Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Removal of antibiotic resistance genes in an anaerobic membrane bioreactor treating primary clarifier effluent at 20 °C
Anthony D.
Kappell
 a,
Lee K.
Kimbell
a,
Lee K.
Kimbell
 a,
Matthew D.
Seib†
a,
Daniel E.
Carey‡
a,
Melinda J.
Choi
b,
Tino
Kalayil
a,
Masanori
Fujimoto§
a,
Daniel H.
Zitomer
a and
Patrick J.
McNamara
a,
Matthew D.
Seib†
a,
Daniel E.
Carey‡
a,
Melinda J.
Choi
b,
Tino
Kalayil
a,
Masanori
Fujimoto§
a,
Daniel H.
Zitomer
a and
Patrick J.
McNamara
 *a
*a
aDepartment of Civil, Construction and Environmental Engineering, Marquette University, P.O. Box 1881, Milwaukee, WI, USA. E-mail: patrick.mcnamara@mu.edu
bDepartment of Biomedical Engineering, Marquette University, P.O. Box 1881, Milwaukee, WI, USA
First published on 17th July 2018
Abstract
Anaerobic membrane bioreactors (AnMBR) play a key role in future plans for sustainable wastewater treatment and resource recovery because they have no energy-intensive oxygen transfer requirements and can produce biomethane for renewable energy. Recent research results show that they can meet relatively stringent discharge limits with respect to BOD5 and TSS when treating municipal wastewater primary effluent. Sustainable used water recovery plans should also consider removal of unregulated pollutants. Antibiotic resistance genes (ARGs) represent an important emerging contaminant due to public health concerns surrounding the spread of infections resistant to common antibiotics. Conventional activated sludge processes have demonstrated mixed results regarding ARG removal. The objective of this research was to determine the impact of an AnMBR on ARG removal when treating municipal primary clarifier effluent at 20 °C. AnMBR treatment resulted in 3.3 to 3.6 log reduction of ARG and the horizontal gene transfer determinate, intI1, copies in filtrate. Membrane treatment significantly decreased the total biomass as indicated by a decrease in 16S rRNA gene concentration. Microbial community analysis via Illumina sequencing revealed that the relative abundance of putative pathogens was higher in membrane filtrate compared to primary effluent although the overall bacterial 16S rRNA gene concentrations was lower in filtrate. Membrane treatment also substantially reduced microbial diversity in filtrate compared to anaerobic reactor contents.
Water impactMainstream anaerobic wastewater treatment is a promising technology for sustainable water resource recovery. Anaerobic membrane bioreactors (AnMBRs) can meet relatively stringent BOD5 and TSS regulatory standards (BOD5 <10 mg L−1, TSS <10 mg L−1) for municipal wastewater at temperatures as low as 10 °C. The research reported herein demonstrated that AnMBR technology can also significantly reduce antibiotic resistance gene copies. |
Introduction
Treating wastewater to only meet discharge limits may be unsustainable due to increasing population, urbanization, water stress, resource consumption and water reuse plans.1 These factors also increase the demand on food and energy. Collectively, these challenges may be better overcome by considering not just wastewater treatment, but the holistic food–energy–water nexus.2 The practice of employing used water reclamation is being more widely embraced as an opportunity to yield value-added products while reducing energy demands and yielding clean water at water resource reclamation facilities (WRRFs).1,3 WRRFs provide a centralized opportunity to recover nutrients (particularly phosphorus), reduce energy demands or produce energy, and provide water for reuse.4–6 To implement resource recovery, WRRFs may need to move beyond conventional activated sludge treatment that requires energy for oxygen transfer and biosolids disposal with limited opportunities for nutrient recovery.3,7Since anaerobic treatment is often a key technology for sustainable water resource recovery management scenarios of the future, anaerobic membrane bioreactors (AnMBRs) have garnered attention for multiple reasons.3,7 AnMBRs produce less solids than aerobic systems and do not require energy for aeration. Additionally, they produce methane that could be recovered for energy.8–10 Recent research results show that AnMBRs can yield effluent BOD5 less than 10 mg L−1 at temperatures as low as 10 °C.11,12 Furthermore, AnMBR filtrate can be treated using processes such as ion-exchange to concentrate nutrients and recover valuable products such as struvite.5,6
In addition to producing valuable products and recovering energy, sustainable water resource recovery must produce water having appropriate quality for reuse. Chemical micropollutants are prevalent in wastewater and pose a variety of ecological risks including fish feminization and the spread of antibiotic resistance.13–18 Results vary in the few existing publications regarding chemical micropollutant removal through AnMBRs.19,20 Additional research is warranted to help determine if pre- or post-treatment is needed to achieve chemical micropollutant removal.21 Removal of biological micropollutants, including antibiotic resistance genes (ARGs), is also important to consider when evaluating treatment options.
Over 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths a year in the US are attributed to infections caused by antibiotic resistant bacteria.22 Health costs associated with antibiotic resistant infections are approximately $1 billion per year.22 The World Health Organization has deemed antibiotic resistance to be a major threat to modern medicine and noted that the role the environment plays in increasing antibiotic resistance is a serious concern.23 Appropriate technical solutions could be applied at hotspots, such as WRRFs,24 to limit ARG dissemination and slow the spread of antibiotic resistance.25 Indeed, ARGs can even be enriched during activated sludge treatment.26,27 Thus, alternative treatment processes to conventional activated sludge could be beneficial to limit dissemination of ARGs in the environment.
000 deaths a year in the US are attributed to infections caused by antibiotic resistant bacteria.22 Health costs associated with antibiotic resistant infections are approximately $1 billion per year.22 The World Health Organization has deemed antibiotic resistance to be a major threat to modern medicine and noted that the role the environment plays in increasing antibiotic resistance is a serious concern.23 Appropriate technical solutions could be applied at hotspots, such as WRRFs,24 to limit ARG dissemination and slow the spread of antibiotic resistance.25 Indeed, ARGs can even be enriched during activated sludge treatment.26,27 Thus, alternative treatment processes to conventional activated sludge could be beneficial to limit dissemination of ARGs in the environment.
Understanding the impact of AnMBRs on the fate of ARGs is important to evaluate sustainable water resource recovery options to remove emerging pollutants while minimizing energy costs and generating value-added products. Membrane treatment is a plausible option to increase ARGs removal. Membrane (100 kDa) filtration of wastewater spiked with plasmid-associated ARGs resulted in 1- to 2-log reduction of ARGs.28 Therefore, a membrane bioreactor could result in ARGs removal due to physical filtration alone. During the biological process, ARGs increased in an aerobic bioreactor in one full-scale study, but were significantly lower in MBR filtrate compared to the wastewater treated.29 The ARGs decrease was correlated to decreased 16S rRNA gene copies.29 In general, aerobic MBR treatment results in higher removal of ARGs than conventional activated sludge treatment,30 but little research is available that describes how AnMBR processes impact ARG fate.
The objective of this research was to determine changes in ARG abundance due to AnMBR treatment of primary clarifier effluent. The most effective mechanism for ARG removal was hypothesized to be membrane filtration rather than biological treatment. To test this hypothesis, a laboratory-scale anaerobic fluidized bed bioreactor followed by an external crossflow membrane was fed primary clarifier effluent from a full-scale water reclamation facility. The abundance of individual ARGs, sul1, erm(B), and tet(O), as well as, the integrase from class 1 integrons (intI1) and 16S rRNA genes were quantified at different points within a lab-scale AnMBR. Furthermore, the microbial communities were analyzed in the membrane bioreactor effluent and the membrane filtrate.
Materials and methods
AnMBR setup, operation, and sampling
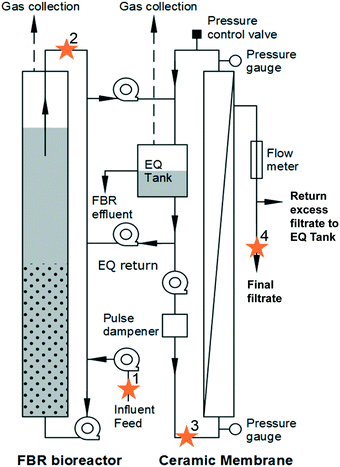
An AnMBR comprised of a fluidized bed bioreactor (FBR) followed by an external, ceramic tubular crossflow membrane with a nominal 0.05 μm pore size (Type1/16, Atech Innovations, Gladbeck, Germany) was operated at 20 °C (Fig. 1) as described by Seib et al.9,12 Operational, performance, and energy demand details can be found elsewhere.9,12 Briefly, the FBR hydraulic residence time (HRT) was 6 h and the nominal solids retention time (SRT) was 49 d. The FBR was initially fed synthetic primary effluent (235 ± 35 mg L−1 BOD5; 115 ± 40 mg L−1 volatile suspended solids [VSS]) for 320 d and then fed actual primary clarifier effluent (160 ± 60 mg L−1 BOD5; 77 ± 25 mg L−1 volatile suspended solids [VSS]) from a full-scale facility (South Shore WRRF, Oak Creek, WI) for 45 d.9 The FBR with external crossflow membrane achieved greater than 90% BOD5 and chemical oxygen demand (COD) removal, with filtrate BOD5 less than 10 mg L−1.Samples were taken on days 348, 349, 352, and 353. Four locations were sampled (Fig. 1): (1) primary influent feed, (2) FBR effluent, (3) liquid in the equalization tank connected to the membrane that served as influent to the membrane (membrane feed), and (4) final filtrate from the membrane. For sample locations 1 through 3, 50 mL were collected and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 15 minutes. The sample pellet was resuspended in 1.0 mL lysis buffer (CLS-TC) and transferred to a 2 mL centrifuge tube. For the final filtrate (4) samples, 500 mL was filtered through a 0.2 μm filter. The filter was placed in a 2 mL centrifuge tube and 1.0 mL lysis buffer was added. The resuspended pellet and filter with lysis buffer were stored at −20 °C until DNA was extracted.
000 × g for 15 minutes. The sample pellet was resuspended in 1.0 mL lysis buffer (CLS-TC) and transferred to a 2 mL centrifuge tube. For the final filtrate (4) samples, 500 mL was filtered through a 0.2 μm filter. The filter was placed in a 2 mL centrifuge tube and 1.0 mL lysis buffer was added. The resuspended pellet and filter with lysis buffer were stored at −20 °C until DNA was extracted.
Molecular methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 or greater per reaction. Each sample was run in triplicate and results averaged. If only two of the three replicates were quantifiable, then the average was taken of the two quantifiable results. Single quantification of three replicates was reported at the quantification limit. Measurements of a no-template control and standards containing target gene DNA between 100 and 108 copies were also performed in triplicate. Cycling conditions were as follows: 2 min at 50 °C to activate the uracil-DNA glycosylase (UDG), 10 min at 95 °C to inactivate UDG and activate the DNA polymerase, 40 cycles of 95 °C for 30 s, 60 °C for 30 s, followed by 72 °C for 30 s. Following amplification, melting curves were acquired. Reactions were carried out using a LightCycler® 96 (Roche Applied Science, Mannheim, Germany). Results were reported as copy number (CN) per L or standardized to total 16S rRNA CN. Specificity of amplification of target genes in samples was confirmed by melt curves consistent with that of the standard. Amplification efficiency was determined by the resulting standard curve and was considered acceptable between 0.9 and 1.1. Efficiency was consistent with previously published ranges for this designed assay.34 Coefficient of determination in the linear regression of the standard curves were greater than 0.99, consistent with the designed assay.34 The qPCR limit of quantification for ARGs and related genes was 5 copy numbers (CNs), which was equivalent to 5 × 104 CN L−1 for final filtrate and 5 × 105 CN L−1 for samples taken at primary influent feed, FBR effluent, and membrane feed (Fig. 1) based upon the volume of sample, resuspension volume following DNA extraction, and volume of DNA used in PCR. The 16S rRNA gene limit of quantification was 500 CN in a reaction.
100 or greater per reaction. Each sample was run in triplicate and results averaged. If only two of the three replicates were quantifiable, then the average was taken of the two quantifiable results. Single quantification of three replicates was reported at the quantification limit. Measurements of a no-template control and standards containing target gene DNA between 100 and 108 copies were also performed in triplicate. Cycling conditions were as follows: 2 min at 50 °C to activate the uracil-DNA glycosylase (UDG), 10 min at 95 °C to inactivate UDG and activate the DNA polymerase, 40 cycles of 95 °C for 30 s, 60 °C for 30 s, followed by 72 °C for 30 s. Following amplification, melting curves were acquired. Reactions were carried out using a LightCycler® 96 (Roche Applied Science, Mannheim, Germany). Results were reported as copy number (CN) per L or standardized to total 16S rRNA CN. Specificity of amplification of target genes in samples was confirmed by melt curves consistent with that of the standard. Amplification efficiency was determined by the resulting standard curve and was considered acceptable between 0.9 and 1.1. Efficiency was consistent with previously published ranges for this designed assay.34 Coefficient of determination in the linear regression of the standard curves were greater than 0.99, consistent with the designed assay.34 The qPCR limit of quantification for ARGs and related genes was 5 copy numbers (CNs), which was equivalent to 5 × 104 CN L−1 for final filtrate and 5 × 105 CN L−1 for samples taken at primary influent feed, FBR effluent, and membrane feed (Fig. 1) based upon the volume of sample, resuspension volume following DNA extraction, and volume of DNA used in PCR. The 16S rRNA gene limit of quantification was 500 CN in a reaction.
Results and discussion
Enumeration of antibiotic resistance and related genes
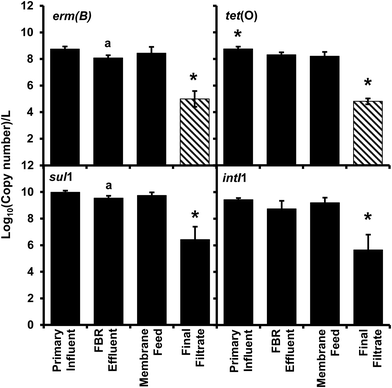
All antibiotic resistance genes (erm(B), tet(O), and sul1) and intI1 abundance values in the final filtrate were between 3 and 4 log units lower than those in primary influent, FBR effluent and the membrane feed (p < 0.01) (Fig. 2). In contrast, ARG and intI1 abundance values were not considerably different among primary influent, FBR effluent or membrane feed water samples; however, the tet(O) and sul1 gene abundance values were greater in the primary influent and membrane feed water compared to the FBR effluent (Fig. 2, p < 0.05).There was a significant 3.94 log reduction of abundance in the final filtrate compared to the other sampled locations in the tet(O) gene (p < 0.01) conferring resistance to tetracycline through protection of the ribosome.45 A significant 3.7 log reduction in erm(B) abundance was determined compared to the other sampled locations (p < 0.01) indicating a loss in resistance to macrolides, lincosamides, and streptogramin B antibiotics through modification of 23S rRNA.46 There was a modest bust significant 0.66 log10 greater abundance of the erm(B) gene within the primary influent compared to FBR effluent (Fig. 2, p < 0.05), suggesting a loss of resistance within the fluidized bed reactor and before membrane filtration. The final filtrate has a significant 3.6 log reduction in sul1 abundance compared to all other sampled locations (Fig. 2, p < 0.01) indicating a loss of sulfonamide resistance by expression of a dihydropteroate synthase with less affinity for the drug.47 The sul1 gene is normally found linked to other resistance genes in class 1 integrons.47 The class 1 integrons have been utilized to quantify the potential of horizontal gene transfer in the bacteria community, specifically in the spread of antibiotic resistance.47,48 The class 1 integron (intI1) had a similar log reduction as the sul1 gene (Fig. 2) indicating a reduction in the potential for horizontal gene transfer in the bacterial community of the final filtrate.
Membrane filtration alone led to a 3.3–3.6 log reduction in ARGs and intI1 in the final filtrate compared to the membrane feed which is consistent with the removal of antibiotic resistant bacteria (ARB) and cell-free ARG-bearing plasmids (extracellular DNA) observed by Cheng and Hong for AnMBRs.49 However, the type of wastewater treated and the temperature employed by Cheng and Hong differed from those employed herein; Cheng and Hong operated an AnMBR at 35 °C and fed synthetic wastewater that was spiked with bacteria containing ARGs.49 Additionally, the methods of sampling in the current study, low-speed centrifugation with disposal of supernatant and filtration reduces the probability of acquiring free-extracellular DNA. However, the methods are not efficient enough to distinguish between bound-extracellular DNA and intracellular DNA.50 Extracellular DNA containing ARGs has the potential for horizontal gene transfer through transformation of bacteria within the environment, including wastewater treatment plants.51–56 The detection of bound-extracellular DNA and intracellular DNA would have been similarly dependent on the ability of amplification, therefore representing a risk of dissemination of antibiotic resistance through vertical or horizontal gene transfer. The results herein demonstrate that AnMBR treating actual wastewater at ambient temperature reduce ARGs.
Enumeration of 16S rRNA genes
There were no significant differences among 16S rRNA gene concentration measurements for primary influent, FBR effluent, and membrane feed samples (p ≥ 0.34; Fig. 3). There was a significant 3-log decrease in biomass indicated by 16S rRNA gene copy number in the final filtrate compared to samples from all other locations within the AnMBR system (p < 0.01). The 16S rRNA gene copy log removal was consistent with bacterial removal via anaerobic digestion as reviewed by Avery et al.57 and an AnMBR operated at 35 °C that also demonstrated a 3-log reduction in total bacteria copy numbers.58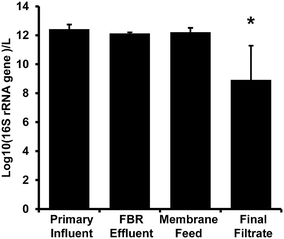 | ||
| Fig. 3 16S rRNA gene abundance per liter from the AnMBR (Fig. 1). Bars represent the average value and error bars represent the standard deviation from four sampling events. The asterisk (*) denotes significant difference from all other sample locations. | ||
Changes in abundance of the ARGs and intI1 normalized to 16S rRNA gene copies (Fig. 4) showed no significant difference (ANOVAs, p > 0.85) suggesting no selective removal of any specific gene over another gene.
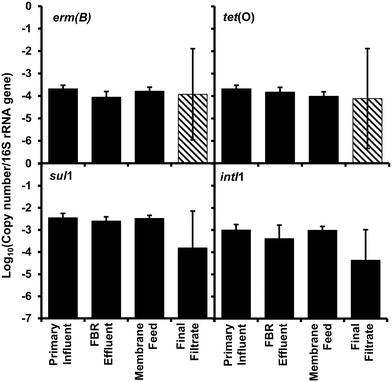 | ||
| Fig. 4 Relative abundance of antibiotic resistance genes erm(B), tet(O), and sul1 and the class 1 associated integrase gene (intI1) per 16S rRNA gene collected from the AnMBR (Fig. 1). Bars represent the average value and error bars represent the standard deviation from four sampling events. Striped bars represent reported results equivalent to the detection limit because only one or two samples out of four were quantifiable. | ||
Membrane feed and membrane filtrate microbial communities
A total of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 730
730![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 988 16S rRNA gene reads were generated, with reads per sample ranging from 970 to 359
988 16S rRNA gene reads were generated, with reads per sample ranging from 970 to 359![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 868. Taxonomic assignments of 12
868. Taxonomic assignments of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 713 unique OTUs designated 47 Phyla, 227 Families, and 588 Genera. Alpha-diversity metrics measuring diversity at each sampling location (Fig. 5), showed significantly greater diversity in sequences in the membrane feed compared to the final filtrate (ANOVA, p < 0.05) in the Chao1, ACE, Shannon, Simpson, Inverse Simpson, and Fisher indices. The change of diversity in sequences suggests that the AnMBR is selective for specific bacteria in the membrane feed through the membrane.
713 unique OTUs designated 47 Phyla, 227 Families, and 588 Genera. Alpha-diversity metrics measuring diversity at each sampling location (Fig. 5), showed significantly greater diversity in sequences in the membrane feed compared to the final filtrate (ANOVA, p < 0.05) in the Chao1, ACE, Shannon, Simpson, Inverse Simpson, and Fisher indices. The change of diversity in sequences suggests that the AnMBR is selective for specific bacteria in the membrane feed through the membrane.
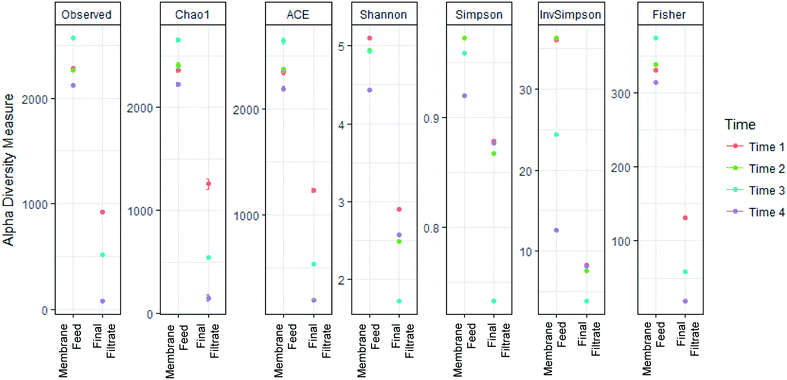 | ||
| Fig. 5 Membrane substantially decreased richness and diversity between the membrane feed and membrane final filtrate. | ||
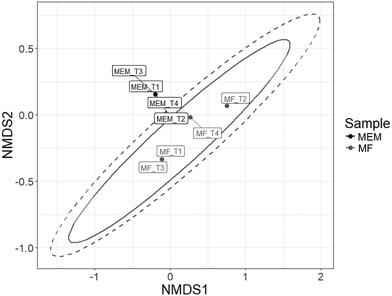
In addition, membrane feed and the membrane final filtrate communities were significantly different as determined using the Horn–Moristia index,41,42 visualized by NMDS (Fig. 6) and calculated by permutational multivariate analysis of variance (p = 0.029). These results are consistent with previous observations by Harb and Hong who reported distinct communities within the influent and effluent from an AnMBR operated at 35 °C treating real wastewater.58
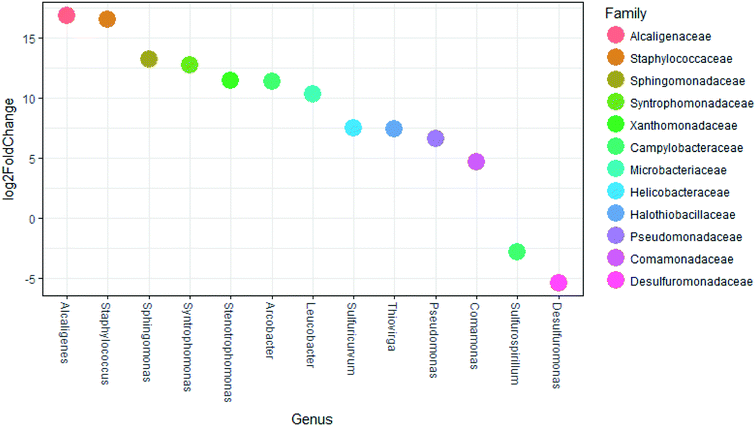
Significant differences (p < 0.01) in dominant genera between membrane feed and membrane final filtrate were determined by DESeq (Fig. 7). Genera that include known opportunistic pathogens and showed increased abundance included: Alcaligenes, Staphylococcus, Sphingomonas, Stenotrophomonas, Arcobacter and Pseudomonas.53,57,59–61,63–67 While the membrane final filtrate showed significant decreases in biomass, the increased relative abundance of these opportunistic pathogens suggests positive selection for these genera. In the US Center for Disease Control and Prevention (CDC) Antibiotic Resistance Threat Report in 2013, methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Staphylococcus aureus (VRSA) were each listed as a serious and concerning level of hazard.22 Interestingly, the Stenotrophomonas genus contains a multidrug resistant opportunistic pathogen.63 Further study by quantification of specific virulence genes associated with these and other opportunistic pathogens would yield information on the potential to select for pathogens.
Of those genera that showed significant increases in relative abundance between the membrane feed and membrane filtrate (Fig. 7), Alcaligenes and Arcobacter are known to perform anaerobic respiration,67,68Staphylococcus, Sulfuricurvum, and Pseudomonas are facultative anaerobes,66,68 while Syntrophomonas are identified as anaerobic.69,70 Similarly, the aerobic genera of Sphingomonas, Stenotrophomonas, Leucobacter, Thiovirga, and Comamonas also showed significant increases in relative abundance in the membrane filtrate compared to the membrane feed.62,63,71–75 Only two genera showed significant decreases in relative abundance between the membrane feed and membrane filtrate, consisting of the genus Sulfurospirillum, able to ferment fumarate under anaerobic conditions68 and the strictly anaerobic Desulfuromonas.76 The decrease in Desulfuromonas relative abundance might have been due to its lack of tolerance to oxygen. However, the increases in both anaerobic and aerobic bacteria suggest that selection based on oxygen tolerance was absent or weak.
The genera Alcaligenes, Sphingomonas, Syntrophomonas, Thiovirga, Stenotrophomonas, Leucobacter, Pseudomonas, Comamonas, and Desulfuromonas have been associated with wastewater treatment and resource recovery processes including anaerobic digestors and AnMBR treatment. Alcaligenes taxa have been identified in anaerobic digestors.77Sphingomonas, while being aerobic, have been identified in anaerobic digesters, though their role remains unclear.71 The Syntrophomonas genus include anaerobic fatty-acid-oxidizing bacteria that are associated with methanogens and are common to anaerobic bioreactors when long-chain fatty-acids are avalaible69,70 indicating they are present in the studied FBR. Thiovirga are chemolithoautotrophic, sulfur-oxidizing bacteria which were first isolated from a microaerobic wastewater biofilm while investigating its sulfur cycle.73Stenotrophomonas, while aerobic, has been previously detected in the effluent of a lab-scale AnMBR treating municipal wastewater.58Leucobacter genus have been isolated from activated sludge. Pseudomonas genera have been detected and the species Pseudomonas aeruginosa has been quantified previously from the filtrate of an AnMBR.58Comamonas are aerobic bacteria which have been identified in AnMBR, activated sludge, and anaerobic digesters.74,75Desulfuromonas were identified in an AnMBR feed with synthetic textile industry wastewater.78
Differences in primary effluent and membrane filtrate microbial communities showed that the AnMBR system selected for specific bacteria. This selection was presumably due to size exclusion, however, the sizes of the bacteria represented in the membrane feed and membrane filtrate are comparable based on published direct observation which are usually taken under ideal growth conditions.79 The bacterial cell wall structure is an important factor in bacteria breakthrough of small-pore-size membranes.80–82 The bacterial cell wall can be elastic and deform under pressure or become less rigid dependent on the environment, such as the presence of antibiotics.83–85 The presence of foulant is known to improve the removal of extracellular DNA containing ARGs and ARB,49 which most likely led to the variability of the microbial community and differential removal of ARGs found in the final filtrate. Optimization of the control of the level of foulant could improve ARB and ARG removal.49
Collectively, the survey of the relative abundance of 16S rRNA genes in membrane final filtrate and membrane feed samples demonstrated a significant increase in the relative abundance of genera containing opportunistic pathogens as well as decreased diversity in the membrane filtrate compared to the membrane feed samples. The changes indicated selective removal at the membrane and/or selective growth or regrowth of bacteria in the membrane filtrate tank.
Conclusions
This research revealed that AnMBR significantly reduce ARGs in primary clarifier effluent. ARGs are an important emerging contaminant as antibiotic resistance is a major public health concern.86 We no longer have the luxury to view drinking water, wastewater, and stormwater as isolated entities, but rather as one water. It is important to develop and implement sustainable wastewater treatment and resource recovery technologies that produce high quality water, recovery energy, and generate value-added products as the population continues to increase and WRRFs receive more stress. AnMBRs produce methane that can be recovered on-site and offer potential for down-stream nutrient recovery. This research revealed that they substantially reduce concentrations of ARGs, likely to a much greater and consistent extent than conventional activated sludge reactors.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was funded in part by National Science Foundation awards OCI-0923037 “MRI: Acquisition of a Parallel Computing Cluster and Storage for the Marquette University Grid (MUGrid)” and CBET-0521602 “Acquisition of a Linux Cluster to Support College-Wide Research & Teaching Activities”. The authors acknowledge the use of the Roche qPCR instrument from Marquette University, funded by the GHR Foundation. Funding from the Marquette University Water Quality Center was provided for A. K. and L. K.References
- G. T. Daigger, Aeesp Lecture: Evolving Urban Water and Residuals Management Paradigms: Water Reclamation and Reuse, Decentralization, Resource Recovery. Proc Water Environ Fed, 2008, vol. 2008(15), pp. 1537–1565, DOI:10.2175/193864708790893378.
- W. Mo and Q. Zhang, Energy-nutrients-water nexus: Integrated resource recovery in municipal wastewater treatment plants, J. Environ. Manage., 2013, 127, 255–267, DOI:10.1016/j.jenvman.2013.05.007.
- P. L. McCarty, J. Bae and J. Kim, Domestic wastewater treatment as a net energy producer - can this be achieved?, Environ. Sci. Technol., 2011, 45(17), 7100–7106, DOI:10.1021/es2014264.
- Z. Liu, P. McNamara and D. Zitomer, Autocatalytic Pyrolysis of Wastewater Biosolids for Product Upgrading, Environ. Sci. Technol., 2017, 51(17), 9808–9816, DOI:10.1021/acs.est.7b02913.
- A. T. Williams, D. H. Zitomer and B. K. Mayer, Ion exchange-precipitation for nutrient recovery from dilute wastewater, Environ. Sci.: Water Res. Technol., 2015, 1(6), 832–838, 10.1039/C5EW00142K.
- Y. Tong, P. J. McNamara and B. K. Mayer, Fate and impacts of triclosan, sulfamethoxazole, and 17β-estradiol during nutrient recovery via ion exchange and struvite precipitation, Environ. Sci.: Water Res. Technol., 2017, 3, 1109–1119, 10.1039/C7EW00280G.
- W. Verstraete and S. Vlaeminck, Short-cycling of Wastewater Resources for Sustainable Cities of the Future, Int. J. Sust. Dev. World, 2011, 18(3), 253–364 CrossRef.
- A. L. Smith, L. B. Stadler, N. G. Love, S. J. Skerlos and L. Raskin, Perspectives on anaerobic membrane bioreactor treatment of domestic wastewater: A critical review, Bioresour. Technol., 2012, 122, 149–159, DOI:10.1016/j.biortech.2012.04.055.
- M. D. Seib, K. J. Berg and D. H. Zitomer, Low energy anaerobic membrane bioreactor for municipal wastewater treatment, J. Membr. Sci., 2016, 514, 450–457, DOI:10.1016/j.memsci.2016.05.007.
- J. Kim, K. Kim and H. Ye, et al., Anaerobic fluidized bed membrane bioreactor for wastewater treatment, Environ. Sci. Technol., 2011, 45(2), 576–581, DOI:10.1021/es1027103.
- C. Shin, P. L. McCarty, J. Kim and J. Bae, Pilot-scale temperate-climate treatment of domestic wastewater with a staged anaerobic fluidized membrane bioreactor (SAF-MBR), Bioresour. Technol., 2014, 159, 95–103, DOI:10.1016/j.biortech.2014.02.060.
- M. D. Seib, K. J. Berg and D. H. Zitomer, Reduced energy demand for municipal wastewater recovery using an anaerobic floating filter membrane bioreactor, Environ. Sci.: Water Res. Technol., 2016, 2(2), 290–297, 10.1039/C5EW00244C.
- B. D. Blair, J. P. Crago, C. J. Hedman and R. D. Klaper, Pharmaceuticals and personal care products found in the Great Lakes above concentrations of environmental concern, Chemosphere, 2013, 93(9), 2116–2123, DOI:10.1016/j.chemosphere.2013.07.057.
- M. V. Alan, L. B. Barber, J. L. Gray, E. M. Lopez, J. D. Woodling and D. O. Norris, Reproductive disruption in fish downstream from an estrogenic wastewater effluent, Environ. Sci. Technol., 2008, 42(9), 3407–3414, DOI:10.1021/es0720661.
- D. E. Carey, D. H. Zitomer, K. R. Hristova, A. D. Kappell and P. J. McNamara, Triclocarban Influences Antibiotic Resistance and Alters Anaerobic Digester Microbial Community Structure, Environ. Sci. Technol., 2016, 50(1), 126–134, DOI:10.1021/acs.est.5b03080.
- D. E. Carey and P. J. McNamara, Altered antibiotic tolerance in anaerobic digesters acclimated to triclosan or triclocarban, Chemosphere, 2016, 163, 22–26, DOI:10.1016/j.chemosphere.2016.07.097.
- D. E. Carey and P. J. McNamara, The impact of triclosan on the spread of antibiotic resistance in the environment, Front. Microbiol., 2014, 5(DEC), 1–11, DOI:10.3389/fmicb.2014.00780.
- D. E. Carey, D. H. Zitomer, A. D. Kappell, M. J. Choi, K. R. Hristova and P. J. McNamara, Chronic exposure to triclosan sustains microbial community shifts and alters antibiotic resistance gene levels in anaerobic digesters, Environ. Sci.: Processes Impacts, 2016, 18(8), 1060–1067, 10.1039/C6EM00282J.
- M. R. Abargues, A. Robles, A. Bouzas and A. Seco, Micropollutants removal in an anaerobic membrane bioreactor and in an aerobic conventional treatment plant, Water Sci. Technol., 2012, 65(12), 2242–2250, DOI:10.2166/wst.2012.145.
- V. M. Monsalvo, J. A. McDonald, S. J. Khan. and P. Le-Clech, Removal of trace organics by anaerobic membrane bioreactors, Water Res., 2014, 49, 103–112, DOI:10.1016/j.watres.2013.11.026.
- Y. Tong, B. K. Mayer and P. J. McNamara, Triclosan adsorption using wastewater biosolids-derived biochar, Environ. Sci.: Water Res. Technol., 2016, 2(4), 761–768, 10.1039/C6EW00127K.
- States U., Antibiotic Resistance Threats in the United States, 2013, US Department of Health and Human Services, 2013, http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf Search PubMed.
- World Health Organization, Global Action Plan on Antimicrobial Resistance, In: Geneva: World Health Organization, 2015, http://www.who.int/antimicrobial-resistance/global-action-plan/en/ Search PubMed.
- T. M. Lapara, T. R. Burch, P. J. McNamara, D. T. Tan, M. Yan and J. J. Eichmiller, Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor, Environ. Sci. Technol., 2011, 45(22), 9543–9549, DOI:10.1021/es202775r.
- A. Pruden, D. G. Joakim Larsson and A. Amézquita, et al., Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment, Environ. Health Perspect., 2013, 121(8), 878–885, DOI:10.1289/ehp.1206446.
- M. Liu, Y. Zhang, M. Yang, Z. Tian, L. Ren and S. Zhang, Abundance and Distribution of Tetracycline Resistance Genes and Mobile Elements in an Oxytetracycline Production Wastewater Treatment System, Environ. Sci. Technol., 2012, 46(14), 7551–7557, DOI:10.1021/es301145m.
- T. Zhang, X.-X. Zhang and L. Ye, Plasmid Metagenome Reveals High Levels of Antibiotic Resistance Genes and Mobile Genetic Elements in Activated Sludge, PLoS One, 2011, 6(10), e26041, DOI:10.1371/journal.pone.0026041.
- M. V. Riquelme Breazeal, J. T. Novak, P. J. Vikesland and A. Pruden, Effect of wastewater colloids on membrane removal of antibiotic resistance genes, Water Res., 2013, 47(1), 130–140, DOI:10.1016/j.watres.2012.09.044.
- J. Du, J. Geng, H. Ren, L. Ding, K. Xu and Y. Zhang, Variation of antibiotic resistance genes in municipal wastewater treatment plant with A2O-MBR system, Environ. Sci. Pollut. Res., 2015, 22(5), 3715–3726, DOI:10.1007/s11356-014-3552-x.
- M. Munir and I. Xagoraraki, Levels of antibiotic resistance genes in manure, biosolids, and fertilized soil, J. Environ. Qual., 2011, 40(1), 248–255, DOI:10.2134/jeq2010.0209.
- P. J. McNamara, T. M. Lapara and P. J. Novak, The impacts of triclosan on anaerobic community structures, function, and antimicrobial resistance, Environ. Sci. Technol., 2014, 48(13), 7393–7400, DOI:10.1021/es501388v.
- D. W. Graham, S. Olivares-Rieumont, C. W. Knapp, L. Lima, D. Werner and E. Bowen, Antibiotic resistance gene abundances associated with waste discharges to the Almendares river near Havana, Cuba, Environ. Sci. Technol., 2011, 45(2), 418–424, DOI:10.1021/es102473z.
- H. Heuer and K. Smalla, Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months, Environ. Microbiol., 2007, 9(3), 657–666, DOI:10.1111/j.1462-2920.2006.01185.x.
- L. K. Kimbell, A. D. Kappell and P. J. McNamara, Effect of pyrolysis on the removal of antibiotic resistance genes and class I integrons from municipal wastewater biosolids, Environ. Sci.: Water Res. Technol., 2018 10.1039/C8EW00141C.
- P. D. Schloss, S. L. Westcott and T. Ryabin, et al., Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 2009, 75(23), 7537–7541, DOI:10.1128/AEM.01541-09.
- J. J. Kozich, S. L. Westcott, N. T. Baxter, S. K. Highlander and P. D. Schloss, Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform, Appl. Environ. Microbiol., 2013, 79(17), 5112–5120, DOI:10.1128/AEM.01043-13.
- C. Quast, E. Pruesse, P. Yilmaz, J. Gerken, T. Schweer and P. Yarza, et al., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res., 2013, 41(Database Issue), D590–D596 Search PubMed.
- R Core Team, R Core Team (2017). R: A language and environment for statistical computing. R Found Stat Comput Vienna, Austria URL http://wwwR-project.org/, 2017:R Foundation for Statistical Computing Search PubMed.
- P. J. McMurdie and S. Holmes, Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 2013, 8(4), e61217 CrossRef PubMed.
- P. J. McMurdie and S. Holmes, Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data, Pac. Symp. Biocomput., 17th, 2012, 235–246, DOI:10.1142/9789814366496_0023.
- H. S. Horn, Measurement of “Overlap” in Comparative Ecological Studies, Am. Nat., 1966, 100(914), 419–424, DOI:10.1086/282436.
- M. Moristita, Measuring of Interspecific Association and Similarity between communities, Memoirs of the Faculty of Sciences, Kyushu University, Series E (Biology), 1959, vol. 13(1), pp. 65–80 Search PubMed.
- M. I. Love, S. Anders and W. Huber, Differential Analysis of Count Data - the DESeq2 Package, 2014, vol. 15, 1 DOI:10.1186/s13059-014-0550-8.
- M. Mariadassou, Phyloseq-extended (https://github.com/mahendra-mariadassou/phyloseq-extended), 2017.
- D. N. Wilson, Ribosome-targeting antibiotics and mechanisms of bacterial resistance, Nat. Rev. Microbiol., 2014, 12(1), 35–48, DOI:10.1038/nrmicro3155.
- M. C. Roberts, J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood and H. Seppala, Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants, Antimicrob. Agents Chemother., 1999, 43(12), 2823–2830 Search PubMed.
- P. P. Antunes, J. Machado, J. C. J. C. Sousa and L. L. Peixe, Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons, Antimicrob. Agents Chemother., 2005, 49(2), 836–839, DOI:10.1128/AAC.49.2.836-839.2005.
- S. Nandi, J. J. Maurer, C. Hofacre and A. O. Summers, Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(18), 7118–7122, DOI:10.1073/pnas.0306466101.
- H. Cheng and P. Y. Hong, Removal of antibiotic-resistant bacteria and antibiotic resistance genes affected by varying degrees of fouling on anaerobic microfiltration membranes, Environ. Sci. Technol., 2017, 51(21), 12200–12209 CrossRef PubMed.
- M. Nagler, S. M. Podmirseg, G. W. Griffith, H. Insam and J. Ascher-Jenull, The use of extracellular DNA as a proxy for specific microbial activity, Appl. Microbiol. Biotechnol., 2018, 102(6), 2885–2898, DOI:10.1007/s00253-018-8786-y.
- S.-S. Liu, H.-M. Qu and D. Yang, et al., Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant, Water Res., 2018, 136, 131–136, DOI:10.1016/J.WATRES.2018.02.036.
- Y. Zhang, Z. Niu, Y. Zhang and K. Zhang, Occurrence of intracellular and extracellular antibiotic resistance genes in coastal areas of Bohai Bay (China) and the factors affecting them, Environ. Pollut., 2018, 236, 126–136, DOI:10.1016/J.ENVPOL.2018.01.033.
- Y. Zhang, D. D. Snow, D. Parker, Z. Zhou and X. Li, Intracellular and Extracellular Antimicrobial Resistance Genes in the Sludge of Livestock Waste Management Structures, Environ. Sci. Technol., 2013, 47(18), 10206–10213, DOI:10.1021/es401964s.
- P. Cai, Q. Huang, X. Zhang and H. Chen, Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol, Soil Biol. Biochem., 2006, 38(3), 471–476, DOI:10.1016/J.SOILBIO.2005.05.019.
- S. Borin, E. Crotti, F. Mapelli, I. Tamagnini, C. Corselli and D. Daffonchio, DNA is preserved and maintains transforming potential after contact with brines of the deep anoxic hypersaline lakes of the Eastern Mediterranean Sea, Saline Syst., 2008, 4(1), 10, DOI:10.1186/1746-1448-4-10.
- D. N. Wang, L. Liu and Z. G. Qiu, et al., A new adsorption-elution technique for the concentration of aquatic extracellular antibiotic resistance genes from large volumes of water, Water Res., 2016, 92, 188–198, DOI:10.1016/j.watres.2016.01.035.
- L. M. Avery, K. Y. Anchang, V. Tumwesige, N. Strachan and P. J. Goude, Potential for Pathogen reduction in anaerobic digestion and biogas generation in Sub-Saharan Africa, Biomass Bioenergy, 2014, 70, 112–124, DOI:10.1016/j.biombioe.2014.01.053.
- M. Harb and P. Y. Hong, Molecular-based detection of potentially pathogenic bacteria in membrane bioreactor (MBR) systems treating municipal wastewater: a case study, Environ. Sci. Pollut. Res., 2017, 24(6), 5370–5380, DOI:10.1007/s11356-016-8211-y.
- H.-J. Busse and G. Auling, Alcaligenes, In: Bergey's Manual of Systematics of Archaea and Bacteria, John Wiley & Sons, Inc., in association with Bergey's Manual Trust, 2015, pp. 1–14 Search PubMed.
- S. R. Gill, D. E. Fouts and G. L. Archer, et al., Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain, J. Bacteriol., 2005, 187(7), 2426–2438, DOI:10.1128/JB.187.7.2426-2438.2005.
- D. C. White, S. D. Suttont and D. B. Ringelberg, The genus Sphingomonas: Physiology and ecology, Curr. Opin. Biotechnol., 1996, 7(3), 301–306, DOI:10.1016/S0958-1669(96)80034-6.
- R. P. Ryan, S. Monchy and M. Cardinale, et al., The versatility and adaptation of bacteria from the genus Stenotrophomonas, Nat. Rev. Microbiol., 2009, 7(7), 514–525, DOI:10.1038/nrmicro2163.
- W. J. Looney, M. Narita and K. Mühlemann, Stenotrophomonas maltophilia: an emerging opportunist human pathogen, Lancet Infect. Dis., 2009, 9(5), 312–323, DOI:10.1016/S1473-3099(09)70083-0.
- S. Ferreira, J. A. Queiroz, M. Oleastro and F. C. Domingues, Insights in the pathogenesis and resistance of Arcobacter: A review, Crit. Rev. Microbiol., 2016, 42(3), 364–383, DOI:10.3109/1040841X.2014.954523.
- A. Peix, M. H. Ramírez-Bahena and E. Velázquez, The current status on the taxonomy of Pseudomonas revisited: An update, Infect., Genet. Evol., 2018, 57, 106–116, DOI:10.1016/j.meegid.2017.10.026.
- N. J. Palleroni, Pseudomonas, In: Bergey's Manual of Systematics of Archaea and Bacteria, 2015, p. 1 Search PubMed.
- U. Warnecke-Eberz and B. Friedrich, Three nitrate reductase activities in Alcaligenes eutrophus, Arch. Microbiol., 1993, 159(5), 405–409, DOI:10.1007/BF00288585.
- F. T. Saia, T. S. O. Souza, R. T. D. Duarte, E. Pozzi, D. Fonseca and E. Foresti, Microbial community in a pilot-scale bioreactor promoting anaerobic digestion and sulfur-driven denitrification for domestic sewage treatment, Bioprocess Biosyst. Eng., 2016, 39(2), 341–352, DOI:10.1007/s00449-015-1520-6.
- Y. Sekiguchi, Syntrophomonas, In: Bergey's Manual of Systematics of Archaea and Bacteria, 2015, pp. 1–11 Search PubMed.
- D. Z. Sousa, M. A. Pereira, J. I. Alves, H. Smidt, A. J. M. Stams and M. M. Alves, Anaerobic microbial LCFA degradation in bioreactors, Water Sci. Technol., 2008, 57(3), 439–444, DOI:10.2166/wst.2008.090.
- J. Kim and C. Lee, Response of a continuous anaerobic digester to temperature transitions: A critical range for restructuring the microbial community structure and function, Water Res., 2016, 89, 241–251, DOI:10.1016/j.watres.2015.11.060.
- L. N. Sun, D. D. Pan, X. W. Wu, E. D. Yang, R. M. Hua and Q. X. Li, Leucobacter triazinivorans sp. nov., a s-triazine herbicide prometryn-degrading bacterium isolated from sludge, Int. J. Syst. Evol. Microbiol., 2018, 68(1), 204–210, DOI:10.1099/ijsem.0.002483.
- T. Ito, K. Sugita, I. Yumoto, Y. Nodasaka and S. Okabe, Thiovirga sulfuroxydans gen. nov., sp. nov., a chemolithoautotrophic sulfur-oxidizing bacterium isolated from a microaerobic waste-water biofilm, Int. J. Syst. Evol. Microbiol., 2005, 55(3), 1059–1064, DOI:10.1099/ijs.0.63467-0.
- W. Zheng, Z. Yu, Y. Xia and X. Wen, Influence of polyaluminum chloride on microbial characteristics in anaerobic membrane bioreactors for sludge digestion, Appl. Microbiol. Biotechnol., 2018, 102(2), 1005–1017, DOI:10.1007/s00253-017-8613-x.
- A. Willems and M. Gillis, Comamonas. Bergey's Man Syst Archaea Bact, 2015, pp. 1–17 Search PubMed.
- J. Kuever, F. A. Rainey and W. Friedrich, Desulfuromonas, In: Bergey's Manual of Systematics of Archaea and Bacteria, 2015, pp. 1–7 Search PubMed.
- S. Xu, C. He, L. Luo, F. Lü, P. He and L. Cui, Comparing activated carbon of different particle sizes on enhancing methane generation in upflow anaerobic digester, Bioresour. Technol., 2015, 196, 606–612, DOI:10.1016/j.biortech.2015.08.018.
- A. Yurtsever, B. Calimlioglu and E. Sahinkaya, Impact of SRT on the efficiency and microbial community of sequential anaerobic and aerobic membrane bioreactors for the treatment of textile industry wastewater, Chem. Eng. J., 2017, 314, 378–387, DOI:10.1016/j.cej.2016.11.156.
- C. Söhngen, B. Bunk, A. Podstawka, D. Gleim and J. Overmann, BacDive—the Bacterial Diversity Metadatabase, Nucleic Acids Res., 2014, 42(Database issue), D592–D599, DOI:10.1093/nar/gkt1058.
- N. Lebleu, C. Roques, P. Aimar and C. Causserand, Role of the cell-wall structure in the retention of bacteria by microfiltration membranes, J. Memb. Sci., 2009, 326(1), 178–185 CrossRef.
- S. B. Sadr Ghayeni, P. J. Beatson, A. J. Fane and R. P. Schneider, Bacterial passage through microfiltration membranes in wastewater applications, J. Memb. Sci., 1999, 153(1), 71–82 CrossRef.
- Y. Wang, F. Hammes, M. Düggelin and T. Egli, Influence of size, shape, and flexibility on bacterial passage through micropore membrane filters, Environ. Sci. Technol., 2008, 42(17), 6749–6754 CrossRef PubMed.
- A. Helling, A. Kubicka, I. A. T. Schaap, M. Polakovic, B. Hansmann and H. Thiess, et al., Passage of soft pathogens through microfiltration membranes scales with transmembrane pressure, J. Memb. Sci., 2017, 522, 292–302 CrossRef.
- A. Gaveau, C. Coetsier, C. Roques, P. Bacchin, E. Dague and C. Causserand, Bacteria transfer by deformation through microfiltration membrane, J. Memb. Sci., 2017, 523, 446–455 CrossRef.
- C. Formosa, M. Grare, R. E. Duval and E. Dague, Nanoscale effects of antibiotics on P. aeruginosa, Nanomedicine, 2012, 8(1), 12–16 CrossRef PubMed.
- A. Pruden, R. E. Alcalde, P. J. Alvarez, N. Ashbolt, H. Bischel, N. L. Caprio, E. Crossette, D. Frigon, K. Grimes, C. N. Haas, K. Ikuma, A. Kappel, T. LaPara, L. Kimbell, M. Li, X. Li, P. McNamara, Y. Seo, M. Sobsey, E. Sozzi, T. Navab-Daneshmand, T. H. Nguyen, L. Raskin, M. V. Riquelme, P. Vikesland, K. Wigginton and Z. Zhou, An Environmental Science and Engineering Framework for Combating Antimicrobial Resistance, Environ. Eng. Sci. DOI:10.1089/ees.2017.0520 , in press.
Footnotes |
| † Madison Metropolitan Sewerage District, 1610 Moorland Rd., Madison, WI, USA |
| ‡ Brown & Caldwell, Charlotte, North Carolina, USA |
| § Department of Soil and Water Sciences, University of Florida, Gainesville, FL, USA |
| This journal is © The Royal Society of Chemistry 2018 |