Extraction of triterpenoids and phenolic compounds from Ganoderma lucidum: optimization study using the response surface methodology†
Taofiq
Oludemi
 abc,
Lillian
Barros
abc,
Lillian
Barros
 a,
M. A.
Prieto
a,
M. A.
Prieto
 ad,
Sandrina A.
Heleno
ad,
Sandrina A.
Heleno
 ab,
Maria F.
Barreiro
ab,
Maria F.
Barreiro
 b and
Isabel C. F. R.
Ferreira
b and
Isabel C. F. R.
Ferreira
 *a
*a
aCentro de Investigação de Montanha (CIMO), Instituto Politécnico de Bragança, Campus de Santa Apolónia, 5300-253 Bragança, Portugal. E-mail: iferreira@ipb.pt; Fax: +351-273-325405; Tel: +351-273-303219
bLaboratory of Separation and Reaction Engineering – Laboratory of Catalysis and Materials (LSRE-LCM), Bragança Polytechnic Institute, 5301-857 Bragança, Portugal
cGIP- USAL, Unidad de Nutrición y Bromatología, Faculty of Pharmacy, University of Salamanca, Campus Miguel de Unamuno, 37007 Salamanca, Spain
dNutrition and Bromatology Group, Faculty of Food Science and Technology, University of Vigo, Ourense Campus, E32004 Ourense, Spain
First published on 23rd November 2017
Abstract
The extraction of triterpenoids and phenolic compounds from Ganoderma lucidum was optimized by using the response surface methodology (RSM), using heat and ultrasound assisted extraction techniques (HAE and UAE). The obtained results were compared with that of the standard Soxhlet procedure. RSM was applied using a circumscribed central composite design with three variables (time, ethanol content, and temperature or ultrasonic power) and five levels. The conditions that maximize the responses (extraction yield, triterpenoids and total phenolics) were: 78.9 min, 90.0 °C and 62.5% ethanol and 40 min, 100.0 W and 89.5% ethanol for HAE and UAE, respectively. The latter was the most effective, resulting in an extraction yield of 4.9 ± 0.6% comprising a content of 435.6 ± 21.1 mg g−1 of triterpenes and 106.6 ± 16.2 mg g−1 of total phenolics. The optimized extracts were fully characterized in terms of individual phenolic compounds and triterpenoids by HPLC-DAD-ESI/MS. The recovery of the above-mentioned bioactive compounds was markedly enhanced using the UAE technique.
1. Introduction
The globalization of the food, cosmetic and pharmaceutical industries, together with consumer's awareness of natural-derived solutions, has led to an increased demand for natural bioactive compounds to be used in the design of functional foods, multifunctional cosmeceutical products and pharmacologically active formulations. Ganoderma lucidum (Curtis) P. Karst. (commonly referred to as lingzhi in China, reishi in Japan and yeongji in South Korea) is a medicinal mushroom that has been consumed for several years due to its many recognized health benefits.1 Mycelium, fruiting bodies and spores have all been reported to reveal different bioactive properties such as antioxidant, antitumor, anti-inflammatory, antimicrobial, antityrosinase, antimalaria, hypoglycemic and anti-HIV-1 activities, and they are being used in the treatment of cancer, hypertension, insomnia, anorexia, dizziness, chronic hepatitis, immunosuppression and hypercholesterolemia.2,3 Their bioactive and nutritional composition includes carbohydrates (mono-, oligo- and polysaccharides), terpenoids (monoterpenes, sesquiterpenes, diterpenes and triterpenes), nucleotides, steroids (ergosterol), fatty acids (saturated, monounsaturated and polyunsaturated fatty acids), vitamins (B1, B2, B6), proteins, minerals (calcium, phosphorus, iron and magnesium), organic acids (malic and citric) and glycol-peptides, among other constituents.4 Phenolic compounds, terpenoids and polysaccharides (mainly β-glucan) are described as major contributors to the above-mentioned bioactive properties.5 In particular, there are several studies reporting the optimization of β-glucan extraction from the fruiting bodies of G. lucidum.6–8Terpenoids are one of the most widespread natural compounds in medicinal plants and mushrooms. Among the various sub-types of terpenoids, triterpenes are the most abundant in G. lucidum. The HPLC-DAD-ESI/MS analyses of the fruiting bodies have revealed the presence of triterpenes such as ganoderiol F, ganoderic acid B, ganoderiol B, lucidenic acid O, lucidenic lactone and cerevisterol.9–11 Because of their well-reported bioactive properties, some studies focused on the optimization of the cultivation practices of this mushroom have been conducted. Among them, the use of submerged fermentation by manipulating parameters such as the medium components, oxygen supply and pH aiming at achieving the biotechnological production of triterpenes was reported.12 However, very few reports are available describing the extraction optimization of these compounds from G. lucidum.13
The phenolic compound composition of G. lucidum has been widely studied with phenolic acids being the most prominent class, which include chlorogenic, cinnamic, gallic, protocatechuic, p-hydroxybenzoic and p-coumaric acids. These compounds have been related to the antioxidant, antimicrobial, anti-tyrosinase and anti-inflammatory activities of G. lucidum.14–18
Generally, solid–liquid extraction using conventional techniques such as Soxhlet extraction (SE) and heat assisted extraction (HAE) requires the consumption of large amounts of solvent and these are time-consuming processes. In this context, several non-conventional extraction techniques are being successively proposed and applied to maximally extract valuable compounds from natural matrices using much more amiable conditions. Among them, ultrasound assisted extraction (UAE) is one of the most widely used alternative extraction methods, offering the advantage of achieving interesting results using, for example, hydroalcoholic mixtures. Moreover, some advantages over other conventional systems include the use of lower extraction times and lower solvent consumption.19–21
Independently of the employed system, the extraction of target compounds is often influenced by several variables, which require individual analysis due to their intrinsic nature and stability. Therefore, it is essential to define the main variables and relevant responses prior to the optimization process in order to determine the values corresponding to response maximization according to the defined objectives (e.g. using minimum time, energy and solvent to achieve a cost-effective and profitable extraction system).22 To effectively carry out an optimization, the influence of each defined variable should be independently assessed. Nevertheless, the application of mathematical models such as the response surface methodology (RSM) is gaining visibility among the scientific community. Through RSM design, the optimization of possible interactions between experimental variables is allowed simultaneously with the prediction of the most efficient conditions. This is achieved by using second-order polynomial models with interactions that are able to describe and maximize the selected response criteria, based on the tested experimental range.23 RSM is utilised to optimise the process conditions from extraction, controlled release from pharmaceutical formulations, and production of microbial enzymes and other metabolites, making it a vital tool to improve the system performance in the food and biopharmaceutical industries.24
The purpose of the present work was to optimize and compare the extraction of phenolic compounds and triterpenes from G. lucidum for application in the food, pharmaceutical and cosmetic industries using SE, HAE and UAE. For this, the extraction yield and the content of triterpenes and total phenolics were maximized by RSM using ethanol solvent proportion (hydroethanolic mixtures), time, temperature and ultrasonic power (in the case of UAE) as independent variables. The extracts corresponding to the optimal conditions were fully characterized in terms of individual phenolic compounds and triterpenoids by HPLC-DAD-ESI/MS analysis.
2. Materials and methods
2.1. Standards and reagents
Perchloric acid 70% and glacial acetic acid were purchased from Fisher Chemicals (Loughborough, UK). Ursolic acid was purchased from Acros Organics (New Jersey, USA), vanillin was obtained from Labkem (Barcelona, Spain), while all the other chemicals and solvents were of analytical grade and purchased from common suppliers. Water was treated in a Milli-Q water purification system (TGI Pure Water Systems, Greenville, SC, USA).2.2. Mushroom material
The fruiting bodies of Ganoderma lucidum were provided by BioReishi, Portugal (reishi producers; Batalha, Portugal) in the form of a dry powdered material. For all the experiments, three samples were analysed and all the assays were carried out in triplicate.2.3. Soxhlet extraction as the standard technique
Soxhlet extraction was chosen as the standard technique, it being also a HAE. The powdered samples (3.0 g) were extracted with 100 mL of ethanol by refluxing in a Soxhlet apparatus. To compare the efficiency of the extraction system, the number of cycles in the Soxhlet system was taken into consideration and up to seven cycles were analysed. After the desired number of cycles, the solvent was evaporated under reduced pressure (rotary evaporator Büchi R-210, Flawil, Switzerland) to obtain the dried ethanolic extracts.2.4. Optimization of the HAE and UAE techniques as alternatives towards process industrialization
Based on a combination of in-house knowledge (previous extractions performed in our laboratory) and a performed bibliographic survey, the relevant variables and appropriate ranges for the studied extraction techniques (HAE and UAE) were selected and tested. A detailed description of the variables and considered ranges to support the RSM design is presented in Table 1. The solid/solvent ratio was kept constant (30 g L−1) for all the techniques (HAE and UAE).| Experimental design | HAE responses | UAE responses | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coded values | HAE conditions | UAE conditions | Residue | Triterpene content | Phenolic content | Residue | Triterpene content | Phenolic content | |||||||||||
| X 1 | X 2 | X 3 | X 1: t (min) | X 2: T (°C) | X 3: S (%) | X 1: t (min) | X 2: P (W) | X 3: S (%) | Yield (%) | Y 1 (mg per g dw) | Y 2 (mg per g R) | Y 1 (mg per g dw) | Y 2 (mg per g R) | Yield (%) | Y 1 (mg per g dw) | Y 2 (mg per g R) | Y 1 (mg per g dw) | Y 2(mg per g R) | |
| 1 | −1 | −1 | −1 | 40 | 35 | 20.3 | 13.5 | 180 | 20.3 | 3.83 | 3.60 | 93.81 | 2.54 | 66.17 | 4.30 | 7.02 | 163.25 | 3.02 | 70.25 |
| 2 | 1 | −1 | −1 | 130 | 35 | 20.3 | 33.5 | 180 | 20.3 | 3.86 | 3.67 | 95.51 | 2.71 | 68.93 | 4.62 | 7.54 | 162.89 | 3.20 | 66.31 |
| 3 | −1 | 1 | −1 | 40 | 75 | 20.3 | 13.5 | 420 | 20.3 | 6.99 | 3.51 | 50.27 | 3.61 | 56.10 | 3.64 | 6.12 | 167.79 | 2.96 | 85.32 |
| 4 | 1 | 1 | −1 | 130 | 75 | 20.3 | 33.5 | 420 | 20.3 | 3.35 | 3.46 | 103.56 | 3.17 | 90.08 | 4.56 | 6.89 | 152.82 | 4.46 | 102.08 |
| 5 | −1 | −1 | 1 | 40 | 35 | 79.7 | 13.5 | 180 | 79.7 | 15.51 | 7.49 | 48.23 | 5.93 | 39.03 | 7.02 | 17.10 | 245.65 | 6.28 | 88.60 |
| 6 | 1 | −1 | 1 | 130 | 35 | 79.7 | 33.5 | 180 | 79.7 | 10.84 | 13.02 | 120.14 | 5.82 | 49.16 | 5.77 | 21.62 | 374.96 | 6.67 | 110.81 |
| 7 | −1 | 1 | 1 | 40 | 75 | 79.7 | 13.5 | 420 | 79.7 | 8.14 | 13.52 | 166.27 | 6.00 | 73.68 | 5.58 | 18.68 | 336.73 | 6.22 | 112.65 |
| 8 | 1 | 1 | 1 | 130 | 75 | 79.7 | 33.5 | 420 | 79.7 | 11.64 | 12.91 | 112.18 | 5.79 | 52.18 | 5.10 | 16.00 | 314.30 | 6.13 | 123.25 |
| 9 | −1.68 | 0 | 0 | 9 | 55 | 50 | 6.7 | 300 | 50 | 5.11 | 11.07 | 219.19 | 5.41 | 105.77 | 8.55 | 13.06 | 152.92 | 7.56 | 93.81 |
| 10 | 1.68 | 0 | 0 | 160 | 55 | 50 | 40.3 | 300 | 50 | 8.50 | 15.21 | 181.41 | 6.21 | 72.92 | 6.04 | 11.43 | 189.26 | 5.93 | 96.17 |
| 11 | 0 | −1.68 | 0 | 85 | 20 | 50 | 23.5 | 100 | 50 | 6.02 | 11.44 | 189.85 | 5.86 | 100.62 | 6.09 | 14.35 | 235.76 | 6.28 | 100.93 |
| 12 | 0 | 1.68 | 0 | 85 | 90 | 50 | 23.5 | 500 | 50 | 4.50 | 14.66 | 325.37 | 7.38 | 160.39 | 8.48 | 10.92 | 128.66 | 6.80 | 84.99 |
| 13 | 0 | 0 | −1.68 | 85 | 55 | 0 | 23.5 | 300 | 0 | 4.97 | 4.67 | 94.41 | 3.27 | 78.27 | 7.19 | 4.75 | 65.07 | 3.43 | 48.74 |
| 14 | 0 | 0 | 1.68 | 85 | 55 | 100 | 23.5 | 300 | 100 | 5.74 | 12.34 | 214.20 | 3.61 | 59.34 | 3.71 | 15.24 | 410.68 | 4.90 | 126.09 |
| 15 | 0 | 0 | 0 | 85 | 55 | 50 | 23.5 | 300 | 50 | 5.52 | 12.29 | 222.66 | 5.28 | 100.74 | 6.59 | 11.01 | 166.99 | 6.04 | 100.37 |
| 16 | 0 | 0 | 0 | 85 | 55 | 50 | 23.5 | 300 | 50 | 5.14 | 11.52 | 224.43 | 5.29 | 103.52 | 6.08 | 10.46 | 172.13 | 6.84 | 114.00 |
| 17 | 0 | 0 | 0 | 85 | 55 | 50 | 23.5 | 300 | 50 | 4.84 | 11.73 | 242.70 | 5.36 | 110.75 | 6.42 | 10.94 | 170.70 | 6.44 | 97.82 |
| 18 | 0 | 0 | 0 | 85 | 55 | 50 | 23.5 | 300 | 50 | 5.26 | 11.83 | 225.40 | 5.49 | 105.00 | 6.63 | 10.58 | 159.78 | 6.39 | 96.54 |
| 19 | 0 | 0 | 0 | 85 | 55 | 50 | 23.5 | 300 | 50 | 4.74 | 10.77 | 228.07 | 5.04 | 104.53 | 6.54 | 10.01 | 153.68 | 6.44 | 99.14 |
| 20 | 0 | 0 | 0 | 85 | 55 | 50 | 23.5 | 300 | 50 | 4.81 | 11.78 | 245.84 | 5.10 | 106.53 | 8.04 | 11.27 | 140.07 | 7.32 | 91.96 |
2.5. Determination of the extraction yield
The obtained extraction solutions were filtered through Whatman paper no. 4 and then evaporated under reduced pressure to remove the solvent. The extraction yield was expressed as a percentage (%, w/w) calculated by dividing the weight of the recovered residue (extract, R) by the weight of the used dry sample (Ganoderma lucidum, dw) and given as response Y3.2.6. Determination of the extract's phytochemical content
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, v/v) and sodium carbonate (Na2CO3; 2 mL, 75 g L−1). The tubes containing the solutions were then vortexed for 15 s and allowed to stand for 30 min at 40 °C for colour development. The absorbance was then measured at 765 nm against a blank (methanol). Gallic acid (chosen as the standard) was used to obtain the calibration curve (0.0094–0.15 mg mL−1), and the results were expressed as mg of gallic acid equivalents (mg equiv. GA).
10, v/v) and sodium carbonate (Na2CO3; 2 mL, 75 g L−1). The tubes containing the solutions were then vortexed for 15 s and allowed to stand for 30 min at 40 °C for colour development. The absorbance was then measured at 765 nm against a blank (methanol). Gallic acid (chosen as the standard) was used to obtain the calibration curve (0.0094–0.15 mg mL−1), and the results were expressed as mg of gallic acid equivalents (mg equiv. GA).
2.7. Experimental design, model analysis and optimization method for the HAE and UAE techniques
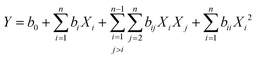 | (1) |
2.8. Chemical characterization of the extracts
For the characterization of the individual compounds of Tr and Ph, the extracts produced under the optimal achieved conditions, for SE, HAE and UAE, were dissolved at a concentration of 10 mg mL−1 in an ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture (20
water mixture (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 v/v), then filtered through a 0.22 μm nylon syringe filter and further analyzed by HPLC-DAD-ESI/MSn on a Dionex Ultimate 3000 UPLC (ThermoScientific, San Jose, CA, USA) system, equipped with a diode array detector coupled with an electrospray ionization mass detector, a quaternary pump, an auto-sampler (kept at 5 °C), a degasser and an automated thermostated column section (kept at 35 °C). Waters Spherisorb S3 ODS-2 C18 (3 μm, 4.6 × 150 mm, Waters, Milford, MA, USA) was used. The solvents were (A) 0.1% formic acid in water and (B) acetonitrile. The applied gradient elution was: 15% B (0–5 min), 15% B to 20% B (5–10 min), 20–25% B (10–20 min), 25–35% B (20–30 min), 35–50% B (30–40 min); the column was then re-equilibrated, using a flow rate of 0.5 mL min−1. The data were collected simultaneously with DAD (280 and 370 nm) and in negative mode detection on a linear ion trap LTQ XL mass spectrometer (ThermoScientific, San Jose, CA, USA), following a procedure reported previously.30 Sheath gas (nitrogen) was kept at 50 psi. Other parameter settings were: source temperature 325 °C, spray voltage 5 kV, capillary voltage −20 V, tube lens offset −66 V, and collision energy 35 arbitrary units. The full scan captured the mass between m/z 100 and 1800. An Xcalibur® data system (ThermoScientific, San Jose, CA, USA) was used for data acquisition. For identification, the retention times, of the UV-VIS and mass spectra, were compared with those of the available standards and the data from the literature were further used to tentatively identify the remaining compounds. To perform quantitative analysis, the calibration curves of the available Ph and Tr standards were constructed based on the UV signal (protocatechuic acid: y = 214168x + 27
80 v/v), then filtered through a 0.22 μm nylon syringe filter and further analyzed by HPLC-DAD-ESI/MSn on a Dionex Ultimate 3000 UPLC (ThermoScientific, San Jose, CA, USA) system, equipped with a diode array detector coupled with an electrospray ionization mass detector, a quaternary pump, an auto-sampler (kept at 5 °C), a degasser and an automated thermostated column section (kept at 35 °C). Waters Spherisorb S3 ODS-2 C18 (3 μm, 4.6 × 150 mm, Waters, Milford, MA, USA) was used. The solvents were (A) 0.1% formic acid in water and (B) acetonitrile. The applied gradient elution was: 15% B (0–5 min), 15% B to 20% B (5–10 min), 20–25% B (10–20 min), 25–35% B (20–30 min), 35–50% B (30–40 min); the column was then re-equilibrated, using a flow rate of 0.5 mL min−1. The data were collected simultaneously with DAD (280 and 370 nm) and in negative mode detection on a linear ion trap LTQ XL mass spectrometer (ThermoScientific, San Jose, CA, USA), following a procedure reported previously.30 Sheath gas (nitrogen) was kept at 50 psi. Other parameter settings were: source temperature 325 °C, spray voltage 5 kV, capillary voltage −20 V, tube lens offset −66 V, and collision energy 35 arbitrary units. The full scan captured the mass between m/z 100 and 1800. An Xcalibur® data system (ThermoScientific, San Jose, CA, USA) was used for data acquisition. For identification, the retention times, of the UV-VIS and mass spectra, were compared with those of the available standards and the data from the literature were further used to tentatively identify the remaining compounds. To perform quantitative analysis, the calibration curves of the available Ph and Tr standards were constructed based on the UV signal (protocatechuic acid: y = 214168x + 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102; R2 = 0.999; p-hydroxybenzoic acid: y = 208604x + 173
102; R2 = 0.999; p-hydroxybenzoic acid: y = 208604x + 173![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056; R2 = 0.999; syringic acid: y = 376056x + 141
056; R2 = 0.999; syringic acid: y = 376056x + 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 329; R2 = 0.999 and ganoderic acid A: y = 2539.7x, R2 = 0.999). The identified compounds with unavailable commercial standards were quantified using the available calibration curve of the most similar standard. The results were expressed in the response format of Y2 (mg per g R).
329; R2 = 0.999 and ganoderic acid A: y = 2539.7x, R2 = 0.999). The identified compounds with unavailable commercial standards were quantified using the available calibration curve of the most similar standard. The results were expressed in the response format of Y2 (mg per g R).
2.9. Fitting procedures and statistical analysis
The statistical analysis of the experimental results and fitting was performed according to the equations for the responses obtained using a Microsoft Excel spreadsheet in three phases:(1) The measurement of the coefficients was achieved using the nonlinear least-squares (quasi-Newton) method provided by the macro ‘Solver’ in Microsoft Office Excel,31 by minimization of the sum of the quadratic differences between the observed and model-predicted values.
(2) The significance of the coefficients was obtained via the ‘SolverAid’ macro in Microsoft Office Excel32 to determine the parametric confidence intervals. The terms that were not statistically significant (p-value > 0.05) were dropped to simplify the model.
(3) Model reliability was confirmed by applying the following standards: (a) the Fisher F-test (α = 0.05) was used to determine the consistency of the constructed models to describe the obtained data;33 (b) the ‘SolverStat’ macro in Microsoft Office Excel was used to make an assessment of the parameter and model prediction uncertainties;34 and (c) R2 was determined to explain the proportion variability of the dependent variable obtained by the model.
3. Results and discussion
3.1. Extraction efficiency by using the Soxhlet technique
The extraction conditions used in the SE technique were selected by applying 7 cycles that correspond to a total extraction time of 4 h. The solvent used was ethanol and the extraction was performed at its boiling point (80 °C). However, depending on the compounds to be extracted, the number of time-cycles can influence the recovered quantity. For the studied target compounds (Tr and Ph), there are many scientific references suggesting a significant degradation rate at ethanol's boiling point as the time-cycles are prolonged.35 Therefore, to optimize the number of cycles and consequently measure the total content of Tr and Ph at its maximum, the time-cycle (note that each cycle accounts approximately for a 30 min period) of SE for G. lucidum was monitored.Fig. 1 shows the effect of the number of time-cycles on the content of Tr (part A) and Ph (part B); the results were obtained in triplicate for each considered time-cycle and confidence intervals were calculated (α = 0.05). For Tr content (Fig. 1, part A), a positive linear dependency was achieved for the responses Y3 and Y1. At the final number of performed time-cycles, the response Y1 shows an asymptotic-decreasing extraction stage (likely to be related to the maximum extraction level of Tr combined with the slight degradation of Tr). However, the increase in Y3 is more prominent than the one observed for Y1, suggesting that other compounds of the sample start to be extracted at higher rates than Tr, which also intensifies the decrease in Y2, causing a sharp decrease in Tr content in R, which is reflected by the decreasing patterns of Y2 (from 410 to 150 mg equiv. UA per g R). Although the Tr content in R is lower, the overall number of optimal time-cycles for Tr extraction was determined to be 5–6 in which ∼14 mg equiv. UA per g dw was obtained. These claims have been supported by other authors36 who reported that the extraction of Tr from G. lucidum is largely influenced in the T range between 80 and 100 °C and if prolonged time-cycles of 2 h (∼4 cycles) are used.
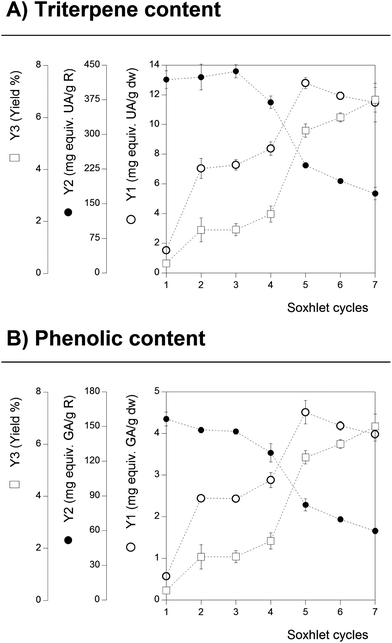 | ||
| Fig. 1 Results obtained for the cycle optimization of the conventional Soxhlet extraction using the response extraction yield and the two format values (Y1 and Y2) of the Tr and Ph content. | ||
Regarding Ph from G. lucidum, a nearly identical pattern to the one described for Tr was observed. The Ph content expressed in terms of Y1 was found to increase significantly up to 5 time-cycles (∼5 mg equiv. GA per g dw) reaching a maximum value before a gradual decrease starts to be observed (likely attributed to the degradation rates of Ph). The Y3 response increases linearly with the increase in the number of time-cycles, presenting also a higher slope when compared with the Y1 response, finishing with a sharp decrease in the Ph content in the dry material. In terms of the Ph content in the extract (Y2 format value), the number of time-cycles maximizing its value was achieved in the early stages of the process (1–3 time-cycles with ∼160 mg equiv. GA per g R), thereafter decreasing significantly as the extraction process is extended. This behaviour is similar to the one reported for the extraction of flavonoids from the flowers of Tabernaemontana heyneana Wall.35 To the authors best knowledge, the effect of the applied time-cycles number on the extraction efficiency of Tr and Ph from G. lucidum, through SE, has not been previously reported.
3.2. Extraction efficiency of HAE and UAE optimized using RSM
The obtained results, according to the proposed RSM design (based on a CCCD), are shown in Table 1 for the optimization of the HAE and UAE techniques using as responses the extraction yield (%) and Tr and Ph in the format values Y1 and Y2. Therefore, by using a non-linear least-squares procedure to fit eqn (1) to the response results, the estimated parametric values of the equation, parametric confidence intervals (α = 0.05) and relevant statistical values to assess the goodness of fit are presented in the first part of Table 2. Those coefficients which showed effects with coefficient interval values higher than the parameter value were considered as non-significant (ns) and were not considered for the model development.
| Parameters | Residue | Triterpene content | Phenolic content | |||
|---|---|---|---|---|---|---|
| Yield | Y 1 | Y 2 | Y 1 | Y 2 | ||
| Heat assisted extraction (HAE) | ||||||
| Intercept | b0 | 5.37 ± 1.0 | 11.62 ± 0.7 | 220.09 ± 25.2 | 5.33 ± 0.3 | 108.07 ± 10.5 |
| Linear effect | b1 | ns | 2.15 ± 0.7 | −14.12 ± 12.6 | 0.16 ± 0.0 | ns |
| b2 | −0.46 ± 0.3 | 0.51 ± 0.7 | 38.37 ± 18.7 | 0.01 ± 0.0 | 10.44 ± 7.7 | |
| b3 | 3.24 ± 0.7 | 3.67 ± 0.7 | ns | 1.21 ± 0.0 | −7.99 ± 7.7 | |
| Quadratic effect | b11 | 1.01 ± 0.7 | −0.28 ± 0.1 | −23.41 ± 18.7 | ns | −12.63 ± 7.9 |
| b22 | ns | ns | ns | 0.05 ± 0.0 | ns | |
| b33 | 0.96 ± 0.7 | −1.73 ± 0.7 | −37.41 ± 18.7 | −1.07 ± 0.0 | −20.01 ± 7.9 | |
| Interactive effect | b12 | 0.35 ± 0.3 | ns | −39.42 ± 18.7 | ns | ns |
| b13 | ns | −0.11 ± 0.3 | 20.62 ± 18.7 | ns | ns | |
| b23 | −1.38 ± 0.9 | 1.27 ± 0.3 | ns | ns | ns | |
| Statistics (R2) | 0.8948 | 0.8978 | 0.8136 | 0.9104 | 0.8210 | |
| Ultrasound assisted extraction (UAE) | ||||||
| Intercept | b0 | 6.64 ± 0.3 | 11.26 ± 0.7 | 178.99 ± 17.7 | 6.45 ± 0.3 | 96.70 ± 1.8 |
| Linear effect | b1 | −0.35 ± 0.3 | ns | ns | ns | 1.95 ± 1.8 |
| b2 | ns | −0.62 ± 0.5 | ns | ns | 3.67 ± 1.8 | |
| b3 | 0.73 ± 0.3 | 4.41 ± 0.5 | 89.57 ± 16.0 | 1.22 ± 0.2 | 18.44 ± 1.8 | |
| Quadratic effect | b11 | ns | ns | ns | −0.23 ± 0.2 | ns |
| b22 | ns | 0.55 ± 0.5 | ns | ns | ns | |
| b33 | −1.36 ± 0.3 | −0.54 ± 0.5 | 30.50 ± 16.3 | −1.17 ± 0.2 | −2.53 ± 1.8 | |
| Interactive effect | b12 | ns | ns | −19.32 ± 9.9 | 0.21 ± 0.2 | 4.55 ± 1.8 |
| b13 | −0.27 ± 0.3 | −0.58 ± 0.3 | ns | −0.23 ± 0.2 | ns | |
| b23 | −0.41 ± 0.3 | ns | ns | −0.19 ± 0.2 | −2.55 ± 1.8 | |
| Statistics (R2) | 0.9222 | 0.9349 | 0.9330 | 0.9223 | 0.9013 | |
Consequently, non-linear equations were built based on the second-order polynomial model of eqn (1). Then, for the HAE system:
| PhHAEY1 = 5.3 + 0.16t + 0.01T + 1.2S + 0.05T2 − 1.1S2 | (2) |
| TrHAEY1 = 11.6 + 2.1t + 0.5T + 3.7S − 0.28t2 − 1.7S2 − 0.11tS + 1.3TS | (3) |
| YieldHAE = 5.3 − 0.46T + 3.2S + 1.1t2 + 0.96S2 − 1.4TS | (4) |
| PhHAEY2 = 108.1 + 10.4T − 7.9S − 12.6t2 − 20.1S2 | (5) |
| TrHAEY2 = 220.1 − 14.1t + 38.4T − 23.4t2 − 37.4S2 − 39.4tT + 20.6TS | (6) |
| PhUAEY1 = 6.45 + 1.22S − 0.23t2 − 1.17S2 + 0.21tP − 0.23tS − 0.19PS | (7) |
| TrUAEY1 = 11.2 − 0.62P + 4.41S + 0.55P2 − 0.54S2 − 0.58tS | (8) |
| YieldUAE = 6.64 − 0.35t + 0.73S − 1.4S2 − 0.27tS − 0.41PS | (9) |
| PhUAEY2 = 96.7 + 1.9t + 3.7P + 18.4S − 2.5S2 + 4.5tP − 2.5PS | (10) |
| TrUAEY2 = 178.9 + 89.5S + 30.5S2 − 19.3tP. | (11) |
These equations translate the patterns for the individual measurement of the assessed response (eqn (2) to (6) for HAE and eqn (7) to (11) for UAE). Note that not all the parameters of eqn (1) were used for building the model since some coefficients were rejected (Table 2). The profile patterns derived from the parametric values of these mathematical models on the assessed response criteria can also be depicted by graphical representation. Fig. 2 and 3 present a 3D graphical analysis of the results concerning the extraction yield and Ph and Tr (expressed as Y1 and Y2) for the studied techniques (HAE and UAE).
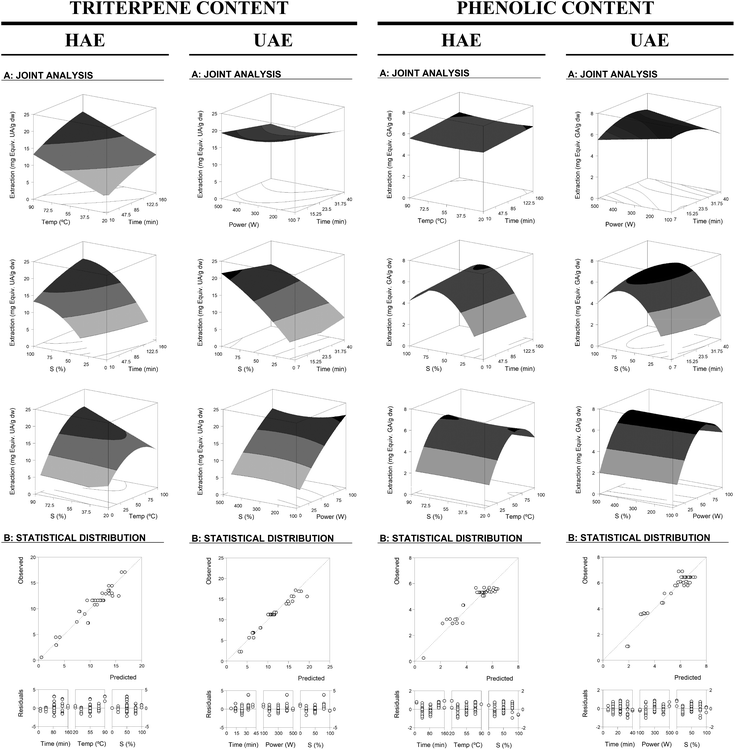 | ||
| Fig. 2 Graphical results in terms of the response surfaces of the format value of Y1 (mg per g dw) of Tr and Ph from the developed equations for the HAE and UAE system optimizations. Part A: Joint graphical 3D analysis as a function of each of the variables involved. Each of the net surfaces represents the theoretical three-dimensional response surface predicted with the second-order polynomial eqn (1) as a function of each one of the involved variables and described by eqn (2) and (3) for HAE and eqn (7) and (8) for UAE. The statistical design and results are described in Table 1. The estimated parametric values are shown in Table 2. The binary actions between the variables are presented when the excluded variable is positioned at the centre of the experimental domain (Table 1). Part B: To illustrate the goodness of fit, two basic graphical statistic criteria are used. The first one, the ability to simulate the changes of the response between the predicted and observed data; and the second one, the residual distribution as a function of each of the variables. Note all the differences in the axes scales. | ||
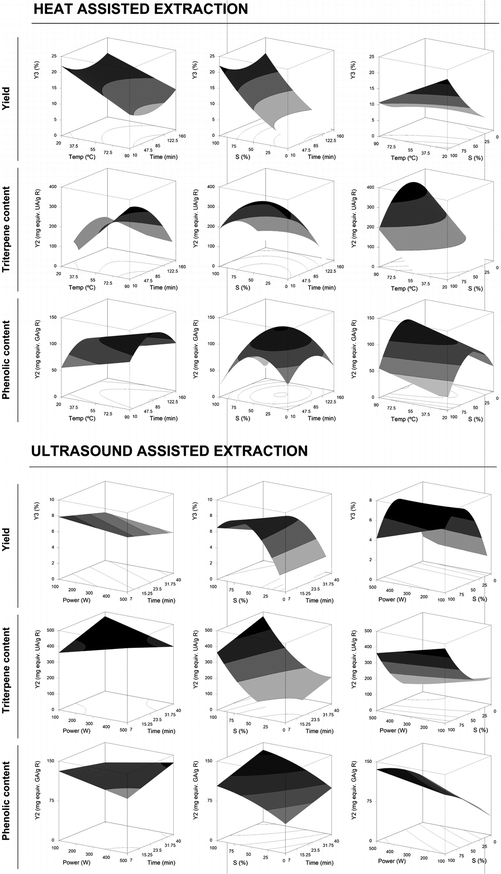 | ||
| Fig. 3 Joint graphical 3D analysis as a function of each of the variables involved for the HAE and UAE systems for the extraction yield of R obtained and the response content of Tr and Ph in the format value of Y2 (mg per g R). Each of the net surfaces represents the theoretical three-dimensional response surface predicted with the second-order polynomial eqn (1) as a function of each one of the involved variables and described by eqn (4), (5) and (6) for HAE and eqn (9), (10) and (11) for UAE. The statistical design and results are described in Table 1. The estimated parametric values are shown in Table 2. The binary actions between the variables are presented when the excluded variable is positioned at the centre of the experimental domain (Table 1). | ||
Regarding the interactive effects for the HAE system in terms of Tr-Y1, there was no significant interaction between t & T but a positive correlation for the pairs t & S and T & S was observed. In terms of Tr-Y2, there was a negative correlation for t & T and a positive correlation for t & S, and no interaction for T & S. In the case of the HAE system in terms of Ph-Y1 and Ph-Y2, no interaction was found among the variables t, T and S. For the UAE system in terms of Tr-Y1, there was no significant interaction between t & S, and P & S while a negative interaction for t & P was observed. In terms of Tr-Y2, there was a negative correlation for t & P, a positive correlation for t & S, and no interaction for P & S. Also in Ph-Y1, there was a negative correlation for t & S and P & S. No significant interaction was observed in terms of Ph-Y1 and Ph-Y2 between t & P, while for P & S it was negative for Ph-Y1 and not significant for Ph-Y2.
The regression coefficients related to the interactive effects of the responses are presented in Table 2 suggesting that the use of a RSM approach to optimize the extraction responses (extraction yield and Tr and Ph (expressed as Y1 and Y2)) for both systems (HAE and UAE) is correctly justified. If approaches based on one-variable-at-a-time were applied, the high level of interactions seen between the variables will make the optimum values difficult to determine. As an illustrative example, Fig. 2 shows the response of Tr in HAE representing the effect of T & S (at constant values of t), in which the related parameter shows a positive value b23 of 1.27 ± 0.3 that causes an increased extraction of Tr as both variables increase (closely related to synergistic effects). However, by looking at their individual profile, at fixed values for the other, such interactions will be nearly impossible to predict without a large investment on experimental analysis.
In terms of the statistical analysis of the fitting of the models to the responses, the lack-of-fit test used to assess the competence of the models showed that the significant parameters in both RSM approaches (Table 2) were highly consistent (p < 0.01) and if any of them was suppressed, the achieved solution would not be acceptable. This was also verified by the achieved high R2 value, indicating the percentage of the variability explained by the model (Table 2). In all cases, R2 was higher than 0.8, with values over ∼0.9 in almost all responses. The residual distribution presented in Fig. 2 was arbitrarily around zero and no groups of values or autocorrelations were observed. Additionally, the agreement between the experimental and predicted values implies an acceptable explanation of the obtained results through the used independent variables. Therefore, the models developed in eqn (2) to (6) for HAE and eqn (7) to (11) for UAE are completely functional and adequate to be used for prediction and process optimization.
Fig. 3 shows an illustrative 3D analysis representing the effect of each one of the involved variables in the HAE and UAE systems on the extraction yield (%) and Tr-Y2 and Ph-Y2 (mg per g R). Each one of the net surfaces represents the response predicted by the second-order polynomial eqn (4), (5) and (6) for HAE and eqn (9), (10) and (11) for UAE. The actions between the two considered variables are presented when the excluded variable is positioned at the centre of the experimental domain (Table 1). The Ph-Y2 for HAE and UAE showed a bell-shaped profile indicating that an increase in t and S increases the content of Ph to a certain point, followed by a significant decrease. These patterns are also found in the Tr-Y2 for HAE and UAE but to a lesser extent. As in SE, this behaviour is likely to be related to the joint action of the two processes. The extraction yield increases at higher rates than the content of Tr and Ph, suggesting that other compounds also start to be extracted from the G. lucidum sample, which causes a decrease in the content of both Tr and Ph since a maximum extraction level is achieved. In addition, degradation of these compounds may also occur justifying the strongest decrease visualized in the produced surfaces.
- For the extraction yield, HAE was the best solution; 21.3 ± 1.7% was achieved at *160.0 min, *20.0 °C and 100.0% ethanol.
- For the Tr-Y1, UAE was the best solution; 21.3 ± 4.1 mg equiv. UA per g dw was achieved at *7 min, *100.0 W and *100.0% ethanol.
- For the Tr-Y2, UAE was the best solution; 470.5 ± 41.7 mg equiv. UA per g extract was achieved at *40 min, *100.0 W and *100.0% ethanol.
- For the Ph-Y1, the UAE technique was the best solution; 6.7 ± 0.7 mg equiv. GA per g dw was achieved at 29.4 min, *100.0 W and 59.7% ethanol.
- For the Ph-Y2, HAE was the best solution; 125.9 ± 12.7 mg equiv. GA per g extract was achieved at 81.3 min, *90.0 °C and 49.1% ethanol.
| Optimal extraction conditions | Response optimum | |||
|---|---|---|---|---|
| X 1 : t (min) | X 2 : T (°C) or P (W) | X 3 : Et (%) | ||
| Individual optimal responses | ||||
| Heat assisted extraction (HAE) | ||||
| Yield | *160.0 | *20.0 | *100.0 | 21.3 ± 1.7 (%) |
| Tr (Y1) | *160.0 | *90.0 | *100.0 | 22.1 ± 2.9 (mg equiv. UA per g dw) |
| Tr (Y2) | *10.0 | *90.0 | 35.4 | 362.1 ± 33.1 (mg equiv. UA per g R) |
| Ph (Y1) | *160.0 | *20.0 | 65.7 | 6.3 ± 0.7 (mg equiv. GA per g dw) |
| Ph (Y2) | 81.3 | *90.0 | 49.1 | 125.9 ± 12.7 (mg equiv. GA per g R) |
| Ultrasound assisted extraction (UAE) | ||||
| Yield | *7.0 | *100.0 | 70.6 | 7.8 ± 0.9 (%) |
| Tr (Y1) | *7.0 | *100.0 | *100.0 | 21.3 ± 4.1 (mg equiv. UA per g dw) |
| Tr (Y2) | *40.0 | *100.0 | *100.0 | 470.5 ± 41.7 (mg equiv. UA per g R) |
| Ph (Y1) | 29.4 | *500.0 | 59.7 | 6.7 ± 0.7 (mg equiv. GA per g dw) |
| Ph (Y2) | *40.0 | *500.0 | *100.0 | 135.65 ± 15.7 (mg equiv. GA per g R) |
| Global optimal responses | ||||
| Heat assisted extraction (HAE) | ||||
| Yield | 78.9 | *90.0 | 62.5 | 5.2 ± 0.6 (%) |
| Tr (Y1) | 14.6 ± 1.9 (mg equiv. UA per g dw) | |||
| Tr (Y2) | 285.7 ± 31.2 (mg equiv. UA per g R) | |||
| Ph (Y1) | 5.8 ± 1.2 (mg equiv. GA per g dw) | |||
| Ph (Y2) | 116.9 ± 13.2 (mg equiv. GA per g R) | |||
| Ultrasound assisted extraction (UAE) | ||||
| Yield | *40 | *100.0 | 89.5 | 4.9 ± 0.6 (%) |
| Tr (Y1) | 17.4 ± 2.9 (mg equiv. UA per g dw) | |||
| Tr (Y2) | 435.6 ± 21.1 (mg equiv. UA per g R) | |||
| Ph (Y1) | 4.6 ± 0.2 (mg equiv. GA per g dw) | |||
| Ph (Y2) | 106.6 ± 16.2 (mg equiv. GA per g R) | |||
Comparing both techniques in terms of extraction efficiency, UAE corresponded to significantly higher values than HAE, probably due to compound's degradation as described previously by other authors.40,41 Regarding the extraction time, UAE was the fastest extraction method for almost all responses. In the second part of Table 3 the global optimal conditions are presented, as well as the relative response values:
- For HAE, the global conditions that maximize the responses were 78.9 min, *90.0 °C and 62.5% of ethanol corresponding to an extraction yield of 5.2 ± 0.6%, Tr content in the dry material and in the extract of 14.6 ± 1.9 mg equiv. UA per g dw and 285.7 ± 31.2 mg equiv. UA per g R, respectively, and Ph content in the dry material and in the extract of 5.8 ± 1.2 mg equiv. GA per g dw and 116.3 ± 13.2 mg equiv. GA per g R, respectively.
- For UAE, the global conditions that maximize the responses were *40 min, *100.0 W and 89.5% of ethanol corresponding to an extraction yield of 4.9 ± 0.6%, Tr content in the dry material and in the extract of 17.4 ± 2.9 mg equiv. UA per g dw and 435.6 ± 41.2 mg equiv. UA per g R, respectively, and Ph content in the dry material and in the extract of 4.6 ± 0.2 mg equiv. GA per g dw and 106.6 ± 16.2 mg equiv. GA per g extract, respectively.
Finally, Fig. 4 shows the summarized individual 2D responses as a function of the defined variables for the HAE and UAE techniques guiding the selection of the most favourable conditions. The line represents the variable response pattern when the other responses are located at the optimal values presented in the third part of Table 3. The dots (⊙) presented alongside the line highlight the location of the optimal value. For all techniques and responses, the conditions that lead to the optimal values were re-checked to ensure the accuracy of the presented results.
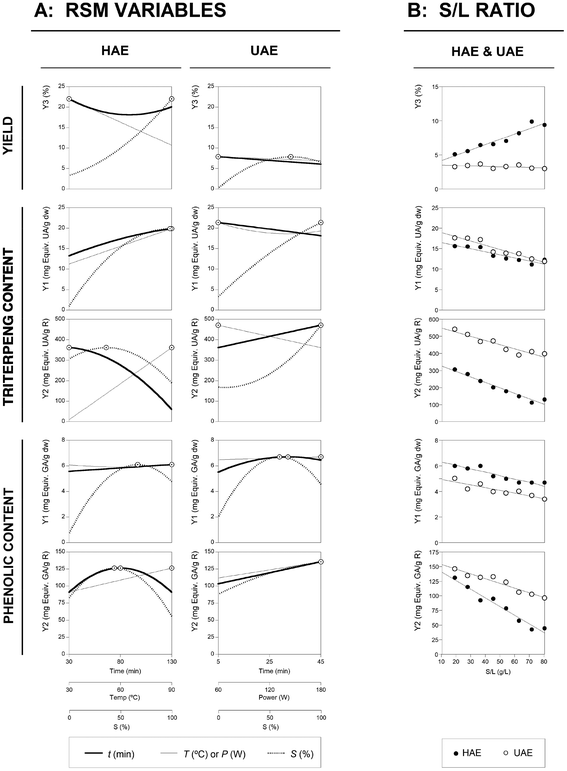 | ||
| Fig. 4 Illustration summarizing the effects of all variables assessed for the HAE and UAE systems. Part A: Individual 2D responses of all studied responses as a function of all the variables assessed. The variables in each of the 2D graphs were positioned at the optimal values of the others (Table 3). The dots (⊙) presented alongside each line highlights the location of the optimum value. The lines and dots are generated by the theoretical second-order polynomial models of eqn (2) to (6) for ME and eqn (7) to (11). Part B: Dose response of the S/L ratio at the optimal values of the other three variables optimized using RSM. The points (● for HAE and ○ for UAE) represent the obtained experimental results, while the line shows the predicted pattern by a simple linear relation. The limit value (80 g L−1) shows the maximum achievable experimental concentration until the sample cannot be physically stirred at the laboratory scale. | ||
The interest to scale up processes to obtain the bioactives of interest, such as the ones proposed in the present work (triterpenes and phenolic compounds), can take advantages from the data reported in Fig. 4. The data were organized in a simple format to allow an easy interpretation of the responses. For the developed models, other variables can be further included in order to consider the minimization of costs (i.e. energy and materials), which can contribute to the economic viability of the process.
3.3. Dose–response analysis of the solid-to-liquid effect under the optimum conditions
The S/L studies for the HAE and UAE techniques, performed under the optimal global conditions predicted by the polynomial eqn (4) to (11), are presented in the third part of Table 3. For HAE, the conditions were: 78.9 min, 90.0 °C and 62.5% of ethanol, whereas for UAE, the conditions were: 40.0 min, 100.0 W and 89.5% of ethanol. The preliminary obtained results showed that the experimental limit value at the lab scale was close to 80 g L−1. Therefore, in both techniques an experimental dose–response test was designed to verify the S/L behavior between 10 g L−1 and 80 g L−1. In all cases, the S/L effect can be described by a simple non-linear relationship with an intercept, and in almost all cases, the linear relationship shows a decreasing pattern as the S/L ratio increases. The increase in the S/L ratio leads to a decrease in the solvent extraction ability and the observed decreasing patterns are expected due to the saturation effects. Consequently, the dose–response is explained by means of the linear parametric values of the slope (m) and the intercept (b). The effects of the S/L dose–response for the HAE and UAE techniques are presented in Fig. 4. In all cases, the obtained responses through the HAE and UAE systems are consistent with the previous results obtained using an S/L ratio of 30 g L−1. Fig. 4 shows the experimental results (dots: ● for HAE and ○ for UAE) and model predictions of the linear relationship (lines) obtained for each technique. The results derived from the linear fitting analysis are summarized as follows:- For the extraction yield, the HAE technique corresponded to a predicted intercept of b = 4.2 ± 0.4% of R and a slope with positive values (m = 0.069 ± 0.008%), which can indicate an increase in the extract with the S/L ratio. This was the only case that presented increased patterns as a function of the rise of the S/L ratio. The reasons behind this may be the high values of T applied (90.0 °C), which may help to dissolve other substances even at high values of the S/L ratio. Meanwhile, for the UAE technique b = 3.5 ± 0.2% of R and m = −0.005 ± 0.004% of R per g per L.
- For the Tr-Y1 response for the HAE and UAE techniques, the b values obtained were 16.4 ± 0.6 and 18.9 ± 0.5 mg equiv. UA per g dw, respectively. The decreasing m effect caused by the S/L ratio increase were −0.066 ± 0.012 and −0.091 ± 0.011 mg equiv. UA per g dw per g per L, respectively.
- For the Tr-Y2 response for the HAE and UAE techniques, the b values obtained were 325.6 ± 12.7 and 547.7 ± 15.3 mg equiv. UA per g R, respectively. The decreasing m effect caused by the S/L ratio increase were −2.80 ± 0.25 and −2.12 ± 0.31 mg equiv. UA per g R per g per L, respectively.
- For the Ph-Y1 response for the HAE and UAE techniques, the b values obtained were 6.3 ± 0.2 and 4.9 ± 0.2 mg equiv. GA per g dw, respectively. The decreasing m effect caused by the S/L ratio increase were −0.023 ± 0.003 and −0.019 ± 0.004 mg equiv. GA per g dw per g per L, respectively.
- For the Ph-Y2 response for the HAE and UAE techniques, the b values obtained were 140.6 ± 5.4 and 153.6 ± 3.7 mg equiv. GA per g R, respectively. The decreasing m effect caused by the S/L ratio increase were −1.299 ± 0.107 and −0.710 ± 0.073 mg equiv. GA per g R per g per L, respectively.
Consequently, the dose–response in terms of all response criteria can be explained by the parametric results derived from the linear relationships and this trend was visually interpreted in Fig. 4, for comparison purposes, in which the modelling predictions obtained for each technique are represented jointly up to the determination of the experimental limit value of 80 g L−1.
3.4. Characterization of the optimized extract in terms of individual triterpenoids and phenolic compounds by HPLC-DAD-ESI/MSn
The global optimum conditions for Tr and Ph extraction by using the HAE and UAE techniques are presented in Table 3. These conditions were used to obtain extracts that were further characterized by using HPLC-DAD-ESI/MSn.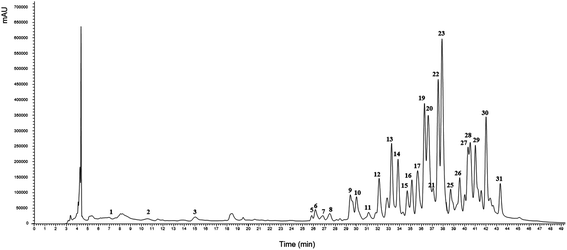 | ||
| Fig. 5 Example of the HPLC profiles derived from G. lucidum with regard to the content of Ph and Tr compounds. A representative case under the optimal global conditions of UAE presented in Table 3. The identification numbers of the compounds are described in detail in Table 4. | ||
| Peak | R t (min) | UV λmax (nm) | [M − H]− & [2M − H] (m/z) | MS2 (m/z) | Tentative identification | Quantification (mg per g R) | ||
|---|---|---|---|---|---|---|---|---|
| HAE | UAE | SE | ||||||
| A: Characterization of phenolic compounds | ||||||||
| 1 | 5.23 | 259![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 293sh 293sh |
153 | 109(100) | Protocatechuic acid | — | — | 1.80± 0.01 |
| 2 | 8.39 | 257 | 137 | 93(100) | p-Hydroxybenzoic acid | — | 0.364 ± 0.01 | 2.98 ± 0.01 |
| 3 | 11.51 | 280 | 197 | 121(100) | Syringic acid | 0.250 ± 0.001 | 0.260 ± 0.001 | 1.51 ± 0.01 |
| Total phenolic acids | 0.250 ± 0.001 | 0.620 ± 0.001 | 6.30 ± 0.01 | |||||
| B: Characterization of triterpenes | ||||||||
| 4 | 29.06 | 259 | 533/1067 | 515(5), 497(12), 404(100), 303(12) | 12-3,7,15-Trihydroxy-4-(hydroxymethyl)-11,23-dioxo-lanost-8-en-26-oic acid | 3.77 ± 0.03 | 7.18 ± 0.05 | 14.21 ± 0.02 |
| 5 | 30.8 | 256 | 533/1067 | 515(5), 497(21), 453(16), 423(11), 319(5), 303(8) | 12-Hydroxyganoderic acid C2 | 5.01 ± 0.03 | 1.43 ± 0.04 | 4.51 ± 0.06 |
| 6 | 31.27 | 252 | 529/1059 | 511(10), 467(100), 449(11), 434(5), 431(15), 419(8), 312(8), 285(5), 263(11) | 20-Hydroxyganoderic acid AM1 | — | 5.41 ± 0.06 | 11.40 ± 0.01 |
| 7 | 31.73 | 251 | 529/1059 | 511(13), 499(100), 481(6), 465(18), 438(28), 419(18), 367(11), 287(11) | 12-Deacetylganoderic acid H | 2.28 ± 0.04 | 3.62 ± 0.06 | 6.1 ± 0.14 |
| 8 | 32.28 | 255 | 547/1095 | 529(50), 511(41), 485(3), 467(100), 449(24), 431(26), 304(11), 265(7) | Ganoderic acid derivative | 3.78 ± 0.03 | 5.27 ± 0.04 | 7.3 ± 0.1 |
| 9 | 33.94 | 258 | 531/1063 | 513(13), 451(3), 401(100), 385(5), 304(19), 301(45), 286(3), 249(27) | Ganoderic acid ɳ | 5.50 ± 0.03 | 14.13 ± 0.08 | 25.41 ± 0.04 |
| 10 | 34.48 | 254 | 511/1023 | 493(23), 449(100), 431(5), 413(15), 405(3) | Ganoderic acid F | 5.34 ± 0.03 | 12.46 ± 0.05 | 17.97 ± 0.02 |
| 11 | 35.37 | 265 | 529/1059 | 511(5), 467(100), 449(20), 437(29), 317(10), 301(5), 263(5) | 12-Hydroxyganoderic acid D | 4.15 ± 0.04 | 7.78 ± 0.05 | 6.86 ± 0.07 |
| 12 | 35.95 | 251 | 515/1031 | 497(100), 453(31), 437(8), 303(19), 287(5), 235(3) | Ganoderic acid derivative | 11.03 ± 0.06 | 21.61 ± 0.06 | 21.20 ± 0.08 |
| 13 | 36.84 | 256 | 517/1035 | 499(100), 481(48), 456(17), 438(52), 407(8), 304(6), 287(35) | Ganoderic acid C2 | 17.92 ± 0.03 | 28.09 ± 0.08 | 38.70 ± 0.15 |
| 14 | 37.51 | 257 | 529/1059 | 511(5), 481(7), 467(100), 451(14), 438(38), 424(3), 319(5), 303(3), 301(5) | Ganoderic acid C6 | 10.50 ± 0.04 | 21.11 ± 0.04 | 29.10 ± 0.08 |
| 15 | 38.16 | 256 | 529/1059 | 511(58), 493(5), 449(10), 399(100), 301(3) | Ganoderic acid derivative | 5.22 ± 0.06 | 11.07 ± 0.06 | 12.77 ± 0.01 |
| 16 | 38.46 | 256 | 531/1063 | 513(11), 498(15), 469(100), 454(29), 452(24), 437(6), 304(5), 302(6), 290(20), 266(7) | Ganoderic acid G | 8.04 ± 0.02 | 14.31 ± 0.29 | 16.84 ± 0.07 |
| 17 | 38.75 | 248 | 513/1027 | 495(10), 480(16), 451(100), 437(14), 433(22), 407(17), 331(5), 315(3), 303(5), 287(5) | Ganoderenic acid B | 13.12 ± 0.01 | 23.05 ± 0.50 | 10.66 ± 0.09 |
| 18 | 39.01 | 259 | 527/1055 | 509(26), 465(6), 397(100), 355(4) | Elfvingic acid derivative | — | — | 10.41 ± 0.09 |
| 19 | 39.35 | 254 | 515/1031 | 497(10), 453(100), 439(5), 409(5), 304(21), 287(12), 263(3), 250(14) | Ganoderic acid B | 18.86 ± 0.07 | 35.72 ± 0.02 | 23.99 ± 0.09 |
| 20 | 39.46 | 250 | 513/1027 | 495(100), 479(27), 462(12), 451(30), 433(31), 381(25), 301(15) | Ganoderic acid derivative | 25.64 ± 0.03 | 44.70 ± 0.16 | 22.44 ± 0.10 |
| 21 | 39.66 | 261 | 513/1027 | 495(21), 480(5), 451(100), 433(24), 381(6), 301(6), 247(3) | Ganoderic acid AM1 | — | 11.56 ± 0.03 | 15.82 ± 0.01 |
| 22 | 40.29 | 254 | 515/1031 | 497(100), 480(5), 454(6), 436(10), 302(8), 301(4), 285(3) | Ganoderic acid A | 28.79 ± 0.05 | 43.46 ± 0.20 | 36.77 ± 0.04 |
| 23 | 40.66 | 261 | 571/1143 | 553(100), 511(8), 481(3), 468(8), 437(3), 423(2) | Ganoderic acid H | 27.32 ± 0.19 | 58.13 ± 0.03 | 41.05 ± 0.39 |
| 24 | 40.87 | 252 | 527/1055 | 509(20), 479(13), 465(100), 435(3), 421(3), 317(3), 301(3) | Elfvingic acid A | — | — | 9.83 ± 0.01 |
| 25 | 41.34 | 254 | 529/1059 | 511(10), 496(22), 493(21), 467(100), 449(61), 434(4), 319(6), 317(8), 301(16), 300(11), 299(12) | Ganoderic acid derivative | 6.12 ± 0.07 | 13.98 ± 0.05 | 11.56± 0.04 |
| 26 | 41.93 | 246 | 511/1023 | 493(12), 478(20), 449(100), 435(15), 431(4), 405(4), 329(3), 301(4), 285(5), 283(3), 261(4) | Ganoderenic acid D | 12.38 ± 0.04 | 23.06 ± 0.04 | 10.27 ± 0.07 |
| 27 | 42.55 | 255 | 513/1027 | 495(16), 451(100), 437(6), 433(3), 407(4), 301(23), 286(3), 284(11), 247(8) | Ganoderic acid D | 15.17 ± 0.09 | 17.61 ± 0.12 | 11.31 ± 0.01 |
| 28 | 42.74 | 245 | 509/1019 | 491(100), 476(18), 461(34), 447(15), 429(3), 417(3), 300(5), 299(4) | Ganoderic acid derivative | 13.37 ± 0.13 | 28.36 ± 0.04 | 12.81 ± 0.08 |
| 29 | 43.15 | 256 | 511/1023 | 493(100), 449(65), 435(3), 300(5), 247(4) | Ganoderic acid E | 12.84 ± 0.03 | 31.53 ± 0.08 | 10.81 ± 0.07 |
| 30 | 43.95 | 255 | 569/1139 | 551(100), 509(35), 508(21), 466(8) | 12-Acetoxyganoderic acid F | 15.91 ± 0.03 | 32.59 ± 0.12 | 12.08 ± 0.07 |
| 31 | 44.92 | 272 | 513/1027 | 451(100), 437(8), 433(3), 422(3), 301(5) | Ganoderic acid J | 8.41 ± 0.16 | 14.05 ± 0.42 | 2.92 ± 0.05 |
| Total triterpenoids | 280.46 ± 0.11 | 531.26 ± 0.24 | 455.01 ± 1.47 | |||||
3.5. Assessment of extraction efficiencies between HAE and UAE
The extraction of bioactive compounds from natural matrices using solid–liquid extraction systems constitutes an important step to obtain ingredients to produce phytochemical-rich products. This is in accordance with the current trends where increasing interest is devoted to the use of natural-derived ingredients in different industrial fields. Over the years, the conventional extraction systems were predominantly applied.47,48 Traditional methods such as SE and HAE have been used everywhere for many different purposes. They are often time-consuming and require large quantities of polluting solvents in comparison with other emerging technologies. However, in terms of efficiency (extraction yield and purity), SE is described as the standard chemical extraction process at the laboratory scale. Nonetheless, by itself, it is an optimized extraction system and in addition, the literature offers a large number of practical examples that report the favourable conditions.49 Additionally, its sister in industrial applications, the repeated heat extraction, has been used by the food processing industries and researchers with the purpose of extracting more efficiently major and minor compounds.However, to maximally recover compounds of interest from natural sources, while also taking into consideration environmental factors, financial feasibility, time and extraction quality, non-conventional technologies have been applied (such as UAE as well as pulsed electric field, enzyme digestion, extrusion, microwave assisted extraction, supercritical fluid extraction, etc.). New extraction techniques are continuously developed and/or modified to properly identify the most suitable technique that maximally increases the recovery of bioactive compounds.
Even though there are reports on the optimization of Tr extraction from natural matrices,37 only Ruan et al.13 (2014) have been able to report the effects of different extraction variables on the extraction efficiency of Tr from G. lucidum. The above authors reported that the Tr yield was 13.23 mg per g dw when the extraction conditions were 100% ethanol, 60.2 °C and 6 h. Wei et al.37 (2015) reported an increase in Tr extraction from Jatropha curcas L. using the UAE system from 14.39 to 19.51 mg per g dw as the solvent proportion increased from 50 to 75%, respectively. Also, as the time increased from 10 to 40 min, the extraction yield increased from 12.50 to 18.93 mg per g dw, respectively. The above authors reported an optimum yield of 26.7 ± 0.2 mg per g dw at 70% ethanol, 50 min and 100 W, values that are consistent with the Tr-Y1 content reported in the present work of 28.4 ± 4.1 mg per g dw under the optimal individual conditions (Table 3, 100% of ethanol, 40 min and 100 W). Studies conducted by Gao et al.36 (2011) revealed an optimal extraction yield of 1.09% for the HAE system at 90% ethanol, 120 min and 80 °C while it was 5.0 ± 0.6% in the present work (Table 3). The dissimilarity in the above response could be due to the influence of the application of mathematical and statistic techniques to maximize the recovery. To the authors' best knowledge, no previous studies have been conducted on the optimization of the extraction of Ph compounds from G. lucidum using the UAE and HAE systems. However, Lin, Yu, & Weng50 (2012) conducted an optimization study using the supercritical fluid extraction technique, obtaining optimum extraction values at lower temperatures.
4. Conclusions
The SE, HAE and UAE extraction techniques have been examined, optimized and compared with regard to the recovery of bioactive compounds from G. lucidum. The results showed that the main variables involved in these processes have significant effects on the extraction yield. The UAE system was found to be the most effective and fastest method for Tr extraction, capable of yielding 17.4 ± 2.9 mg equiv. UA per g dw and producing 4.9% R (49 mg R per g dw) with a purity of 435.6 ± 21.1 mg equiv. UA per g R under its optimal extraction conditions. Meanwhile, HAE was found to offer a better result for the Ph extraction showing 5.8 ± 1.2 mg equiv. GA per g dw and producing 5.2% R (52 mg R per g dw) with a purity of 116.9 ± 13.2 mg equiv. GA per g R under its optimal extraction conditions. In global terms, applying the RSM conditions by using the UAE technique leads to higher amounts of compounds but also higher extraction yields than those found in HAE and SE, but the costs involving the use of this technique at the industrial scale may not cover the investment needed. This work offers an overview through environmentally compatible extraction processes, in which G. lucidum is subjected to ‘clean’ technologies able to integrate a potential industrial sector in a sustainable approach. The obtained results indicate the viability of using G. lucidum as a productive source of an enriched extract with bioactive compounds. Under the optimum conditions, HPLC-DAD-ESI/MSn confirmed the validity of the model with a total of 24, 26 and 28 Tr correctly identified in the HAE, UAE and SE extracts, respectively. Protocatechuic, p-hydroxybenzoic and syringic acids were identified in the SE ethanolic extract. The HAE system showed the presence of only syringic acid while p-hydroxybenzoic acid was additionally present in the UAE extract. In addition, the present work can reinforce the potential of G. lucidum to serve as a source of bioactive compounds to be used as natural additives in functional foods, as cosmeceutical ingredients and as novel compounds with pharmaceutical potential. Because of the widespread presence of Tr and Ph compounds in G. lucidum, further studies should be conducted using the optimal extraction conditions determined in the present work.Conflicts of interest
No conflict of interest.Acknowledgements
The authors are grateful to the Foundation for Science and Technology (FCT, Portugal) and the FEDER under Programme PT2020 for financial support to the CIMO (UID/AGR/00690/2013), the REQUIMTE (UID/QUI/50006/2013-POCI/01/0145/FEDER/007265) and L. Barros; to POCI-01-0145-FEDER-006984 (LA LSRE-LCM), funded by the FEDER, through POCI-COMPETE2020 and the FCT and Project NORTE-01-0145-FEDER-000006, funded by NORTE2020 under PT2020; and to the Xunta de Galicia for financial support for the post-doctoral research of M.A. Prieto. This work was also funded by the European Structural and Investment Funds (FEEI) through the Regional Operational Program North 2020, within the scope of Project ValorNatural®.References
- S. Baby, A. J. Johnson and B. Govindan, Secondary metabolites from Ganoderma, Phytochemistry, 2015, 114, 66–101 CrossRef PubMed.
- X. R. Zhao, B. J. Zhang, S. Deng, H. L. Zhang, S. S. Huang, X. K. Huo, C. Wang, F. Liu and X. C. Ma, Isolation and identification of oxygenated lanostane-type triterpenoids from the fungus Ganoderma lucidum, Phytochem. Lett., 2016, 16, 87–91 CrossRef.
- Z. Liu, J. Xing, Y. Huang, R. Bo, S. Zheng, L. Luo, Y. Niu, Y. Zhang, Y. Hu, J. Liu, Y. Wu and D. Wang, Activation effect of Ganoderma lucidum polysaccharides liposomes on murine peritoneal macrophages, Int. J. Biol. Macromol., 2016, 82, 973–978 CrossRef PubMed.
- K. S. Bishop, C. H. J. Kao, Y. Xu, M. P. Glucina, R. R. M. Paterson and L. R. Ferguson, From 2000 years of Ganoderma lucidum to recent developments in nutraceuticals, Phytochemistry, 2015, 114, 56–65 CrossRef PubMed.
- F. Hennicke, Z. Cheikh-Ali, T. Liebisch, J. G. MacIá-Vicente, H. B. Bode and M. Piepenbring, Distinguishing commercially grown Ganoderma lucidum from Ganoderma lingzhi from Europe and East Asia on the basis of morphology, molecular phylogeny, and triterpenic acid profiles, Phytochemistry, 2016, 127, 29–37 CrossRef PubMed.
- C. Wah Ma, M. Feng, X. Zhai, M. Hu, L. You, W. Luo and M. Zhao, Optimization for the extraction of polysaccharides from Ganoderma lucidum and their antioxidant and antiproliferative activities, J. Taiwan Inst. Chem. Eng., 2013, 44, 886–894 CrossRef.
- Y. Kan, T. Chen, Y. Wu, J. wu and J. Wu, Antioxidant activity of polysaccharide extracted from Ganoderma lucidum using response surface methodology, Int. J. Biol. Macromol., 2015, 72, 151–157 CrossRef PubMed.
- Z. Liu, X. Ma, B. Deng, Y. Huang, R. Bo, Z. Gao, Y. Yu, Y. Hu, J. Liu, Y. Wu and D. Wang, Development of liposomal Ganoderma lucidum polysaccharide: Formulation optimization and evaluation of its immunological activity, Carbohydr. Polym., 2015, 117, 510–517 CrossRef PubMed.
- N. Benkeblia, Ganoderma lucidum Polysaccharides and Terpenoids: Profile and Health Benefits, J. Food Nutr. Diet., 2015, 1, 1–6 Search PubMed.
- R. A. S. Thabit, X. Cheng, N. Q. Al-hajj, H. Shi, X. Tang and G. Le, Optimization of Extraction of Triterpense from Geum japonicum using Response Surface Methodology, Int. J. Pharma Res. Rev., 2014, 3, 31–40 Search PubMed.
- O. Taofiq, S. A. Heleno, R. C. Calhelha, M. J. Alves, L. Barros, A. M. González-Paramás, M. F. Barreiro and I. C. F. R. Ferreira, The potential of Ganoderma lucidum extracts as bioactive ingredients in topical formulations, beyond its nutritional benefits, Food Chem. Toxicol., 2017, 108, 139–147 CrossRef PubMed.
- P. Xu, Z. Y. Ding, Z. Qian, C. X. Zhao and K. C. Zhang, Improved production of mycelial biomass and ganoderic acid by submerged culture of Ganoderma lucidum SB97 using complex media, Enzyme Microb. Technol., 2008, 42, 325–331 CrossRef.
- W. Ruan, A. H. H. Lim, L. G. Huang and D. G. Popovich, Extraction optimisation and isolation of triterpenoids from Ganoderma lucidum and their effect on human carcinoma cell growth, Nat. Prod. Res., 2014, 28, 2264–2272 CrossRef PubMed.
- M.-Y. Kim, P. Seguin, J.-K. Ahn, J.-J. Kim, S.-C. Chun, E.-H. Kim, S.-H. Seo, E.-Y. Kang, S.-L. Kim, Y.-J. Park, H.-M. Ro and I.-M. Chung, Phenolic Compound Concentration and Antioxidant Activities of Edible and Medicinal Mushrooms from Korea, J. Agric. Food Chem., 2008, 56, 7265–7270 CrossRef PubMed.
- I. A. Sheikh, D. Vyas, M. A. Ganaie, K. Dehariya and V. Singh, HPLC determination of phenolics and free radical scavenging activity of ethanolic extracts of two polypore mushrooms, Int. J. Pharm. Pharm. Sci., 2014, 6, 679–684 Search PubMed.
- S. A. Heleno, L. Barros, A. Martins, M. J. R. P. Queiroz, C. Santos-Buelga and I. C. F. R. Ferreira, Fruiting body, spores and in vitro produced mycelium of Ganoderma lucidum from Northeast Portugal: A comparative study of the antioxidant potential of phenolic and polysaccharidic extracts, Food Res. Int., 2012, 46, 135–140 CrossRef.
- O. Taofiq, A. M. González-Paramás, A. Martins, M. F. Barreiro and I. C. F. R. Ferreira, Mushrooms extracts and compounds in cosmetics, cosmeceuticals and nutricosmetics-A review, Ind. Crops Prod., 2016, 90, 38–48 CrossRef.
- O. Taofiq, A. Martins, M. F. Barreiro and I. C. F. R. Ferreira, Anti-inflammatory potential of mushroom extracts and isolated metabolites, Trends Food Sci. Technol., 2016, 50, 193–210 CrossRef.
- F. Chemat, Zill-E-Huma and M. K. Khan, Applications of ultrasound in food technology: Processing, preservation and extraction, Ultrason. Sonochem., 2011, 18, 813–835 CrossRef PubMed.
- P. Garcia-Salas, A. Morales-Soto, A. Segura-Carretero and A. Fernández-Gutiérrez, Phenolic-compound-extraction systems for fruit and vegetable samples, Molecules, 2010, 15, 8813–8826 CrossRef PubMed.
- L. Wang and C. L. Weller, Recent advances in extraction of nutraceuticals from plants, Trends Food Sci. Technol., 2006, 17, 300–312 CrossRef.
- J. Dai and R. J. Mumper, Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties, Molecules, 2010, 15, 7313–7352 CrossRef PubMed.
- B. R. Albuquerque, M. A. Prieto, M. F. Barreiro, A. Rodrigues, T. P. Curran, L. Barros and I. C. F. R. Ferreira, Catechin-based extract optimization obtained from Arbutus unedo L. fruits using maceration/microwave/ultrasound extraction techniques, Ind. Crops Prod., 2016, 95, 404–415 CrossRef.
- M. Yolmeh and S. M. Jafari, Applications of Response Surface Methodology in the Food Industry Processes, Food Bioprocess Technol., 2017, 10, 413–433 CrossRef.
- C. L. Chang, C. S. Lin and G. H. Lai, Phytochemical characteristics, free radical scavenging activities, and neuroprotection of five medicinal plant extracts, J. Evidence-Based Complementary Altern. Med., 2012 Search PubMed , 984295.
- G. Box and J. Hunter, Multi-factor experimental designs for exploring response surfaces, Ann. Math. Stat., 1957, 28, 195–241 CrossRef.
- G. E. P. Box, J. S. Hunter and W. G. Hunter, Statistics for Experimenters: Design, Innovation, and Discovery, in Statistics for Experimenters: Design, Innovation, and Discovery, 2nd edn, 2005 Search PubMed.
- J. Pinela, M. A. Prieto, M. F. Barreiro, A. M. Carvalho, M. B. P. P. Oliveira, J. A. Vázquez and I. C. F. R. Ferreira, Optimization of microwave-assisted extraction of hydrophilic and lipophilic antioxidants from a surplus tomato crop by response surface methodology, Food Bioprod. Process., 2016, 98, 283–298 CrossRef.
- S. A. Heleno, P. Diz, M. A. Prieto, L. Barros, A. Rodrigues, M. F. Barreiro and I. C. F. R. Ferreira, Optimization of ultrasound-assisted extraction to obtain mycosterols from Agaricus bisporus L. by response surface methodology and comparison with conventional Soxhlet extraction, Food Chem., 2016, 197, 1054–1063 CrossRef PubMed.
- S. M. F. Bessada, J. C. M. Barreira, L. Barros, I. C. F. R. Ferreira and M. B. P. P. Oliveira, Phenolic profile and antioxidant activity of Coleostephus myconis (L.) Rchb.f.: An underexploited and highly disseminated species, Ind. Crops Prod., 2016, 89, 45–51 CrossRef.
- M. A. Prieto, J. A. Vázquez and M. A. Murado, Crocin bleaching antioxidant assay revisited: Application to microplate to analyse antioxidant and pro-oxidant activities, Food Chem., 2015, 1, 299–310 CrossRef PubMed.
- M. A. Prieto, T. P. Curran, A. Gowen and J. A. A. Vázquez, An efficient methodology for quantification of synergy and antagonism in single electron transfer antioxidant assays, Food Res. Int., 2015, 67, 284–298 CrossRef.
- J. Pinela, M. A. P. Lage, A. L. Antonio, A. M. Carvalho, M. B. P. P. Oliveira, L. Barros and I. C. F. R. Ferreira, Ellagitannin-rich bioactive extracts of Tuberaria lignosa: Insights into the radiation-induced effects in the recovery of high added-value compounds, Food Funct., 2017, 8, 2485 Search PubMed.
- C. Comuzzi, P. Polese, A. Melchior, R. Portanova and M. Tolazzi, SOLVERSTAT: a new utility for multipurpose analysis. An application to the investigation of dioxygenated Co(II) complex formation in dimethylsulfoxide solution, Talanta, 2003, 59, 67–80 CrossRef PubMed.
- T. Sathishkumar, R. Baskar, M. Aravind, S. Tilak, S. Deepthi and V. M. Bharathikumar, Simultaneous Extraction Optimization and Analysis of Flavonoids from the Flowers of Tabernaemontana heyneana by High Performance Liquid Chromatography Coupled to Diode Array Detector and Electron Spray Ionization/Mass Spectrometry, ISRN Biotechnol., 2013, 2013, 1–10 CrossRef PubMed.
- Y. Gao, R. Zhang, J. Zhang, S. Gao, W. Gao, H. Zhang, H. Wang and B. Han, Study of the extraction process and in vivo inhibitory effect of ganoderma triterpenes in oral mucosa cancer, Molecules, 2011, 16, 5315–5332 CrossRef PubMed.
- L. Wei, W. Zhang, L. Yin, F. Yan, Y. Xu and F. Chen, Extraction optimization of total triterpenoids from jatropha curcas leaves using response surface methodology and evaluations of their antimicrobial and antioxidant capacities, Electron. J. Biotechnol., 2015, 18, 88–95 CrossRef.
- W. H. Wong, W. X. Lee, R. N. Ramanan, L. H. Tee, K. W. Kong, C. M. Galanakis, J. Sun and K. N. Prasad, Two level half factorial design for the extraction of phenolics, flavonoids and antioxidants recovery from palm kernel by-product, Ind. Crops Prod., 2015, 63, 238–248 CrossRef.
- E. Roselló-Soto, C. M. Galanakis, M. Brnčić, V. Orlien, F. J. Trujillo, R. Mawson, K. Knoerzer, B. K. Tiwari and F. J. Barba, Clean recovery of antioxidant compounds from plant foods, by-products and algae assisted by ultrasounds processing. Modeling approaches to optimize processing conditions, Trends Food Sci. Technol., 2015, 42, 134–149 CrossRef.
- Y. Li, A. S. Fabiano-Tixier, V. Tomao, G. Cravotto and F. Chemat, Green ultrasound-assisted extraction of carotenoids based on the bio-refinery concept using sunflower oil as an alternative solvent, Ultrason. Sonochem., 2013, 20, 12–18 CrossRef PubMed.
- M. Jacotet-Navarro, N. Rombaut, S. Deslis, A.-S. Fabiano-Tixier, F.-X. Pierre, A. Bily and F. Chemat, Towards a ‘dry’ bio-refinery without solvents or added water using microwaves and ultrasound for total valorization of fruit and vegetable by-products, Green Chem., 2016, 18, 3106–3115 RSC.
- G. S. Marques, W. F. Leão, M. A. M. Lyra, M. S. Peixoto, R. P. M. Monteiro, L. A. Rolim, H. S. Xavier, P. J. R. Neto and L. A. L. Soares, Comparative evaluation of UV/VIS and HPLC analytical methodologies applied for quantification of flavonoids from leaves of Bauhinia forficata, Braz. J. Pharmacog., 2013, 23, 51–57 CrossRef.
- M. Popova, V. Bankova, D. Butovska, V. Petkov, B. Nikolova-Damyanova, A. G. Sabatini, G. L. Marcazzan and S. Bogdanov, Validated methods for the quantification of biologically active constituents of poplar-type propolis, Phytochem. Anal., 2004, 15, 235–340 CrossRef PubMed.
- M. Hadda, C. Djamel and O. Akila, Production and Qualitative Analysis of Triterpenoids and Steroids of Ganoderma Species Harvested from Cork Oak Forest of North-Eastern Algeria, Res. J. Microbiol., 2015, 10, 366–376 CrossRef.
- M. Yang, X. Wang, S. Guan, J. Xia, J. Sun, H. Guo and D. An Guo, Analysis of Triterpenoids in Ganoderma lucidum Using Liquid Chromatography Coupled with Electrospray Ionization Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2007, 18, 927–939 CrossRef CAS PubMed.
- X.-Y. Guo, J. Han, M. Ye, X.-C. Ma, X. Shen, B.-B. Xue and Q.-M. Che, Identification of major compounds in rat bile after oral administration of total triterpenoids of Ganoderma lucidum by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry, J. Pharm. Biomed. Anal., 2012, 63, 29–39 CrossRef CAS PubMed.
- G. Joana Gil-Chávez, J. A. Villa, J. Fernando Ayala-Zavala, J. Basilio Heredia, D. Sepulveda, E. M. Yahia and G. A. González-Aguilar, Technologies for Extraction and Production of Bioactive Compounds to be Used as Nutraceuticals and Food Ingredients: An Overview, Compr. Rev. Food Sci. Food Saf., 2013, 12, 5–23 CrossRef.
- N. N. Azwanida, A Review on the Extraction Methods Use in Medicinal Plants, Principle, Strength and Limitation, Med. Aromat. Plants, 2015, 4, 3–8 Search PubMed.
- J. Azmir, I. S. M. Zaidul, M. M. Rahman, K. M. Sharif, A. Mohamed, F. Sahena, M. H. A. Jahurul, K. Ghafoor, N. A. N. Norulaini and A. K. M. Omar, Techniques for extraction of bioactive compounds from plant materials: A review, J. Food Eng., 2013, 117, 426–436 CrossRef CAS.
- M. S. Lin, Z. R. Yu and Y. M. Weng, Study of Continuous Extraction Process Utilizing Supercritical Fluid for Ganoderma lucidum, Adv. Mater. Res., 2012, 524–527, 2310–2315 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7fo01601h |
| This journal is © The Royal Society of Chemistry 2018 |
