Open Access Article
Open Access ArticleGreen synthesis and biotransformation of amorphous Se nanospheres to trigonal 1D Se nanostructures: impact on Se mobility within the concept of radioactive waste disposal†
Miguel A.
Ruiz Fresneda
 ,
Josemaría
Delgado Martín
,
Jaime
Gómez Bolívar
,
María V.
Fernández Cantos
,
Josemaría
Delgado Martín
,
Jaime
Gómez Bolívar
,
María V.
Fernández Cantos
 ,
Germán
Bosch-Estévez
,
Marcos F.
Martínez Moreno
,
Germán
Bosch-Estévez
,
Marcos F.
Martínez Moreno
 and
Mohamed L.
Merroun
and
Mohamed L.
Merroun
 *
*
Department of Microbiology, University of Granada, Granada, Spain. E-mail: merroun@ugr.es
First published on 27th July 2018
Abstract
Nuclear waste containing radionuclides including selenium isotopes, Se79, will be disposed of in future deep geological repositories. Due to the long lifetime of these radioisotopes, studies on the impact of microbial processes on their chemical speciation would contribute significantly to understanding the risks associated with these repositories. Here we report a green method for biogenic reduction of Se(IV), production of amorphous Se(0) (a-Se) nanospheres and their subsequent transformation to one-dimensional (1D) trigonal selenium (t-Se) nanostructures using a combination of methods (XRD, STEM/HAADF, HRTEM/EDX, ESEM, etc.). The bacterial strain used, Stenotrophomonas bentonitica, was isolated from Spanish bentonites considered as artificial barriers for future Spanish repositories. After 24 h of incubation, 30–200 nm sized biogenic individual a-Se nanospheres were synthesized and then started to coalesce, forming aggregates after 48 and 72 h of incubation. The 144 h sample presented a mixture of single crystal and polycrystalline 1D t-Se nanostructures with different shapes (e.g. nanowires, hexagonal, polygonal, etc.) and diameters of 30–400 nm, in addition to a-Se nanospheres. The HRTEM analysis showed that the 1D nanostructures presented different lattice spacings corresponding to the (100), (101) and (111) planes of t-Se. Thus, a-Se nanospheres were initially synthesized and then would transform into t-Se nanostructures. STEM/HAADF and ESEM revealed that the cells and their extracellular proteins play an important role in this transformation process. Due to the low solubility of t-Se nanostructures compared to that of a-Se nanospheres and Se(IV), the mobility of selenium in future repositories may be significantly reduced.
Environmental significanceSelenium is a common component of radioactive waste which will be disposed of in future geological repositories using bentonites as artificial barriers. A bentonite bacterial isolate, Stenotrophomonas bentonitica, is demonstrated here to reduce Se(IV) to Se(0), forming initially amorphous Se(0) (a-Se) nanospheres which subsequently transform to one-dimensional (1D) trigonal selenium (t-Se) nanostructures with diverse crystallinities, morphologies and sizes under ambient conditions. Due to the low solubility of t-Se nanostructures compared to that of a-Se nanospheres and Se(IV), the mobility of selenium in the environment may be significantly reduced. The potential environmental significance of this study includes understanding the impact of microbial processes on the transport of selenium in future radioactive repositories which in turn will help to support the implementation of planned repositories. |
Introduction
Radioactive waste produced mainly by the nuclear energy industry contains materials contaminated with radionuclides (e.g. uranium, curium, selenium, etc.). Due to its toxicity, nuclear waste will be disposed of for a long period of time until its toxicity decreases to non-hazardous levels. For this purpose, deep geological repositories (DGRs), consisting of the encapsulation of radioactive waste in metal containers surrounded by artificial and natural barriers, have been proposed as the safest option for its disposal in the near future.1 In different countries, bentonite clay formations have been selected as reference materials for safety barriers in a DGR. In Spain, bentonite formations located in Cabo de Gata Natural Park (Almeria) were selected for their geochemical and mineralogical properties as artificial barrier reference materials.2 The high microbial diversity of these Spanish clays was shown by means of culture dependent and independent techniques.3,4 In addition, more than 100 microbial strains were isolated from these bentonites including a novel species of the genus Stenotrophomonas (Stenotrophomonas bentonitica BII-R7).5The microbial communities of the selected artificial barriers can affect the safety and performance of future DGRs at 3 different levels by: i) transformation of clay minerals; ii) corrosion of metal canisters; and iii) mobilization of radionuclides through different interaction processes.6 Microorganisms are able to interact with typical elements of radioactive waste like selenium (Se), uranium (U) or curium (Cm) through different processes like biomineralization, biosorption or reduction leading to their mobilization or immobilization.7–9
Selenium is considered a common component of high-level radioactive waste (HLRW), particularly the Se79 isotope produced by nuclear fission reactions. Different studies have indicated the Se79 isotope to be one of the critical radionuclides for the geological disposal of HLRW due to its ability to emit β particles.10 Selenium can exist in the environment in different oxidation states {selenate [Se(VI)], selenite [Se(IV)], elemental selenium [Se(0)] and selenide [Se(-II)]}, whose toxicity is mainly related to their solubility.11 Selenate and selenite are the most soluble and hence the most toxic forms of Se, being dominant species under mildly and strongly oxidizing conditions, respectively.12 On the other hand, elemental selenium has a relatively low toxicity because of its low solubility, and it is considered the dominant species under reducing conditions.12 Therefore, it is important to investigate the speciation of Se associated with microbial populations occurring in different barriers in future DGRs to evaluate the safety of this kind of disposal. Different physico-chemical and biological methods were applied to reduce selenate and selenite to produce elemental Se nanoparticles (SeNPs).13,14 Several bacterial strains have been described for their ability to reduce toxic selenium [Se(VI) and Se(IV)] to less toxic forms [Se(0)], producing NPs under different physiological conditions.15–17 In general, the solubility and mobility of SeNPs seem to be affected by their physicochemical properties (size, shape, morphology, crystallinity, etc.), and also by the action of microbial processes through the synthesis of coating agents (proteins, polysaccharides, etc.).18–20
Selenium could exist in three different allotropic forms: amorphous, monoclinic and trigonal phase.13 The trigonal selenium (t-Se) phase is one of the most thermodynamically stable forms of selenium. This phase is characterized by its anisotropic crystal structure where the Se atoms are coordinated to each other through covalent bonds forming infinite chains.13 Several studies have previously reported that bacteria can produce intracellular and extracellular amorphous Se (a-Se) nanospheres with a size ranging from 100 to 200 nm.14,21 To a lesser extent, few studies have also described the formation of biogenic trigonal and monoclinic selenium (t-Se and m-Se).8,22 Ho et al.23 reported the control of organic solvents like DMSO in the transformation of biogenic a-Se nanospheres to 1D t-Se nanostructures (e.g. nanowires, nanoribbons, nanorods, etc.). In addition, the anaerobic biotransformation of a-Se nanospheres to t-Se nanorods has been shown for microbial granular activated sludge only under thermophilic (55 °C) conditions.18 Biogenic Se nanorods were characterized by their low colloidal stability and high settling efficiency in comparison with a-Se nanospheres, making them highly attractive for bioremediation of Se contaminated waters.18 However, no Se nanorods were produced under mesophilic conditions (30 °C). Here we report the first study on the synthesis of Se nanostructures with different shapes (spherical, hexagonal, polygonal, and nanowires) and crystallographic properties under mesophilic conditions (30 °C) by the cells of S. bentonitica, isolated from Spanish bentonite formations.5 A scanning transmission electron microscope equipped with a high angle annular field detector (STEM/HAADF) showed the time-dependent transformation process of a-Se nanospheres to t-Se nanostructures with different morphologies (hexagonal, polygonal, and nanowires). Environmental scanning electron microscopy (ESEM) analysis revealed the impact of extracellular flagella-like proteins in this transformation process since the a-Se nanospheres aggregate along the axis of the flagella-like proteins whose monomers seem to embed these nanomaterials. This work provides new molecular insights into the impact of biogenic Se nanostructures on the mobility of this element and the effect of microbial processes on the safety of deep geological repository systems. In addition, this diversity of Se nanostructure shapes will play a basic role in the development of nanoscience and technology.
Experimental
Bacterial strain and growth conditions
The bacterial strain used in the present work was recently described by our research group as a novel species named S. bentonitica BII-R7T.5 This strain was isolated from Spanish bentonites collected from the Cabo de Gata Natural Park (Almeria, Spain).3 The cells were grown aerobically in Luria-Bertani (LB) broth medium (tryptone 10 g l−1, yeast extract 5 g l−1 and NaCl 10 g l−1, pH 7.0 ± 0.2) at 28 °C and 180 rpm in a rotary shaker.Metal solution preparation
Sodium selenite (Na2SeO3) (Sigma-Aldrich) was prepared as a 1 M stock solution by dissolving appropriate quantities in distilled water. Finally, the solution was sterilized by filtration using 0.22 μm syringe filters (Sartorius®).Determination of the minimum inhibitory concentration (MIC) of selenite for the bacterial growth
The minimum inhibitory concentrations (MICs) of Se(IV) were determined in triplicate. Cells of the isolate were grown to the late exponential phase to a final optical density (O.D) of 0.9 (at 600 nm) in LB broth and washed twice with 0.9% NaCl. Finally, 10 μl of the cell suspension was inoculated to LB agar supplemented with increasing concentrations of Se(IV) from 1 to 400 mM. Afterwards, the plates were incubated at 28 °C for 48 h. The MIC was defined as the lowest concentration of the element at which complete inhibition of colony formation is observed.24X-ray diffraction
XRD analysis was carried out to determine the size and crystalline phase of biogenic Se nanoparticles. S. bentonitica cells treated with 2 mM Se(IV) for 144 h were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and the resultant pellet was washed with double distilled water. The washed samples were dried at 28 °C for 24 h. X-ray patterns of the biogenic SeNPs were obtained with a Bruker D8 Advanced diffractometer equipped with a LINXEYE detector available at the University of Granada. The obtained diffractograms were analysed using the software DIFFRAC PLUS. In order to calculate the average size of the nanoparticles, Scherrer's equation (D = kλ/β
000 rpm for 10 min and the resultant pellet was washed with double distilled water. The washed samples were dried at 28 °C for 24 h. X-ray patterns of the biogenic SeNPs were obtained with a Bruker D8 Advanced diffractometer equipped with a LINXEYE detector available at the University of Granada. The obtained diffractograms were analysed using the software DIFFRAC PLUS. In order to calculate the average size of the nanoparticles, Scherrer's equation (D = kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ) was used, where k is a constant whose value is approximately 0.9, λ is the wavelength of the X-ray, β is the width in radians of the peak due to the size effect and θ is Bragg's diffraction angle. Specifically, the particle size of the sample was estimated from the line width of the (101) XRD peak.
θ) was used, where k is a constant whose value is approximately 0.9, λ is the wavelength of the X-ray, β is the width in radians of the peak due to the size effect and θ is Bragg's diffraction angle. Specifically, the particle size of the sample was estimated from the line width of the (101) XRD peak.
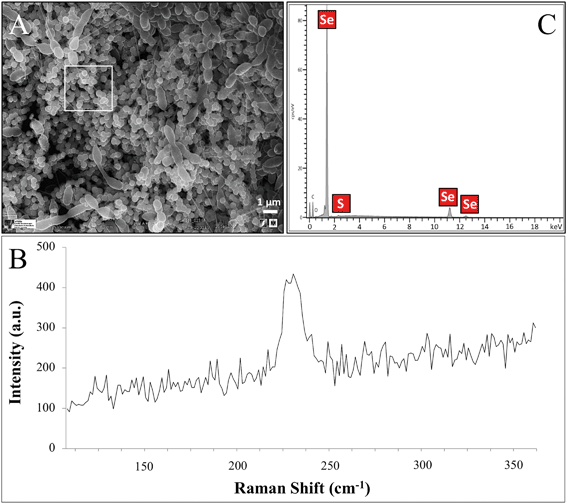
VP-FESEM and Raman spectroscopy analysis
A variable pressure field emission scanning electron microscope (VP-FESEM) equipped with an X-ray detector Raman spectroscopy system enabled in situ 3-D structural and elemental characterization of the SeNPs produced by the cells. Cell suspensions supplemented with 2 mM Se(IV) for 144 h were fixed with 3% glutaraldehyde in 0.05 M sodium cacodylate buffer (pH 7.2) for 24 h at 4 °C. Afterwards, the samples were washed three times with the same buffer. The resultant pellets were fixed with 1% osmium tetroxide solution (OsO4) in cacodylate buffer before dehydration in graded ethanol solutions in water. The critical point drying method was also used to dehydrate the samples. Finally, they were coated with carbon and stored in a desiccator. The samples were analysed under a variable pressure field emission scanning electron microscope (Zeiss SUPRA 40VP).STEM/HAADF analysis
The morphology, elemental composition and cellular location of reduced Se were analysed by using a scanning-transmission electron microscope (STEM) equipped with an energy dispersive X-ray (EDX) spectrometer. EDX analysis was performed at 300 kV using a spot size of 4 Å and a live counting time of 50 s. The structural characterization of Se nanostructures was performed by using selected-area electron diffraction (SAED) and high-resolution TEM combined with fast Fourier transform (FFT).STEM samples consisting of Se(IV)-treated cells (2 mM) were prepared as described in Merroun et al.25 after 24, 48, 72 and 144 h of incubation. Finally, the samples were examined under a high-angle annular dark field scanning transmission electron microscope (HAADF-STEM, FEI TITAN G2 80-300). STEM specimen holders were cleaned with plasma prior to STEM analysis to minimize contamination.
FEG-ESEM
A field emission gun environmental scanning electron microscope (FEG-ESEM) equipped with secondary and circular backscatter electron detectors (ETD and CBS) enabled the determination of the presence of organic matter surrounding the biogenic SeNPs. For this purpose, the samples described above for VP-FESEM were also analysed by using this technique. In addition, this technique was also applied to explain the biotransformation of a-Se nanospheres to t-Se nanostructures. Thus, the cells were brought into contact for 24 h with solutions containing two different types of a-Se nanospheres: 1) chemically produced (CheSeNPs); and 2) organically produced SeNPs (OrgSeNPs). Chemical SeNPs were produced by reduction of sodium selenite (5 mM) by reduced glutathione (GSH) (20 mM) at 28 °C. OrgSeNPs were synthesized using proteins (Bovine Serum Albumin; BSA) (0.01 g l−1) in addition to GSH (20 mM) and Se(IV) (2 mM). Finally, the samples were analysed under a FEG-SEM microscope (FEI QEMSCAN 650F).Effect of selenite on the bacterial growth
The potential of S. bentonitica to tolerate selenite was assayed by growing the cells in LB liquid medium (30 ml) supplemented with 2 mM Na2SeO3. Untreated (control) and Se(IV)-treated cells were incubated at 28 °C by shaking at 180 rpm. In addition, dead cells obtained by heating the biomass at 90 °C and LB broth containing Se(IV) (abiotic) were used as controls. The growth was evaluated by quantifying the total protein content in bacterial cell extracts using a modification of the method of Dhanjal and Cameotra.26 The total protein content was correlated with the increase in the cell growth. A 1 ml aliquot of bacterial culture was taken at different time intervals to measure the growth based on the protein content of the cells by using the Bradford reagent (Bio-Rad®). Bovine serum albumin (BSA) was used as a standard. All the measurements were performed in triplicate.Results and discussion
Bacterial reduction of Se(IV)
The ability of S. bentonitica to reduce aerobically Se(IV) to Se(0) was tested in LB liquid medium at 2 mM selenite. The colour of the Se(IV)-treated cultures of S. bentonitica turned intense orange-red after 24 h of incubation, in contrast to untreated cultures (blanks) (Fig. 1). This colour change reveals the capacity of the strain to reduce Se(IV) to Se(0).27 The Se(IV) reduction is mediated by a biological rather than a chemical process since no colour change was observed in abiotic and dead cell control samples supplemented with Se(IV) (Fig. 1). The MIC of Se(IV) for the growth of the studied strain in solid medium was 400 mM as red precipitates and cell growth were observed in all assayed Se(IV) concentrations ranging from 1 to 200 mM (Fig. 2). The absence of both cell growth and red precipitates was appreciated at 400 mM Se(IV) (data not shown). This indicated the high Se tolerance of S. bentonitica compared to other species of the same genus like S. maltophilia SeITE02 and S. maltophilia Sm777, both of which exhibited a tolerance to Se(IV) of up to 50 mM.11,28 The high tolerance of S. bentonitica to selenite can also be appreciated by comparing its MIC with those of strains belonging to different bacterial genera. For example, the MIC of Se(IV) for the growth of Rhizobium selenitireducens strain B1 and Comamonas testosteroni S44 is between 8–16 mM and 100 mM, respectively.29,30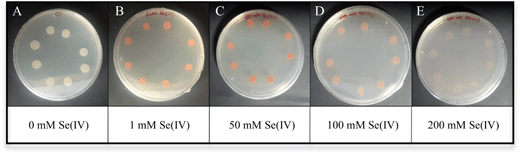 | ||
| Fig. 2 Growth of Stenotrophomonas bentonitica in LB solid medium without (A) or with 1 (B), 50 (C), 100 (D) and 200 mM (E) Se(IV) after 48 h of incubation. | ||
Recent studies indicated the presence of Se in the tetravalent oxidation state in high-level nuclear waste.31 The bioreduction of this soluble Se species to insoluble elemental Se would reduce the mobility of Se in future DGR systems. This fact makes S. bentonitica an excellent candidate to improve the safety of these future repositories of radioactive waste.
Effect of Se on the bacterial growth
In order to evaluate the biological effect of Se(IV) on S. bentonitica, the growth profile of the cells was studied and expressed as the total protein content as a function of time to avoid optical interferences of Se(0) red accumulations in spectrophotometric measurements. In comparison with the control (0 mM Se(IV)), the microbial growth was negatively affected at an initial concentration of 2 mM Se(IV) (Fig. 3). It is important to note that the growth of S. bentonitica cells presented a longer lag phase of about 48 hours in the presence of Se. Vogel et al.32 detected that the growth of Azospirillum brasilense also exhibited a lag phase whose duration depends on the initial concentration of Se(IV): 2 days at 1 mM Se(IV) and up to 6 days when the cells were brought into contact with Se(IV) concentrations ranging from 1 to 5 mM. Furthermore, Ralstonia metallidurans also presented a prolonged lag phase (around 30 h) at 2 mM Se(IV), suggesting the adaptation of the cells to Se(IV) stress.33 In Se(IV)-treated cells of S. bentonitica, this stress could be associated with the presence of Se(IV) which was determined to be about 50% of the initial Se(IV) concentration at 48 h (data not shown). The optimum growth reached by the cells treated with Se(IV) was at 144 h of incubation where the Se(IV) concentration was about 0% (100% reduction of Se(IV) to Se(0)) (data not shown). It was shown that 15, 50, 60 and 100% of Se(IV) was reduced to Se(0) after a period of 24, 48, 72 and 144 h time incubation, respectively (data not shown).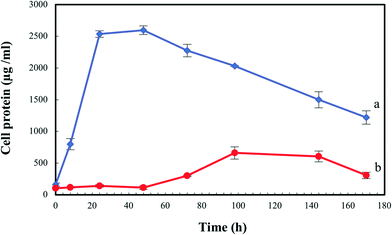 | ||
| Fig. 3 Growth profile of Stenotrophomonas bentonitica in the absence (a) and the presence of 2 mM Se(IV) (b). Selenite was added at time zero. Standard errors are given correspondingly. | ||
The lag phase observed in the present and other studies could be probably associated with differential expressions of enzymes involved in Se(IV) tolerance. The whole genome analysis of S. bentonitica revealed the presence of genes codifying enzymes described for their ability to reduce Se(IV) to Se(0).34 Glutathione reductase and thioredoxin reductase as well as NADH-dependent enzymes such as NADH:flavin oxidoreductase (OYE family) previously reported to be implicated in the reduction of this oxy-anion35 have been identified in the genome of S. bentonitica. Several mechanisms have been proposed for the reduction of selenite in microorganisms including a Painter-type reaction,36 thioredoxin reductase system,37 sulphide mediated reduction,38 dissimilatory reduction,39etc.
X-ray diffraction (XRD) analysis
The nature of the crystalline phase and the size of reduced SeNPs produced by cells treated with 2 mM Se(IV) for 144 h were determined by using X-ray diffraction. The XRD patterns obtained show the main peaks characteristic of crystalline t-Se (COD-9008579) at 2θ values of 23.5°, 29.7°, 41.4°, 43.7° and 45.4° (Fig. 4) corresponding to the crystal planes (100), (101), (110), (102) and (111), respectively. The average crystalline size of biogenic Se nanostructures measured by Scherrer's equation was about 34 nm.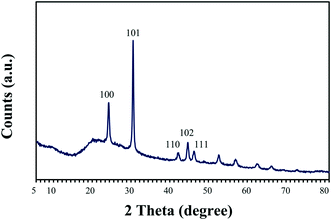 | ||
| Fig. 4 X-ray diffraction pattern of selenium nanoparticles synthesized by Stenotrophomonas bentonitica after 144 h of incubation. | ||
These results agree well with literature data from Borghese et al.40 and Srivastava and Mukhopadhyay,41 who reported the production of crystalline nanostructures by Rhodobacter capsulatus and Zooglea ramigera, respectively. The diffraction patterns corresponding to these nanoparticles show the main peaks belonging to trigonal Se(0) crystals. The production of crystalline Se nanostructures by the cells of S. bentonitica could be of great significance in DGR systems since the mobility of Se through the surrounding environment may be reduced due to the low solubility of these Se nanostructures.18 Zhang et al.22 could not detect in the XRD pattern the presence of t-Se nanorods previously detected by microscopic techniques after 48 h of incubation with Pseudomonas alcaliphila. They attributed it to the shortage of Se nanorods and the attachment of Se nanospheres onto the surface of nanorods. Se nanospheres produced by Bacillus sp. SN3 have been previously reported for their amorphous nature since sharp Bragg reflections were not observed in their diffraction pattern.8 As a consequence, amorphous nanospheres could interfere with XRD results. In our case, the XRD peak clearly showed the presence of crystalline t-Se.
Microscopic characterization of Se(IV) reduction by the S. bentonitica cells
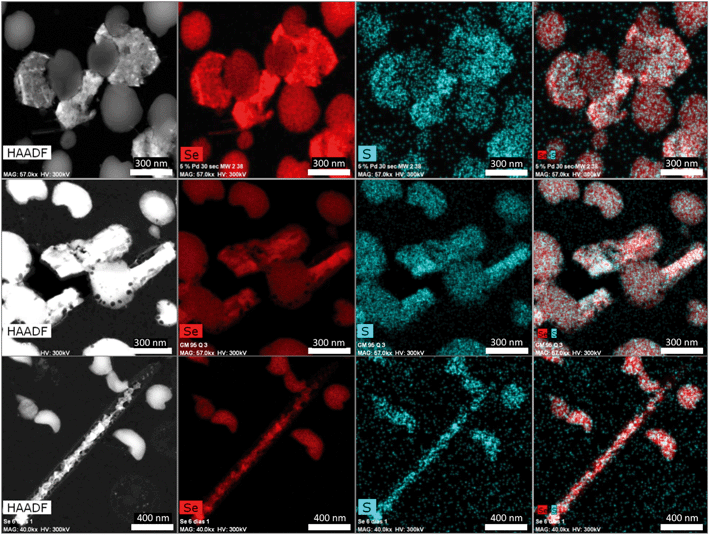
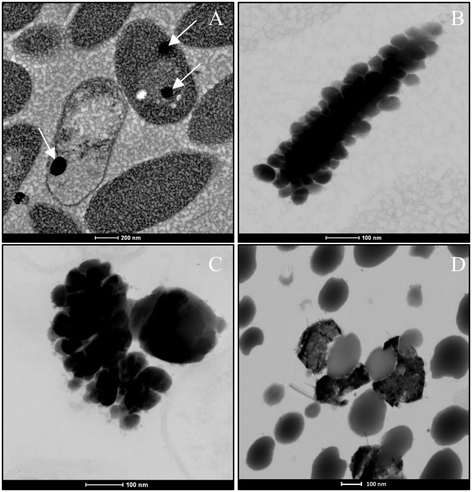
The Se nanostructures identified by STEM exhibited different shapes: spheres, nanowires, hexagons, polygons, etc. Energy dispersive X-ray (EDX) elemental mapping derived from these accumulations showed that they are mainly composed of Se, in addition to S (Fig. 6). Many investigations reported previously the formation of biogenic SeNPs.14,44 However, very few authors described the presence of S in these nanostructures. For instance, Vogel et al.32 detected S in Se nanospheres synthesized by Azospirillum brasilense by using EDX analysis. Combining Raman and EXAFS spectroscopy they could determine the local coordination of selenium within the biogenic SeNPs where Se is coordinated to S atoms forming Se8−nSn structured SeNPs forming 8-rings typical for amorphous Se(0) with a partial S-for-Se substitution in the 8-ring. The presence of S suggests the involvement of this element in the formation of the nanospheres. In this sense, extensive studies on the mechanisms of Se(IV) reduction have indicated the involvement of molecules containing thiol groups (–SH) such as reduced glutathione (GSH) in the reduction of Se(IV) to Se(0).36,47 GSH is proposed to act as an electron donor and reacts with selenite to form selenodiglutathione (GS–Se–SG). Selenodiglutathione reduction by glutathione reductase (GR) leads to the formation of glutathione selenopersulfide (GS–Se−), which dismutates into Se(0) and GSH.
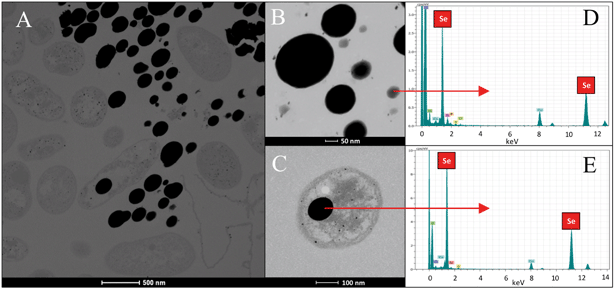
In the case of Se nanospheres, extensive accumulations of this morphology with two considerable different sizes were detected. Specifically, smaller nanospheres with diameters of 20 to 30 nm and bigger NPs around 200 nm were found (Fig. 7A–C). EDX microanalysis indicated that they are mainly composed of Se and S (Fig. 7D and E). In addition, the selected area electron diffraction (SAED) pattern of the nanospheres indicated their amorphous nature (Fig. 8A and B). Production of biogenic spherical SeNPs through the reduction of Se(IV) to Se(0) by several bacterial strains has been previously reported. The strain Rhodopseudomonas palustris N produces nanospheres between 80 to 200 nm as indicated by TEM analysis when cells grew on Se(IV) after 8 days.48 On the other hand, Srivastava and Mukhopadhyay41 reported the formation of spherical SeNPs ranging from 30 to 150 nm by Zooglea ramigera after 48 h. Benko et al.49 demonstrated that biologically produced spherical SeNPs are less toxic than Se oxyanions such as Se(IV) and Se(VI). However, other studies revealed that the toxicity of SeNPs can vary widely among different species.50 In their experiments, Li et al.50 found a higher toxicity of SeNPs compared to Se(IV) in the fish species Oryzias latipes.
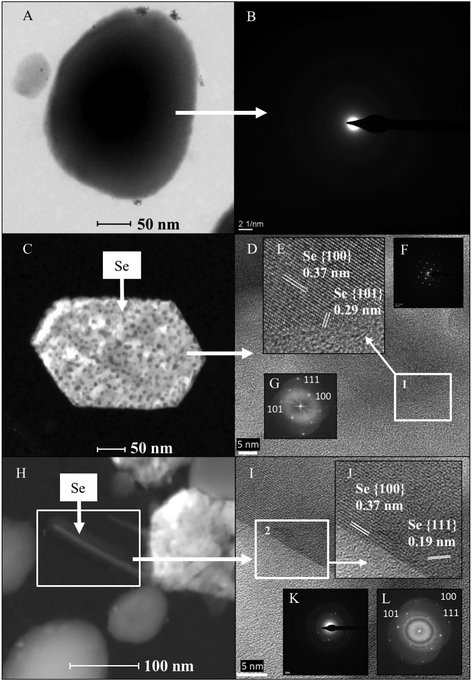
To a lesser extent, hexagonal-shaped nanostructures from 100 to 400 nm in size were produced (Fig. 8C). Their crystal structure was investigated using SAED and high-resolution scanning transmission electron microscopy (HRSTEM) combined with Fast Fourier Transform (FFT). The HRTEM image of an individual hexagonal-shaped nanoparticle shows 2 distinct lattice spacings of 0.37 and 0.29 nm, corresponding to the (100) and (101) planes of t-Se, respectively (Fig. 8D and E). The SAED pattern indicated that the hexagonal-shaped nanostructures are single crystals, which can be well indexed to t-Se (Fig. 8F). In addition, the lattice spacing values obtained for all the diffraction rings from the FFT of the image (Fig. 8G) are in agreement with the trigonal phase of Se. The production of SeNPs with a hexagonal shape has rarely been reported. Ho et al.23 demonstrated the formation of Se nanoribbons with a hexagonal cross section from biogenic Se nanospheres in the presence of 80% dimethyl sulfoxide (DMSO). As far as we know, the present work describes for the first time the biological production of crystalline hexagonal-shaped SeNPs without the addition of organic solvents under mesophilic conditions. Consequently, here we report a simpler method for the preparation of crystalline SeNPs than those previously described.23,51 We provide an economic, environmentally-friendly and promising method for the synthesis of SeNPs, well known to be useful in many industrial and medical applications. The SAED patterns and HRTEM images of the polygonal forms also evidenced the trigonal phase of Se (data not shown).
The last type of SeNP shape detected consisted of nanowires. In the same way as the hexagonal and polygonal-shaped nanostructures, Se nanowires around 30 nm in diameter were found (Fig. 8H) to a lesser extent compared with the nanospheres. The HRTEM image of an individual nanowire shows 2 distinct lattice spacings of 0.37 and 0.19 nm, corresponding to the (100) and (111) planes of t-Se (Fig. 8I and J). The SAED spots revealed that the Se nanowires are polycrystalline (Fig. 8K). In addition, the FFT of the image shown in Fig. 8L further confirms the trigonal phase of selenium. These results match very well with the XRD diffraction pattern described in the section above, indicating the presence of t-Se ranging from 30 to 45 nm in size.
The shape of Se nanostructures greatly affects their colloidal stability and surface charge, which in turn influences their mobility and the effectiveness of bioremediation.18 Jain et al.18 demonstrated the lower colloidal stability and mobility of biogenic Se nanorods compared with biogenic Se nanospheres produced by anaerobic granular sludge. For this reason, the formation Se nanowires would be beneficial for Se immobilization in future disposal of radioactive waste. In addition to environmental purposes, SeNPs have acquired major relevance in the field of medicine owing to their antioxidative and anticancer properties. They are also extensively used in industrial applications (solar cells, photocopiers, electronics, optics, etc.) due to their semiconductor, photoelectrical and catalytic properties.16
Careful observation of TEM micrographs revealed that some nanowires are protruding out of the hexagonal and polygonal-shaped Se nanostructures (Fig. 8H). This could indicate that the nanowires aggregate, forming Se hexagons and polygon-shaped nanostructures. This hypothesis is supported by the discontinuous distribution of Se within these nanostructures as indicated by elemental mapping analysis, and also by the fact that the hexagons, polygons and nanowires have the same crystalline structure (trigonal). It has been previously reported that crystallization of SeNPs increases their settleability and hence immobility through the environment.52 For this reason, the formation of crystalline Se nanostructures may have a positive effect on the safety of DGRs due to their lower solubility and mobility.
Biotransformation of amorphous Se nanospheres to trigonal Se nanostructures
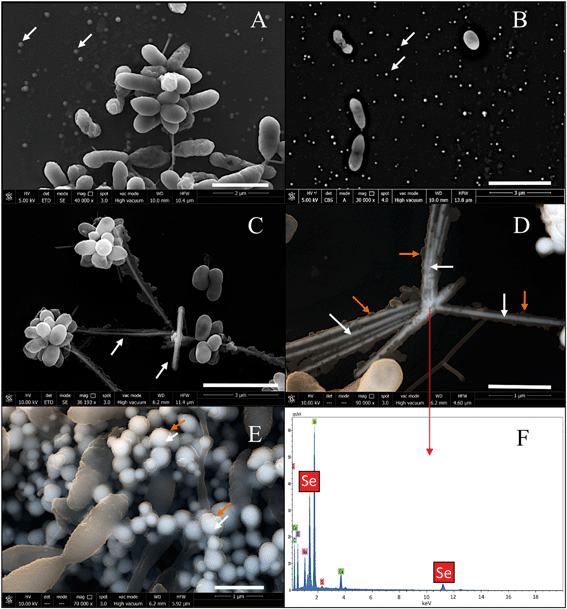
The production of different Se nanostructure shapes by the cells is a time dependent process. After an incubation period of 24 h, 30–200 nm sized biogenic individual a-Se nanospheres were synthesized (Fig. 9A). The latter start to coalesce, forming aggregates after 48 and 72 h (Fig. 9B and C). However, the cells were able to produce a mixture of single crystal and polycrystalline one-dimensional (1D) t-Se nanostructures with different shapes (e.g. nanowires, hexagons, polygons, etc.) and diameters of 30–400 nm, in addition to a-Se nanospheres after 144 h of incubation (Fig. 9D). These results suggested the probable transformation of a-Se nanospheres to t-Se nanostructures with different shapes. This hypothesis is supported by VP-FESEM and Raman analysis results showing the aggregation and crystallization of SeNPs (see Fig. 4). Li et al.53 and Chen et al.13 evidenced the synthesis of Se nanospheres and t-Se nanotubes and nanowires with HRSTEM analysis by using different chemical processes. They both suggested that spherical SeNPs are initially generated and then they transform into the more stable t-Se nanowires and nanotubes.In order to determine the impact that bacterial cells and their components (e.g. proteins) can have on the transformation of a-Se NPs to t-Se NPs, we performed a series of control experiments where S. bentonitica cells were brought into contact for 24 h with chemically (CheSeNPs) and organically produced SeNPs (OrgSeNPs). The organic SeNPs were synthesized using proteins (i.e. BSA) as the template and represent the early stage of biogenic formation of amorphous SeNPs. The CheSeNPs are the SeNPs produced chemically and were used as the negative control (the absence of proteins and bacteria).
When the experiment was conducted without the presence of bacteria, TEM images revealed the presence of only amorphous spherical SeNPs with a homogenous size distribution in both CheSeNPs and OrgSeNPs samples (Fig. S1†). However, in the presence of bacteria, whilst FEG-ESEM showed a high number of individual CheSeNPs distributed in the extracellular space without any contact with the cells (Fig. 10A and B), both Se nanowires and nanospheres associated with cells were detected (Fig. 10C–E) when proteins (BSA) were present. EDX analysis confirmed that both nanostructures are composed of Se (Fig. 10F). In the latter sample, spherical SeNP aggregates are formed by a high number of individual Se nanospheres which seem to be embedded in an organic matrix. This aggregation has been previously reported to be a crucial step during the transformation process to Se nanowires.43 The aggregation takes place along an axis which could correspond to the flagella-like protein structures characteristic of this bacterium. Flagella-like proteins of S. maltophilia are composed of several thousand copies of flagellin subunits,54 and are involved in the formation of biofilms on abiotic surfaces such as glass, Teflon, polystyrene, and stainless steel,54,55 as well as on biotic surfaces.56 Bacterial flagella are also used as templates for formation of different nanomaterials including pearl-necklace-like 1D silica nanotubes in aqueous solution.57 Flagellin monomers from Pseudomonas fluorescens were reported for their ability to form a monolayer thick protein film on the Au(111) surface through hydrophobic interactions.58 These results suggested that organic matter (mainly proteins capping the NPs and flagella-like proteins) produced by S. bentonitica (Fig. S2†) plays an important role in the aggregation and hence in the transformation from nanospheres to nanowires. We proposed that the transformation to t-Se nanostructures occurs once the nanospheres have been released from the cells, probably through the process of cell lysis. This is supported by the appearance of some SeNPs in the proximity of the lysed cells (Fig. S3†). The transport of Se nanospheres across the membrane seems unlikely due to their large size and no detection of vesicles in HAADF-STEM analysis. A transformation process has been proposed where the released Se nanospheres from the intracellular space start to coalesce and probably grow on the axis of flagella-like proteins (Fig. 11). Finally, they crystallize forming different t-Se nanostructures. However, more experimental data are needed to prove this proposed transformation mechanism model. The different Se(0) allotropes should eventually be converted to one of the most thermodynamically stable forms such as t-Se. Minaev et al.59 reported that from a thermodynamic point of view, the transformation of amorphous Se to crystalline Se allotropes (monoclinic and trigonal Se) requires high temperatures. Specifically, the amorphous form is transformed into monoclinic Se at temperatures around 70 °C and into the hexagonal form at higher temperatures. However, in this work, the reduction of Se(IV) to Se(0) and the transformation to t-Se nanostructures by Stenotrophomonas bentonitica is considered as detoxification mechanisms used by this bacterium to cope with the toxicity of Se(IV) and it is not governed by a thermodynamic law. This process takes place at a temperature of about 28 °C for 6 days.
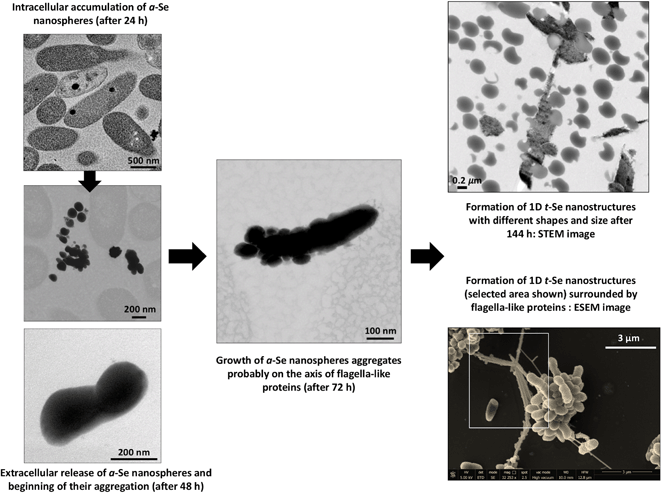 | ||
| Fig. 11 Proposed mechanism of time-dependent transformation of a-Se nanospheres to different 1D t-Se nanostructures. | ||
Many authors have reported the synthesis of SeNPs coated by organic layers mainly composed of proteins, lipids and polysaccharides.18,60 Ni et al.61 reported the first 3D electron tomographic analysis of Se(IV) reduction by the cells of Pseudomonas moraviensis stanleyae providing new insights into the interactions at the nanoparticle/protein interface. They identified the first polypeptide playing a major role in the reduction of Se(IV) and formation of intracellular of SeNPs. This polypeptide is also involved in retaining the produced Se(0)NPs forming complexes with the latter in the reduction site, and controlling their size. In addition, the authors demonstrated that this biopolymer layer is acquired after intracellular NP formation, either during extracellular transport or when the NPs are outside the cell. However, the impact of bioorganic layers during the formation of SeNPs remains to be fully elucidated. In particular, the role of proteins in the transformation of SeNPs as well as in controlling the size distribution has been previously suggested.43,62,63 Dobias et al.63 demonstrated the role of AdhP, a single purified protein, in decreasing the average size of chemically produced SeNPs. On the other hand, Wang et al.43 reported the polyvalency of proteins during the transformation from Se nanospheres to stable Se nanowires. According to them, proteins are involved in trapping on the surface and reducing the Se(IV) ions. In addition, they suggested that proteins excreted by Bacillus subtilis serve as templates hindering the further growth of monoclinic-Se and accelerating its transformation to trigonal nanowires. Although the exact mechanism of the transformation of spherical SeNPs to t-Se 1D structures in S. bentonitica is still unclear, the directing role of cells and their proteins is undoubtedly significant.
Future studies are in progress to elucidate the molecular mechanisms of interaction between the SeNPs on the surface of purified flagella and the impact of these proteins in the transformation of spherical SeNPs to trigonal ones.
Conclusions
In summary, this study described the synthesis of biogenic Se nanostructures with different shapes (spherical, hexagonal, polygonal and nanowires) and distinct crystallographic properties (amorphous and trigonal structure) by selenite-tolerant Stenotrophomonas bentonitica, isolated from Spanish bentonites. Time dependent SeNP experiments showed that the cells of the strain S. bentonitica and their proteins are able to transform amorphous Se(0) nanospheres (a-Se) to one-dimensional (1D) trigonal selenium (t-Se) nanostructures (hexagons, polygons and nanowires) under mesophilic conditions. However, more studies are needed to determine the exact mechanism of the transformation process. The Se(IV) reduction and production of crystalline t-Se would be of great significance in DGR systems since the mobility of Se through the surrounding environment may be reduced. The potential environmental significance of this study includes understanding of the impact of microbial processes on the transport of selenium in future radioactive repositories which in turn will help to support the implementation of planned repositories.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the Euratom research and training programme 2014-2018 under grant agreement no. 661880. The authors acknowledge the assistance of Maria del Mar Abad Ortega, Isabel Sánchez Almazo, Isabel Guerra Tschuschke and Concepción Hernández Castillo (Centro de Instrumentación Científica, University of Granada, Spain) for their help with microscopy measurements. The authors also thank José Romero Garzón (Centro de Instrumentación Científica, University of Granada, Spain) for his assistance with the XRD measurements.References
- IAEA, Scientific and technical basis for geological disposal of radioactive wastes, Vienna, International Atomic Energy Agency, 2003, Technical reports series, n. 413. STI/DOC/010/413. Available from: http://www-pub.iaea.org/MTCD/Publications/PDF/TRS413_web.pdf Search PubMed.
- M. V. Villar, L. Pérez del Villar, P. L. Martín, M. Pelayo, A. M. Fernández and A. Garralon, et al. The study of Spanish clays for their use as sealing materials in nuclear waste repositories : 20 years of progress, Journal of Iberian Geology, 2006, 32(1), 15–36 Search PubMed.
- M. López-Fernández, O. Fernández-Sanfrancisco, A. Moreno-García, I. Martín-Sánchez, I. Sánchez-Castro and M. L. Merroun, Microbial communities in bentonite formations and their interactions with uranium, Appl. Geochem., 2014, 49, 77–86 CrossRef.
- M. López-Fernández, A. Cherkouk, R. Vilchez-Vargas, R. Jauregui, D. Pieper and N. Boon, et al. Bacterial Diversity in Bentonites, Engineered Barrier for Deep Geological Disposal of Radioactive Wastes, Microb. Ecol., 2015, 70(4), 922–935 CrossRef PubMed.
- I. Sánchez-Castro, M. A. Ruiz-Fresneda, M. Bakkali, P. Kämpfer, S. P. Glaeser and H. J. Busse, et al. Stenotrophomonas bentonitica sp. nov., isolated from bentonite formations, Int. J. Syst. Evol. Microbiol., 2017, 67(8), 2779–2786 CrossRef PubMed.
- A. Meleshyn, Microbial Processes Relevant for Long-Term Performance of Radioactive Waste Repositories in Clays. Gesellschaftfür Anlagen-und Reaktorsicherheit (GRS) mbH, 2011. GRS – 291. ISBN 978-3-939355-67-0. Availablefrom: https://www.grs.de/sites/default/files/pdf/GRS-291.pdf Search PubMed.
- M. L. Merroun, M. Nedelkova, J. J. Ojeda, T. Reitz, M. L. Fernández and J. M. Arias, et al. Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X-ray absorption spectroscopic analyses, J. Hazard. Mater., 2011, 197, 1–10 CrossRef PubMed.
- N. T. Prakash, N. Sharma, R. Prakash, K. K. Raina, J. Fellowes and C. I. Pearce, et al. Aerobic microbial manufacture of nanoscale selenium: Exploiting nature's bio-nanomineralization potential, Biotechnol. Lett., 2009, 31(12), 1857–1862 CrossRef PubMed.
- H. Moll, L. Lütke, V. Bachvarova, A. Cherkouk, S. Selenska-Pobell and G. Bernhard, Interactions of the Mont Terri Opalinus Clay Isolate Sporomusa sp. MT-2.99 with Curium(III) and Europium(III), Geomicrobiol. J., 2014, 31(8), 682–696 CrossRef.
- G. Jörg, R. Bühnemann, S. Hollas, N. Kivel, K. Kossert and S. Van Winckel, et al. Preparation of radiochemically pure 79Se and highly precise determination of its half-life, Appl. Radiat. Isot., 2010, 68(12), 2339–2351 CrossRef PubMed.
- S. Di Gregorio, S. Lampis and G. Vallini, Selenite precipitation by a rhizospheric strain of Stenotrophomonas sp. isolated from the root system of Astragalus bisulcatus: a biotechnological perspective, Environ. Int., 2005, 31(2), 233–241 CrossRef PubMed.
- E. Breynaert, A. C. Scheinost, D. Dom, A. Rossberg, J. Vancluysen and E. Gobechiya, et al. Reduction of Se(IV) in boom clay: XAS solid phase speciation, Environ. Sci. Technol., 2010, 44(17), 6649–6655 CrossRef PubMed.
- H. Chen, D. W. Shin, J. G. Nam, K. W. Kwon and J. B. Yoo, Selenium nanowires and nanotubes synthesized via a facile template-free solution method, Mater. Res. Bull., 2010, 45(6), 699–704 CrossRef.
- D. B. Li, Y. Y. Cheng, C. Wu, W. W. Li, N. Li and Z. C. Yang, et al. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm, Sci. Rep., 2014, 4, 1–7 CrossRef.
- R. S. Dungan, S. R. Yates and W. T. Frankenberger, Transformations of selenate and selenite by Stenotrophomonas maltophilia isolated from a seleniferous agricultural drainage pond sediment, Environ. Microbiol., 2003, 5(4), 287–295 CrossRef PubMed.
- S. A. Wadhwani, U. U. Shedbalkar, R. Singh and B. A. Chopade, Biogenic selenium nanoparticles: current status and future prospects, Appl. Microbiol. Biotechnol., 2016, 100(6), 2555–2566 CrossRef PubMed.
- S. Lampis, E. Zonaro, C. Bertolini, P. Bernardi, C. S. Butler and G. Vallini, Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SeITE01 as a consequence of selenite reduction under aerobic conditions, Microb. Cell Fact., 2014, 13(1), 1–14 CrossRef PubMed.
- R. Jain, N. Jordan, S. Tsushima, R. Hübner, S. Weiss and P. N. L. Lens, Shape change of biogenic elemental selenium nanomaterials from nanospheres to nanorods decreases their colloidal stability, Environ. Sci.: Nano, 2017, 4(5), 1054–1063 RSC.
- B. Buchs, M. W. H. Evangelou, L. H. E. Winkel and M. Lenz, Colloidal properties of nanoparticular biogenic selenium govern environmental fate and bioremediation effectiveness, Environ. Sci. Technol., 2013, 47(5), 2401–2407 CrossRef PubMed.
- A. A. Keller, H. Wang, D. Zhou, H. S. Lenihan, G. Cherr and B. J. Cardinale, et al. Stability and aggregation of metal oxide nanoparticles in natural aqueous matrices, Environ. Sci. Technol., 2010, 44(6), 1962–1967 CrossRef PubMed.
- A. S. Eswayah, T. J. Smith, A. C. Scheinost, N. Hondow and P. H. E. Gardiner, Microbial transformations of selenite by methane-oxidizing bacteria, Appl. Microbiol. Biotechnol., 2017, 101(17), 6713–6724 CrossRef PubMed.
- W. Zhang, Z. Chen, H. Liu, L. Zhang, P. Gao and D. Li, Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonas alcaliphila, Colloids Surf., B, 2011, 88(1), 196–201 CrossRef PubMed.
- C. T. Ho, J. W. Kim, W. B. Kim, K. Song, R. A. Kanaly and M. J. Sadowsky, et al. Shewanella-mediated synthesis of selenium nanowires and nanoribbons, J. Mater. Chem., 2010, 20(28), 5899–5905 RSC.
- S. Rossbach, T. L. Wilson, M. L. Kukuk and H. A. Carty, Elevated zinc induces siderophore biosynthesis genes and a znt A-like gene in Pseudomonas fluorescens, FEMS Microbiol. Lett., 2000, 191(1), 61–70 CrossRef PubMed.
- M. L. Merroun, J. Raff, A. Rossberg, C. Hennig, T. Reich and S. Selenska-Pobell, Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12, Appl. Environ. Microbiol., 2005, 71(9), 5532–5543 CrossRef PubMed.
- S. Dhanjal and S. S. Cameotra, Aerobic biogenesis of selenium nanospheres by Bacillus cereus isolated from coalmine soil, Microb. Cell Fact., 2010, 9, 52 CrossRef PubMed.
- M. Sabaty, C. Avazeri, D. Pignol and A. Vermeglio, Characterization of the Reduction of Selenate and Tellurite by Nitrate Reductases, Appl. Environ. Microbiol., 2001, 67(3–12), 5122–5126 CrossRef PubMed.
- D. Pages, J. Rose, S. Conrod, S. Cuine, P. Carrier and T. Heulin, et al. Heavy metal tolerance in Stenotrophomonas maltophilia, PLoS One, 2008, 3(2), e1539, DOI:10.1371/journal.pone.0001539.
- W. J. Hunter and L. D. Kuykendall, Reduction of selenite to elemental red selenium by Rhizobium sp. strain B1, Curr. Microbiol., 2007, 55, 344–349 CrossRef PubMed.
- S. Zheng, J. Su, L. Wang, R. Yao, D. Wang and Y. Deng, et al. Selenite reduction by the obligate aerobic bacterium Comamonas testosteroni S44 isolated from a metal-contaminated soil, BMC Microbiol., 2014, 14, 204 CrossRef PubMed.
- K. Dardenne, E. González-Robles, J. Rothe, N. Müller, G. Christill and D. Lemmer, et al. XAS and XRF investigation of an actual HAWC glass fragment obtained from the Karlsruhe vitrification plant (VEK), J. Nucl. Mater., 2015, 460, 209–215 CrossRef.
- M. Vogel, S. Fischer, A. Maffert, R. Hübner, A. C. Scheinost and C. Franzen, et al. Biotransformation and detoxification of selenite by microbial biogenesis of selenium-sulfur nanoparticles, J. Hazard. Mater., 2018, 344, 749–757 CrossRef PubMed.
- M. Roux, G. Sarret, I. Pignot-Paintrand, M. Fontecave and J. Coves, Mobilization of selenite by Ralstonia metallidurans CH34, Appl. Environ. Microbiol., 2001, 67(2), 769–773 CrossRef PubMed.
- I. Sánchez-Castro, M. Bakkali and M. L. Merroun, Draft genome sequence of Stenotrophomonas bentonitica BII-R7, a selenite-reducing bacterium isolated from Spanish bentonites, Genome Announc., 2017, 5(31), e00719, DOI:10.1128/genomeA.00719-17.
- W. J. Hunter, A Rhizobium selenitireducens protein showing selenite reductase activity, Curr. Microbiol., 2014, 68(3), 311–316 CrossRef PubMed.
- J. Kessi and K. W. Hanselmann, Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli, J. Biol. Chem., 2004, 279(49), 50662–50669 CrossRef PubMed.
- A. Yamada, M. Miyashita, K. Inoue and T. Matsunaga, Extracellular reduction of selenite by a novel marine photosynthetic bacterium, Appl. Microbiol. Biotechnol., 1997, 48(3), 367–372 CrossRef PubMed.
- A. M. Zawadzka, R. L. Crawford and A. J. Paszczynski, Pyridine-2,6-bis(thiocarboxylic acid) produced by Pseudomonas stutzeri KC reduces and precipitates selenium and tellurium oxyanions, Appl. Environ. Microbiol., 2006, 72(5), 3119–3129 CrossRef PubMed.
- M. Basaglia, A. Toffanin, E. Baldan, M. Bottegal, J. P. Shapleigh and S. Casella, Selenite-reducing capacity of the copper-containing nitrite reductase of Rhizobium sullae, FEMS Microbiol. Lett., 2007, 269(1), 124–130 CrossRef PubMed.
- R. Borghese, C. Baccolini, F. Francia, P. Sabatino and R. J. Turner, Zannoni D. Reduction of chalcogen oxyanions and generation of nanoprecipitates by the photosynthetic bacterium Rhodobacter capsulatus, J. Hazard. Mater., 2014, 269, 24–30 CrossRef PubMed.
- N. Srivastava and M. Mukhopadhyay, Biosynthesis and structural characterization of selenium nanoparticles mediated by Zooglea ramigera, Powder Technol., 2013, 244, 26–29 CrossRef.
- B. Gates, B. Mayers, B. Cattle and Y. Xia, Synthesis and characterization of uniform nanowires of trigonal selenium, Adv. Funct. Mater., 2002, 12(3), 219–227 CrossRef.
- T. Wang, L. Yang, B. Zhang and J. Liu, Extracellular biosynthesis and transformation of selenium nanoparticles and application in H2O2 biosensor, Colloids Surf., B, 2010, 80(1), 94–102 CrossRef PubMed.
- S. Lampis, E. Zonaro, C. Bertolini, D. Cecconi, F. Monti and M. Micaroni, et al. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles, J. Hazard. Mater., 2017, 324, 3–14 CrossRef PubMed.
- A. J. McBroom and M. J. Kuehn, Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response, Mol. Microbiol., 2007, 63(2), 545–558 CrossRef PubMed.
- H. Fernandez-Llamosas, L. Castro, M. L. Blazquez, E. Diaz and M. Carmona, Biosynthesis of selenium nanoparticles by Azoarcus sp. CIB, Microb. Cell Fact., 2016, 15(1), 1–10 CrossRef PubMed.
- Y. Tan, R. Yao, R. Wang, D. Wang, G. Wang and S. Zheng, Reduction of selenite to Se(0) nanoparticles by filamentous bacterium Streptomyces sp. ES2-5 isolated from a selenium mining soil, Microb. Cell Fact., 2016, 15, 157 CrossRef PubMed.
- B. Li, N. Liu, Y. Li, W. Jing, J. Fan and D. Li, et al. Reduction of selenite to red elemental selenium by Rhodopseudomonas palustris strain N, PLoS One, 2014, 9(4), e95955, DOI:10.1371/journal.pone.0095955.
- I. Benko, G. Nagy, B. Tanczos, E. Ungvari, A. Sztrik and P. Eszenyi, et al. Subacute toxicity of nano-selenium compared to other selenium species in mice, Environ. Toxicol. Chem., 2012, 31(12), 2812–2820 CrossRef PubMed.
- H. Li, J. Zhang, T. Wang, W. Luo, Q. Zhou and G. Jiang, Elemental selenium particles at nano-size (Nano-Se) are more toxic to Medaka (Oryzias latipes) as a consequence of hyper-accumulation of selenium: A comparison with sodium selenite, Aquat. Toxicol., 2008, 89(4), 251–256 CrossRef PubMed.
- Y. Shin, J. M. Blackwood, I. T. Bae, B. W. Arey and G. J. Exarhos, Synthesis and stabilization of selenium nanoparticles on cellulose nanocrystal, Mater. Lett., 2007, 61(21), 4297–4300 CrossRef.
- M. Lenz, A. C. van Aelst, M. Smit, P. F. X. Corvini and P. N. L. Lens, Biological production of selenium nanoparticles from wastewaters, Adv. Mater. Res., 2009, 71–73, 721–724 Search PubMed.
- X. Li, Y. Li, S. Li, W. Zhou, H. Chu and W. Chen, et al. Single crystalline trigonal selenium nanotubes and nanowires synthesized by sonochemical process, Cryst. Growth Des., 2005, 5(3), 911–916 CrossRef.
- B. A. Jucker, H. Harms and A. J. B. Zehnder, Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to Glass and Teflon, J. Bacteriol., 1996, 178(18), 5472–5479 CrossRef PubMed.
- N. D. Friedman, T. M. Korman, C. K. Fairley, J. C. Franklin and D. W. Spelman, Bacteraemia due to Stenotrophomonas maltophilia: An analysis of 45 episodes, J. Infect., 2002, 45(1), 47–53 CrossRef PubMed.
- S. Chhibber and A. K. Zgair, Involvement of Stenotrophomonas maltophilia flagellin in bacterial adhesion to airway biotic surfaces : an in vitro study, Am. J. Biomed. Sci., 2009, 1(3), 188–195 Search PubMed.
- F. Wang, D. Li and C. Mao, Genetically modifiable flagella as templates for silica fibers: From hybrid nanotubes to 1D periodic nanohole arrays, Adv. Funct. Mater., 2008, 18(24), 4007–4013 CrossRef.
- A. González-Orive, D. E. Pissinis, C. Diaz, A. Miñán, G. A. Benítez and A. Rubert, et al. Self-assembly of flagellin on Au(111) surfaces, J. Colloid Interface Sci., 2014, 433, 86–93 CrossRef PubMed.
- V. S. Minaev, S. P. Timoshenkov and V. V. Kalugin, Structural and Phase Transformation in Condensed Selenium, J. Optoelectron. Adv. Mater., 2005, 7(4), 1717–1741 Search PubMed.
- A. A. Kamnev, P. V. Mamchenkova, Y. A. Dyatlova and A. V. Tugarova, FTIR spectroscopic studies of selenite reduction by cells of the rhizobacterium Azospirillum brasilense Sp7 and the formation of selenium nanoparticles, J. Mol. Struct., 2017, 1140, 106–112 CrossRef.
- T. W. Ni, L. C. Staicu, R. S. Nemeth, C. L. Schwartz, D. Crawford and J. D. Seligman, et al. Progress toward clonable inorganic nanoparticles, Nanoscale, 2015, 7(41), 17320–17327 RSC.
- G. Kaur, M. Iqbal and M. S. Bakshi, Biomineralization of fine selenium crystalline rods and amorphous spheres, J. Phys. Chem. C, 2009, 113(31), 13670–13676 CrossRef.
- J. Dobias, E. I. Suvorova and R. Bernier-Latmani, Role of proteins in controlling selenium nanoparticle size, Nanotechnology, 2011, 22(19), 195605 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8en00221e |
| This journal is © The Royal Society of Chemistry 2018 |