Z. Wang and M. A. Koffas, Recent advances in modular co-culture engineering for synthesis of natural products, Curr. Opin. Biotechnol., 2020, 62, 65–71 CrossRef CAS.
- L. Huo, et al., Heterologous expression of bacterial natural product biosynthetic pathways, Nat. Prod. Rep., 2019, 36, 1412–1436 RSC.
- D. Mao, B. K. Okada, Y. Wu, F. Xu and M. R. Seyedsayamdost, Recent advances in activating silent biosynthetic gene clusters in bacteria, Curr. Opin. Microbiol., 2018, 45, 156–163 CrossRef CAS PubMed.
- J. J. Zhang, X. Tang and B. S. Moore, Genetic platforms for heterologous expression of microbial natural products, Nat. Prod. Rep., 2019, 36, 1313–1332 RSC.
- Y. Ahmed, et al., Engineering of Streptomyces lividans for heterologous expression of secondary metabolite gene clusters, Microb. Cell Fact., 2020, 19, 5 CrossRef CAS PubMed.
- C. Meyer, et al., Analysis of the Innovation Trend in Cell-Free Synthetic Biology, Life, 2021, 11, 551 CrossRef CAS PubMed.
- A. D. Silverman, A. S. Karim and M. C. Jewett, Cell-free gene expression: an expanded repertoire of applications, Nat. Rev. Genet., 2020, 21, 151–170 CrossRef CAS.
- S. J. Moore, Enzyme alchemy: cell-free synthetic biochemistry for natural products, Emerging Top. Life Sci., 2019, 3, 529–535 CrossRef PubMed.
- J. Li, L. Zhang and W. Liu, Cell-free synthetic biology for in vitro biosynthesis of pharmaceutical natural products, Synth. Syst. Biotechnol., 2018, 3, 83–89 CrossRef.
- J. Garamella, R. Marshall, M. Rustad and V. Noireaux, The All E. coli TX-TL Toolbox 2.0: a Platform for Cell-Free Synthetic Biology, ACS Synth. Biol., 2016, 5, 344–355 CrossRef CAS.
- M. C. Jewett and J. R. Swartz, Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis, Biotechnol. Bioeng., 2004, 86, 19–26 CrossRef CAS.
- S. J. Moore, et al., Rapid acquisition and model-based analysis of cell-free transcription-translation reactions from nonmodel bacteria, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E4340–E4349 CrossRef CAS.
- D. Garenne, S. Thompson, A. Brisson, A. Khakimzhan and V. Noireaux, The all-E. coliTXTL toolbox 3.0: new capabilities of a cell-free synthetic biology platform, Synth. Biol., 2021, 6, ysab017 CrossRef.
- I. Siebels, et al., Cell-Free Synthesis of Natural Compounds from Genomic DNA of Biosynthetic Gene Clusters, ACS Synth. Biol., 2020, 9, 2418–2426 CrossRef CAS PubMed.
- J. Li, H. Wang and M. C. Jewett, Expanding the palette of Streptomyces-based cell-free protein synthesis systems with enhanced yields, Biochem. Eng. J., 2018, 130, 29–33 CrossRef CAS.
- H. Xu, et al., Regulatory part engineering for high-yield protein synthesis in an all-Streptomyces-based cell-free expression system, ACS Synth. Biol., 2022, 11, 570–578 CrossRef CAS PubMed.
- S. J. Moore, H.-E. Lai, H. Needham, K. M. Polizzi and P. S. Freemont, Streptomyces venezuelae TX-TL - a next generation cell-free synthetic biology tool, Biotechnol. J., 2017, 12, 1600678 CrossRef.
- L. K. Nagappa, et al., A ubiquitous amino acid source for prokaryotic and eukaryotic cell-free transcription-translation systems, Front. Bioeng. Biotechnol., 2022, 992708 CrossRef PubMed.
- M. Toh, K. Chengan, T. Hanson, P. S. Freemont and S. J. Moore, A High-Yield Streptomyces Transcription-Translation Toolkit for Synthetic Biology and Natural Product Applications, J. Visualized Exp., 2021, 175, e63012 Search PubMed.
- S. J. Moore, et al., A Streptomyces venezuelae Cell-Free Toolkit for Synthetic Biology, ACS Synth. Biol., 2021, 10, 402–411 CrossRef CAS.
- H. Xu, W.-Q. Liu and J. Li, Translation Related Factors Improve the Productivity of a Streptomyces-Based Cell-Free Protein Synthesis System, ACS Synth. Biol., 2020, 9, 1221–1224 CrossRef CAS PubMed.
- J. Li, H. Wang, Y.-C. Kwon and M. C. Jewett, Establishing a high yielding Streptomyces-based cell-free protein synthesis system, Biotechnol. Bioeng., 2017, 114, 1343–1353 CrossRef CAS PubMed.
- W. Wang, et al., An engineered strong promoter for streptomycetes, Appl. Environ. Microbiol., 2013, 79, 4484–4492 CrossRef CAS.
- Y. Jeong, J.-N. Kim, M. W. Kim, G. Bucca, S. Cho, Y. J. Yoon, B.-G. Kim, J.-H. Roe, S. C. Kim, C. P. Smith and B.-K. Cho, The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2), Nat. Commun., 2016, 7, 11605 CrossRef CAS PubMed.
- J.-E. Kim, J.-S. Choi, J.-S. Kim, Y.-H. Cho and J.-H. Roe, Lysine acetylation of the housekeeping sigma factor enhances the activity of the RNA polymerase holoenzyme, Nucleic Acids Res., 2020, 48, 2401–2411 CrossRef CAS PubMed.
- J. Park, S. S. Yim and H. H. Wang, High-Throughput Transcriptional Characterization of Regulatory Sequences from Bacterial Biosynthetic Gene Clusters, ACS Synth. Biol., 2021, 10, 1859–1873 CrossRef CAS.
- K. D. Bauman, et al., Refactoring the Cryptic Streptophenazine Biosynthetic Gene Cluster Unites Phenazine, Polyketide, and Nonribosomal Peptide Biochemistry, Cell Chem. Biol., 2019, 26, 724–736 CrossRef CAS PubMed.
- C. Bai, et al., Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12181–12186 CrossRef CAS.
- X. Ji, W.-Q. Liu and J. Li, Recent advances in applying cell-free systems for high-value and complex natural product biosynthesis, Curr. Opin. Microbiol., 2022, 67, 102142 CrossRef CAS.
- X. Tian, et al., Cell-free expression of NO synthase and P450 enzyme for the biosynthesis of an unnatural amino acid L-4-nitrotryptophan, Synth. Syst. Biotechnol., 2022, 7, 775–783 CrossRef CAS PubMed.
- R. Liu, et al., A cell-free platform based on Nisin biosynthesis for discovering novel lanthipeptides and guiding their overproduction in vivo, Adv. Sci., 2020, 7, 2001616 CrossRef CAS PubMed.
- Y. Si, A. M. Kretsch, L. M. Daigh, M. J. Burk and D. A. Mitchell, Cell-free biosynthesis to evaluate lasso peptide formation and enzyme-substrate tolerance, J. Am. Chem. Soc., 2021, 143, 5917–5927 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2023 |