Solid-phase extraction as sample preparation of water samples for cell-based and other in vitro bioassays†
Peta A.
Neale
 a,
Werner
Brack
bc,
Selim
Aït-Aïssa
d,
Wibke
Busch
b,
Juliane
Hollender
a,
Werner
Brack
bc,
Selim
Aït-Aïssa
d,
Wibke
Busch
b,
Juliane
Hollender
 ef,
Martin
Krauss
b,
Emmanuelle
Maillot-Maréchal
d,
Nicole A.
Munz
ef,
Rita
Schlichting
b,
Tobias
Schulze
ef,
Martin
Krauss
b,
Emmanuelle
Maillot-Maréchal
d,
Nicole A.
Munz
ef,
Rita
Schlichting
b,
Tobias
Schulze
 b,
Bernadette
Vogler
e and
Beate I.
Escher
b,
Bernadette
Vogler
e and
Beate I.
Escher
 *abg
*abg
aAustralian Rivers Institute, School of Environment and Science, Griffith University, Southport, QLD 4222, Australia
bUFZ – Helmholtz Centre for Environmental Research, 04318 Leipzig, Germany. E-mail: beate.escher@ufz.de; Tel: +49 341 235 1244
cRWTH Aachen University, Institute for Environmental Research, 52074 Aachen, Germany
dInstitut National de l'Environnement Industriel et des Risques INERIS, 60550 Verneuil-en-Halatte, France
eEawag, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, Switzerland
fInstitute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, 8092 Zürich, Switzerland
gEberhard Karls University Tübingen, Environmental Toxicology, Center for Applied Geosciences, 72074 Tübingen, Germany
First published on 15th February 2018
Abstract
In vitro bioassays are increasingly used for water quality monitoring. Surface water samples often need to be enriched to observe an effect and solid-phase extraction (SPE) is commonly applied for this purpose. The applied methods are typically optimised for the recovery of target chemicals and not for effect recovery for bioassays. A review of the few studies that have evaluated SPE recovery for bioassays showed a lack of experimentally determined recoveries. Therefore, we systematically measured effect recovery of a mixture of 579 organic chemicals covering a wide range of physicochemical properties that were spiked into a pristine water sample and extracted using large volume solid-phase extraction (LVSPE). Assays indicative of activation of xenobiotic metabolism, hormone receptor-mediated effects and adaptive stress responses were applied, with non-specific effects determined through cytotoxicity measurements. Overall, effect recovery was found to be similar to chemical recovery for the majority of bioassays and LVSPE blanks had no effect. Multi-layer SPE exhibited greater recovery of spiked chemicals compared to LVSPE, but the blanks triggered cytotoxicity at high enrichment. Chemical recovery data together with single chemical effect data were used to retrospectively estimate with reverse recovery modelling that there was typically less than 30% effect loss expected due to reduced SPE recovery in published surface water monitoring studies. The combination of targeted experiments and mixture modelling clearly shows the utility of SPE as a sample preparation method for surface water samples, but also emphasizes the need for adequate controls when extraction methods are adapted from chemical analysis workflows.
Environmental significanceSolid-phase extraction (SPE) is commonly applied for sample enrichment prior to bioanalysis. While many studies have assessed recovery of targeted chemicals, much less is known about effect recovery using common SPE methods. Using a complex mixture of chemicals spiked into a pristine surface water sample, the current study shows acceptable effect recovery for a range of bioassays after enrichment using large-volume SPE. Reverse recovery modelling was applied to predict effect loss by SPE in previously published water quality monitoring studies, with no substantial loss of effect by SPE found in most cases. With effect recovery similar to chemical recovery, the current study provides support for the application of bioassays for water quality monitoring. |
1. Introduction
There is increasing interest in applying bioanalytical tools complementary to chemical analysis for water quality monitoring.1,2 While targeted chemical analysis provides information about the presence of known chemicals in a sample, bioanalysis yields information about the mixture effects of the known and unknown bioactive chemicals in the sample. This complementary approach has been applied to a range of water samples including wastewater, surface water and drinking water,3–5 with studies showing that many more chemicals than those quantified contribute to the biological effects for many endpoints. As the concentration of chemicals in environmental waters is typically in the nanogram per litre to microgram per litre range, sample preparation prior to bioanalysis is required, with solid-phase extraction (SPE) commonly applied to enrich water samples.6–10 As bioassays are increasingly applied to cleaner matrices, such as surface water and drinking water, samples often need to be enriched up to 100 times to detect an effect.11 For practical purposes, the extracts are diluted in the bioassays, hence the initial enrichment of the water sample by SPE is typically 1000 to 2000 fold. Many studies have evaluated the recovery or the fraction of individual chemicals retained by SPE based on chemical analysis in a range of water matrices,12–15 with recovery dependent on the physicochemical properties of the target chemical, the matrix, the SPE material and the extraction conditions. To capture a broad range of chemicals, including very polar chemicals, combinations of SPE materials, such as reverse-phase materials with ion-exchange materials, are used.12,16,17 However, there is considerably less work on understanding the recovery of biological effects by typically applied enrichment techniques, but this is essential for the application of bioassays for water quality monitoring and for regulatory acceptance of these tools.The aim of the current study was to review the different approaches applied to evaluate effect recovery by SPE from the literature and to propose a new approach to experimentally determine effect recovery for bioassays. This approach will be applied to assess the recovery of a complex mixture of 579 chemicals spiked into surface water prior to large volume solid-phase extraction (LVSPE) using a combination of chemical analysis and bioassays. For water quality monitoring, bioassays covering different stages of the cellular toxicity pathway, as well as apical effects, are recommended.18 Therefore, we applied nine cell-based bioassays indicative of xenobiotic metabolism, hormone receptor-mediated effects and adaptive stress responses, as well as the fish embryo toxicity test with Danio rerio as a representative for an in vitro assay covering apical effects in whole organisms. A single bioassay will not be able to detect all potential effects, but by using a test battery with assays that target specific modes of action, as well as assays that detect more integrative effects, such as adaptive stress responses and apical effects in whole organisms, we are able to detect the effects of a wide range of chemicals.
Another potential issue associated with the application of SPE extracts to bioassays is effects caused by impurities captured during the extraction process. The SPE material and solvents used for high enrichment of many different chemicals with diverse physicochemical properties might lead to unwanted blank effects. Therefore, in addition to high recovery of individual chemicals and effects, a low blank effect is a prerequisite for sample preparation with SPE for bioanalytical assessment. Potential blank effects from two different SPE methods recently used for water quality monitoring, LVSPE and multi-layer SPE, were evaluated in the current study using the bioassay test battery described above.
This study represents the most comprehensive experimental evaluation of effect recovery by SPE to date. In addition, we also applied chemical recovery data from two SPE methods and reverse recovery modelling to estimate how much the measured effects underestimate predicted effects by back-calculating measured effects to expected effects for 100% chemical recovery using water quality monitoring case studies from the literature. Three of the case studies focused on samples collected from European rivers19–21 extracted with a LVSPE method using the same neutral HR-X sorbent applied in the current study. The fourth case study on Swiss effluent-impacted streams used a multi-layer SPE method with multiple layers of solid phases, namely Oasis HLB, a mixture of Strata-X-CW, Strata-X-AW and Isolute Env+, and Supelclean EnviCarb, for maximum chemical recovery.22
2. Current state of knowledge on effect recovery
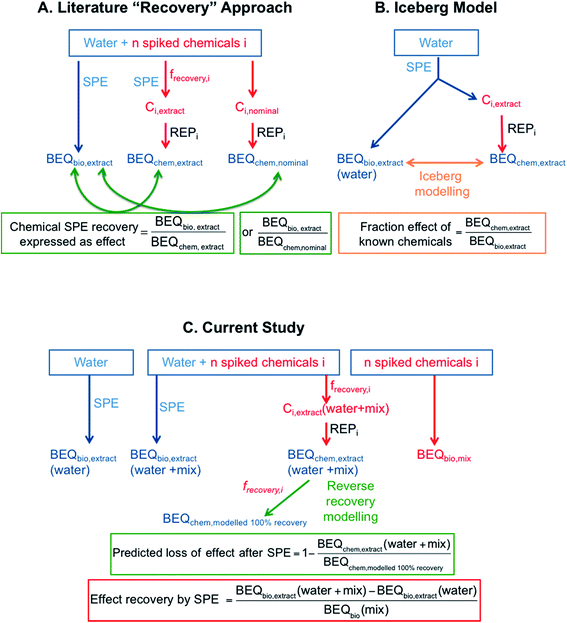
While studies on recovery of individual chemicals with SPE in the preparation of chemical analysis are abundant, very little systematic work has been performed on effect recovery by SPE. Effect recovery for bioassays in the literature is typically assessed by spiking a cocktail of chemicals into a water matrix before enrichment by SPE. Since it is most often not possible to measure the water sample prior to SPE directly in the bioassays, the effect of the extract expressed as a bioanalytical equivalent concentration from bioanalysis (BEQbio,extract) is often compared to the predicted mixture effect using the BEQ approach, which assumes that the spiked chemicals are acting in a concentration additive manner. BEQ for chemical analysis can be calculated based on either the concentration of individual chemicals detected in the extract (BEQchem,extract) or the nominal concentration of spiked chemicals (BEQchem,nominal), along with the potency of the individual chemicals in the assay.23,24 This type of mixture modelling and comparison between BEQbio and BEQchem has been applied extensively to quantify the effect triggered by unknown chemicals in environmental samples5,19,21,22 but can also be used to quantify effect recovery in SPE, provided that the effect is dominated by the spiked chemicals. The ratio of BEQbio,extract/BEQchem,nominal or BEQbio,extract/BEQchem,extract is a measure of the spiked chemical SPE recovery expressed as effect and assumes that the water sample receiving the spiked chemicals does not contribute to the effect and that the spiked chemicals act concentration-additive in mixtures (Fig. 1A).The comparison of BEQbio,extract with BEQchem,extract is mathematically similar to iceberg modelling (Fig. 1B), which is often applied in water quality monitoring to quantify the fraction of unknown bioactive chemicals in a water sample by calculating BEQchem,extract/BEQbio,extract.19 The difference between iceberg modelling used for water quality monitoring and the current approach using the BEQbio,extract/BEQchem,extract ratio is that for chemical SPE recovery expressed as effect we assume that we know all chemicals in the sample. In this application of mixture modelling, it is assumed that the spiked chemicals dominate the effect in the water sample and hence the evaluation of the BEQbio,extract/BEQchem,extract ratio can be a proxy for effect recovery. However, because this approach compares chemical analysis and bioanalysis after extraction only, it is, strictly speaking, a measure of quality/applicability of mixture toxicity models based on concentration addition rather than an effect recovery.
Studies that have determined the BEQbio,extract/BEQchem,extract ratio are summarised in Table S1 of the ESI.† Leusch et al.23 spiked eight estrogenic compounds to various water types and reported BEQbio,extract/BEQchem,extract of 0.3 to 1.64 for five different estrogen receptor (ER) assays. Kolkman et al.25 spiked a surface water sample with a mixture of 39 chemicals including natural and synthetic hormones, pesticides and pharmaceuticals and determined BEQbio,extract and BEQchem,extract for a suite of assays indicative of different hormone receptor-mediated effects. The BEQbio,extract/BEQchem,extract ratio ranged from 0.02 and 1.06, with the low BEQbio,extract/BEQchem,extract ratio in the assays indicative of activation of the androgen receptor (AR) and activation of the progesterone receptor (PR) attributed to the spiked mixture containing both agonists and antagonists. Using the BEQbio,extract/BEQchem,extract ratio one can relate the extracted chemicals to the observed effects against a background water matrix, but this is not a recovery of the biological effect in the true sense. Rather these studies compare the predicted effects in the extracts based on known bioactive chemicals with the measured effects of the extracts, similar to iceberg modelling.
In contrast, the BEQbio,extract/BEQchem,nominal ratio is more useful for determining effect recovery by SPE as the effect in the bioassay is related to the predicted effect based on the nominal concentration, rather than the concentration measured in the extract. Studies that have applied this approach are summarised in Table S2.† For example, Neale and Escher26 found a BEQbio,extract/BEQchem,nominal ratio of 0.91 for six spiked herbicides in treated wastewater in the combined algae assay. Further, Kunz et al.24 found a BEQbio,extract/BEQchem,nominal ratio of 0.27 to 1.38 for spiked estrogenic compounds in assays indicative of estrogenic activity and a similar study using four estrogenic chemicals spiked into wastewater reported a ratio of 1.13 to 1.24 for the yeast estrogen screen (YES).27
One issue with comparing BEQbio,extract with BEQchem,extract or BEQchem,nominal based on the spiked chemicals alone is that the spiked water matrix may have an effect itself in the bioassay. This is especially likely for complex matrices, such as wastewater. Therefore, it is important to consider the effect of the matrix itself when assessing effect recovery for bioassays. By adding a chemical cocktail to a urine sample, which was selected as a representative for a matrix-rich water, and testing both urine alone and urine spiked with the cocktail, Escher et al.28 were able to confirm good effect recovery by SPE, with between 75 and 148% recovery for YES and the bioluminescence inhibition test.
To truly assess effect recovery by SPE one would need to spike water prior to SPE and compare the effects before and after SPE, which is technically challenging. As a proxy, Escher et al.29 previously extracted spiked and unspiked wastewater with Lichrolut Env/C18 SPE cartridges and SDC Empore Disks and compared the resulting effects. Full bioassay recovery was achieved for spiked estradiol in YES, spiked parathion in the acetylcholinesterase inhibition assay and spiked diuron in the combined algae assay, confirming high extraction efficacy as well as concentration-additive mixture effects of the wastewater matrix and spiked chemicals.29 A limitation of this study was that concentrations in the samples were not chemically verified.
In summarising the available literature, there is a lack of experimentally determined effect recoveries for bioassays using commonly applied SPE techniques. To fill this knowledge gap, the current study evaluated the effect recovery of a mixture of micropollutants by SPE using a combination of bioanalysis and chemical analysis (Fig. 1C). Spiked and unspiked water samples were enriched using LVSPE, and chemical analysis was performed on the spiked and unspiked SPE extracts. Effect recovery was calculated by applying mixture modelling based on the assumption that the chemical mixture and the unspiked water extract would act in a concentration additive manner. Effect recovery was hence defined as the ratio of the difference in BEQbio between the spiked and unspiked extract to the BEQbio of the spiked chemical mixture. Thus all parameters of the recovery calculations are derived from experimentally quantified effects. In addition, reverse recovery modelling was applied to determine how much greater the predicted effect would be if all chemicals had been completely recovered by SPE (Fig. 1C). This was termed BEQchem,modelled 100% recovery and was also calculated for existing iceberg modelling studies from the literature and compared with the reported BEQchem,extract values.
3. Materials and methods
3.1 Chemical mixture
The spiked chemical mixture (sample “mix”) contained 579 chemicals in methanolic solution. The spiked mixture contained chemical classes commonly detected in environmental waters and wastewater30 including pharmaceuticals, pesticides, industrial compounds and natural and synthetic hormones. This set of chemicals covers a wide range of physicochemical properties, including acids and bases as well as multiprotic chemicals to explore the applicability domain of SPE. The test set of 579 chemicals includes and expands our previous study of the chemical recovery of 251 organic chemicals.12 The concentrations of 532 compounds in the mix stock solution were 800 ng mL−1, though the concentrations of the 47 steroidal hormones were 20 ng mL−1 to account for their high bioactivity. A list of the spiked chemicals is provided in Table S3† along with selected chemical properties, such as octanol–water partition constant (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow) and the ionisation-corrected octanol–water distribution ratio (log
Kow) and the ionisation-corrected octanol–water distribution ratio (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Dow).
Dow).
3.2 Sample collection and extraction
Surface water from Wormsgraben, a pristine creek in the Harz Mountains, Germany, was used as the water matrix for the effect recovery experiments. Ninety litres of the water were collected using a submersible rotary pump (Comet, Pfaffschwende, Germany) equipped with polytetrafluoroethylene tubing and stored in three solvent-cleaned stainless steel drums. The flow rate of the pump was 20 L min−1. Therefore, it can be assumed that the water condition of the creek was not altered during the short sampling period (approximately 5 min) and thus the water composition was similar in all drums. The samples were stored at 4 °C in a cooling chamber for three weeks until performance of the spiking experiments. The mix stock solution was diluted with methanol by a factor of five prior to spiking, with 10 mL of the diluted mix stock solution spiked into 20 L Wormsgraben water to give final concentrations of 80 ng L−1 for the majority of compounds and 2 ng L−1 for steroidal hormones. The spiked water sample was enriched using LVSPE with the neutral HR-X sorbent (sample “water + mix”), with further information about the LVSPE method available in Schulze et al.12 A modified elution procedure with neutral, acidic and basic elution steps was used as detailed in Välitalo et al.31 The final extract had a volume of 20 mL, giving an enrichment factor of 1000 based on the water volume. Twenty litres of unspiked Wormsgraben water were also extracted by the same LVSPE method (sample “water”). Five litres of ultrapure water (LCMS grade water) were extracted using LVSPE by circulating the water four times to obtain a process blank containing possible impurities from the extraction process (e.g., leachates from machine materials or residues from SPE sorbent) as described in Schulze et al.12 Both the unspiked Wormsgraben water and the process blank had a final enrichment factor of 1000. In addition to the process blank, a methanol solvent blank was also included.Recovery of a suite of chemicals spiked in surface water from the Rhine River was also evaluated using multi-layer SPE cartridges. These multi-layer SPE cartridges have been previously applied to extract wastewater and surface water samples for bioanalysis22 and the recovery data were used for reverse recovery modelling in the current study. Briefly, Rhine water was filtered with a glass microfiber filter (GF/F, 47 mm, Whatman) and adjusted to pH 6.5. Three different sample types were prepared, a background sample with no chemicals spiked and recovery samples where 193 chemicals were spiked before SPE and after elution, respectively. Internal standards were also spiked into samples to account for possible analyte loss.
One litre of water was enriched using the multi-layer SPE cartridge, which was composed of 200 mg of Oasis HLB (Waters, U.S), 350 mg of a mixture of Strata-X-CW, Strata-X-AW (Phenomenex, USA) and Isolute Env+ (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5) (Separtis, Germany) and 200 mg of Supelclean EnviCarb (Sigma-Aldrich, Germany). The cartridges were conditioned with 5 mL methanol and 10 mL nanopure water, then the samples were loaded onto the cartridges and dried completely by pumping air through the cartridge. Elution occurred in back flush mode with ethyl acetate/methanol (1
1.5) (Separtis, Germany) and 200 mg of Supelclean EnviCarb (Sigma-Aldrich, Germany). The cartridges were conditioned with 5 mL methanol and 10 mL nanopure water, then the samples were loaded onto the cartridges and dried completely by pumping air through the cartridge. Elution occurred in back flush mode with ethyl acetate/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 6 mL) containing ammonium (0.5%), followed by ethyl acetate/methanol (1
1, 6 mL) containing ammonium (0.5%), followed by ethyl acetate/methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 mL) containing formic acid (1.7%) and then pure methanol (2 mL), which resulted in a final neutral elution volume of 11 mL. The samples were then concentrated to a volume of 100 μL under a gentle nitrogen flow, diluted with nanopure water (100 μl) and filtered (4 mm Cronus Filter, regenerated cellulose, 0.45 μm, Infochroma, Switzerland). The vial and filter were rinsed with nanopure water (800 μL), giving a final volume of 1 mL and thus an enrichment factor of 1000.
1, 3 mL) containing formic acid (1.7%) and then pure methanol (2 mL), which resulted in a final neutral elution volume of 11 mL. The samples were then concentrated to a volume of 100 μL under a gentle nitrogen flow, diluted with nanopure water (100 μl) and filtered (4 mm Cronus Filter, regenerated cellulose, 0.45 μm, Infochroma, Switzerland). The vial and filter were rinsed with nanopure water (800 μL), giving a final volume of 1 mL and thus an enrichment factor of 1000.
3.3 Chemical analysis
Analysis of all spiked compounds was performed using liquid chromatography (LC) coupled to tandem mass spectrometry (MS/MS) or high-resolution tandem mass spectrometry (HRMS/MS). For the LVSPE recovery experiment, 561 compounds were analysed by a LC-HRMS/MS target screening method in positive and negative electrospray ionization (ESI+/ESI−) using a QExactive Plus instrument (Thermo). An additional 18 compounds (phenols and steroids) were analysed by LC-MS/MS in ESI-mode on a QTrap 6500 instrument (ABSciex), as the sensitivity of the LC-HRMS screening method was not sufficient. Details on the analytical method used can be found in Section S1.† Analysis of 193 chemicals in the multi-layer SPE extracts was also conducted using LC coupled to a QExactive HRMS. Further information is provided in Section S2.†3.4 Bioanalysis
Ten bioassays covering 9 different endpoints were selected in the current study (Table 1). The assays were indicative of activation of the aryl hydrocarbon receptor (AhR, AhR CALUX), activation of the pregnane X receptor (PXR, HG5LN-hPXR), binding to peroxisome proliferator-activated receptor gamma (PPARγ, PPARγ GeneBLAzer), activation of ER (ER GeneBLAzer, MELN), activation of AR (AR GeneBLAzer), activation of the glucocorticoid receptor (GR, GR GeneBLAzer), activation of PR (PR GeneBLAzer), oxidative stress response (AREc32) and fish embryo toxicity (FET). Cell viability was assessed in parallel for all assays indicative of non-apical effects. Detailed information about the studied assays can be found in König et al.,21 Neale et al.18 and Nivala et al.32| Bioassay | Endpoint | Method reference | Positive reference compound | EC value | Positive reference compound EC value (M) |
|---|---|---|---|---|---|
| AhR CALUX | Activation of aryl hydrocarbon receptor (AhR) | Brennan et al.38 | 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) | EC10 | (5.68 ± 0.17) × 10−13 |
| HG5LN-hPXR | Activation of pregnane X receptor (PXR) | Lemaire et al.39 | SR 12813 | EC10 | (1.41 ± 0.15) × 10−8 |
| PPARγ GeneBLAzer | Binding to the peroxisome proliferator-activated receptor gamma (PPARγ) | Neale et al.18 | Rosiglitazone | EC10 | (9.87 ± 0.14) × 10−10 |
| MELN | Activation of estrogen receptor (ER) | Balaguer et al.40 | 17β-Estradiol | EC10 | (2.42 ± 0.06) × 10−12 |
| ER GeneBLAzer | Activation of ER | König et al.21 | 17β-Estradiol | EC10 | (2.50 ± 0.08) × 10−11 |
| AR GeneBLAzer | Activation of androgen receptor (AR) | König et al.21 | Metribolone (R1881) | EC10 | (2.37 ± 0.07) × 10−10 |
| GR GeneBLAzer | Activation of glucocorticoid receptor (GR) | König et al.21 | Dexamethasone | EC10 | (8.49 ± 0.36) × 10−10 |
| PR GeneBLAzer | Activation of progesterone receptor (PR) | König et al.21 | Promegestone | EC10 | (1.52 ± 0.06) × 10−10 |
| AREc32 | Oxidative stress response | Wang et al.41, Escher et al.42 | tert-Butylhydroquinone (tBHQ) | ECIR1.5 | (1.93 ± 0.04) × 10−6 |
| Fish embryo toxicity (FET) | Mortality | OECD43 | 3,4-Dichloroaniline | — | — |
The mix stock solution and the five times diluted mix stock solution were also analysed in the bioassays in their original methanolic form and were equivalent to an enrichment factor of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 2000, respectively, of a water sample that had 100% recovery. As both the mix stock solution and the diluted mix stock solution gave consistent concentration–effect curves in all bioassays they were evaluated together as sample “mix”.
000 and 2000, respectively, of a water sample that had 100% recovery. As both the mix stock solution and the diluted mix stock solution gave consistent concentration–effect curves in all bioassays they were evaluated together as sample “mix”.
SPE process blank samples from LVSPE and multi-layer SPE were also tested in all assays, with the exception of HG5LN-hPXR and MELN in the case of multi-layer SPE. In addition, blank samples from different materials used in multi-layer SPE (e.g., Oasis HLB, Oasis HLB + Strata-X-AW, Strata-X-CW and Isolute ENV+) were tested, as well as different conditioning solvents.
3.5 Data evaluation
The concentration causing 10% effect (EC10) was derived from linear concentration–effect curves for the assays indicative of xenobiotic metabolism and hormone receptor-mediated effects, while the effect concentration causing an induction ratio of 1.5 (ECIR1.5) was derived from linear concentration–effect curves for the AREc32 assay. Log-sigmoidal concentration–effect curves were applied to the FET assay to determine the concentration causing 50% effect (EC50). Further information about the applied data evaluation methods can be found in Escher et al.33 and Neale et al.18 The EC values for all samples were expressed in units of relative enrichment factor (REF), which was calculated based on the SPE enrichment factor, or equivalent enrichment factor in the case of sample “mix”, and the dilution factor in the bioassay.6 The EC values for the assay positive reference compounds were expressed in molar units.To relate the effect of the sample in a bioassay in units of REF to the concentration of a reference compound (ref) in molar units that would elicit the same effect the EC values were converted to BEQbio using eqn (1).
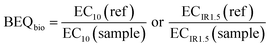 | (1) |
Effect recovery for the bioassays was calculated for each assay using eqn (2) with the BEQbio value of the spiked Wormsgraben water extract (BEQbio,extract (water + mix)), the BEQbio value of the unspiked Wormsgraben water extract (BEQbio,extract (water)) and the BEQbio of the mix stock solution (BEQbio (mix)).
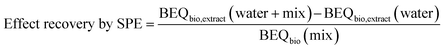 | (2) |
The effect based on spiked chemicals was modelled using BEQchem,extract based on the concentration of the individual chemical in the extract (Ci) and its relative effect potency (REPi) in the studied bioassay (eqn (3)). REPi was calculated using eqn (4), with effect concentrations of the individual chemicals collected from the peer reviewed literature or the US EPA ToxCast database.34 As the data in the ToxCast database were expressed as 50% activity concentrations (AC50), EC10,absolute was calculated using the reported AC50 value and the maximum of the concentration–effect curve based on the approach described in Neale et al.22
 | (3) |
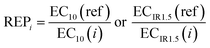 | (4) |
To evaluate how much effect would be overlooked due to loss of chemicals during SPE, we predicted the biological effect if the recovery of all chemicals by SPE were 100%, BEQchem,modelled 100% recovery, using eqn (5), where frecovery,i is the fraction of each chemical i recovered by SPE. frecovery,i was calculated using eqn (6), where Cextract (water + mix) is the measured chemical concentration in the spiked water extract (ng L−1), Cextract (water) is the measured chemical concentration in the unspiked water extract (ng L−1) and Ci,nominal is the nominal chemical concentration spiked into the water (ng L−1).
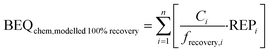 | (5) |
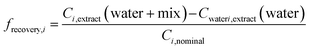 | (6) |
Reverse recovery modelling was also applied to existing iceberg modelling studies from the literature.19–22 The predicted loss of effect by SPE was calculated using eqn (7).
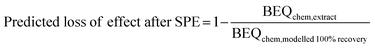 | (7) |
4. Results and discussion
4.1 Recovery of individual chemicals
The concentration of each chemical measured in the spiked water extract, along with the calculated frecovery,i values, is provided in Table S4.† Of the 579 chemicals spiked, 29 were not detected at all after LVSPE, while a further 88 were not measureable as no calibration was obtained, either due to lack of ionization or high background noise. The majority of the 29 chemicals that were not detected after LVSPE were hydrophilic or charged compounds, with predicted log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Dow values less than 0.5. Another three compounds, 4-n-octylphenol, benzyldimethyldodecylammonium and lauramidopropylbetaine, were detected in the unspiked water extract at similar concentrations as in the spiked water extract due to background contamination, resulting in negative frecovery,i values. Of the remaining 459 compounds, frecovery,i ranged from 0.01 for the insecticide ethion to 3.08 for the pharmaceutical metabolite canrenone, with an average frecovery,i of 0.70. As can be seen from Fig. 2, frecovery,i for the majority of chemicals was between 0.75 and 1.25. The low recovery of ethion fits with previous studies, with Schulze et al.12 finding no recovery of hydrophobic ethion by LVSPE using HR-X, the weak anion exchanger HR-XAW and weak cation exchanger HR-XCW. Thirty six chemicals had a frecovery,i greater than 1. The higher recovery is likely to be due to the presence of an isobaric compound or a mismatch of the internal standard used for quantification, as only 40 isotope-labelled compounds were available, with the one with the closest retention time used for quantification. frecovery,i was set to 1 for reverse recovery modelling for these chemicals.
Dow values less than 0.5. Another three compounds, 4-n-octylphenol, benzyldimethyldodecylammonium and lauramidopropylbetaine, were detected in the unspiked water extract at similar concentrations as in the spiked water extract due to background contamination, resulting in negative frecovery,i values. Of the remaining 459 compounds, frecovery,i ranged from 0.01 for the insecticide ethion to 3.08 for the pharmaceutical metabolite canrenone, with an average frecovery,i of 0.70. As can be seen from Fig. 2, frecovery,i for the majority of chemicals was between 0.75 and 1.25. The low recovery of ethion fits with previous studies, with Schulze et al.12 finding no recovery of hydrophobic ethion by LVSPE using HR-X, the weak anion exchanger HR-XAW and weak cation exchanger HR-XCW. Thirty six chemicals had a frecovery,i greater than 1. The higher recovery is likely to be due to the presence of an isobaric compound or a mismatch of the internal standard used for quantification, as only 40 isotope-labelled compounds were available, with the one with the closest retention time used for quantification. frecovery,i was set to 1 for reverse recovery modelling for these chemicals.
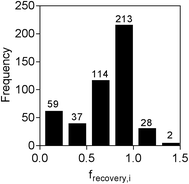 | ||
| Fig. 2 Distribution of frecovery,i for spiked chemicals (n = 459) in LVSPE. Six chemicals had a frecovery,i greater than 1.5 (not shown). | ||
The ratio of frecovery,i for LVSPE from the current study to frecovery,i from Schulze et al.12 for HR-X only was calculated to compare recovery between studies (Fig. S1†). For 79% of common chemicals (165 out of 208 chemicals), the ratio was within a factor of 2, indicating that the results are generally reproducible. While the same sorbent, HR-X, was used, neutral, acidic and basic elution steps were undertaken in the current study, while only a neutral elution step was applied in Schulze et al.12 As a result, greater recovery of some chemicals, such as positively charged quaternary ammonium compounds, was achieved in the current study.
To compare chemical recovery in the current study with a mixed sorbent SPE cartridge designed to capture a wide range of neutral and charged chemicals, the ratio of frecovery,i for LVSPE from the current study to frecovery,i for multi-layer SPE was determined (Fig. S1†). The calculated frecovery,i values for multi-layer SPE are provided in Table S5.† Despite using different sorbents, frecovery,i for 87% of common chemicals (153 out of 175 chemicals) was within a factor of 2 of the LVSPE method. Many of the compounds with greater than two times higher recovery in multi-layer SPE were either charged hydrophilic compounds with log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Dow less than 0 or hydrophobic compounds with a log
Dow less than 0 or hydrophobic compounds with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Dow greater than 4.
Dow greater than 4.
4.2 SPE process blank effects in bioassays
Bioassays cannot differentiate between the effects of micropollutants in a water sample and the effects associated with impurities from the extraction process, but their benefit is that they can provide information about the mixture effects of all bioactive chemicals. Solvent traces in bioassays, non-volatile residues from solvents, leachates from the LVSPE device, residues from SPE material and dirty glassware can potentially cause blank effects. Consequently, it is important to include procedural blanks as part of bioassay quality control. In the current study with LVSPE two procedural blanks were included, namely a process blank, where 5 L of ultrapure water was extracted by circulating through LVSPE four times, and a methanol blank to act as a solvent control. The volume of the ultrapure water in the process blank was restricted to 5 L to prevent potential effects from any trace level contamination in the water itself, rather than from the LVSPE material, glassware or solvents used.12 Neither of the process nor solvent blanks induced a response in the bioassays, though cytotoxicity was observed at high sample enrichment (REF 306) in the AhR CALUX assay. For the other assays, no induction or cytotoxicity was observed up to the maximum tested REF, which ranged from 50 (MELN, HG5LN-hPXR) to 150 (GeneBLAzer assays) (Table S6, Fig. S2 to S11†).In contrast, the multi-layer SPE produced higher blank effects, with the cytotoxicity 10% inhibitory concentration (IC10) around REF 20 for several of the assays (Table S7 and Fig. S12†). We also tested the SPE blanks with the EnviCarb layer removed and with Oasis HLB only, as well as the effect of using methanol only or ethyl acetate and methanol for conditioning. The different sorbents and conditioning solvents did not have a significant influence on cytotoxicity (Table S7 and Fig. S12†). In addition to cytotoxicity, some of the SPE blanks induced a response in the xenobiotic metabolism assays and the oxidative stress response assay. None of the SPE blanks had an effect at 24 or 48 h in the FET assay, though all blanks induced mortality after 120 h, with EC50 values ranging from REF 29 to 59. While the multi-layer SPE has the highest chemical recovery, this comparison also demonstrates that when optimising a SPE method not only maximum chemical recovery but also effect recovery and bioassay blank effects must be considered.
We recommend that samples are tested up to enrichments where no blank effects occur. In exceptional situations and at very high enrichments, when the process blank has an effect, the blank effect concentration in units of REF cannot be simply subtracted, but the BEQ of the blank can be subtracted by applying eqn (8). For this equation to be valid, the process blank should be prepared using the same SPE extraction and elution conditions as the sample.
| BEQbio(blank-corrected sample) = BEQbio(sample) − BEQbio(blank) | (8) |
4.3 Pristine water effects in bioassays
A pristine surface water sample was used as the matrix to assess effect recovery by SPE in the current study. Chemical analysis revealed that 43 chemicals were present at low concentrations in the unspiked water extract. Consequently, the effect of the water alone, BEQbio,extract (water), was included in eqn (2). In any case, it is still important to consider the effect of the water matrix alone even if no chemicals are detected as chemicals may still be present at concentrations below the analytical limit of detection. The BEQbio,extract (water) correction was zero for ER GeneBLAzer, AR GeneBLAzer, GR GeneBLAzer and PR GeneBLAzer as no effect was observed in the unspiked water extract in these assays, though it had a minor effect in the other assays (Table 2) and this effect was subtracted using eqn (2). Some of the chemicals detected in the unspiked water extract, including bisphenol A, estriol and propylparaben, are active in the studied bioassays (Tables S8 and S9†) and may have contributed to the effect observed in the unspiked water. It should be noted that the 43 chemicals found in the unspiked water extract were also detected at similar concentrations in the process blank (Table S4†), suggesting that they may have originated from the LVSPE material or solvent residues, rather than from Wormsgraben. Interestingly, the process blank did not induce a response in any of the assays, suggesting that other undetected chemicals may be contributing to the effect observed in the unspiked water extract.| Assay | BEQbio,extract (water) (M) ± standard error | BEQbio (mix) (M) ± standard error | BEQbio,extract (water + mix) (M) ± standard error | Effect recovery (%) ± standard error |
|---|---|---|---|---|
| AhR CALUX | (2.39 ± 0.11) × 10−14 | (1.56 ± 0.08) × 10−13 | (1.20 ± 0.07) × 10−13 | 61.2 ± 5.6 |
| HG5LN-hPXR | (3.90 ± 0.69) × 10−10 | (2.59 ± 0.51) × 10−10 | (3.76 ± 0.86) × 10−9 | 1300 ± 420 |
| PPARγ GeneBLAzer | (2.95 ± 0.27) × 10−11 | (1.83 ± 0.09) × 10−10 | (3.10 ± 0.29) × 10−10 | 153 ± 18 |
| MELN | (6.68 ± 0.96) × 10−14 | (2.38 ± 0.31) × 10−11 | (2.94 ± 0.34) × 10−11 | 124 ± 21 |
| ER GeneBLAzer | <8.33 × 10−13 | (4.27 ± 0.17) × 10−11 | (1.49 ± 0.07) × 10−11 | 34.9 ± 2.1 |
| AR GeneBLAzer | <2.63 × 10−12 | (3.46 ± 0.12) × 10−11 | (4.33 ± 0.16) × 10−11 | 125 ± 6.4 |
| GR GeneBLAzer | <2.84 × 10−11 | (1.76 ± 0.08) × 10−10 | (1.24 ± 0.07) × 10−10 | 70.5 ± 5.4 |
| PR GeneBLAzer | <5.07 × 10−12 | (4.47 ± 0.18) × 10−11 | (2.96 ± 0.15) × 10−11 | 66.2 ± 4.3 |
| AREc32 | (8.43 ± 0.27) × 10−8 | (1.49 ± 0.05) × 10−8 | (1.19 ± 0.04) × 10−7 | 236 ± 29 |
4.4 Effect recovery by SPE
BEQbio values for the unspiked water extract (BEQbio,extract (water)), mix stock solution (BEQbio (mix)) and spiked water extract (BEQbio,extract (water + mix)) are provided in Table 2, along with effect recovery (eqn (2)). All EC values are provided in Table S6,† along with the full concentration–effect curves in Fig. S2 to S11.† Effect recovery could not be calculated for FET as the unspiked water extract, mix stock solution and spiked water extract all resulted in similar concentration–effect curves (Fig. S11†).If all bioactive chemicals spiked in the water extract were 100% recovered by SPE, BEQbio (mix) and BEQbio,extract (water + mix) (after subtraction of BEQbio,extract (water)) should be the same. Effect recovery for the studied bioassays was calculated using eqn (2) and ranged from 35% for ER GeneBLAzer to 236% for AREc32, with one extreme value of 1300% in HG5LN-hPXR. The effect recovery in HG5LN-hPXR is not likely to be representative, but is instead related to the small and rather variable effect of the mix stock solution in the assay, which is in the denominator of eqn (2) and therefore is strongly driving the effect recovery. Similarly, the variable response in the mix stock solution in ER GeneBLAzer may also explain the low recovery reported.
Effect recovery was within a factor of two of the optimal 100% effect recovery for AhR CALUX, PPARγ GeneBLAzer, MELN, AR GeneBLAzer, GR GeneBLAzer and PR GeneBLAzer, suggesting that LVSPE is suitable to capture bioactive chemicals for the majority of applied assays.
It must be noted that the concentration axis in bioassays is typically on a logarithmic scale so a small variation on the concentration–effect curve might have quite dramatic effects on the calculated effect recovery. Most likely the samples where the BEQbio,extract (water) was below the detection limit are more robust than those where the BEQbio,extract (water) was subtracted from the BEQbio,extract (water + mix). Also, the BEQ addition and subtraction assumes that chemicals and samples act concentration additive, which is conceptually likely and recommended for mixture risk assessment of environmental mixtures35,36 but there might still be some variability due to the contribution of antagonistically or independently acting chemicals (i.e., chemicals that act according to different modes of action).
This experimental case study demonstrates the difficulty in assessing recoveries directly with experimental data and the number of replicates would need to be increased to increase the power of the experiment. While a recovery range of 80–120% is desirable and within the range of uncertainty for chemical analysis, this range must likely be expanded for bioassays to a range of a factor of 2, i.e., from 50% to 200%.
To get a better feeling of how much of the effect we would overlook by chemical losses incurred during SPE, we did reverse recovery modelling for the experimental data (Section 4.5) and also for case studies from the literature (Section 4.6).
4.5 Reverse recovery modelling of LVSPE extracts
The BEQchem,extract was calculated using the detected chemical concentration in the LVSPE extract and available REPi values from the literature or the US EPA ToxCast database. REPi values were available for between 4 and 45 chemicals in the different assays, with the EC and REPi values provided in Tables S8 and S9.† Of the chemicals with REPi values, five (4-n-octylphenol, acrylamide, amitraz, flufenoxuron and iopamidol) were not detected after LVSPE and could not be included in the BEQchem,extract calculations. No effect data could be found for the individual spiked chemicals in PR GeneBLAzer, so it was not possible to calculate BEQchem,extract for this assay, though some of the spiked chemicals, such as progesterone and canrenone, are active in other assays indicative of activation of PR.4BEQchem,extract was compared to BEQchem,modelled 100% recovery predicted by reverse recovery modelling using frecovery,i data from the current study to determine how much greater the effect would be if all chemicals were completely recovered by SPE. The predicted loss of effect after SPE ranged from 13% for the AR GeneBLAzer assay to 61% for the AREc32 assay (Table 3). For most assays, the predicted loss of BEQ by SPE was around 40%, which is less than a factor of two, i.e., relatively small in relation to the variability of effect concentrations in bioassays. A ratio of two in effect concentrations such as EC50 or EC10 is a factor of ±0.3 on a log scale and a standard deviation of ±0.3 is more than typical for a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EC value derived from a log-sigmoidal concentration–effect curve.
EC value derived from a log-sigmoidal concentration–effect curve.
| Assay | BEQbio,extract (water + mix) (M) | BEQchem,extract (M) | BEQchem,modelled 100% recovery (M) | % BEQbio,extract (water + mix) explained by BEQchem,extract | % BEQbio,extract (water + mix) explained by BEQchem,modelled 100% recovery | % predicted loss by SPE |
|---|---|---|---|---|---|---|
| AhR CALUX | 1.20 × 10−13 | 4.31 × 10−17 | 7.22 × 10−17 | 0.04% | 0.06% | 40.3% |
| HG5LN-hPXR | 3.76 × 10−9 | 1.07 × 10−10 | 2.06 × 10−10 | 2.84% | 5.48% | 48.1% |
| PPARγ GeneBLAzer | 3.10 × 10−10 | 1.31 × 10−12 | 2.13 × 10−12 | 0.42% | 0.69% | 38.6% |
| MELN | 2.94 × 10−11 | 1.90 × 10−11 | 3.29 × 10−11 | 64.7% | 112% | 42.1% |
| ER GeneBLAzer | 1.49 × 10−11 | 8.47 × 10−12 | 1.29 × 10−11 | 56.9% | 86.8% | 34.4% |
| AR GeneBLAzer | 4.33 × 10−11 | 8.31 × 10−11 | 9.62 × 10−11 | 192% | 222% | 13.6% |
| GR GeneBLAzer | 1.24 × 10−10 | 1.68 × 10−10 | 2.91 × 10−10 | 135% | 234% | 42.4% |
| AREc32 | 1.19 × 10−7 | 1.28 × 10−10 | 3.33 × 10−10 | 0.11% | 0.28% | 61.6% |
4.6 Reverse recovery modelling of literature data
Recently, iceberg modelling using the BEQ concept (Fig. 1B) has been applied to determine the contribution of detected chemicals to the biological effect in surface water and wastewater.19,21,22 These studies all use the detected chemical concentrations after SPE to calculate BEQchem,extract, but some chemicals may be poorly recovered by SPE, meaning BEQchem,extract may underestimate the true effect potential in the water sample. Since concentration–effect curves are only linear at low effect levels we cannot just assume that an 80% average chemical recovery results in 80% effect recovery. In addition the composition might change in the extract and we do not expect any correlation between recovery and potency but this remains to be proven. Therefore, BEQchem,modelled 100% recovery was calculated for three studies that have previously applied LVSPE, Neale et al.,19 König et al.21 and Tousova et al.,20 using frecovery,i determined in the current study. Neale et al.19 used a LVSPE device with three sorbents in a row, HR-X, HR-XAW and HR-XCW, to extract surface water samples, but previous studies have shown that the majority of chemicals are primarily extracted using the neutral HR-X.12 Despite the different elution steps used, results presented in Section 4.1 indicate that the majority of frecovery,i values in the current study were similar to those obtained for HR-X by Schulze et al.,12 therefore using frecovery,i values from the current study for reverse recovery modelling for Neale et al.19 is still acceptable.BEQchem,extract and BEQchem,modelled 100% recovery from the current study and the literature19–21 were compared for assays indicative of activation of PXR, activation of ER and oxidative stress response in Fig. 3, with literature data shown for the FET assay. Comparisons for assays indicative of binding to PPARγ, activation of AR and p53 response are provided in Fig. S13.† Generally, BEQchem,extract was within a factor of 2 of BEQchem,modelled 100% recovery, indicating less than 50% loss of effect after LVSPE. The exceptions included one sample from Neale et al.19 (JDS 59) in the activation of PXR and FET assays, with 60% and 64% predicted loss of effect after LVSPE, respectively. Similarly, up to 92% loss by LVSPE was predicted for several samples in Tousova et al.20 for the FET assay. In all examples this could be attributed to the poor recovery of triclosan, which had a frecovery,i of 0.06. Similarly, low recovery of triclosan was also observed previously,12,37 and may be related to the hydrophobicity of triclosan, which has a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow of 4.98 (Chemaxon, Table S3†). Therefore, strong sorption of triclosan to the HR-X sorbent and/or LVSPE materials is likely, with the solvents used for elution seemingly unable to completely desorb triclosan from the LVSPE device.
Kow of 4.98 (Chemaxon, Table S3†). Therefore, strong sorption of triclosan to the HR-X sorbent and/or LVSPE materials is likely, with the solvents used for elution seemingly unable to completely desorb triclosan from the LVSPE device.
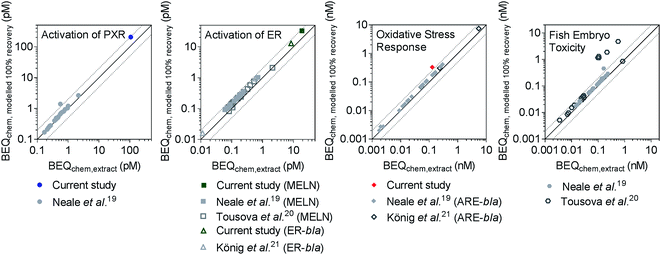 | ||
| Fig. 3 Comparison of BEQchem,extract and BEQchem,modelled 100% recovery for activation of PXR, activation of ER (both ER GeneBLAzer (ER-bla) and MELN), oxidative stress response and fish embryo toxicity for the current study, Neale et al.,19 König et al.21 and Tousova et al.20 The dotted lines indicate a factor of 2 difference between BEQchem,extract and BEQchem,modelled 100% recovery. NB: two different oxidative stress response assays based on the same endpoint were included, with AREc32 applied in the current study and ARE GeneBLAzer (ARE-bla) applied in Neale et al.19 and König et al.21 | ||
BEQchem,modelled 100% recovery was also calculated for Neale et al.,22 where a suite of bioassays were applied to surface water extracts collected from streams in Switzerland upstream and downstream of wastewater treatment discharges. As multi-layer SPE was used for sample enrichment prior to bioanalysis, multi-layer SPE frecovery,i values measured in the current study were applied for reverse recovery modelling. The studied assays were indicative of activation of AhR, activation of AR, oxidative stress response, photosystem II inhibition and algal growth inhibition. No information was available on the recovery of 4-nonylphenol, alfuzosin, bisphenol A, estrone, etodolac and ritonavir by multi-layer SPE. Therefore, frecovery,i data for LVSPE were used for 4-nonylphenol, bisphenol A and estrone, given similar recoveries in the multi-layer SPE and LVSPE (Fig. S1†), while a frecovery,i of 1 was assumed for the other chemicals. BEQchem,extract was within a factor of 2 of BEQchem,modelled 100% recovery for all assays (Fig. 4), indicating a good agreement and a minor loss of effect after SPE.
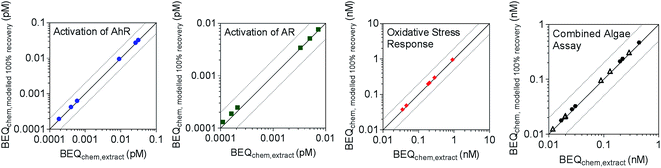 | ||
| Fig. 4 Comparison of BEQchem,extract and BEQchem,modelled 100% recovery for activation of AhR, activation of AR, oxidative stress response and the combined algae assay (open symbols indicate PSII inhibition and closed symbols indicate algal growth inhibition) from Neale et al.22 derived from the recovery data of the multi-layer SPE (Table S5†). The dotted lines indicate a factor of two difference between BEQchem,extract and BEQchem,modelled 100% recovery. | ||
While the reverse recovery modelling approach has some limitations, such as the lack of recovery data for some of the detected chemicals and lack of effect data for some others, it suggests that there are no substantial losses of effect equivalents due to SPE. This indicates that the current method of iceberg modelling for environmental water samples is meaningful.
5. Conclusions
A complementary chemical analysis and bioanalysis approach was applied in the current study to assess the chemical and effect recovery of a complex mixture of chemicals by SPE. Overall, comparison with other studies and different extraction processes indicates that chemical recovery by LVSPE in the current study is within an acceptable range. The majority of chemicals were well recovered by LVSPE, with 79% and 87% of spiked chemicals having frecovery,i values within a factor of 2 of previously measured recovery values for LVSPE and for multi-layer SPE from the current study, respectively. Effect recovery was determined from experimentally quantified effects in the spiked water extract, the unspiked water extract and mix stock solution. For the majority of assays, experimental effect recovery was within a factor of 2 of the expected 100% recovery, though small variations in the concentration–effect curve may have implications for the calculated effect recovery. Reverse recovery modelling of existing published studies that applied LVSPE and multi-layer SPE for bioanalysis indicated that in most cases there was no substantial loss of effect by SPE. Further, the theoretical correction for chemical losses using reverse recovery modelling is a useful approach to predict effect when chemical recovery is less than 100%. Overall, the current study found that available SPE methods for bioanalysis are appropriate, with effect recovery similar to recovery by chemical analysis and that we can confidently apply bioassays after SPE extraction without fear of substantial loss of effect due to incomplete SPE recovery. This provides support for the use of current SPE methods with low blank effects for bioanalytical assessment and the application of bioassays for water quality monitoring and for assessing treatment efficacy in natural and engineered treatment systems.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The project SOLUTIONS is supported by the European Union Seventh Framework Programme (FP7-ENV-2013-two-stage Collaborative project) under grant agreement number 603437. Neale was supported by the National Health and Medical Research Council (NHMRC) – European Union Collaborative Research Grant (APP1074775). We are grateful to Liza-Marie Beckers (UFZ), Margit Petre (UFZ) and Jörg Ahlheim (UFZ) for sampling and sample processing. We thank Janet Krüger (UFZ), Juliane Riedel (University of Tübingen), Maria König (UFZ), Christin Kühnert (UFZ) and Lisa Glauch (UFZ) for experimental assistance. We thank Heinz Singer (Eawag) for supervision of the master study on the multi-layer SPE cartridge recovery. We thank Patrick Balaguer (INSERM) for provision of the PXR cell line and Michael Denison (UC Davis) for provision of the AhR CALUX cell line. A free academic licence of ChemAxon Calculator Plugins 2017 (http://www.chemaxon.com) was used to calculate physicochemical properties.References
- K. A. Maruya, N. G. Dodder, A. C. Mehinto, N. D. Denslow, D. Schlenk, S. A. Snyder and S. B. Weisberg, Integr. Environ. Assess. Manage., 2016, 12, 540–547 CrossRef CAS PubMed.
- A.-S. Wernersson, M. Carere, C. Maggi, P. Tusil, P. Soldan, A. James, W. Sanchez, V. Dulio, K. Broeg, G. Reifferscheid, S. Buchinger, H. Maas, E. Van Der Grinten, S. O'Toole, A. Ausili, L. Manfra, L. Marziali, S. Polesello, I. Lacchetti, L. Mancini, K. Lilja, M. Linderoth, T. Lundeberg, B. Fjällborg, T. Porsbring, D. J. Larsson, J. Bengtsson-Palme, L. Förlin, C. Kienle, P. Kunz, E. Vermeirssen, I. Werner, C. D. Robinson, B. Lyons, I. Katsiadaki, C. Whalley, K. den Haan, M. Messiaen, H. Clayton, T. Lettieri, R. N. Carvalho, B. M. Gawlik, H. Hollert, C. Di Paolo, W. Brack, U. Kammann and R. Kase, Environ. Sci. Eur., 2015, 27, 1–11 CrossRef CAS.
- P. A. Neale, A. Antony, M. Bartkow, M. Farre, A. Heitz, I. Kristiana, J. Y. M. Tang and B. I. Escher, Environ. Sci. Technol., 2012, 46, 10317–10325 CAS.
- N. Creusot, S. Aït-Aïssa, N. Tapie, P. Pardon, F. Brion, W. Sanchez, E. Thybaud, J.-M. Porcher and H. Budzinski, Environ. Sci. Technol., 2014, 48, 3649–3657 CrossRef CAS PubMed.
- J. Y. M. Tang, F. Busetti, J. W. A. Charrois and B. I. Escher, Water Res., 2014, 60, 289–299 CrossRef CAS PubMed.
- B. I. Escher and F. D. L. Leusch, Bioanalytical tools in water quality assessment, IWA Publishing, London, 2012 Search PubMed.
- N. Reineke, K. Bester, H. Huhnerfuss, B. Jastorff and S. Weigel, Chemosphere, 2002, 47, 717–723 CrossRef CAS PubMed.
- K. V. Thomas, J. Balaam, M. R. Hurst and J. E. Thain, Environ. Toxicol. Chem., 2004, 23, 1156–1163 CrossRef CAS PubMed.
- K. V. Thomas, M. R. Hurst, P. Matthiessen and M. J. Waldock, Environ. Toxicol. Chem., 2001, 20, 2165–2170 CrossRef CAS PubMed.
- S. Weigel, K. Bester and H. Huhnerfuss, J. Chromatogr. A, 2001, 912, 151–161 CrossRef CAS PubMed.
- F. D. L. Leusch, P. A. Neale, A. Hebert, M. Scheurer and M. C. M. Schriks, Environ. Int., 2017, 99, 120–130 CrossRef CAS PubMed.
- T. Schulze, M. Ahel, J. Ahlheim, S. Ait-Aissa, F. Brion, C. Di Paolo, J. Froment, A. O. Hidasi, J. Hollender, H. Hollert, M. Hu, A. Klolss, S. Koprivica, M. Krauss, M. Muz, P. Oswald, M. Petre, J. E. Schollee, T. B. Seiler, Y. Shao, J. Slobodnik, M. Sonavane, M. J. F. Suter, K. E. Tollefsen, Z. Tousova, K. H. Walz and W. Brack, Sci. Total Environ., 2017, 581, 350–358 CrossRef PubMed.
- A. L. Batt, M. S. Kostich and J. M. Lazorchak, Anal. Chem., 2008, 80, 5021–5030 CrossRef CAS PubMed.
- B. J. Vanderford, R. A. Pearson, D. J. Rexing and S. A. Snyder, Anal. Chem., 2003, 75, 6265–6274 CrossRef CAS PubMed.
- P. Välitalo, N. Perkola, T. B. Seiler, M. Sillanpää, J. Kuckelkorn, A. Mikola, H. Hollert and E. Schultz, Water Res., 2016, 88, 740–749 CrossRef PubMed.
- S. Huntscha, H. P. Singer, C. S. McArdell, C. E. Frank and J. Hollender, J. Chromatogr. A, 2012, 1268, 74–83 CrossRef CAS PubMed.
- S. Kern, K. Fenner, H. P. Singer, R. P. Schwarzenbach and J. Hollender, Environ. Sci. Technol., 2009, 43, 7039–7046 CrossRef CAS PubMed.
- P. A. Neale, R. Altenburger, S. Aït-Aïssa, F. Brion, W. Busch, G. de Aragão Umbuzeiro, M. S. Denison, D. Du Pasquier, K. Hilscherová, H. Hollert, D. A. Morales, J. Novák, R. Schlichting, T.-B. Seiler, H. Serra, Y. Shao, A. J. Tindall, K. E. Tollefsen, T. D. Williams and B. I. Escher, Water Res., 2017, 123, 734–750 CrossRef CAS PubMed.
- P. A. Neale, S. Ait-Aissa, W. Brack, N. Creusot, M. S. Denison, B. Deutschmann, K. Hilscherova, H. Hollert, M. Krauss, J. Novak, T. Schulze, T. B. Seiler, H. Serra, Y. Shao and B. I. Escher, Environ. Sci. Technol., 2015, 49, 14614–14624 CrossRef CAS PubMed.
- Z. Tousova, P. Oswald, J. Slobodnik, L. Blaha, M. Muz, M. Hu, W. Brack, M. Krauss, C. Di Paolo, Z. Tarcai, T. B. Seiler, H. Hollert, S. Koprivica, M. Ahel, J. E. Schollee, J. Hollender, M. J. F. Suter, A. O. Hidasi, K. Schirmer, M. Sonavane, S. Ait-Aissa, N. Creusot, F. Brion, J. Froment, A. C. Almeida, K. Thomas, K. E. Tollefsen, S. Tufi, X. Y. Ouyang, P. Leonards, M. Lamoree, V. O. Torrens, A. Kolkman, M. Schriks, P. Spirhanzlova, A. Tindall and T. Schulze, Sci. Total Environ., 2017, 601, 1849–1868 CrossRef PubMed.
- M. König, B. I. Escher, P. A. Neale, M. Krauss, K. Hilscherova, J. Novak, I. Teodorovic, T. Schulze, S. Seidensticker, M. A. K. Hashmi, J. Ahlheim and W. Brack, Environ. Pollut., 2017, 220, 1220–1230 CrossRef PubMed.
- P. A. Neale, N. A. Munz, S. Ait-Aissa, R. Altenburger, F. Brion, W. Busch, B. I. Escher, K. Hilscherova, C. Kienle, J. Novak, T. B. Seiler, Y. Shao, C. Stamm and J. Hollender, Sci. Total Environ., 2017, 576, 785–795 CrossRef CAS PubMed.
- F. D. L. Leusch, C. De Jager, Y. Levi, R. Lim, L. Puijker, F. Sacher, L. A. Tremblay, V. S. Wilson and H. F. Chapman, Environ. Sci. Technol., 2010, 44, 3853–3860 CrossRef CAS PubMed.
- P. Y. Kunz, E. Simon, N. Creusot, B. S. Jayasinghe, C. Kienle, S. Maletz, A. Schifferli, C. Schonlau, S. Ait-Aissa, N. D. Denslow, H. Hollert, I. Werner and E. L. M. Vermeirssen, Water Res., 2017, 110, 378–388 CrossRef CAS PubMed.
- A. Kolkman, M. Schriks, W. Brand, P. S. Bauerlein, M. M. E. van der Kooi, R. H. van Doorn, E. Emke, A. A. Reus, S. C. van der Linden, P. de Voogt and M. B. Heringa, Environ. Toxicol. Pharmacol., 2013, 36, 1291–1303 CrossRef CAS PubMed.
- P. A. Neale and B. I. Escher, Chemosphere, 2014, 108, 281–288 CrossRef CAS PubMed.
- K. L. Thorpe, M. Gross-Sorokin, I. Johnson, G. Brighty and C. R. Tyler, Environ. Health Perspect., 2006, 114, 90–97 CrossRef PubMed.
- B. I. Escher, N. Bramaz, M. Maurer, M. Richter, D. Sutter, C. von Kanel and M. Zschokke, Environ. Toxicol. Chem., 2005, 24, 750–758 CrossRef CAS PubMed.
- B. I. Escher, N. Bramaz, P. Quayle, S. Rutishauser and E. L. M. Vermeirssen, J. Environ. Monit., 2008, 10, 622–631 RSC.
- W. Busch, S. Schmidt, R. Kühne, T. Schulze, M. Krauss and R. Altenburger, Environ. Toxicol. Chem., 2016, 35, 1887–1899 CrossRef CAS PubMed.
- P. Välitalo, R. Massei, I. Heiskanen, P. Behnisch, W. Brack, A. J. Tindall, D. Du Pasquier, E. Küster, A. Mikola, T. Schulze and M. Sillanpää, Water Res., 2017, 126, 153–163 CrossRef PubMed.
- J. Nivala, P. A. Neale, T. Haasis, S. Kahl, M. König, R. A. Müller, T. Reemtsma, R. Schlichting and B. I. Escher, Environ. Sci.: Water Res. Technol., 2018, 4, 206–217 CAS.
- B. I. Escher, M. Allinson, R. Altenburger, P. A. Bain, P. Balaguer, W. Busch, J. Crago, N. D. Denslow, E. Dopp, K. Hilscherova, A. R. Humpage, A. Kumar, M. Grimaldi, B. S. Jayasinghe, B. Jarosova, A. Jia, S. Makarov, K. A. Maruya, A. Medvedev, A. C. Mehinto, J. E. Mendez, A. Poulsen, E. Prochazka, J. Richard, A. Schifferli, D. Schlenk, S. Scholz, F. Shiraish, S. Snyder, G. Y. Su, J. Y. M. Tang, B. van der Burg, S. C. van der Linden, I. Werner, S. D. Westerheide, C. K. C. Wong, M. Yang, B. H. Y. Yeung, X. W. Zhang and F. D. L. Leusch, Environ. Sci. Technol., 2014, 48, 1940–1956 CrossRef CAS PubMed.
- US EPA, Interactive Chemical Safety for Sustainability (iCSS) Dashboard v2, http://actor.epa.gov/dashboard/, accessed 26th Aug 2017.
- T. Backhaus and M. Faust, Environ. Sci. Technol., 2012, 46, 2564–2573 CrossRef CAS PubMed.
- T. Backhaus and M. Karlsson, Water Res., 2014, 49, 157–165 CrossRef CAS PubMed.
- S. Weigel, R. Kallenborn and H. Huhnerfuss, J. Chromatogr. A, 2004, 1023, 183–195 CrossRef CAS PubMed.
- J. C. Brennan, G. C. He, T. Tsutsumi, J. Zhao, E. Wirth, M. H. Fulton and M. S. Denison, Environ. Sci. Technol., 2015, 49, 11903–11912 CrossRef CAS PubMed.
- G. Lemaire, W. Mnif, J. M. Pascussi, A. Pillon, F. Rabenoelina, H. Fenet, E. Gomez, C. Casellas, J. C. Nicolas, V. Cavailles, M. J. Duchesne and P. Balaguer, Toxicol. Sci., 2006, 91, 501–509 CrossRef CAS PubMed.
- P. Balaguer, F. Francois, F. Comunale, H. Fenet, A. M. Boussioux, M. Pons, J. C. Nicolas and C. Casellas, Sci. Total Environ., 1999, 233, 47–56 CrossRef CAS PubMed.
- X. J. Wang, J. D. Hayes and C. R. Wolf, Cancer Res., 2006, 66, 10983–10994 CrossRef CAS PubMed.
- B. I. Escher, M. Dutt, E. Maylin, J. Y. M. Tang, S. Toze, C. R. Wolf and M. Lang, J. Environ. Monit., 2012, 14, 2877–2885 RSC.
- OECD, Test No. 236: Fish Embryo Acute Toxicity (FET) Test, OECD Guidelines for the Testing of Chemicals, Section 2, OECD Publishing, Paris, 2013 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7em00555e |
| This journal is © The Royal Society of Chemistry 2018 |