Quercetin induces the selective uptake of HDL-cholesterol via promoting SR-BI expression and the activation of the PPARγ/LXRα pathway†
Kun
Ren
 ab,
Ting
Jiang
a and
Guo-Jun
Zhao
*a
ab,
Ting
Jiang
a and
Guo-Jun
Zhao
*a
aDepartment of Histology and Embryology, Guilin Medical University, Guilin, Guangxi 541004, China. E-mail: zzhcsu@163.com; Tel: +86 -773-3680365
bInstitute of Cardiovascular Research, Key Laboratory for Atherosclerology of Hunan Province, University of South China, Hengyang, 421001, China
First published on 8th December 2017
Abstract
Reverse cholesterol transport (RCT) is the process to deliver cholesterol to the liver for further excretion and involves scavenger receptor class B type I (SR-BI)-mediated selective lipid uptake (SLU) from high-density lipoprotein cholesterol (HDL-C). The up-regulation of hepatic SR-BI expression accelerates HDL-C clearance in circulation and impedes the development of atherosclerosis (AS). In the present study, we explored the modulation of hepatic SR-BI expression and SR-BI-mediated SLU by quercetin, a natural flavonoid compound in the diet with a favorable role in cardiovascular disorders. We found that quercetin significantly increased the expression level of SR-BI in HepG2 cells in a concentration- and time-dependent manner. Besides, quercetin had stimulatory effects on the binding of 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Dil)-labeled HDL to hepatocytes and 125I/3H-CE-HDL association. Treatment with small interfering RNA (siRNA) or SR-BI specific inhibitor, BLT-1, inhibited quercetin-induced Dil-HDL binding and selective HDL-C uptake. Treatment with quercetin increased both proliferator-activated receptor γ (PPARγ) and liver X receptor α (LXRα) levels. Additionally, the quercetin-induced expression of SR-BI, Dil-HDL binding and the selective uptake of HDL-C were significantly attenuated by treatment with PPARγ siRNA, LXRα siRNA, and their antagonists, respectively. In C57BL/6 mice, quercetin administration potently increased SR-BI, PPARγ and LXRα levels and lipid accumulation in the liver. Altogether, our results suggest that quercetin-induced up-regulation of SR-BI and subsequent lipid uptake in hepatocytes might contribute to its beneficial effects on cholesterol homeostasis and atherogenesis.
1. Introduction
Atherosclerosis (AS) is the fundamental pathology of cardiovascular disease (CVD),1 one of the most common causes of mortality around the world, especially in advanced countries. It is characterized by the accumulation of lipoprotein cholesterol and the formation of macrophage-derived foam cells within the blood vessel wall. In the bloodstream, high-density lipoprotein (HDL) serves as a vehicle for lipid transport. Reverse cholesterol transport (RCT) is the process responsible for cholesterol delivery from peripheral cells and tissues to the liver for secretion, representing a key mechanism by which HDL exerts anti-atherogenic properties. RCT is initialized by ATP-binding cassette transporter A1 (ABCA1)-mediated removal of excess cholesterol from peripheral cells to lipid-free apolipoprotein A-I (apoA-I), resulting in nascent preβ-HDL formation.2,3 Consequently, mature α-HDL conveys its cholesterol load to hepatocytes for further excretion into bile and feces.4 Scavenger receptor class B type I (SR-BI) is a physiological high-affinity receptor for HDL.5 It mediates the uptake of cholesteryl esters (CE) from HDL-C to hepatocytes, a process termed selective lipid uptake (SLU), thus playing a crucial role in the later stage of RCT.6 Studies with mice showed that hepatic SR-BI over-expression results in decreased levels of total cholesterol (TC), HDL cholesterol (HDL-C) as well as free cholesterol/TC (FC/TC) ratio in the plasma, ameliorating atherogenesis. In sharp contrast, down-regulation of hepatic SR-BI leads to elevated plasma cholesterol levels and aggravation of AS.7 Most importantly, other research has identified that humans who carry the loss-of-function variants of SCARB1 (a gene for coding human SR-BI) presented relatively higher plasma HDL-C levels and increased risk of CVD.8,9 These observations demonstrated a critical role for hepatic SR-BI in rodent and human cholesterol metabolism.Quercetin is one of the widely distributed flavonoids in the plant kingdom. It exists richly and extensively in human diet, including fruits, vegetables, tea, red wine and diverse food supplements.10 It is estimated that quercetin and its relevant derivatives account for nearly 60%–70% of flavonoids intake in human daily life.11 Recently, the medicinal properties of quercetin have captured much attention for its powerful biological activities, including anti-inflammation, anti-oxidation, anti-virus and anti-cancer.12–15 Besides, studies showed that quercetin could also protect against AS by reducing low-density lipoprotein (LDL) oxidation, lowering blood pressure, enhancing blood capillary elasticity and reversing endothelial dysfunction.16–18 Although this phytomedicine can exert many anti-atherogenic activities, the underlying mechanisms have not been fully unraveled. This study was designed to evaluate the effects of quercetin on hepatic SR-BI expression and SR-BI-mediated SLU from HDL-C, as well as its potential mechanisms. Considering that proliferator-activated receptor γ (PPARγ)/liver X receptor α (LXRα) pathway is involved in the modulation of SR-BI expression and cholesterol homeostasis,19–23 we hypothesized that quercetin might impede the atherogenic process by enhancing hepatic SR-BI-mediated selective HDL-C uptake in a PPARγ/LXRα pathway-dependent manner.
2. Materials and methods
2.1 Cell culture
Human HepG2 cell line was bought from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in 6-well plates under standard culture conditions (5% CO2, 37 °C). Before cells were used for the experiments, they were grown in DMEM-high glucose without FBS and antibiotics overnight to synchronize cell growth.2.2 Western blot analysis
After indicated treatments, cells were harvested and total proteins were extracted for the detection of SR-BI, PPARγ and LXRα, as described previously.24 A BCA protein assay kit (CWBIO, Peking, China) was used to measure protein concentrations. Approximately 20 μg proteins (per lane) were loaded on a 10% SDS-polyacrylamide electrophoresis gel (SDS-PAGE, Solarbio Co, Peking, China). After electrophoresed and separated at 120 V for 1.5 h in gel running buffer, proteins were transferred to 0.45 μm polyvinylidene fluoride membranes (PVDF, Merck Millipore, Darmstadt, Germany). The success of protein transfer was judged using Coomassie Brilliant Blue staining (CWBIO, Peking, China). Subsequently, membranes were blocked using 5% fat-free milk in Tween-20 Tris-buffered saline (TBS-T) at least 4 h at room temperature and then immunoblotted overnight at 4 °C with a primary antibody (rabbit anti-human; diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) against SR-BI, PPARγ, LXRα and β-actin (Santa Cruz, CA, USA) at 4 °C with soft shaking. After being rinsed three times in TBS-T (10 min each), membranes were further incubated with a goat anti-rabbit peroxidase-conjugated secondary antibody (diluted 1
1000) against SR-BI, PPARγ, LXRα and β-actin (Santa Cruz, CA, USA) at 4 °C with soft shaking. After being rinsed three times in TBS-T (10 min each), membranes were further incubated with a goat anti-rabbit peroxidase-conjugated secondary antibody (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000) in 5% TBS-T for 2 h at room temperature, followed by another series of rinses in TBS-T. Finally, the β-actin, SR-BI, PPARγ and LXRα proteins were visualized by enhanced chemiluminescence (ECL; Merck Millipore, Darmstadt, Germany), and their concentrations were assessed using LabWorks image analysis software. The β-Actin level was used as an internal control.
4000) in 5% TBS-T for 2 h at room temperature, followed by another series of rinses in TBS-T. Finally, the β-actin, SR-BI, PPARγ and LXRα proteins were visualized by enhanced chemiluminescence (ECL; Merck Millipore, Darmstadt, Germany), and their concentrations were assessed using LabWorks image analysis software. The β-Actin level was used as an internal control.
2.3 Total RNA extraction and quantitative real-time PCR (qPCR) analysis
HepG2 cells (1 × 106 cells per well) were cultured in DMEM supplemented with 10% FBS with or without the indicated treatments. Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Then, approximately 2 μg of cDNA for each sample was obtained by the reverse transcription of RNA (1 μg) using a TaqMan Reverse Transcription Reagents Kit (Applied Biosystems). Afterwards, PCR was conducted on a real-time qPCR machine (Real-Time PCR System, Bio-Rad, USA) using a 2 × Taq PCR master mix (Sangon Biotech, Shanghai, China) and specific primers for SR-BI, PPARγ, LXRα and GADPH (see below). The PCR protocols were as follows: denaturation at 92 °C for 3 min, followed by amplification at 94 °C for 28 s, 55 °C for 30 s, 75 °C for 48 s, and 70 °C for 9 min for 42 cycles. The primers were designed using Primer Premier Six software 6.0 (Applied Biosystems, Foster City, CA, USA). The GADPH level was used as an internal control. The sequences of the primers used were as shown below: SR-BI forward, 5′-GAGAGGCTCGTCAACAAG-3′; SR-BI reverse, 5′-GTCCATAGGATGATGTCAGTT-3′; LXRα sense, 5′-ACACCTACATGCGTCGCAAG-3′ and anti-sense, 5′-GACGAGCTTCTCGATCATGCC-3′; PPARγ sense, 5′-TGGAATTAGATGACAGCGACTTGG-3′ and anti-sense, 5′-CTGGAGCAGCTTGGCAAACA-3′; GADPH forward, 5′-AGTGAGATTGCTTCTTACCT-3′; and reverse, 5′-TCCATTCTTCCTGCTCCT-3′. Melt curve analysis was used to assess all qPCR products, in which DNA duplexes were identified. The measurements were evaluated using ΔΔCt method with software in the qPCR apparatus (SDS 2.3).252.4 Cell transfection
HepG2 cells, when reached approximately 80% confluence, were transfected with small-interfering RNA (siRNA) specific for human SR-BI (80 nM, Santa Cruz Biotechnology, sense, 5′-GGACAAGUUCGGAUUAUUUdTdT-3′; and anti-sense, 5′-AAAUAAUCCGAACUUGUCCTdTd-3′), LXRα (50 nM, Santa Cruz Biotechnology, sense, 5′-GGCUGCAAGUGGAAUUCAUTT-3′; anti-sense, 5′-AUGAAUUCCACUUGCAGCCTT-3′) and PPARγ (100 nM, Santa Cruz Biotechnology, sense, 5′-GGAUGCAAGGGUUUCUUCCTT-3′; anti-sense, 5′-GGAAGAAACCCUUGCAUCCTT-3′) using lipofectamine 2000 (Invitrogen) according to the instructions of manufacturers. After transfection for 48 h, cells were harvested for western blot analysis. Western blot analysis was used to evaluate the silencing efficiency.2.5 HDL labeling procedure
The apolipoprotein constituent of human HDL (Yiyuan Biotechnologies, Guangzhou, China) was radiolabeled with sodium 125iodide (125I; Hartmann Analytic, Germany) using the Pierce IODO-BEADS test kit (ThermoScientific, Rockford, IL, USA), and then purified from unincorporated label by gel filtration. Subsequently, 125I-HDL was further labeled in its lipid moiety (125I/3H-CE-HDL) with the following procedures: approximately 50 μCi [cholesteryl-1,2–3H(N)]-oleate (PerkinElmer, Waltham, MA, USA) were evaporated in a small glass tube and then resuspended in DMSO. After incubation in rocking water for 1 h, 125I-HDL (1 mg per 500 μl PBS) was added and then purified, as described above.262.6 SLU assay with radiolabeled HDL
After HDL was double-radiolabeled, SLU was assessed as described previously.26 Briefly, cells were treated with 10 μg ml−1 125I-HDL or 125I/3H-CE-HDL (∼585 cpm ng−1 for 125I and ∼785 cpm ng−1 for 3H-CE) for 2 h. A 50-fold excess of unlabeled HDL was added to culture medium every fourth data point. Cells were rinsed several times in the mixed solution of cold 0.9% NaCl, Tris HCl (pH ≈ 7.4), and without or with 0.2% bovine serum albumin (BSA), and then lysed using 0.1 M NaOH. Cell protein concentration was determined using a BCA protein assay kit (CWBIO, Peking, China). The radioactivity of 125I-HDL and 125I/3H-CE-HDL was determined utilizing a γ-ray counter and a β-ray counter, respectively. Cell association was assessed by deducting the intensity of radioactivity detected in the 50-fold excess from the total activity, and HDL uptake was presented as ng HDL per mg cell protein.2.7 Dil-HDL uptake assay
Dil-HDL binding assay was performed to further evaluate cholesterol uptake induced by quercetin. After pre-treatment with 15 μM quercetin (Cayman Chemicals, AnnArbor, MI) for 24 h, cells were incubated with 10 μg ml−1 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (Dil)-labeled HDL (Yiyuan Biotechnologies, Guangzhou, China) for an additional 4 h at 37 °C. Adherent cells were then collected and rinsed three times with PBS. Analysis was conducted on a FACScalibur Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ, USA) with Cell Quest Pro software (Becton Dickinson Biosciences).2.8 Mice and treatments
C57BL/6 mice (male, six-week old) from Shanghai Laboratory Animal Center, Chinese Academy of Sciences were maintained on a 12 h light/dark cycle. Animals were randomly divided into two groups (n = 8 per group), housed at a constant temperature (24 ± 2 °C) with 60% humidity and fed a chow diet. Mice in the quercetin group were gavaged with quercetin (50 mg kg−1, dissolved in 50% propylene glycol) every day for 8 weeks. The control group was orally administered with an equal bulk of 50% propylene glycol as a vehicle. Then, mice were euthanized, and the livers were isolated and prepared as described previously.27 SR-BI, LXRα and PPARγ protein levels in the liver homogenate were measured using western blot analysis. Hepatic lipid accumulation was evaluated using Oil Red O staining and hematoxylin–eosin (HE) staining.The animal studies were performed according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication no. 85–23, revised in 1996) and Care and Use guidelines for experimental animals of Guilin Medical University. The investigation procedure was approved by the Animal Ethics Committee of Guilin Medical University. To minimize sufferings, sodium pentobarbital anesthesia was performed prior to all surgeries.
2.9 Statistical analysis
All results are expressed as the mean ± standard deviation (SD) obtained from at least three independent experiments. Values were processed by one-way ANOVA, followed by the Student–Newman–Keuls (SNK) post hoc multiple comparison tests using Graphpad Prism 6.0 software. A p value of less than 0.05 was considered statistically significant.3. Results
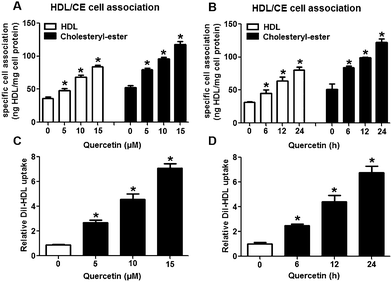
3.1 Effects of quercetin on SLU and Dil-HDL binding to HepG2 cells
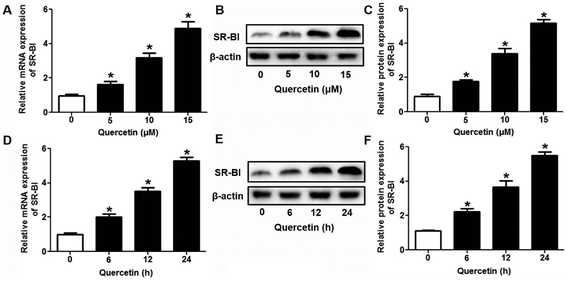
The selective uptake of HDL-C into hepatocytes and steroidogenic cells is the last step of RCT, a process that promotes HDL-C clearance in the plasma. Low clearance of plasma HDL-C aggravates the development of AS while high clearance of plasma HDL-C impedes atherogenesis.28,29 Firstly, we explored whether quercetin influences SLU and the binding of Dil-HDL to HepG2 cells using the method of double-radiolabeled HDL particles (125I/3H-CE-HDL) and flow cytometry, respectively. HepG2 cells (1 × 106 cells per well) were incubated with different concentrations of quercetin (0, 5, 10, 15 μM, respectively) for 24 h. The treatment of cells with quercetin resulted in increases in both the HDL holo-particle association (as measured by the 125I activity) and CE association (as determined by the 3H activity) as well as Dil-HDL binding. HDL, CE association and Dil-HDL binding to HepG2 cells were gradually enhanced with increasing quercetin concentrations (Fig. 1A and C). Subsequently, cells were pre-treated with 15 μM quercetin for different times (0, 6, 12, 24 h, respectively) to evaluate whether quercetin promotes SLU and Dil-HDL binding in a time-dependent fashion. The results showed that 6 h incubation with quercetin remarkably increased HDL and CE association as well as Dil-HDL binding (Fig. 1B and D). These outcomes suggest that quercetin promotes the selective uptake of HDL-C in HepG2 cells in both concentration- and time-dependent patterns.3.2 The role of SR-BI in the effects of quercetin on SLU and Dil-HDL binding to HepG2 cells
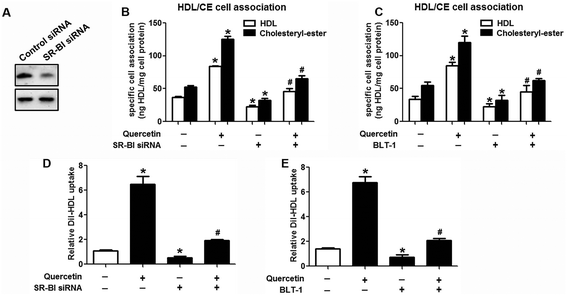
Research has identified that hepatic SLU from HDL-C is largely mediated by the HDL physiological receptor, SR-BI. To determine whether SR-BI is involved in the effects of quercetin on SLU and Dil-HDL binding, cells (1 × 106 cells per well) were pre-treated with different concentrations of quercetin (0, 5, 10, 15 μM, respectively) for 24 h or 15 μM quercetin for different durations (0, 6, 12, 24 h, respectively), and then SR-BI mRNA and protein levels were measured using qPCR and western blot analyses, respectively. The results showed that quercetin potently amplified the expression of SR-BI at both the mRNA and protein levels in a concentration- and time-dependent manner (Fig. 2). Next, cells were transfected with SR-BI siRNA for 48 h or pre-treated with 10 μM BLT-1 (SR-BI specific inhibitor) for 2 h, followed by the addition of quercetin into the media. Data showed that pre-incubation with SR-BI siRNA or BLT-1 significantly attenuated the positive effects of quercetin on SLU and Dil-HDL binding to HepG2 cells (Fig. 3). These results suggested that enhancement of SLU and Dil-HDL binding to HepG2 cells by quercetin is mediated by SR-BI.3.3 The role of PPARγ/LXRα signaling in the effects of quercetin on SR-BI expression, SLU and Dil-HDL binding to HepG2 cells
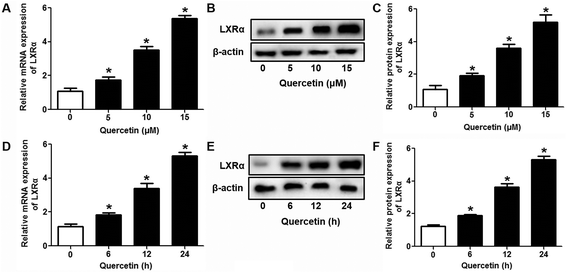
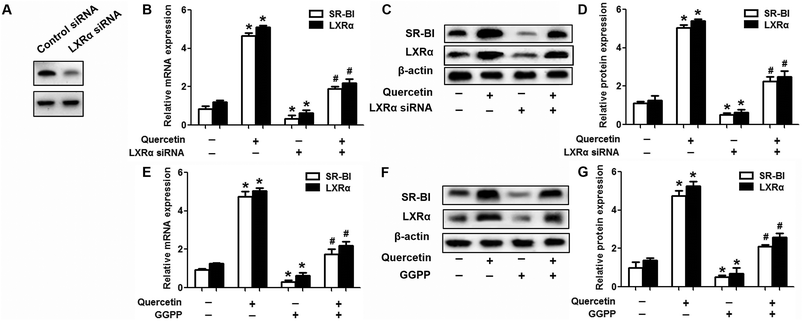
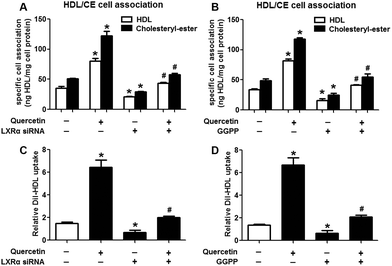
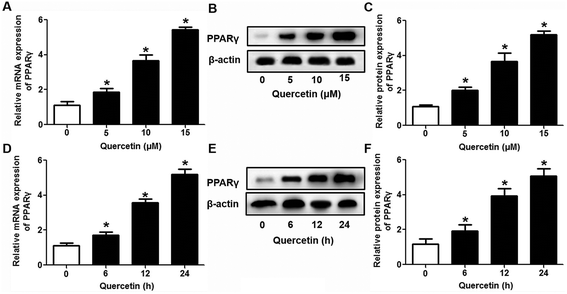
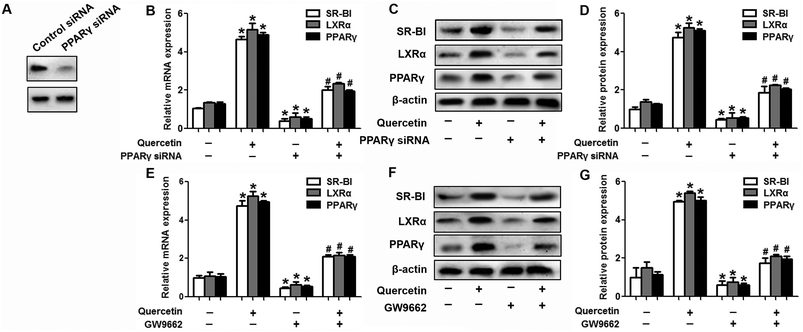
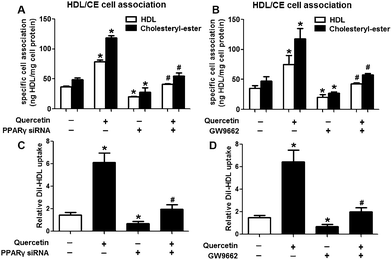
Evidence has shown that LXRs can modulate SR-BI expression, a process that controls lipid and cholesterol homeostasis in hepatocytes and macrophages.22,30,31 LXRα is abundantly expressed in liver and is involved in regulating the expression of several genes responsible for HDL metabolism. LXRα-induced signaling can be modulated by one of its common upstream factors, PPARγ.19,32,33 Next, we investigated whether the PPARγ/LXRα pathway participates in the effects of quercetin on hepatic SR-BI expression and SR-BI-mediated selective HDL-C uptake. The results showed that quercetin significantly enhanced the mRNA and protein levels of LXRα in HepG2 cells in a concentration- and time-dependent fashion (Fig. 4). Thereafter, cells were transfected with LXRα siRNA for 48 h, followed by adding quercetin. As shown in Fig. 5A–D, transfection with LXRα siRNA reduced the mRNA and protein levels of LXRα and SR-BI, and almost abolished the amplification of quercetin on LXRα and SR-BI levels. At the same time, SR-BI-mediated SLU and Dil-HDL binding were remarkably down-regulated in cells treated with both LXRα siRNA and quercetin when compared with those treated with quercetin alone (Fig. 6A and C). Thereafter, we explored the effects of LXRα-specific inhibitor GGPP (Sigma-Aldrich, CA, USA) on the enhancement of SR-BI expression. Cells were pre-incubated with 10 μM GGPP for 2 h and subsequently treated with 15 μM quercetin for another 24 h. As indicated in Fig. 5E, the treatment of GGPP decreased LXRα and SR-BI mRNA levels when compared with the control group. Meanwhile, the enhanced SR-BI and LXRα mRNA levels induced by quercetin were potently down-regulated by the addition of GGPP. Similar observations were obtained for the LXRα and SR-BI protein levels as detected using western blot analysis (Fig. 5F and G). Moreover, GGPP also inhibited the quercetin-induced SLU and Dil-HDL binding to HepG2 cells (Fig. 6B and D).Next, we assessed the effects of PPARγ siRNA and its inhibitor GW9662 (Santa Cruz Biotechnology, Inc. CA, USA) on LXRα and SR-BI expression, selective HDL-C uptake as well as Dil-HDL binding. HepG2 cells were pre-treated with PPARγ siRNA for 48 h or 10 μM GW9662 for 2 h, followed by the incubation of quercetin for 24 h. As shown in Fig. 7, quercetin can likewise enhance the expression of PPARγ at both mRNA and protein levels in a concentration- and time-dependent manner. In addition, the transfection of HepG2 cells with PPARγ siRNA potently reduced the mRNA and protein levels of PPARγ, LXRα and SR-BI, and almost abolished the effects of quercetin on the expression of PPARγ, LXRα and SR-BI (Fig. 8A–D). In line with these results, the promotion of SLU and Dil-HDL binding by quercetin was abolished by PPARγ siRNA incubation (Fig. 9A and C). Similarly, the treatment of GW9662 and quercetin also markedly down-regulated the expressions of PPARγ, LXRα and SR-BI (Fig. 8E–G), and selective HDL-C uptake as well as Dil-HDL binding compared with the groups treated with quercetin alone (Fig. 9B and D). These outcomes suggested that PPARγ/LXRα signaling is involved in the positive effects of quercetin on SR-BI expression, SR-BI-mediated SLU and Dil-HDL binding to HepG2 cells. As for other target genes of the PPARγ/LXRα pathway associated with lipid and fat metabolism, we found that quercetin significantly promoted the expression of ABCA1/G1 and sterol regulatory element binding transcription factor-1c (SREBP-1c), while poorly activating steroyl CoA desaturase-1 (SCD-1) or fatty acid synthase (FASN) (Fig. S1†).
3.4 Effects of quercetin on SR-BI, PPARγ and LXRα levels as well as lipid accumulation in the liver of C57BL/6 mice
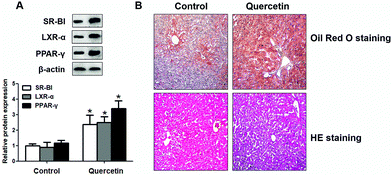
To confirm the effects of quercetin in liver, we detected whether quercetin could regulate SR-BI, PPARγ and LXRα expressions in vivo. As shown in Fig. 10A, quercetin markedly increased SR-BI, PPARγ and LXRα protein levels in liver homogenate compared with the counterpart group. Lastly, we evaluated effects of quercetin on hepatic lipid accumulation in vivo and found that quercetin administration remarkably increased the amounts of lipid droplets in mouse liver tissue (Fig. 10B). These observations are consistent with our in vitro results that quercetin can enhance hepatic SR-BI expression and lipid uptake.4. Discussion
The central role of HDL in athero-protection is to facilitate RCT, a critical process to prevent the onset and development of AS.34,35 The initial stage of AS is featured by the formation and deposition of macrophage-derived foam cells within vessel walls due to excessive cholesterol accumulation. ABCA1-mediated cholesterol efflux from lipid-laden macrophages is the first step of RCT to transfer cellular FC to apoA-I, resulting in nascent HDL formation. Finally, circular spherical HDL delivers its cholesterol cargo to the liver for further metabolism and secretion. This process is dependent on SR-BI-mediated selective HDL-C uptake. Accordingly, hepatic SR-BI is critical for plasma HDL-C clearance.6,36,37 Additionally, increasing studies have suggested that quercetin administration correlates with some anti-atherogenic properties.38,39 Our study showed that quercetin can up-regulate hepatic ABCA1/G1 and SREBP-1c expressions, which were in line with the previous observations that the macrophage ABCA1/G1 level and RCT can be magnified by quercetin.40 However, expression levels of lipogenic genes including SCD-1 and FASN were not significantly increased by quercetin treatment, indicating that the regulating effects of quercetin on cholesterol metabolism are stronger than that of fat metabolism in hepatocytes. Importantly, we extend the proof that quercetin may increase hepatic SR-BI expression and SR-BI-mediated SLU through activation of the PPARγ/LXRα signaling pathway.Epidemiologic research has revealed a reverse correlation between flavonoid uptake and cardiovascular risks.41,42 It has been demonstrated that quercetin, the most abundant flavonoid in human daily diet, exerts many beneficial effects on the development of CVD and obesity-related disorders.43,44 Hitherto, many studies have concentrated on its properties of anti-inflammation, anti-oxidation, anti-apoptosis as well as anti-proliferation.17,45,46 Boots et al.45 showed that quercetin is effective in eliminating reactive oxygen species (ROS) and nitric oxide (NO), thus protecting against aging. Quercetin can repress the activity of inducible NO synthase and reduce the injury of experimental ischemia-reperfusion and production of damaging peroxynitrite from superoxide anions and NO.45 Besides, quercetin decreases the level of LDL oxidation through interference with neutrophil myeloperoxidase activity, therefore potentially ameliorating the risks of heart diseases. In hypertensive rats, quercetin administration significantly lowered blood pressure (BP) and improved endothelial dysfunction by suppressing endothelin-1 (ET-1)-induced enhancement of protein kinase C (PKC) activity.47 Moreover, quercetin can down-regulate the expression of some pro-inflammatory factors, activation of NF-κB signaling as well as accumulation of lipid rafts in macrophages, suggesting its capacity to attenuate inflammatory processes. In addition to these beneficial effects related to the cardiovascular system, quercetin has been found to be associated with atherogenesis through conferring other biological activities. Juzwiak et al.17 found that the supplementation of quercetin can significantly reduce plasma TC, LDL cholesterol (LDL-C) and very low density lipoprotein cholesterol (VLDL-C) levels in rabbits fed with a high-fat diet, thus preventing AS development. Chang et al.48 observed that quercetin can potently enhance the ABCA1-mediated cholesterol efflux from lipid-loading macrophages to apoA-I, preventing foam cell formation. Taken together, these results have demonstrated that quercetin has the hypolipemic and anti-atherogenic properties. To the best of our knowledge, the effects of quercetin on SR-BI-mediated selective HDL-C uptake and its potential mechanisms were not previously investigated. Here, our studies demonstrated that quercetin-treated hepatocytes displayed a potent increase in SLU and HDL binding mediated by SR-BI. The amplification in SR-BI-mediated SLU and HDL association is consistent with an increase of SR-BI at both the transcriptional and translational levels in response to incubation with quercetin in HepG2 cells. These findings elucidate the notion that quercetin could enhance SLU from HDL-C in hepatocytes. In C57BL/6 mice, Oil Red O and HE staining shows that quercetin administration significantly enhanced SR-BI expression and liver lipid accumulation. As neutral lipids, such as triglycerides, were also stained with Oil Red O, whether quercetin-induced hepatic lipid accumulation in vivo is the result of SR-BI upregulation is still uncertain. Besides, other studies identified a decrease in lipid accumulation in mouse liver upon dietary quercetin administration.49,50 We speculated that the difference in feeding conditions (high fat diet vs. chow diet) may account for this discrepancy. However, more data are still needed to clarify the mechanisms responsible for the effects of quercetin in vivo.
Evidence has shown that treatment with both PPARγ agonist rosiglitazone and LXRα agonist GW3965 can substantially increase SR-BI expression.51 However, transfection with PPARγ siRNA or LXRα siRNA decreased the SR-BI level.23 We therefore explored the effects of quercetin on PPARγ/LXRα signaling to ascertain whether the activation levels changed in the process of quercetin-induced SR-BI up-regulation. Quercetin was found to remarkably increase the expression of PPARγ and LXRα at both mRNA and protein levels in a concentration- and time-dependent manner. Subsequent studies using PPARγ/LXRα siRNA and their specific inhibitors further confirmed that the PPARγ/LXRα pathway was implicated in SR-BI magnification induced by quercetin. The quercetin-induced up-regulation of SR-BI mRNA and protein was almost abolished by PPARγ/LXRα siRNA transfection or pre-treatment with their inhibitors. At the same time, quercetin-induced increase in SLU and Dil-HDL binding was also fully ruled out by PPARγ/LXRα siRNA or their inhibitors. These results suggest that the PPARγ/LXRα pathway is implicated in SR-BI up-regulation and SR-BI-mediated selective uptake of HDL-C induced by quercetin in HepG2 cells.
Interestingly, when the PPARγ/LXRα pathway was inhibited, quercetin still showed a positive effect on SR-BI expression, which indicates that there exist other ways involved in quercetin-induced SR-BI up-regulation. SR-BI expression can be positively regulated by SREBPs.52 As shown, quercetin can significantly enhance the SREBP-1c level in the liver, which may contribute to its amplification in SR-BI expression. Moreover, Wang et al.53 reported that compared to its counterpart group, quercetin-administered mice exhibited lower levels of pro-inflammatory cytokines in liver tissue samples, including IL-1β, IL-6 and TNF-α. Since SR-BI expression can be negatively regulated by LPS, IL-1β and TNF-α,54 quercetin may enhance the SR-BI level through its anti-inflammatory effects. Notably, another study by Hu et al.55 observed that the transfection of HepG2 cells with Na+/H+ exchanger regulatory factors 1/2 (NHERF1/2) cDNA significantly decreased the SR-BI protein level by specifically interacting with SR-BI, suggesting the critical role of NHERF1/2 in SR-BI regulation. Whether these possible mechanisms are implicated in quercetin-induced SR-BI up-regulation still warrants further investigation.
Furthermore, we also noticed that quercetin can exert apoptotic effects on HepG2 cells in a dose-dependent manner,56,57 which may interfere with our conclusions. However, this effect only becomes significant when quercetin concentrations are over 25 μM.57 Treatment with 40 μM quercetin for 24 h can induce 19.1% HepG2 cell apoptosis.56 In the present study, the maximum dose of quercetin used is 15 μM. Thus, we believe that these doses are not apoptosis-inducing obviously, and the interference is limited.
Many in vivo studies have revealed that the oral administration of quercetin is non-carcinogenic and non-genotoxic in rats.58 Particularly in humans, quercetin can also be safely used for food additives.59 Furthermore, quercetin can promote mental and physical performance of human subjects,60 probably through improving exercise-induced inflammation and oxidative damage. Other studies have identified some advantageous effects of quercetin on the progression of tumors, including prostate and breast cancers.61 These pieces of evidence revealed the feasibility of therapeutic application of this flavonoid compound in diverse unfavorable circumstances. In the present study, we emphasized the missing connection between quercetin treatment and increased SLU by providing proof that quercetin incubation positively correlated with SR-BI expression, which likely results from quercetin-induced increase in PPARγ/LXRα expression. Therefore, this research offers a potential explanation for the underlying mechanisms by which quercetin promotes SLU from HDL-C in hepatocytes. Since the natural high-affinity receptor for HDL, SR-BI, and nuclear receptors PPARγ/LXRα are the key factors to modulate lipid and cholesterol homeostasis, this phytomedicine might be considered as a promising therapeutic agent for the prevention and treatment of AS by promoting the activity of this cholesterol-uptake pathway.
5. Conclusion
In conclusion, our studies have revealed that quercetin enhances SR-BI expression by stimulating the PPARγ/LXRα pathway in HepG2 cells, and promotes SR-BI-mediated SLU as well as HDL binding, suggesting a novel mechanism underlying its protection against AS.Conflicts of interest
The authors declared no conflicts of interest in this paper.Acknowledgements
This work is financially sponsored by the National Natural Sciences Foundation of China (81660082), the Guangxi Natural Sciences Foundation (AA139209), the Autonomous region College Students’ innovation and entrepreneurship training program (201610601047), the Hunan Provincial Innovation Foundation For Postgraduate (CX2017B503), the Hunan Provincial Natural Science Foundation of China (14JJ2091), and the Zhengxiang scholar program (prof. Xiangyang Tang) of the University of South China.References
- C. Weber and H. Noels, Atherosclerosis: current pathogenesis and therapeutic options, Nat. Med., 2011, 17, 1410–1422 CrossRef CAS PubMed.
- Z. C. Mo, K. Ren, X. Liu, Z. L. Tang and G. H. Yi, A high-density lipoprotein-mediated drug delivery system, Adv. Drug Delivery Rev., 2016, 106, 132–147 CrossRef CAS PubMed.
- B. Shao, C. Tang, A. Sinha, P. S. Mayer, G. D. Davenport, N. Brot, M. N. Oda, X. Q. Zhao and J. W. Heinecke, Humans with atherosclerosis have impaired ABCA1 cholesterol efflux and enhanced high-density lipoprotein oxidation by myeloperoxidase, Circ. Res., 2014, 114, 1733–1742 CrossRef CAS PubMed.
- D. J. Rader and A. Daugherty, Translating molecular discoveries into new therapies for atherosclerosis, Nature, 2008, 451, 904–913 CrossRef CAS PubMed.
- T. Eberhart, K. Eigner, Y. Filik, S. Fruhwurth, H. Stangl and C. Rohrl, The unfolded protein response is a negative regulator of scavenger receptor class B, type I (SR-BI) expression, Biochem. Biophys. Res. Commun., 2016, 479, 557–562 CrossRef CAS PubMed.
- S. Acton, A. Rigotti, K. T. Landschulz, S. Xu, H. H. Hobbs and M. Krieger, Identification of scavenger receptor SR-BI as a high density lipoprotein receptor, Science, 1996, 271, 518–520 CAS.
- T. Huby, C. Doucet, C. Dachet, B. Ouzilleau, Y. Ueda, V. Afzal, E. Rubin, M. J. Chapman and P. Lesnik, Knockdown expression and hepatic deficiency reveal an atheroprotective role for SR-BI in liver and peripheral tissues, J. Clin. Invest., 2006, 116, 2767–2776 CAS.
- L. R. Brunham, I. Tietjen, A. E. Bochem, R. R. Singaraja, P. L. Franchini, C. Radomski, M. Mattice, A. Legendre, G. K. Hovingh, J. J. Kastelein and M. R. Hayden, Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans, Clin. Genet., 2011, 79, 575–581 CrossRef CAS PubMed.
- M. Vergeer, S. J. Korporaal, R. Franssen, I. Meurs, R. Out, G. K. Hovingh, M. Hoekstra, J. A. Sierts, G. M. Dallinga-Thie, M. M. Motazacker, A. G. Holleboom, T. J. Van Berkel, J. J. Kastelein, M. Van Eck and J. A. Kuivenhoven, Genetic variant of the scavenger receptor BI in humans, N. Engl. J. Med., 2011, 364, 136–145 CrossRef CAS PubMed.
- Y. Marunaka, Actions of quercetin, a flavonoid, on ion transporters: its physiological roles, Ann. N. Y. Acad. Sci., 2017, 1398, 142–151 CrossRef CAS PubMed.
- W. Liu, M. Zhang, J. Feng, A. Fan, Y. Zhou and Y. Xu, The Influence of Quercetin on Maternal Immunity, Oxidative Stress, and Inflammation in Mice with Exposure of Fine Particulate Matter during Gestation, Int. J. Environ. Res. Public Health, 2017, 14 Search PubMed , pii: E592.
- C. Nishimura Fde, A. C. de Almeida, B. A. Ratti, T. Ueda-Nakamura, C. V. Nakamura, V. F. Ximenes and O. Silva Sde, Antioxidant effects of quercetin and naringenin are associated with impaired neutrophil microbicidal activity, J. Evidence-Based Complementary Altern. Med., 2013, 2013, 795916 Search PubMed.
- L. Wang, B. Wang, H. Li, H. Lu, F. Qiu, L. Xiong, Y. Xu, G. Wang, X. Liu, H. Wu and H. Jing, Quercetin, a flavonoid with anti-inflammatory activity, suppresses the development of abdominal aortic aneurysms in mice, Eur. J. Pharmacol., 2012, 690, 133–141 CrossRef CAS PubMed.
- W. Wu, R. Li, X. Li, J. He, S. Jiang, S. Liu and J. Yang, Quercetin as an Antiviral Agent Inhibits Influenza A Virus (IAV) Entry, Viruses, 2015, 8 CrossRef PubMed , pii: E6.
- A. Rivera Rivera, L. Castillo-Pichardo, Y. Gerena and S. Dharmawardhane, Anti-Breast Cancer Potential of Quercetin via the Akt/AMPK/Mammalian Target of Rapamycin (mTOR) Signaling Cascade, PLoS One, 2016, 11, e0157251 Search PubMed.
- M. Garelnabi, H. Mahini and T. Wilson, Quercetin intake with exercise modulates lipoprotein metabolism and reduces atherosclerosis plaque formation, J. Int. Soc. Sports Nutr., 2014, 11, 22 CrossRef PubMed.
- S. Juzwiak, J. Wojcicki, K. Mokrzycki, M. Marchlewicz, M. Bialecka, L. Wenda-Rozewicka, B. Gawronska-Szklarz and M. Drozdzik, Effect of quercetin on experimental hyperlipidemia and atherosclerosis in rabbits, Pharmacol. Rep., 2005, 57, 604–609 CAS.
- F. Perez-Vizcaino, J. Duarte and R. Andriantsitohaina, Endothelial function and cardiovascular disease: effects of quercetin and wine polyphenols, Free Radical Res., 2006, 40, 1054–1065 CrossRef CAS PubMed.
- A. Chawla, W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans and P. Tontonoz, A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis, Mol. Cell, 2001, 7, 161–171 CrossRef CAS PubMed.
- A. Majdalawieh and H. S. Ro, PPARgamma1 and LXRalpha face a new regulator of macrophage cholesterol homeostasis and inflammatory responsiveness, AEBP1, Nucl. Recept. Signaling, 2010, 8, e004 Search PubMed.
- Y. W. Hu, X. Ma, J. L. Huang, X. R. Mao, J. Y. Yang, J. Y. Zhao, S. F. Li, Y. R. Qiu, J. Yang, L. Zheng and Q. Wang, Dihydrocapsaicin Attenuates Plaque Formation through a PPARgamma/LXRalpha Pathway in apoE(-/-) Mice Fed a High-Fat/High-Cholesterol Diet, PLoS One, 2013, 8, e66876 CAS.
- Y. W. Hu, Q. Wang, X. Ma, X. X. Li, X. H. Liu, J. Xiao, D. F. Liao, J. Xiang and C. K. Tang, TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor alpha signaling pathway in THP-1 macrophage-derived foam cells, J. Atheroscler. Thromb., 2010, 17, 493–502 CrossRef CAS PubMed.
- X. Ma, S.-F. Li, Z.-S. Qin, J. Ye, Z.-L. Zhao, H.-H. Fang, Z.-W. Yao, M.-N. Gu and Y.-W. Hu, Propofol up-regulates expression of ABCA1, ABCG1, and SR-B1 through the PPARγ/LXRα signaling pathway in THP-1 macrophage-derived foam cells, Cardiovasc. Pathol., 2015, 24, 230–235 CrossRef CAS PubMed.
- K. Ren, Z. C. Mo, X. Liu, Z. L. Tang, Y. Jiang, X. S. Peng, Q. H. Zhang, J. F. Shi and G. H. Yi, TGF-beta Down-regulates Apolipoprotein M Expression through the TAK-1-JNK-c-Jun Pathway in HepG2 Cells, Lipids, 2017, 52, 109–117 CrossRef CAS PubMed.
- K. Ren, Y. J. Lu, Z. C. Mo, X. Liu, Z. L. Tang, Y. Jiang, X. S. Peng, L. Li, Q. H. Zhang and G. H. Yi, ApoA-I/SR-BI modulates S1P/S1PR2-mediated inflammation through the PI3K/Akt signaling pathway in HUVECs, J. Physiol. Biochem., 2017, 73, 287–296 CrossRef CAS PubMed.
- C. Rohrl, K. Eigner, S. Fruhwurth and H. Stangl, Bile acids reduce endocytosis of high-density lipoprotein (HDL) in HepG2 cells, PLoS One, 2014, 9, e102026 Search PubMed.
- Y. I. Kim, S. Hirai, H. Takahashi, T. Goto, C. Ohyane, T. Tsugane, C. Konishi, T. Fujii, S. Inai, Y. Iijima, K. Aoki, D. Shibata, N. Takahashi and T. Kawada, 9-oxo-10(E),12(E)-Octadecadienoic acid derived from tomato is a potent PPAR alpha agonist to decrease triglyceride accumulation in mouse primary hepatocytes, Mol. Nutr. Food Res., 2011, 55, 585–593 CAS.
- N. Wang, T. Arai, Y. Ji, F. Rinninger and A. R. Tall, Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein ApoB, low density lipoprotein ApoB, and high density lipoprotein in transgenic mice, J. Biol. Chem., 1998, 273, 32920–32926 CrossRef CAS PubMed.
- S. Kuhnast, M. Fiocco, J. W. van der Hoorn, H. M. Princen and J. W. Jukema, Innovative pharmaceutical interventions in cardiovascular disease: Focusing on the contribution of non-HDL-C/LDL-C-lowering versus HDL-C-raising: A systematic review and meta-analysis of relevant preclinical studies and clinical trials, Eur. J. Pharmacol., 2015, 763, 48–63 CrossRef PubMed.
- L. Malerod, M. Sporstol, L. K. Juvet, S. A. Mousavi, T. Gjoen, T. Berg, N. Roos and W. Eskild, Bile acids reduce SR-BI expression in hepatocytes by a pathway involving FXR/RXR, SHP, and LRH-1, Biochem. Biophys. Res. Commun., 2005, 336, 1096–1105 CrossRef CAS PubMed.
- X. Ma, S. F. Li, Z. S. Qin, J. Ye, Z. L. Zhao, H. H. Fang, Z. W. Yao, M. N. Gu and Y. W. Hu, Propofol up-regulates expression of ABCA1, ABCG1, and SR-B1 through the PPARgamma/LXRalpha signaling pathway in THP-1 macrophage-derived foam cells, Cardiovasc. Pathol., 2015, 24, 230–235 CrossRef CAS PubMed.
- S. Bastias-Candia, A. N. Garrido, J. M. Zolezzi and N. C. Inestrosa, Recent Advances in Neuroinflammation Therapeutics: PPARs/LXR as Neuroinflammatory Modulators, Curr. Pharm. Des., 2016, 22, 1312–1323 CrossRef CAS PubMed.
- X. Xu, Q. Li, L. Pang, G. Huang, J. Huang, M. Shi, X. Sun and Y. Wang, Arctigenin promotes cholesterol efflux from THP-1 macrophages through PPAR-gamma/LXR-alpha signaling pathway, Biochem. Biophys. Res. Commun., 2013, 441, 321–326 CrossRef CAS PubMed.
- S. Ganjali, A. A. Momtazi, M. Banach, P. T. Kovanen, E. A. Stein and A. Sahebkar, HDL abnormalities in familial hypercholesterolemia: Focus on biological functions, Prog. Lipid Res., 2017, 67, 16–26 CrossRef CAS PubMed.
- W. Marz, M. E. Kleber, H. Scharnagl, T. Speer, S. Zewinger, A. Ritsch, K. G. Parhofer, A. von Eckardstein, U. Landmesser and U. Laufs, HDL cholesterol: reappraisal of its clinical relevance, Clin. Res. Cardiol., 2017, 106, 663–675 CrossRef PubMed.
- D. Calvo and M. A. Vega, Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family, J. Biol. Chem., 1993, 268, 18929–18935 CAS.
- A. Rigotti, B. L. Trigatti, M. Penman, H. Rayburn, J. Herz and M. Krieger, A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 12610–12615 CrossRef CAS.
- W. Lin, W. Wang, D. Wang and W. Ling, Quercetin protects against atherosclerosis by inhibiting dendritic cell activation, Mol. Nutr. Food Res., 2017, 61 Search PubMed.
- L. Xiao, L. Liu, X. Guo, S. Zhang, J. Wang, F. Zhou, L. Liu, Y. Tang and P. Yao, Quercetin attenuates high fat diet-induced atherosclerosis in apolipoprotein E knockout mice: A critical role of NADPH oxidase, Food Chem. Toxicol., 2017, 105, 22–33 CrossRef CAS PubMed.
- Y. Cui, P. Hou, F. Li, Q. Liu, S. Qin, G. Zhou, X. Xu, Y. Si and S. Guo, Quercetin improves macrophage reverse cholesterol transport in apolipoprotein E-deficient mice fed a high-fat diet, Lipids Health Dis., 2017, 16, 9 CrossRef PubMed.
- M. G. Hertog, E. J. Feskens, P. C. Hollman, M. B. Katan and D. Kromhout, Dietary antioxidant flavonoids and risk of coronary heart disease: the Zutphen Elderly Study, Lancet, 1993, 342, 1007–1011 CrossRef CAS.
- M. G. Hertog, D. Kromhout, C. Aravanis, H. Blackburn, R. Buzina, F. Fidanza, S. Giampaoli, A. Jansen, A. Menotti and S. Nedeljkovic, et al., Flavonoid intake and long-term risk of coronary heart disease and cancer in the seven countries study, Arch. Intern. Med., 1995, 155, 381–386 CrossRef CAS PubMed.
- R. L. Edwards, T. Lyon, S. E. Litwin, A. Rabovsky, J. D. Symons and T. Jalili, Quercetin reduces blood pressure in hypertensive subjects, J. Nutr., 2007, 137, 2405–2411 CAS.
- J. Ahn, H. Lee, S. Kim, J. Park and T. Ha, The anti-obesity effect of quercetin is mediated by the AMPK and MAPK signaling pathways, Biochem. Biophys. Res. Commun., 2008, 373, 545–549 CrossRef CAS PubMed.
- A. W. Boots, G. R. Haenen and A. Bast, Health effects of quercetin: from antioxidant to nutraceutical, Eur. J. Pharmacol., 2008, 585, 325–337 CrossRef CAS PubMed.
- S. C. Bischoff, Quercetin: potentials in the prevention and therapy of disease, Curr. Opin. Clin. Nutr. Metab. Care, 2008, 11, 733–740 CrossRef CAS PubMed.
- M. Romero, R. Jimenez, M. Sanchez, R. Lopez-Sepulveda, M. J. Zarzuelo, F. O'Valle, A. Zarzuelo, F. Perez-Vizcaino and J. Duarte, Quercetin inhibits vascular superoxide production induced by endothelin-1: Role of NADPH oxidase, uncoupled eNOS and PKC, Atherosclerosis, 2009, 202, 58–67 CrossRef CAS PubMed.
- Y. C. Chang, T. S. Lee and A. N. Chiang, Quercetin enhances ABCA1 expression and cholesterol efflux through a p38-dependent pathway in macrophages, J. Lipid Res., 2012, 53, 1840–1850 CrossRef CAS PubMed.
- C. H. Jung, I. Cho, J. Ahn, T. I. Jeon and T. Y. Ha, Quercetin reduces high-fat diet-induced fat accumulation in the liver by regulating lipid metabolism genes, Phytother. Res., 2013, 27, 139–143 CrossRef CAS PubMed.
- M. Kobori, S. Masumoto, Y. Akimoto and H. Oike, Chronic dietary intake of quercetin alleviates hepatic fat accumulation associated with consumption of a Western-style diet in C57/BL6J mice, Mol. Nutr. Food Res., 2011, 55, 530–540 CAS.
- R. Yu, Y. Lv, J. Wang, N. Pan, R. Zhang, X. Wang, H. Yu, L. Tan, Y. Zhao and B. Li, Baicalin promotes cholesterol efflux by regulating the expression of SR-BI in macrophages, Exp. Ther. Med., 2016, 12, 4113–4120 CrossRef PubMed.
- A. Leiva, H. Verdejo, M. L. Benitez, A. Martinez, D. Busso and A. Rigotti, Mechanisms regulating hepatic SR-BI expression and their impact on HDL metabolism, Atherosclerosis, 2011, 217, 299–307 CrossRef CAS PubMed.
- R. Wang, H. Zhang, Y. Wang, F. Song and Y. Yuan, Inhibitory effects of quercetin on the progression of liver fibrosis through the regulation of NF-small ka, CyrillicB/Ismall ka, CyrillicBalpha, p38 MAPK, and Bcl-2/Bax signaling, Int. Immunopharmacol., 2017, 47, 126–133 CrossRef CAS PubMed.
- W. Khovidhunkit, A. H. Moser, J. K. Shigenaga, C. Grunfeld and K. R. Feingold, Regulation of scavenger receptor class B type I in hamster liver and Hep3B cells by endotoxin and cytokines, J. Lipid Res., 2001, 42, 1636–1644 CAS.
- Z. Hu, J. Hu, Z. Zhang, W. J. Shen, C. C. Yun, C. H. Berlot, F. B. Kraemer and S. Azhar, Regulation of expression and function of scavenger receptor class B, type I (SR-BI) by Na+/H+ exchanger regulatory factors (NHERFs), J. Biol. Chem., 2013, 288, 11416–11435 CrossRef CAS PubMed.
- J. Tan, B. Wang and L. Zhu, Regulation of survivin and Bcl-2 in HepG2 cell apoptosis induced by quercetin, Chem. Biodiversity, 2009, 6, 1101–1110 CAS.
- P. Zhao, J. M. Mao, S. Y. Zhang, Z. Q. Zhou, Y. Tan and Y. Zhang, Quercetin induces HepG2 cell apoptosis by inhibiting fatty acid biosynthesis, Oncol. Lett., 2014, 8, 765–769 CAS.
- T. Okamoto, Safety of quercetin for clinical application (Review), Int. J. Mol. Med., 2005, 16, 275–278 CAS.
- M. Harwood, B. Danielewska-Nikiel, J. F. Borzelleca, G. W. Flamm, G. M. Williams and T. C. Lines, A critical review of the data related to the safety of quercetin and lack of evidence of in vivo toxicity, including lack of genotoxic/carcinogenic properties, Food Chem. Toxicol., 2007, 45, 2179–2205 CrossRef CAS PubMed.
- S. Teixeira, Bioflavonoids: proanthocyanidins and quercetin and their potential roles in treating musculoskeletal conditions, J. Orthop. Sports Phys. Ther., 2002, 32, 357–363 CrossRef PubMed.
- S. R. McAnulty, L. S. McAnulty, D. C. Nieman, J. C. Quindry, P. A. Hosick, M. H. Hudson, L. Still, D. A. Henson, G. L. Milne, J. D. Morrow, C. L. Dumke, A. C. Utter, N. T. Triplett and A. Dibarnardi, Chronic quercetin ingestion and exercise-induced oxidative damage and inflammation, Appl. Physiol., Nutr., Metab., 2008, 33, 254–262 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7fo01107e |
| This journal is © The Royal Society of Chemistry 2018 |