Open Access Article
Open Access ArticleImpact of copper and iron binding properties on the anticancer activity of 8-hydroxyquinoline derived Mannich bases†
Veronika F. S.
Pape‡
 a,
Nóra V.
May
a,
Nóra V.
May
 b,
G. Tamás
Gál
b,
István
Szatmári
c,
Flóra
Szeri§
b,
G. Tamás
Gál
b,
István
Szatmári
c,
Flóra
Szeri§
 a,
Ferenc
Fülöp
c,
Gergely
Szakács
a,
Ferenc
Fülöp
c,
Gergely
Szakács
 *ad and
Éva A.
Enyedy
*ad and
Éva A.
Enyedy
 *e
*e
aInstitute of Enzymology, Research Centre for Natural Sciences, Hungarian Academy of Sciences, Magyar Tudósok körútja 2, H-1117 Budapest, Hungary
bResearch Centre for Natural Sciences, Hungarian Academy of Sciences, Magyar Tudósok körútja 2, H-1117 Budapest, Hungary
cInstitute of Pharmaceutical Chemistry and Stereochemistry Research Group of Hungarian Academy of Sciences, University of Szeged, Eötvös u. 6, H-6720 Szeged, Hungary
dInstitute of Cancer Research, Medical University of Vienna, Borschkegasse 8a, A-1090 Vienna, Austria. E-mail: gergely.szakacs@meduniwien.ac.at
eDepartment of Inorganic and Analytical Chemistry, Interdisciplinary Excellence Centre, University of Szeged, Dóm tér 7, H-6720 Szeged, Hungary. E-mail: enyedy@chem.u-szeged.hu
First published on 7th November 2018
Abstract
The anticancer activity of 8-hydroxyquinolines relies on complex formation with redox active copper and iron ions. Here we employ UV-visible spectrophotometry and EPR spectroscopy to compare proton dissociation and complex formation processes of the reference compound 8-hydroxyquinoline (Q-1) and three related Mannich bases to reveal possible correlations with biological activity. The studied derivatives harbor a CH2–N moiety at position 7 linked to morpholine (Q-2), piperidine (Q-3), and chlorine and fluorobenzylamino (Q-4) substituents. Solid phase structures of Q-3, Q-4·HCl·H2O, [(Cu(HQ-2)2)2]·(CH3OH)2·Cl4·(H2O)2, [Cu(Q-3)2]·Cl2 and [Cu(HQ-4)2(CH3OH)]·ZnCl4·CH3OH were characterized by single-crystal X-ray diffraction analysis. In addition, the redox properties of the copper and iron complexes were studied by cyclic voltammetry, and the direct reaction with physiologically relevant reductants (glutathione and ascorbic acid) was monitored. In vitro cytotoxicity studies conducted with the human uterine sarcoma MES-SA/Dx5 cell line reveal the significant cytotoxicity of Q-2, Q-3, and Q-4 in the sub- to low micromolar range (IC50 values 0.2–3.3 μM). Correlation analysis of the anticancer activity and the metal binding properties of the compound series indicates that, at physiological pH, weaker copper(II) and iron(III) binding results in elevated toxicity (e.g.Q4: pCu = 13.0, pFe = 6.8, IC50 = 0.2 μM vs.Q1: pCu = 15.1, pFe = 13.0 IC50 = 2.5 μM). Although the studied 8-hydroxyquinolines preferentially bind copper(II) over iron(III), the cyclic voltammetry data revealed that the more cytotoxic ligands preferentially stabilize the lower oxidation state of the metal ions. A linear relationship between the pKa (OH) and IC50 values of the studied 8-hydroxyquinolines was found. In summary, we identify Q-4 as a potent and selective anticancer candidate with significant toxicity in drug resistant cells.
Introduction
The broad pharmacological activity of 8-hydroxyquinoline (Q-1, Chart 1) and its derivatives has been exploited in several medicinal applications ranging from antineurodegenerative, anti-inflammatory and antimicrobial to anticancer therapies.1,2 In all of these applications, the therapeutic effect of 8-hydroxyquinolines is believed to rely on metal chelation. Indeed, the (N,O) donor set of the planar, heterocyclic scaffold possesses a high affinity for various transition metal ions. For example, clioquinol (5-chloro-7-iodo-8-hydroxyquinoline, CQ), a lipophilic derivative of Q-1, has been explored as a treatment for Alzheimer's disease based on its ability to disrupt the interaction between zinc(II) and copper(II) ions and the Aβ peptide in the brain.3,4 CQ and numerous other 8-hydroxyquinoline derivatives were reported to exhibit a remarkable anticancer effect. The anticancer activity of 8-hydroxyquinolines is also related to complexation with endogenous metals, above all the redox active copper and iron ions.5–7 Despite continuous interest in novel 8-hydroxyquinolines as candidate anticancer agents, relatively little is known about their exact mechanism of toxicity. Studies have suggested several alternative models, including lysosomal disruption, inhibition of proteasome or histone deacetylase, inhibition of angiogenesis, or the inhibition of ribonucleotide reductase.8–10 Labile iron and copper pools in the cytoplasm represent only rather low concentrations under healthy conditions, as most of these metal ions are bound to proteins.11,12 However, the concentration of labile metal ions may be significantly increased in cancer cells.13 CQ can also act as an ionophore, inducing the transfer of metal ions across biological membranes, either in or out of cells.10 The antitumor activity of 8-hydroxyquinolines may also be linked to the toxicity of their metal complexes, as shown for copper(II),6,14 ternary ruthenium(II),15 half-sandwich organoruthenium,16–19 organorhodium,16 platinum(II)20 and gallium(III).21,22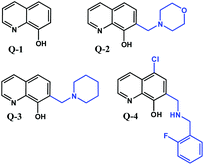 | ||
| Chart 1 Structures of the reference compound 8-hydroxyquinoline (Q-1) and its derivatives (Q-2, Q-3, and Q-4) reported in this work in their neutral form. | ||
Mannich bases of 8-hydroxyquinolines were reported to possess high potency against human cancer cells,23,24 and our meta-analysis of the cytotoxicity of various 8-hydroxyquinolines revealed the importance of the CH2–N subunit at position 7 for cytotoxicity.25–27 In this study we have chosen three 8-hydroxyquinoline derivatives in which the crucial CH2–N building block is retained. Morpholine (7-(morpholinomethyl)quinolin-8-ol, Q-2) and piperidine (7-(piperidin-1-ylmethyl)quinolin-8-ol, Q-3) moieties are introduced at position 7 (Chart 1) to decrease the hydrophobic character of the reference compound Q-1. To include a compound with a more lipophilic character to the study, Q-4 was developed, bearing a chlorine at position 5 and a fluorobenzylamino substituent at position 7 (5-chloro-7-((2-fluorobenzylamino)methyl)quinolin-8-ol, Q-4, Chart 1).
In this work we characterize the effect of these derivatizations on the cytotoxicity, proton dissociation processes, lipophilicity, copper(II) and iron(III) binding abilities and also study the redox properties of the relevant metal complexes to establish the relationship between the determined thermodynamic parameters and the biological activity of the compounds. Correlation analysis exploring the relationship between physico-chemical and biological properties provides an opportunity to better understand the mechanism of action of these compounds. Based on quantitative relationship analyses between chemical structures and bioactivity, between a set of thermodynamic parameters and toxicity, more reliable structure activity predictions can be made. Elegant examples of these types of correlation studies were shown for e.g. polyoxometalates regarding their antibacterial activity,28 or for the antitumor and antiangiogenic effect of half-sandwich Ru(η6-p-cymene) and Ir(η5-C5Me5) benzimidazole complexes.29
Results and discussion
Synthesis and characterization of the 8-hydroxyquinoline derived Mannich bases
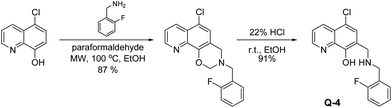
Q-1 is a commercially available compound, while Q-2 and Q-3 were obtained from the National Cancer Institute's Developmental Therapeutics Program (Chart 1).27 As shown in Scheme 1, Q-4 was synthesized by coupling 5-chloro-8-hydroxyquinoline with 2-fluorobenzylamine in the presence of paraformaldehyde via a modified Mannich reaction under microwave conditions. The formed [1,3]oxazino[5,6-h]quinolone derivative was isolated by crystallization from an n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate mixture and converted to the desired product as Q-4·HCl in ethanolic hydrogen chloride in excellent yield. NMR spectra were in agreement with the expected structures, enabling the assignment of all 1H and 13C resonances. The purity of Q-4·HCl was further confirmed by elemental analyses.
ethyl acetate mixture and converted to the desired product as Q-4·HCl in ethanolic hydrogen chloride in excellent yield. NMR spectra were in agreement with the expected structures, enabling the assignment of all 1H and 13C resonances. The purity of Q-4·HCl was further confirmed by elemental analyses.
The structures of Q-3 and Q-4·HCl·H2O were established by single crystal X-ray diffraction (Fig. 1). Crystal data and structure refinement parameters of the crystals are given in Table S1;† selected bond distances and angles are listed in Tables S2 and S4.†Q-3 crystallized in its neutral form in the orthorhombic crystal system, in the space group P212121, without solvent inclusion.
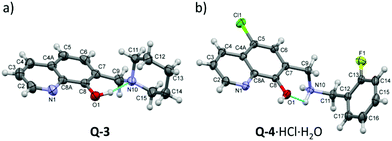 | ||
| Fig. 1 ORTEP views of Q-3 (a) and Q-4·HCl·H2O (b) at the 30% probability level of displacement ellipsoids. Blue lines indicate the suggested intramolecular hydrogen bonds. Selected bond distances and bond angles are listed in Tables S2 and S4.† | ||
The molecular structure is stabilized by the O1–H1O⋯N10 intramolecular hydrogen bond. The crystal structure is stabilized by weak C–H⋯O and C–H⋯π interactions with the participation of the piperidine protons (Fig. S1 and Tables S2, S3†). Q-4 crystallized in its protonated form with Cl− counter ions and a water molecule in the triclinic crystal system, in the space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) . The conformation is stabilized by a weak intermolecular interaction between O1 and one of the amino protons. The neighboring molecules are connected by O–H⋯N and C–H⋯O hydrogen bonds, and π⋯π (off-centered parallel stacking) interactions (Fig. S1, S2 and Tables S3, S5†).
. The conformation is stabilized by a weak intermolecular interaction between O1 and one of the amino protons. The neighboring molecules are connected by O–H⋯N and C–H⋯O hydrogen bonds, and π⋯π (off-centered parallel stacking) interactions (Fig. S1, S2 and Tables S3, S5†).
In vitro toxicity of Q-1 and its Mannich base derivatives against a cancer cell line and primary hepatocytes
The in vitro cytotoxic activity of compounds Q-1 to Q-4 was determined in the human uterine sarcoma MES-SA/Dx5 cell line and in primary hepatocytes (Table 1). The compounds demonstrated marked cytotoxic activity in the micromolar and submicromolar concentration range in the drug resistant human uterine sarcoma MES-SA/Dx5 cell line, increasing in the following order Q-2 < Q-1 < Q-3 < Q-4. Notably, Q-3 and Q-4 possess even stronger cytotoxic activity than the FDA approved anticancer agent doxorubicin. To exclude non-specific toxic effects related to the multiple targets of metal chelators,30 primary hepatocytes were also included in the study. Compared to doxorubicin, the newly investigated compounds displayed a manifold higher selectivity towards cancer cells (Table 1). Due to the high proliferation rate, malignant cancer cells show increased reliance on metal ions, which are required e.g. as co-factors in enzymes catalyzing essential steps in DNA-synthesis, as well as for cell cycle progression and cellular growth.31–34 For example, the rate limiting step in DNA synthesis is the reduction of ribonucleotides, which is catalyzed by the iron dependent enzyme ribonucleotide reductase (RR).35,36 Thus, selective toxicity of the studied compounds to the human uterine sarcoma cells over hepatocytes might be the result of the altered metal homeostasis of the cancerous cells.| IC50/μM | Q-1 | Q-2 | Q-3 | Q-4 | Doxorubicin |
|---|---|---|---|---|---|
| a Determined by means of the MTT assay after exposure for 72 h. Values are means ± standard deviations obtained from at least three independent experiments. | |||||
| MES-SA/Dx5 | 2.46 ± 0.69 | 3.27 ± 0.39 | 0.86 ± 0.26 | 0.20 ± 0.06 | 2.13 ± 0.65 |
| Primary hepatocytes | 115 ± 18 | 243 ± 69 | 163 ± 35 | 40.4 ± 8.9 | 2.14 ± 1.11 |
Proton dissociation processes of the studied 8-hydroxyquinolines
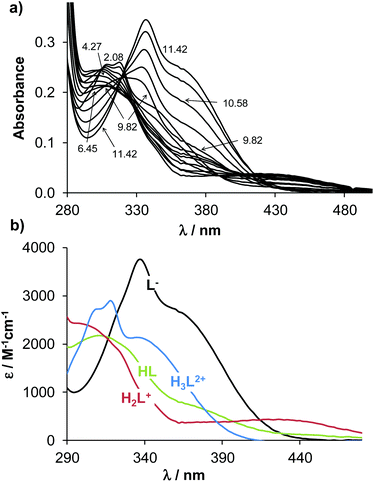
The proton dissociation constant (Ka) is a key physico-chemical parameter influencing the pharmacokinetic properties of the compounds. Determination of the pKa value allows the estimation of the distribution of the chemical forms of the molecule in the various protonation states at a given pH. As the pKa values of the reference compounds Q-1 and Q-3 are available, although for Q-3 under different conditions,16,37 we determined the pKa values of Q-2, Q-3 and Q-4 by UV-visible (UV-Vis) spectrophotometric titrations in an aqueous solution. This method requires lower concentrations as compared to pH-potentiometry, which are compatible with the limited water-solubility of the compounds. The UV-Vis spectra of all ligands recorded in the pH range 2 to 11.5 show characteristic spectral changes upon deprotonation processes.Representative spectra for Q-2, as well as the molar absorbance spectra of individual ligand species in the different protonated forms are shown in Fig. 2. The recorded absorbance spectra were deconvoluted resulting in the pKa values of the compounds and the molar absorbance spectra of the various species (Table S6†). The pKa values were assigned to the particular moieties by careful analysis of the spectral changes (Scheme 2). The deprotonation processes of Q-1 are well-described in the literature,16,37 and pK1 and pK2 values correspond to the deprotonation of the quinolinium-NH+ and the phenolic-OH moieties, respectively, as indicated in Scheme 2.
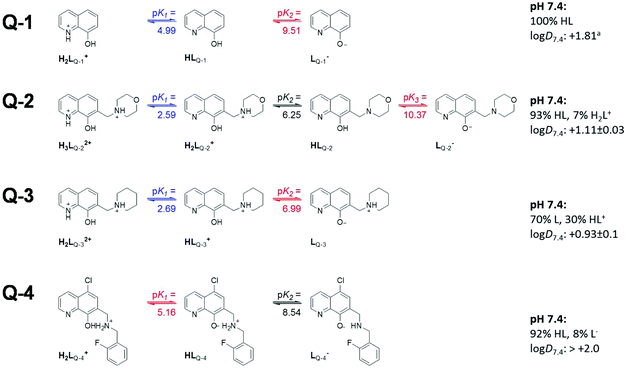 | ||
Scheme 2 Proton dissociation steps of the studied 8-hydroxyquinolines and their pKa values. Predominant species and n-octanol/water distribution coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4) determined at pH 7.4. (T = 25 °C, I = 0.20 M (KCl)) aData taken from ref. 43. D7.4) determined at pH 7.4. (T = 25 °C, I = 0.20 M (KCl)) aData taken from ref. 43. | ||
Values obtained for Q-3 are in good agreement with those reported in the presence of KNO3 instead of KCl.16
The most pronounced spectral changes are linked to the deprotonation of the hydroxyl functional group. The emerging strong bands with higher λmax values originate from the more extended conjugated π-electron system in the deprotonated form.16,37 In comparison with the 8-hydroxyquinoline scaffold (Scheme 2), the morpholino-substituent in the 7-position in Q-2 increases the pKa value of the phenolic-OH by almost one order of magnitude, possibly due to the formation of a hydrogen bond between the OH and the morpholine-N in the HL form of the ligand, which hinders deprotonation. In contrast, the protonated (thus positively charged) amine moieties of the piperidine of Q-3 and of the benzylamine in the case of Q-4 decrease the proton dissociation constants of the OH group by 2.5 and 4.4 orders of magnitude, respectively, as compared to Q-1. These results are consistent with the lower pKa values reported for numerous hydroxamic and carboxylic acids due to the electron withdrawing effect of the protonated piperidinium moiety.38 As confirmed by single crystal X-ray diffraction analysis of the solid compounds Q-3 and Q-4·HCl·H2O (vide supra), possible hydrogen bond formations between the phenolate and the piperidinium moiety of Q-3 (in the zwitterionic L form) can additionally contribute to the diminished pKa values due to the stabilization of the conjugate base. Intramolecular hydrogen bonding is also probable in the HL form of Q-4 between the phenolate and the protonated benzylamine moieties. The pKa of the OH group in Q-4 (pK1) is also affected by the electron withdrawing effects of the chloro-substituent.
pKa values assigned to the deprotonation of the quinolinium-NH+ moiety of the R7-substituted ligands (pK1 for Q-2 and Q-3) were found to be considerably lower compared to that of Q-1 as a consequence of the electron withdrawing effect of the protonated methylene-amine groups in the Mannich bases. In Q-4 the deprotonation of this moiety could not be observed under the applied experimental conditions, since it most probably takes place at lower pH (<2) due to the additional negative inductive effect of the halogen atom (the predicted pKa value for the quinolinium-NH+ moiety in Q-4 is 0.98).39
Based on smaller spectral alterations further pKa values could be assigned corresponding to the deprotonation processes of the non-coordinating nitrogens, such as the morpholinium-NH+ of Q-2 and the benzylamino-NH+ of Q-4. Deprotonation of the piperidinium-NH+ of Q-3 most probably takes place only at strongly basic pH (pKa > 11.5).
On the basis of the obtained pKa values, species distribution was calculated at pH 7.4 (Scheme 2). Thus, the neutral forms of these compounds predominate at physiological pH; however Q-3 and Q-4 are mostly present in their zwitterionic L and HL forms, respectively. The lipophilicity of the compounds was characterized by the n-octanol/water distribution coefficients at pH 7.4 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 values in Scheme 2). These data revealed that while the introduction of the aliphatic methylene amines in position R7 of Q-2 and Q-3 increases the hydrophilic character of the ligands in comparison with the unsubstituted scaffold Q-1, the additional benzyl moiety on the methylene amine in R7 and the halogen substituent in R5 significantly increase the lipophilicity of Q-4.
D7.4 values in Scheme 2). These data revealed that while the introduction of the aliphatic methylene amines in position R7 of Q-2 and Q-3 increases the hydrophilic character of the ligands in comparison with the unsubstituted scaffold Q-1, the additional benzyl moiety on the methylene amine in R7 and the halogen substituent in R5 significantly increase the lipophilicity of Q-4.
Complex formation of the studied 8-hydroxyquinolines with iron(III) and copper(II) in solution
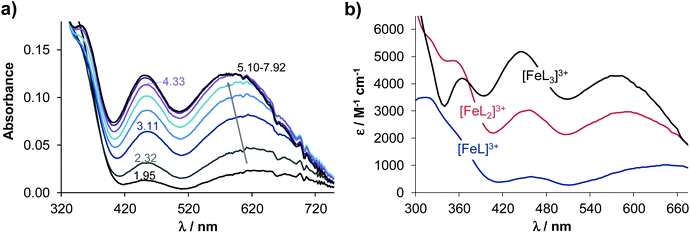
The anticancer activity of 8-hydroxyquinolines may be related to the formation of (redox active) metal complexes with essential ions such as iron and copper7,40 Therefore, we investigated complex formation of Q-2 to Q-4 with two physiologically relevant metal ions (iron(III) and copper(II)), in order to reveal a possible relationship between the metal binding ability of these ligands and their cytotoxicity. Results were compared to data reported for Q-1.41,42Complexation of the 8-hydroxyquinoline derived Mannich bases with iron(III) and copper(II) was studied by UV-Vis spectrophotometry. During titrations with both metal ions the absorbance values tend to decrease at pH ≳ 8. In the case of iron(III), hydrolysis of the metal ion may suppress complex formation at alkaline pH values, whereas neutral copper(II) complexes precipitate at pH ≳ 9 (Fig. S3† for Q-2 and Q-4 complexes). Given these possible artefacts, evaluation was restricted to data collected at pH < 8. Spectral changes upon complexation with iron(III) detected in the 350–750 nm wavelength range were the most useful for the calculations of the overall stability constants, since bands located in this range are relatively well-separated from the ligand bands (see the representative spectra for Q-3 in Fig. 3).
The stoichiometries of the metal complexes and the overall stability constants (β) furnishing the best fits to the experimental data are shown in Table 2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β) of the copper(II) and iron(III) complexes of ligands Q-2 to Q-4 determined by UV-vis spectrophotometric titrations and pM values calculated at pH 7.4. Data for Q-1 are also shown for comparison. (Charges of the complexes are omitted for clarity. L is neutral for Q-3 and singly negatively charged for Q-1, Q-2 and Q-4.) (T = 25 °C, I = 0.20 M (KCl))
β) of the copper(II) and iron(III) complexes of ligands Q-2 to Q-4 determined by UV-vis spectrophotometric titrations and pM values calculated at pH 7.4. Data for Q-1 are also shown for comparison. (Charges of the complexes are omitted for clarity. L is neutral for Q-3 and singly negatively charged for Q-1, Q-2 and Q-4.) (T = 25 °C, I = 0.20 M (KCl))
| Q-1 | Q-2 | Q-3 | Q-4 | |
|---|---|---|---|---|
| a Data taken from ref. 41. b Data taken from ref. 42. c pM = −log[M] calculated at pH = 7.4, cM = 1 μM; cL = 10 μM. d pM* = −log([Fe] + i × ∑[Fei(OH)j]) calculated at pH = 7.4, cM = 1 μM; cL = 10 μM. | ||||
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeLH] β [FeLH] |
— | 19.80 ± 0.04 | — | 18.46 ± 0.04 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeL] β [FeL] |
13.69a | 16.90 ± 0.04 | 11.11 ± 0.02 | 14.31 ± 0.05 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeL2H2] β [FeL2H2] |
— | — | — | 34.44 ± 0.04 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeL2H] β [FeL2H] |
— | 35.18 ± 0.04 | — | 28.50 ± 0.06 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeL2] β [FeL2] |
26.3a | — | 20.32 ± 0.03 | — |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeL3H] β [FeL3H] |
— | 49.03 ± 0.03 | — | — |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [FeL3] β [FeL3] |
36.9a | 42.49 ± 0.03 | 25.42 ± 0.07 | — |
| pMc | 21.1 | 21.1 | 16.0 | 14.8 |
| pM*d | 13.0 | 13.0 | 8.0 | 6.8 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [CuLH] β [CuLH] |
— | — | — | 16.87 ± 0.03 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [CuL] β [CuL] |
12.1b | 16.54 ± 0.04 | 10.6 ± 0.1 | 10.96 ± 0.07 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [CuL2H2] β [CuL2H2] |
— | — | — | 32.54 ± 0.06 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [CuL2H] β [CuL2H] |
— | 34.67 ± 0.04 | — | 26.94 ± 0.06 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β [CuL2] β [CuL2] |
23.0b | 27.5 ± 0.1 | 19.4 ± 0.1 | — |
| pMc | 15.1 | 17.4 | 14.9 | 13.0 |
The molar absorbance spectra of the individual metal complexes were also computed (see λmax and molar absorptivity values in Table S7†). As expected, mono and bis complexes are formed with both metal ions, while tris complexes could also be detected with iron(III). In the case of Q-2 and Q-4, further protonated complexes were formed, in which the protons were attributed to the non-coordinating protonated morpholinium and secondary amine moieties, respectively (Table 2).
Based on the overall stability constants (Table 2), concentration distribution diagrams were computed at various metal-to-ligand ratios (exemplarily diagrams are shown for 5 μM ligand concentration at pH 7.4 in Fig. S4†). These data reveal the different speciation of the four ligands at physiological pH. Complexation of iron(III) with Q-1 and Q-2 predominantly results in tris-ligand complexes, while with Q-3, the formation of the bis-complex is more pronounced. In the iron(III)−Q-4 system, mono and protonated bis complexes co-exist at pH 7.4. Notably, the formation of tris complexes is not observed even at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 metal-to-ligand ratio, most probably due to steric reasons. In the case of copper(II) complexation, at a 1
3 metal-to-ligand ratio, most probably due to steric reasons. In the case of copper(II) complexation, at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 metal-to-ligand ratio, the coordination sphere of the metal ion is likely to be saturated in the physiologically relevant pH range, as bis complexes are formed almost exclusively under these conditions.
2 metal-to-ligand ratio, the coordination sphere of the metal ion is likely to be saturated in the physiologically relevant pH range, as bis complexes are formed almost exclusively under these conditions.
The overall stability constants of the complexes cannot be compared directly owing to the different basicities of the ligands and the number of dissociable protons. However, the stability constants for the iron(III) species are generally higher than those of the corresponding copper(II) complexes, the significantly different extent of hydrolysis of these metal ions should also be taken into consideration when the solution stabilities are compared. Therefore, in order to compare the metal binding ability of the four investigated 8-hydroxyquinolines adequately, pM values (negative decadic logarithm of the equilibrium concentration of the unbound metal ion) were computed at pH 7.4 (Table 2). In the case of iron(III), the formation of hydroxido species was also taken into account (pM* values in Table 2). At pH 7.4, the iron binding capacity of the investigated ligands follows the order Q-1 ∼ Q-2 > Q-3 > Q-4, and a similar trend (Q-2 > Q-1 > Q-3 > Q-4) was obtained for the copper(II) complexes. A comparison of the pM (of copper(II)) and pM* (of iron(III)) values indicates that at pH 7.4, the studied ligands preferentially bind copper(II) over iron(III).
To further investigate the coordination modes of the copper(II) complexes in solution, electron paramagnetic resonance (EPR) spectroscopic measurements were performed.
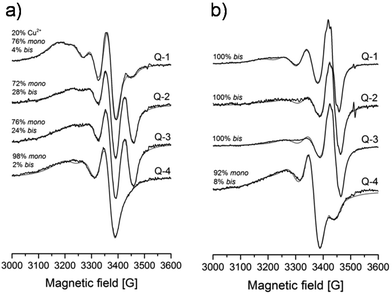
EPR spectra were recorded at room temperature for samples containing copper(II) and the investigated ligands (deprotonated with one equivalent of NaOH) in equimolar concentrations and at two-fold ligand excess (Fig. 4). Dimethyl sulfoxide (DMSO) was used as a solvent in which the complexes are soluble in the mM concentration range. The measured spectra could be deconvoluted into the individual spectra of mono- and bis complexes (and free copper(II)). Notably, the [CuL2] and [CuL2H] complexes could not be distinguished, as side chain deprotonations have a negligible impact on the EPR spectra. The EPR data indicate that the formation of the mono complexes seems to be more pronounced in DMSO than in the aqueous solution in case of Q-1 and Q-4. Additionally, the fraction of the bis complex is much lower for Q-4 as compared to the other ligands, most probably due to steric reasons. The isotropic EPR parameters obtained by the simulation of room temperature spectra are shown in Table 3.
| Ligand | Complex | g o | A o/G | a N/G |
|---|---|---|---|---|
| a The experimental error is ±0.001 for go, ±1 G for Ao and aN. | ||||
| DMSO | Cu(DMSO) | 2.170 | 35 | — |
| Q-1 | Mono | 2.144 | 59 | 9 |
| Bis | 2.116 | 73 | 11, 11 | |
| Q-2 | Mono | 2.142 | 57 | 10 |
| Bis | 2.113 | 71 | 11, 11 | |
| Q-3 | Mono | 2.144 | 59 | 8 |
| Bis | 2.114 | 72 | 11, 11 | |
| Q-4 | Mono | 2.152 | 68 | 9 |
| Bis | 2.113 | 63 | 11, 11 | |
The go values of the copper(II) complexes support a similar bidentate (N,O−) 8-quinolinolato coordination mode in all complexes. It is worth noting that the go constant of the mono complex of Q-4 is higher compared to those of the complexes of the other 3 ligands, indicating a somewhat weaker ligand field.
EPR spectra were also recorded at 77 K in methanol and in toluene. Interestingly, these EPR spectra revealed that while for Q-1 and Q-3 the bis complexes form monomers, for Q-2 and Q-4, additional dimeric species were detectable (Fig. S5, Table S8†) in the frozen samples. A doublet peak detected at around 3000 and 3500 G and the half-field signal at 1500 G unambiguously show the appearance of a coupled-spin system (S = 1), which can be established by two neighboring copper(II) centers. From the simulation of the spectra the orientation of the copper(II) centers can be calculated (polar angles: χ = 28.5°, ψ = −5.0°), showing that one center is above the other. A copper–copper distance of 3.88 Å was calculated from the dipolar coupling by using the point-dipole approach. These results are in good agreement with the obtained X-ray crystallographic results (vide infra) where χ = 35.1° and ψ = 0° could be measured for Q-2 and χ = 30.6° and ψ = 0° for Q-4, and the copper–copper distances are 3.3935(7) Å and 3.8354(8) Å respectively. From this result we conclude that the bis complexes of Q-2 and Q-4 preferably form dimers in frozen solution as well as in the solid state.
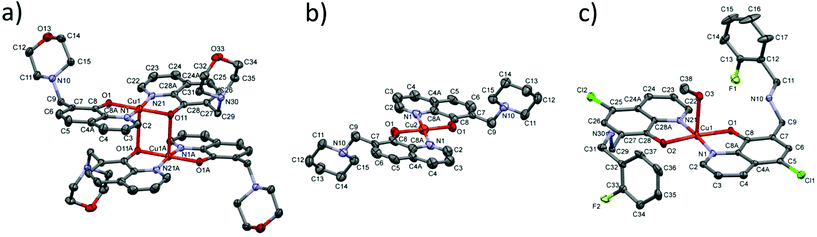
Solid phase characterization of the copper(II) complexes of the studied 8-hydroxyquinolines
Single crystals suitable for X-ray crystallography were obtained from methanolic solution in the case of the copper(II) complexes of Q-2, Q-3 and Q-4 as [(Cu(HQ-2)2)2]·(CH3OH)2·Cl4·(H2O)2, [Cu(Q-3)2]·Cl2 and [Cu(HQ-4)2(CH3OH)]·ZnCl4·CH3OH (Fig. 5). Crystal data and structure refinement parameters are given in Table S1.† Selected bond distances and angles are specified in the legend of Fig. 5, while packing arrangements are shown in Fig. S6–8.† As expected based on their structural similarity, the X-ray diffraction studies reveal a similar bidentate (N,O−) 8-quinolinolato coordination mode for copper(II) complexes of Q-3 and Q-4 and the reference compound Q-1.44 Despite a coordination with the same donor atom set, the copper(II) complex of Q-2 adopts a cyclic dimer [(Cu(HQ-2)2)2]4+ form, in which the phenolato oxygen acts as a bridging atom between two metal centers of the neighboring complexes (Fig. S6†). However, the presence of long intermolecular contacts Cu1⋯O11A or Cu1A⋯O11 (Å) strongly suggests the weak association of complexes into dimers, which most probably dissociate in aqueous solution with the formation of monomeric species.[(Cu(HQ-2)2)2]·(CH3OH)2·Cl4·(H2O)2 crystallized in the monoclinic C2/c space group (Fig. 5 and S6†). The ligand in this complex is in the neutral zwitterionic HL form (with phenolate-O− and morpholinium-NH+). The metal ion is coordinated in a square-pyramidal mode, with the basal plane defined by atoms O1 and N1 from one Q-2 ligand and O11 and N21 from another Q-2 ligand in the trans position to each other, while atom O11 from another bis complex occupies the axial position of the metal ion (and vice versa) with a long Cu1–O11 distance (2.681(3) Å). The complex has an inversion centre symmetry, half of the molecule can be found in the asymmetric unit. The charge of the complex is neutralized by chloride counter ions. It is worth mentioning that the O33 atom of a neighbouring morpholine ring comes closer at a distance of 2.810(3) Å to the second axial position of the copper atom (Fig. S6†). The copper–copper distance is 3.3935(7) Å in the dimeric complex. The complex structure is further stabilized by two intramolecular hydrogen bonds C22–H22⋯O1 and C29–H29B⋯O1, and the main intermolecular interactions are listed in Table S9.†
[Cu(Q-3)2]·Cl2 crystallized in the monoclinic crystal system, in the space group P21/n (Fig. 5 and S7†). The ligand shows a bidentate (N,O−) coordination mode forming a square planar type geometry around the copper center. The planar arrangement of the two quinolinolato rings is also secured by the intramolecular hydrogen bond of C2–H2⋯O1. In addition to a N–H⋯Cl bond and three weak C–H⋯Cl hydrogen bonds, π⋯π intermolecular interactions stabilize the crystal lattice between the neighboring quinoline moieties (Fig. S7 and Table S10†). The closest copper–copper distance is 6.534(5) Å in this structure.
[Cu(HQ-4)2(CH3OH)]·ZnCl4·CH3OH crystallized in the triclinic crystal system, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) (Fig. 5 and S8†). The two HL ligands are coordinated via the (N,O−) donor set resulting in a square planar geometry. The fifth (axial) coordination position is occupied by a methanol-OH group with a Cu1–O3 distance of 2.314(4) Å. Although the copper–copper distance (3.8354(8) Å) is significantly larger than in the crystal [Cu(Q-3)2]·Cl2 the arrangement of the pair of complexes is very similar to it, and the complexes are placed face to face with their parallel basal planes such as O2 is arranged in the axial position of a neighboring copper ion (and vice versa, see Fig. S8†). Intra- and intermolecular interactions can be found in Table S11.†
(Fig. 5 and S8†). The two HL ligands are coordinated via the (N,O−) donor set resulting in a square planar geometry. The fifth (axial) coordination position is occupied by a methanol-OH group with a Cu1–O3 distance of 2.314(4) Å. Although the copper–copper distance (3.8354(8) Å) is significantly larger than in the crystal [Cu(Q-3)2]·Cl2 the arrangement of the pair of complexes is very similar to it, and the complexes are placed face to face with their parallel basal planes such as O2 is arranged in the axial position of a neighboring copper ion (and vice versa, see Fig. S8†). Intra- and intermolecular interactions can be found in Table S11.†
Correlations between solution equilibrium constants and in vitro cytotoxicity
While the low number of the compounds did not allow statistically valid conclusions, we attempted to correlate the solution equilibrium properties of the studied 8-hydroxyquinolines with their biological activity. Correlations between the in vitro cytotoxicity (pIC50) obtained in the MES-SA/Dx5 cancer cell line, lipophilic character (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4), proton dissociation constants (pKa) of the compounds and their metal binding abilities (pM values) were analyzed (Fig. 6). No relationship was found between the pIC50 and log
D7.4), proton dissociation constants (pKa) of the compounds and their metal binding abilities (pM values) were analyzed (Fig. 6). No relationship was found between the pIC50 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4 values (Table 1 and Scheme 2) suggesting that lipophilicity as a single parameter is not a predictor of cytotoxicity of the studied ligands, despite the often well-established correlations between the biological activity and lipophilicity of cytotoxic drug molecules, as the latter strongly affects membrane permeability. Whereas there was no strong correlation between pIC50 and pKa (NquinolinidiumH+) values, cytotoxicity seemed to increase with lower pKa values of the hydroxyl moiety (Fig. 6a and b). Furthermore, compounds with weaker copper(II) and iron(III) binding abilities (at physiological pH) seemed to be more toxic, as suggested by the relationship of pM and pIC50 values (Fig. 6c and d). The more cytotoxic Q-4 and Q-3 depending on the metal-to-ligand ratio form positively charged complexes at pH 7.4 with both metal ions (Q-4: [CuL2H]+, [CuL]+ and [FeL2H]2+; Q-3: [CuL2]2+, [CuL]2+ and [FeL2]3+), while the less active compounds (Q-1 and Q-1) mostly form neutral complexes (Q-1: [CuL2], [CuL]+ and [FeL3]; Q-2: [CuL2], [CuL2H]+, [CuL]+ and [FeL3]).
D7.4 values (Table 1 and Scheme 2) suggesting that lipophilicity as a single parameter is not a predictor of cytotoxicity of the studied ligands, despite the often well-established correlations between the biological activity and lipophilicity of cytotoxic drug molecules, as the latter strongly affects membrane permeability. Whereas there was no strong correlation between pIC50 and pKa (NquinolinidiumH+) values, cytotoxicity seemed to increase with lower pKa values of the hydroxyl moiety (Fig. 6a and b). Furthermore, compounds with weaker copper(II) and iron(III) binding abilities (at physiological pH) seemed to be more toxic, as suggested by the relationship of pM and pIC50 values (Fig. 6c and d). The more cytotoxic Q-4 and Q-3 depending on the metal-to-ligand ratio form positively charged complexes at pH 7.4 with both metal ions (Q-4: [CuL2H]+, [CuL]+ and [FeL2H]2+; Q-3: [CuL2]2+, [CuL]2+ and [FeL2]3+), while the less active compounds (Q-1 and Q-1) mostly form neutral complexes (Q-1: [CuL2], [CuL]+ and [FeL3]; Q-2: [CuL2], [CuL2H]+, [CuL]+ and [FeL3]).
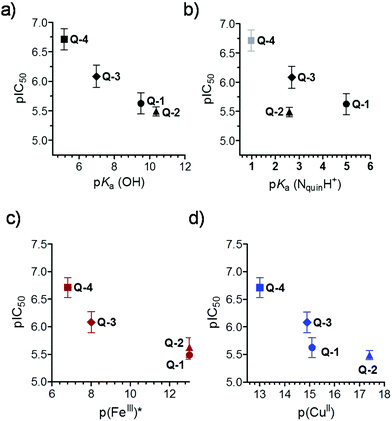 | ||
| Fig. 6 Correlations between pIC50 values measured in MES-SA/Dx5 cell lines (IC50: mol L−1) and pKa (OH) (a), pKa (NquinolinidiumH+) (b), pFe(III)* (c) and pCu(II) (d) of the studied 8-hydroxyquinolines. Data are taken from Tables 1 and 2 and Scheme 2. | ||
Redox properties of the studied 8-hydroxyquinolines with iron(III) and copper(II)
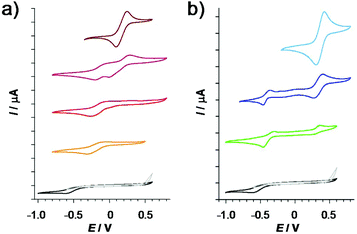
In addition to solution stability, the toxicity of metal complexes is also influenced by their redox properties. We therefore characterized the redox features of the copper(II) and iron(III) complexes of the studied 8-hydroxyquinoline ligands. First, the redox activity of iron(III) and copper(II) complexes was investigated by cyclic voltammetry (CV). The CV measurements were carried out in pure DMSO due to the low water solubility of these metal complexes (c ≪ 0.5 mM). However, aprotic organic solvents can modify complex formation processes and redox properties. Reliable data could be collected only for the complexes formed with Q-1 and Q-3, when quasi-reversible redox processes were observed.Representative voltammograms recorded for the complexes formed with Q-3 at different metal-to-ligand ratios are shown in Fig. 7 and for the complexes formed with the other ligands in Fig. S9.† The corresponding electrochemical data are collected in Table S12.† Based on the voltammograms it could be concluded that upon complex formation with Q-1 and Q-3, the half-wave potentials (E1/2) are shifted to more negative potentials compared to those of the corresponding metal salts. (Ligands Q-3 and Q-2 were found to be redox active as well, but their identified cathodic peaks appear at much lower potentials than the peaks belonging to the metal complexes (Fig. S9 and Table S12†).) On the other hand, redox processes of the copper(II/I) systems occur in lower potential ranges compared to the iron containing systems. Interestingly, the observed shifts from the half-wave potentials of the metal salts were much larger in the case of ligand Q-1 as compared to Q-3 for both metal ions. Thus, Q-3, the more cytotoxic ligand, stabilizes the lower oxidation states of iron and copper more than Q-1.
In comparison with the formal potential of −0.26 V reported for the redox couple of oxidized glutathione/glutathione (GSSG/GSH) at physiological pH,45 the determined E1/2 value of the iron-Q-3 complex is more positive (Table S12†), suggesting that GSH might be able to reduce this iron(III) complex, while the reduction of the iron(III) complex of Q-1 by GSH seems unlikely based on these data. However, since the CV data were obtained in DMSO solutions, care is warranted in extrapolating the obtained results to the intracellular milieu. Therefore, the direct reactions of the iron(III) and copper(II) complexes of Q-1 to Q-4 with two physiologically relevant reducing agents GSH and ascorbate (ASC) were followed by UV-Vis spectrophotometry under strictly anaerobic conditions. Absorbance spectra were analyzed at λ > 310 nm, where neither the reduced nor the oxidized forms of GSH and ASC absorb light.46,47
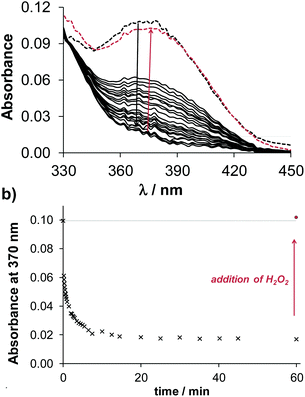
No significant spectral changes were observed upon reaction of the iron(III) complexes with 2.5 equivalents of GSH (in HEPES buffer, pH 7.4) within a time frame of 90 min. It was reported that the reduction of some iron(III)47 and vanadium(V)46,48 complexes by GSH might be kinetically hindered, while the dehydro-L-ascorbic acid/ASC redox pair (E′° at physiological pH = +0.05 V),49 reacts considerably faster. Redox reactions of the iron(III) complexes with ASC were observed at pH 6.87 as shown in Fig. S10,† although the spectral changes were still minor under the conditions, especially in the case of Q-3 and Q-4. Addition of H2O2 to the samples reverses these changes.
Notably, the copper complexes have lower redox potentials than the iron compounds (Table S12†), thus the copper(II) complexes are less powerful oxidizing agents. Nevertheless, copper(II) complexes of Q-1 to Q-4 show characteristic spectral changes upon reaction with GSH, as displayed exemplarily for the complex of Q-2 in Fig. 8. The significant decrease of the absorbance values at the λmax of the copper(II) complex occurred relatively fast, indicating a possible ligand release from the reduced copper(I) complex. In order to assess whether the observed reduction process of the copper(II) complex is reversible, H2O2 was added to the samples at the end of the reaction. Upon addition of the oxidizing agent the spectrum of the original copper(II) complex reappeared, indicating the reversibility of the redox process and the possibility of an intracellular redox-cycling reaction. The time courses of the spectral changes induced by the reaction of the various copper(II) complexes with GSH are compared in Fig. S11.† The observed rate constants (kobs) of the redox reactions, calculated based on the initial slope of the ln(A/A0) vs. t plots (not shown), were as follows: 0.15 min−1 for Q-1, 0.21 min−1 for Q-2, 0.20 min−1 for Q-3, and 0.02 min−1 for Q-4. This trend indicates that a stronger copper(II) binding ability of the ligands is accompanied by the faster redox reaction of their complex with GSH. Despite the trend in the kinetics of the reduction of the copper complexes, showing the slowest reaction for the most toxic ligand, a clear quantitative relationship could not be established between the redox chemical characteristics and the cytotoxic activity of the compounds.
Conclusions
8-Hydroxyquinoline derived Mannich bases were shown to exhibit high potency against human cancer cell lines, and the importance of a CH2–N building block at position 7 has also been described.27 As the biological activity of a chelator can be affected by the lipophilicity, pKa values, metal binding abilities and redox properties of the forming complexes, the possible relationship between these parameters was monitored in detail in the case of 8-hydroxyquinoline (Q-1) and its three derivatives containing the crucial CH2–N moiety. Due to the protonated tertiary amine nitrogens in 7-(morpholinomethyl)quinolin-8-ol (Q-2) and 7-(piperidin-1-ylmethyl)quinolin-8-ol (Q-3), the hydrophilicity is increased as compared to Q-1, while the newly synthesized and characterized 5-chloro-7-((2-fluorobenzylamino)methyl)quinolin-8-ol (Q-4) possesses a more lipophilic character. In addition, the various substitutions of the Q-1 scaffold altered the size, the acid–base character, charges and the metal ion chelation of the compounds. To evaluate the effect of these alterations on the in vitro anticancer activity of the compounds, cytotoxicity of Q-1 to Q-4 was tested against the drug resistant human uterine sarcoma MES-SA/Dx5 cell line as well as against primary hepatocytes. All compounds showed marked cytotoxic activity (IC50 values 0.20–3.27 μM) in the cancer cells increasing in the following order Q-2 < Q-1 < Q-3 < Q-4. IC50 values obtained in primary hepatocytes revealed a much better selectivity of the studied compounds towards cancer cells than the chemotherapy agent doxorubicin. Even though the most lipophilic derivative Q-4 was found to be the most toxic, no clear relationship was apparent between the lipophilicity of the set of derivatives and their toxicity, suggesting that the cytotoxicity of the compound series is not strictly dependent on membrane permeability.Relationship between proton dissociation constants and cytotoxicity
Based on the determined pKa values, the compounds predominate in their neutral forms at physiological pH, although Q-3 and Q-4 are mostly present in their zwitterionic L and HL forms, respectively. As compared to Q-1, the pKa values of the quinolinium-NH+ decrease due to the electron withdrawing effect of the various substituents in Q-2 to Q-4, and the pKa of the phenolic-OH moiety is reduced in Q-3 and Q-4. On the other hand, the morpholino substituent in the 7-position increases the pKa (OH) as a consequence of an intramolecular hydrogen bond between the OH and the morpholine-N in the HL form of the ligand. Interestingly, formation of intramolecular hydrogen bonding is also feasible in the solid Q-3 and Q-4·HCl·H2O according to their single crystal X-ray diffraction analysis. It was concluded that compounds with lower pKa (OH) values possess lower IC50 values in the MES-SA/Dx5 cancer cell lines showing a linear relationship.Solution speciation and solid structures of the complexes
Formation of mono and bis complexes with copper(II) and iron(III) was found based on the UV-visible spectrophotometric titrations, whereas tris iron(III) complexes could also be detected for two of the ligands. At physiological pH, the copper(II) binding strength of the studied compounds follows the order Q-2 > Q-1 > Q-3 > Q-4 and a similar trend was determined for the iron(III) complexes (Q-1 ∼ Q-2 > Q-3 > Q-4). The studied 8-hydroxyquinolines have a binding preference to copper(II) over iron(III) at pH 7.4. Results of the EPR spectroscopic measurements and the solid phase structures of [(Cu(HQ-2)2)2]·(CH3OH)2·Cl4·(H2O)2, [Cu(Q-3)2]·Cl2 and [Cu(HQ-4)2(CH3OH)]·ZnCl4·CH3OH characterized by single-crystal X-ray diffraction analysis confirmed the coordination via the (N,O) donor set in the copper(II) complexes.Relationship between solution stability, redox properties and cytotoxicity
In the investigated set of compounds, those possessing weaker copper(II) and iron(III) binding abilities at physiological pH were found to be more toxic against the cancer cell line. Based on the cyclic voltammetric data of the iron and copper complexes, the more cytotoxic Q-3 stabilizes the lower oxidation states of these metal ions stronger than Q-1. Glutathione was not able to reduce the iron(III) complexes of the tested 8-hydroxyquinolines at five-fold excess within a time frame of 90 min, while copper(II) complexes could be reduced much faster, and in a reversible manner. The compounds with stronger copper(II) binding abilities showed faster redox reactions and were found to be less cytotoxic.In all, the established property–activity relationship for the studied compounds can help in understanding the mechanism of action of 8-hydroxyquinolines and can contribute to the development of anticancer compounds. We found that the introduction of the morpholine moiety at position 7 slightly reduced the cytotoxicity of 8-hydroxyquinoline in the MES-SA/Dx5 cell line, at the same time the piperidine and fluorobenzylamino substituents (with chlorine at position 5) increased the activity. The morpholine derivative has slightly higher, while the latter two compounds have much lower pKa (OH) than the reference compound Q-1. The more cytotoxic ligands possess weaker iron(III) and copper(II) binding abilities than Q-1. Based on these conclusions, Q-4 was selected as a candidate for undergoing preclinical in vivo experiments.
Experimental section
Chemicals
Solid KOH, 8-hydroxyquinoline (Q-1), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), tetrabutylammonium chloride (TBACl), ferrocene, 5-chloroquinolin-8-ol, 2-fluorobenzylamine, paraformaldehyde, ascorbic acid (ASC) and glutathione (GSH) were purchased from Sigma-Aldrich and HCl, KCl, CuCl2, FeCl3, and n-octanol were Reanal products (Budapest, Hungary). Q-2 (NSC662298) and Q-3 (NSC57969) were acquired from the drug repository of the Developmental Therapeutics Program of the National Cancer Institute. Iron(III) and copper(II) stock solutions were prepared by dissolving appropriate amounts of metal chlorides in known amounts of HCl. Their concentrations were determined by complexometry via EDTA complexes. Accurate strong acid content of the metal stock solutions was determined by pH-potentiometric titrations. The studied 8-hydroxyquinolines were poorly soluble in water; therefore stock solutions (0.01 M; 1.4–3.7 g L−1) were in pure DMSO. Solubility in water was not determined experimentally, but based on the determined log D7.4 values we can conclude that Q-2 and Q-3 have somewhat better, while Q-4 has definitely a worse (∼50 μM) water solubility than Q-1 (S ∼ 0.56 g L−1, ∼4 mM). (The solubility of these compounds strongly depends on the pH.)Chemicals used for the synthesis were of reagent grade quality or better, obtained from commercial suppliers and used without further purification. Solvents were used as received or dried.
Synthesis and characterization of 5-chloro-7-((2-fluorobenzylamino)methyl)quinolin-8-ol hydrochloride (Q-4·HCl)
Q-4·HCl was obtained by the acidic cleavage of 5-chloro-3-(2-fluorobenzyl)-3,4-dihydro-2H-[1,3]oxazino[5,6-h]quinoline, which was prepared by the reaction of 5-chloroquinolin-8-ol (0.50 g, 2.78 mmol), 2-fluorobenzylamine (0.52 g, 4.15 mmol) and paraformaldehyde (0.208 g, 6.95 mmol) using ethanol (17 mL) as the solvent in a 35 mL pressurized reaction vial. The mixture was heated by microwave irradiation at 100 °C for 45 min. The solvent was then evaporated off and the residue was crystallized with n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate (5
ethyl acetate (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 30 mL) and recrystallized from 20 mL of diisopropyl ether. Yield: 0.795 g (87%); mp.: 150–152 °C. 1H NMR (DMSO-d6, Fig. S12†): δ = 3.96 (2H, s); 4.08 (2H, s); 5.11 (2H, s); 7.12–7.26 (2H, m); 7.36 (1H, t, J = 7.6 Hz); 7.41–7.52 (2H, m); 7.65 (1H, dd, J = 8.12 Hz, 3.84 Hz); 8.46 (1H, d, J = 8.5 Hz); 8.92 (1H, s); 13C NMR (DMSO-d6, Fig. S13†): δ = 48.3; 48.7; 61.7; 115.2; 115.4; 118.6; 120.1; 122.5; 124.4; 124.9; 125.0; 126.4; 129.4; 131.1; 132.2; 139.4; 148.8; 149.9. Anal. calcd for C18H14ClFN2O (328.77): C, 65.76; H, 4.29; N, 8.52. Found: C, 66.02; H, 4.31; N, 8.49.
1; 30 mL) and recrystallized from 20 mL of diisopropyl ether. Yield: 0.795 g (87%); mp.: 150–152 °C. 1H NMR (DMSO-d6, Fig. S12†): δ = 3.96 (2H, s); 4.08 (2H, s); 5.11 (2H, s); 7.12–7.26 (2H, m); 7.36 (1H, t, J = 7.6 Hz); 7.41–7.52 (2H, m); 7.65 (1H, dd, J = 8.12 Hz, 3.84 Hz); 8.46 (1H, d, J = 8.5 Hz); 8.92 (1H, s); 13C NMR (DMSO-d6, Fig. S13†): δ = 48.3; 48.7; 61.7; 115.2; 115.4; 118.6; 120.1; 122.5; 124.4; 124.9; 125.0; 126.4; 129.4; 131.1; 132.2; 139.4; 148.8; 149.9. Anal. calcd for C18H14ClFN2O (328.77): C, 65.76; H, 4.29; N, 8.52. Found: C, 66.02; H, 4.31; N, 8.49.
To obtain Q-4·HCl 0.15 mL (1.0 mmol) of HCl/ethanol (22%) was added to a stirred solution of 5-chloro-3-(2-fluorobenzyl)-3,4-dihydro-2H-[1,3]oxazino[5,6-h]quinoline (0.20 g; 0.6 mmol) in ethanol (15 mL). The mixture was stirred for 30 min at room temperature. After the evaporation of the solvent and crystallization with diethyl ether (10 mL), the product was obtained in 91% yield (0.19 g); mp.: 211–213 °C. 1H NMR (DMSO-d6, Fig. S14†): δ = 4.25 (2H, s); 4.37 (2H, s); 7.22–7.32 (2H, m); 7.43–7.53 (1H, m); 7.75 (1H, t, J = 7.2 Hz); 7.81 (1H, dd, J = 8.24 Hz, 3.97 Hz ); 8.00 (1H, s); 8.56 (1H, d, J = 8.2 Hz); 9.02 (1H, s); 9.85 (2H, brs); 13C NMR (DMSO-d6, Fig. S15†): δ = 43.0; 44.1; 114.9; 115.6; 118.4; 119.0; 119.1; 123.8; 124.6; 126.2; 129.4; 131.5; 132.5; 133.0; 138.4; 149.3; 151.9. Anal. calcd for C17H15Cl2FN2O (353.22): C, 57.81; H, 4.28; N, 7.93. Found: C, 57.92; H, 4.27; N, 7.95.
1H and 13C NMR spectra were recorded in deuterated solvents on a 400 (1H: 400 MHz, 13C: 100.6 MHz) MHz spectrometer at room temperature. Chemical shifts δ are expressed in ppm values using the residual solvent peaks as internal standards (DMSO-d6 2.50; 39.52 ppm).50 Melting points were determined on a Hinotek X-4 type melting point apparatus and are uncorrected. Elemental analyses were performed with a PerkinElmer 2400 CHNS elemental analyzer. Merck Kieselgel 60F254 plates were used for TLC. Microwave reactions were performed by using a CEM LabMate microwave reactor.
Crystallization, X-ray data collection, structure solution and refinement
Single crystals suitable for X-ray crystallography (SXRD) (Q-3, Q-4·HCl·H2O, [(Cu(HQ-2)2)2]·(CH3OH)2·Cl4·(H2O)2, [Cu(Q-3)2]·Cl2 and [Cu(HQ-4)2(CH3OH)]·ZnCl4·CH3OH) were grown from slow evaporation of a methanolic solution at room temperature. In the case of the crystallisation of the copper complexes the solution contained the metal ion and the ligand at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio. [Cu2(Q-2)4]·(CH3OH)2·(Cl)4·(H2O)2 and [Cu(Q-3)]·(Cl)2 were crystallized from methanolic solutions applying vapour diffusion of diethyl ether to reduce the solubility. The complex of Q-4 could not be crystallized under similar conditions, however when ZnCl2 was also present in the solution the crystals of [Cu(Q-4)]·(ZnCl4)·(CH3OH) could be obtained. As Cu and Zn could not be unambiguously distinguished by SXRD, elemental analysis (ICP-OES) has been performed. This crystal contains 2.6(1) mg kg−1 of Cu and 4.0(2) mg kg−1 of Zn that is a 60/40% Zn/Cu ratio (with an error of ±5%). If we suppose that in this crystal, Zn is possibly in a tetrahedral ZnCl4 form (40%) and Cu is complexed by the Q-4 ligand (40%), there is still 20% of Zn shared, half in ZnCl4 and the other half possibly occupies the position of copper in the Q-4 complex. It means an 80% copper(II) and 20% zinc(II) occupation of the central metal position of the organic ligand complex cation, which may be the reason for the poor crystal quality, which prevented the refinement of the metal occupancy on the weak quality SXRD data.
2 ratio. [Cu2(Q-2)4]·(CH3OH)2·(Cl)4·(H2O)2 and [Cu(Q-3)]·(Cl)2 were crystallized from methanolic solutions applying vapour diffusion of diethyl ether to reduce the solubility. The complex of Q-4 could not be crystallized under similar conditions, however when ZnCl2 was also present in the solution the crystals of [Cu(Q-4)]·(ZnCl4)·(CH3OH) could be obtained. As Cu and Zn could not be unambiguously distinguished by SXRD, elemental analysis (ICP-OES) has been performed. This crystal contains 2.6(1) mg kg−1 of Cu and 4.0(2) mg kg−1 of Zn that is a 60/40% Zn/Cu ratio (with an error of ±5%). If we suppose that in this crystal, Zn is possibly in a tetrahedral ZnCl4 form (40%) and Cu is complexed by the Q-4 ligand (40%), there is still 20% of Zn shared, half in ZnCl4 and the other half possibly occupies the position of copper in the Q-4 complex. It means an 80% copper(II) and 20% zinc(II) occupation of the central metal position of the organic ligand complex cation, which may be the reason for the poor crystal quality, which prevented the refinement of the metal occupancy on the weak quality SXRD data.
The single crystals were mounted on loops. X-ray diffraction data were collected on a Rigaku RAXIS-RAPID II diffractometer at room temperature (293 K) in the case of Q-3, Q-4·HCl·H2O, [(Cu(HQ-2)2)2]·(CH3OH)2·Cl4·(H2O)2, and [Cu(Q-3)2]·Cl2, while the temperature was 103 K during the measurement of [Cu(HQ-4)2(CH3OH)]·ZnCl4·CH3OH. Numerical absorption correction was carried out, except the last crystal where multi-scan absorption correction has been applied using the software CrystalClear.51
Sir201452 and SHELXL53 under WinGX54 software were used for crystal structure solution and refinement, respectively. The structures were solved by direct methods. The models were refined by full-matrix least squares on F2. Hydrogen atoms were included in structure factor calculations but they were not refined, and their isotropic displacement parameters were approximated from the U(eq) value of the atom they were bonded to. Refinement of non-hydrogen atoms was carried out with anisotropic temperature factors. Selected bond lengths and angles of compounds were calculated using PLATON software.55 The graphical representations were done by Mercury,56 and the editing of CIF files was performed using PublCif57 software, respectively.
Crystallographic data for the crystal structures were deposited at the Cambridge Crystallographic Data Centre as supplementary publication numbers CCDC 1575107–1575111† for the five crystals listed above.
UV-Vis spectrophotometry and determination of the distribution coefficients
The pH-metric measurements for determination of the exact concentrations of HCl and KOH stock solutions used for the spectrophotometric titrations were carried out at 25.0 ± 0.1 °C in aqueous solutions and at an ionic strength of 0.20 M KCl in order to keep the activity coefficients constant. All the titrations were performed with carbonate-free KOH solutions of a known concentration (0.10 M) in the presence of 0.1 M KCl. An Orion 710A pH-meter equipped with a Metrohm combined electrode (type 6.0234.100) and a Metrohm 665 Dosimat burette were used for titrations. The electrode system was calibrated to the pH = −log[H+] scale by the method suggested by Irving et al.58 The average water ionization constant, pKwater, is 13.76 ± 0.01 at 25 °C. The samples were deoxygenated by bubbling purified argon for ca. 10 min prior to the measurements and argon was also passed over the solutions during further titrations. A Hewlett Packard 8452A diode array spectrophotometer was used to record the UV-Vis spectra in the 200–800 nm interval. The path length was 1 or 2 cm. The spectrophotometric titrations were performed on the samples of the ligands Q-3, Q-4 and Q-2 alone or with iron(III) or copper(II). The concentration of the ligand was usually 50 μM (10 μM for Q-4 due to its worse water solubility) and the metal ion-to-ligand ratios were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 over the pH range between 2 and 11.5 at an ionic strength of 0.20 M (KCl) at 25.0 ± 0.1 °C. The initial volume of the samples was 10.00 mL. The samples contained 0.5% (v/v) DMSO. The overall protonation constants of the ligands (from which the pKa values were calculated), the overall stability constants of their metal complexes and the individual spectra of the species were computed using the program PSEQUAD59 and literature data were used for iron(III) hydroxido species.60
3 over the pH range between 2 and 11.5 at an ionic strength of 0.20 M (KCl) at 25.0 ± 0.1 °C. The initial volume of the samples was 10.00 mL. The samples contained 0.5% (v/v) DMSO. The overall protonation constants of the ligands (from which the pKa values were calculated), the overall stability constants of their metal complexes and the individual spectra of the species were computed using the program PSEQUAD59 and literature data were used for iron(III) hydroxido species.60
β(MpLqHr) is defined for the general equilibrium pM + qL + rH ⇌ MpLqHr as β(MpLqHr) = [MpLqHr]/[M]p[L]q[H]r where M denotes the metal ion and L the completely deprotonated ligand.
The n-octanol/water distribution coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D7.4) of the ligands Q-3, Q-4 and Q-2 were determined by the shake-flask method in n-octanol/water solutions at 25.0 ± 0.2 °C at pH 7.40 as described previously.16
D7.4) of the ligands Q-3, Q-4 and Q-2 were determined by the shake-flask method in n-octanol/water solutions at 25.0 ± 0.2 °C at pH 7.40 as described previously.16
EPR measurements
The EPR spectra were recorded at room temperature and at 77 K (in a Dewar flask containing liquid nitrogen) using a BRUKER EleXsys E500 spectrometer (microwave frequency 9.81 GHz, microwave power 13 mW, modulation amplitude 5 G, and modulation frequency 100 kHz). All EPR spectra were simulated by a spectral decomposition method using the “EPR” program.61 Since the copper(II) salt used to make the stock solution was a natural mixture of isotopes, the spectrum of each species was calculated as the sum of spectra containing 63Cu and 65Cu weighted by their natural abundances. The copper and ligand coupling constants are given in units of gauss (1 G = 10−4 T). The EPR spectra of the dimeric complex was simulated using a new module of the ‘EPR’ program61 developed for calculating the EPR spectra and dynamic nuclear polarization (DNP) in coupled spin systems (biradicals and paramagnetic dimers). The EPR spectrum is calculated by the complete diagonalization of the Hamiltonian of a two-spin system. The details of the simulation was published previously.62Cyclic voltammetric measurements
All cyclic voltammograms of the iron(III) and copper(II) complexes were measured in DMSO (with a H2O content of up to 1.6% for copper samples and 3.7% for iron samples) at 25.0 ± 0.1 °C and at an ionic strength of 10 mM (TBAClO4). Samples contained 1.0 mM metal ion or 1.0 mM ligand (Q-1, Q-2, Q-3, Q-4) alone or together while the cL/cM ratios were 1, 1.5, 2 or 3. Measurements were performed on a conventional three-electrode system under an argon atmosphere and a PC controlled Autolab-PGSTAT 204 potentiostat. Samples were purged for 15 min with argon before recording the cyclic voltammograms. A glassy carbon electrode was used the working electrode, a platinum electrode as the auxiliary electrode and a Ag/AgCl/KCl (3 M) as the reference electrode. Electrochemical potentials were converted into the normal hydrogen electrode (NHE) scale by adding 0.210 V.63 The electrochemical measuring system was calibrated with ferrocene in 95% (v/v) DMSO–water (E1/2 = +0.50 ± 0.03 V vs. Ag/AgCl/KCl (3 M) in our setup). Redox potentials were obtained at 5 mV s−1 scan rate in the range of −1.2 to +1.0 V.Reduction by GSH and ASC
Reduction of the copper(II) and iron(III) complexes of the ligands Q-1 to Q-4 by GSH and ASC, respectively, was studied by UV-Vis spectrophotometry using a Hewlett Packard 8452A diode array spectrophotometer. A special, tightly closed tandem cuvette (Hellma Tandem Cell, 238-QS, path length: 1 cm) was used and the reactants were separated until the reaction was triggered. Both isolated pockets of the cuvette were completely deoxygenated by bubbling a stream of argon for 10 min before mixing the reactants. Spectra were recorded before and immediately after mixing, and changes were followed for 90 min. One of the isolated pockets contained the reducing agent at five times (GSH) and 2.5 times (ASC) higher concentrations than that of the metal ions. The other pocket contained the metal ions (Cu(II); Fe(III): 100 μM) and ligands at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 metal-to-ligand ratio. The pH of the solutions was adjusted to 7.40 by 50 mM HEPES buffer, or to 6.87 by a mixed acetate
1 metal-to-ligand ratio. The pH of the solutions was adjusted to 7.40 by 50 mM HEPES buffer, or to 6.87 by a mixed acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS buffer (ratio 1
PBS buffer (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), and an ionic strength of 0.1 M (KCl) was applied at 25.0 ± 0.1 °C. At the end of the measurements cc. H2O2 (50 μL to 1.4 mL sample) was added to the samples.
4), and an ionic strength of 0.1 M (KCl) was applied at 25.0 ± 0.1 °C. At the end of the measurements cc. H2O2 (50 μL to 1.4 mL sample) was added to the samples.
Cell lines and culture conditions
The doxorubicin selected human uterine sarcoma MES-SA-Dx5 cell line was obtained from ATCC (MES-SA/Dx5: No. CRL-1977™). The cells were cultivated in Dulbecco's Modified Eagle's Medium (DMEM, Sigma Aldrich, Hungary), supplemented with 10% fetal bovine serum, 5 mM glutamine, and 50 unit per mL penicillin and streptomycin (Life Technologies). All cell lines were cultivated at 37 °C, 5% CO2.For the preparation of primary mouse hepatocytes mouse handling and experimental procedures were conducted in accordance with the institutional guidelines for animal care and use. 16 weeks old male C57BL/6 mice were anaesthetized (20 mg kg−1 tiletamine, 20 mg kg−1 zolazepam, 12.5 mg kg−1 xylazine and 3 mg kg−1 butorphanol) and livers were perfused with 75 mL oxygenized perfusion buffer (120 mmol L−1 NaCl, 5.4 mmol L−1 KCl, 0.9 mmol L−1 NaH2PO4, 26 mmol L−1 NaHCO3, 5.6 mmol L−1 glucose, pH 7.4) supplemented with EGTA (0.5 mmol L−1), followed by a second perfusion with 75 mL perfusion buffer without EGTA. Collagenase digestion was performed by 75 mL perfusion buffer supplemented with 2.5 mmol L−1 CaCl2 and 0.2 mg mL−1 collagenase (C5138, Sigma). The digested liver was taken out of the abdominal cavity and minced using tweezers, thereby releasing the hepatocytes. The cells were washed in ice-cold sterile suspension buffer (10 mmol L−1 HEPES, 142 mmol L−1 NaCl, 7 mmol L−1 KCl, pH 7.4), filtered through a 100 μm mesh membrane and centrifuged for 4 min at 4 °C at 80 g. After one additional washing step, dead cells were removed by Percoll (Sigma) centrifugation. After checking their viability by trypan blue exclusion staining, the cells were seeded on 96-well plates pre-coated with 5 μg cm−2 collagen I (BD, 356234) at a density of 1.35 × 104 cells per well in Williams E medium (Gibco) supplemented with hepatocyte thawing/plating supplement (Gibco) and 10% FBS. The cells that did not attach were removed by refreshing the culture medium after 1 h. The medium was renewed every 24 h during the culture as well as during the experiment (containing the dilutions of the tested compounds).
MTT and PrestoBlue viability assay
MTT viability assays were performed as described earlier with minor modifications.47,64,65 Briefly, cells were seeded into 96-well tissue culture plates (Sarstedt, Newton, USA/Orange, Braine-l'Alleud, Belgium) at a density of 5000 cells per well and allowed to attach overnight. Test compounds were added to achieve the required final concentration in a final volume of 100 μL per well. After an incubation period of 72 h, the supernatant was removed and fresh medium supplemented with the MTT reagent (0.083 mg mL−1) was added. Incubation with MTT at 37 °C was terminated after 1 h by removing the supernatants and lysing the cells with 100 μL DMSO per well. Viability of the cells was measured spectrophotometrically by absorbance at 540 nm using either a PerkinElmer Victor X3 or an EnSpire microplate reader, respectively. Data were background corrected by subtraction of the signal obtained from unstained cell lysates and normalized to untreated cells.On primary mouse hepatocytes, Q-1 to Q-4 were measured using the PrestoBlue assay (Life Technologies) as described earlier.27 Briefly, after the incubation with the compounds for 72 h and removal of the supernatants, a diluted solution of the PrestoBlue reagent (10%) was added to each well. After incubation with the reagent for one hour at 37 °C emission was measured at 579 nm upon excitation at 540 nm using a PerkinElmer EnSpire microplate reader.
Curves were fitted using Prism software66 using the sigmoidal dose–response model (comparing variable and fixed slopes). Curve fit statistics were used to determine the concentration of the test compound that resulted in 50% toxicity (IC50).
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Research, Development and Innovation Office-NKFIH through project GINOP-2.3.2-15-2016-00038, FK124240, K115762, Ministry of Human Capacities, Hungary grant 20391-3/2018/FEKUSTRAT and the J. Bolyai Research Scholarship of the Hungarian Academy of Sciences (E. A. E., N. V. M.). G. S. was supported by a Momentum grant of the Hungarian Academy of Sciences and an ERC Starting Grant (StG-260572). A grant from the Fulbright Visiting Scholar Program sponsored by the U.S. Department of State is also acknowledged (F. S.).References
- Y. Song, H. Xu, W. Chen, P. Zhan and X. Liu, MedChemComm, 2015, 6, 61–74 RSC.
- V. Oliveri and G. Vecchio, Eur. J. Med. Chem., 2016, 120, 252–274 CrossRef CAS PubMed.
- A. Budimir, Acta Pharm., 2011, 61, 1–14 CAS.
- A. Budimir, N. Humbert, M. Elhabiri, I. Osinska, M. Biruš and A.-M. Albrecht-Gary, J. Inorg. Biochem., 2011, 105, 490–496 CrossRef CAS PubMed.
- A. Barilli, C. Atzeri, I. Bassanetti, F. Ingoglia, V. Dall'Asta, O. Bussolati, M. Maffini, C. Mucchino and L. Marchiò, Mol. Pharm., 2014, 11, 1151–1163 CrossRef CAS PubMed.
- S. Tardito, A. Barilli, I. Bassanetti, M. Tegoni, O. Bussolati, R. Franchi-Gazzola, C. Mucchino and L. Marchiò, J. Med. Chem., 2012, 55, 10448–10459 CrossRef CAS PubMed.
- V. Oliveri, V. Lanza, D. Milardi, M. Viale, I. Maric, C. Sgarlata and G. Vecchio, Metallomics, 2017, 9, 1439–1446 RSC.
- D. Chen, Q. C. Cui, H. Yang, R. A. Barrea, F. H. Sarkar, S. Sheng, B. Yan, G. P. V. Reddy and Q. P. Dou, Cancer Res., 2007, 67, 1636–1644 CrossRef CAS PubMed.
- O. N. F. King, X. S. Li, M. Sakurai, A. Kawamura, N. R. Rose, S. S. Ng, A. M. Quinn, G. Rai, B. T. Mott, P. Beswick, R. J. Klose, U. Oppermann, A. Jadhav, T. D. Heightman, D. J. Maloney, C. J. Schofield and A. Simeonov, PLoS One, 2010, 5, e15535 CrossRef PubMed.
- W.-Q. Ding and S. E. Lind, IUBMB Life, 2009, 61, 1013–1018 CrossRef CAS PubMed.
- M. Kruszewski, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2003, 531, 81–92 CrossRef CAS PubMed.
- M. C. Linder, Biochemistry of Copper, Springer, US, 1991 Search PubMed.
- A. Gupte and R. J. Mumper, Cancer Treat. Rev., 2009, 35, 32–46 CrossRef CAS PubMed.
- C. Duncan and A. R. White, Metallomics, 2012, 4, 127–138 RSC.
- D. Havrylyuk, B. S. Howerton, L. Nease, S. Parkin, D. K. Heidary and E. C. Glazer, Eur. J. Med. Chem., 2018, 156, 790–799 CrossRef CAS PubMed.
- O. Dömötör, V. F. S. Pape, N. V. May, G. Szakács and É. A. Enyedy, Dalton Trans., 2017, 46, 4382–4396 RSC.
- M. Kubanik, H. Holtkamp, T. Söhnel, S. M. F. Jamieson and C. G. Hartinger, Organometallics, 2015, 34, 5658–5668 CrossRef CAS.
- R. Schuecker, R. O. John, M. A. Jakupec, V. B. Arion and B. K. Keppler, Organometallics, 2008, 27, 6587–6595 CrossRef CAS.
- S. Movassaghi, M. Hanif, H. U. Holtkamp, T. Söhnel, S. M. F. Jamieson and C. G. Hartinger, Dalton Trans., 2018, 47, 2192–2201 RSC.
- Q.-P. Qin, Z.-F. Chen, J.-L. Qin, X.-J. He, Y.-L. Li, Y.-C. Liu, K.-B. Huang and H. Liang, Eur. J. Med. Chem., 2015, 92, 302–313 CrossRef CAS PubMed.
- A. R. Timerbaev, Metallomics, 2009, 1, 193–198 RSC.
- P. Collery, F. Lechenault, A. Cazabat, E. Juvin, L. Khassanova, A. Evangelou and B. Keppler, Anticancer Res., 2000, 20, 955–958 CAS.
- A.-Y. SHEN, S.-N. WU and C.-T. CHIU, J. Pharm. Pharmacol., 1999, 51, 543–548 CrossRef CAS PubMed.
- A. Y. Shaw, C.-Y. Chang, M.-Y. Hsu, P.-J. Lu, C.-N. Yang, H.-L. Chen, C.-W. Lo, C.-W. Shiau and M.-K. Chern, Eur. J. Med. Chem., 2010, 45, 2860–2867 CrossRef CAS PubMed.
- R. H. Shoemaker, Nat. Rev. Cancer, 2006, 6, 813–823 CrossRef CAS PubMed.
- D. Türk, M. D. Hall, B. F. Chu, J. A. Ludwig, H. M. Fales, M. M. Gottesman and G. Szakács, Cancer Res., 2009, 69, 8293–8301 CrossRef PubMed.
- A. Füredi, S. Tóth, K. Szebényi, V. F. S. Pape, D. Türk, N. Kucsma, L. Cervenak, J. Tóvári and G. Szakács, Mol. Cancer Ther., 2017, 16, 45–56 CrossRef PubMed.
- A. Bijelic, M. Aureliano and A. Rompel, Chem. Commun., 2018, 54, 1153–1169 RSC.
- J. Yellol, S. A. Pérez, A. Buceta, G. Yellol, A. Donaire, P. Szumlas, P. J. Bednarski, G. Makhloufi, C. Janiak, A. Espinosa and J. Ruiz, J. Med. Chem., 2015, 58, 7310–7327 CrossRef CAS PubMed.
- J. Baell and M. A. Walters, Nat. News, 2014, 513, 481 CrossRef CAS PubMed.
- D. S. Kalinowski and D. R. Richardson, Pharmacol. Rev., 2005, 57, 547–583 CrossRef CAS PubMed.
- A. M. Merlot, D. S. Kalinowski and D. R. Richardson, Antioxid. Redox Signaling, 2013, 18, 973–1006 CrossRef CAS PubMed.
- M. Whitnall, J. Howard, P. Ponka and D. R. Richardson, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 14901–14906 CrossRef CAS PubMed.
- P. Heffeter, V. F. S. Pape, É. A. Enyedy, B. K. Keppler, G. Szakacs and C. R. Kowol, Antioxid. Redox Signaling DOI:10.1089/ars.2017.7487.
- H. Eklund, U. Uhlin, M. Färnegårdh, D. T. Logan and P. Nordlund, Prog. Biophys. Mol. Biol., 2001, 77, 177–268 CrossRef CAS PubMed.
- N. T. V. Le and D. R. Richardson, Biochim. Biophys. Acta, Rev. Cancer, 2002, 1603, 31–46 CrossRef CAS.
- É. A. Enyedy, O. Dömötör, E. Varga, T. Kiss, R. Trondl, C. G. Hartinger and B. K. Keppler, J. Inorg. Biochem., 2012, 117, 189–197 CrossRef PubMed.
- SCQuery, The IUPAC Stability Constants Database, Academic Software, Royal Society of Chemistry, 2005 Search PubMed.
- L. ChemAxon, Instant J Chem/MarvinSketch, ChemAxon Ltd., Budapest, Hungary, 2012 Search PubMed.
- P. Leanderson and C. Tagesson, Carcinogenesis, 1996, 17, 545–550 CrossRef CAS PubMed.
- T. D. Turnquist and E. B. Sandell, Anal. Chim. Acta, 1968, 42, 239–245 CrossRef CAS.
- R. M. Smith, A. E. Martell and R. J. Motekaitis, NIST critically selected stability constants of metal complexes database, National Institute of Standards and Technology, U.S. Dept. of Commerce, Gaithersburg, MD, 2004 Search PubMed.
- S. Tsakovski, K. Benkhedda, E. Ivanova and F. C. Adams, Anal. Chim. Acta, 2002, 453, 143–154 CrossRef CAS.
- R. C. Hoy and R. H. Morriss, Acta Crystallogr., 1967, 22, 476–482 CrossRef CAS.
- F. Q. Schafer and G. R. Buettner, Free Radical Biol. Med., 2001, 30, 1191–1212 CrossRef CAS PubMed.
- T. Jakusch, É. A. Enyedy, K. Kozma, Z. Paár, A. Bényei and T. Kiss, Inorg. Chim. Acta, 2014, 420, 92–102 CrossRef CAS.
- V. F. S. Pape, D. Türk, P. Szabó, M. Wiese, E. A. Enyedy and G. Szakács, J. Inorg. Biochem., 2015, 144, 18–30 CrossRef CAS PubMed.
- B. Song, N. Aebischer and C. Orvig, Inorg. Chem., 2002, 41, 1357–1364 CrossRef CAS PubMed.
- C. G. Hartinger, S. Zorbas-Seifried, M. A. Jakupec, B. Kynast, H. Zorbas and B. K. Keppler, J. Inorg. Biochem., 2006, 100, 891–904 CrossRef CAS PubMed.
- G. R. Fulmer, A. J. M. Miller, N. H. Sherden, H. E. Gottlieb, A. Nudelman, B. M. Stoltz, J. E. Bercaw and K. I. Goldberg, Organometallics, 2010, 29, 2176–2179 CrossRef CAS.
- CrystalClear SM 1.4.0, Rigaku/MSC Inc., 2008 Search PubMed.
- M. C. Burla, R. Caliandro, B. Carrozzini, G. L. Cascarano, C. Cuocci, C. Giacovazzo, M. Mallamo, A. Mazzone and G. Polidori, J. Appl. Crystallogr., 2015, 48, 306–309 CrossRef CAS.
- SHELXL-2013 Program for Crystal Structure Solution, University of Göttingen, Germany, 2013 Search PubMed.
- L. J. Farrugia, J. Appl. Crystallogr., 2012, 45, 849–854 CrossRef CAS.
- A. L. Spek, J. Appl. Crystallogr., 2003, 36, 7–13 CrossRef CAS.
- C. F. Macrae, P. R. Edgington, P. McCabe, E. Pidcock, G. P. Shields, R. Taylor, M. Towler and J. van de Streek, J. Appl. Crystallogr., 2006, 39, 453–457 CrossRef CAS.
- S. P. Westrip, J. Appl. Crystallogr., 2010, 43, 920–925 CrossRef CAS.
- H. M. Irving, M. G. Miles and L. D. Pettit, Anal. Chim. Acta, 1967, 38, 475–488 CrossRef CAS.
- L. Zékány and I. Nagypál, in Computational Methods for the Determination of Formation Constants, ed. D. J. Leggett, 1985, pp. 291–353 Search PubMed.
- C. F. Baes and R. E. Mesmer, The Hydrolysis of Cations, Wiley, New York, 1976 Search PubMed.
- A. Rockenbauer and L. Korecz, Appl. Magn. Reson., 1996, 10, 29–43 CrossRef CAS.
- F. Bacher, É. A. Enyedy, N. V. Nagy, A. Rockenbauer, G. M. Bognár, R. Trondl, M. S. Novak, E. Klapproth, T. Kiss and V. B. Arion, Inorg. Chem., 2013, 52, 8895–8908 CrossRef CAS PubMed.
- A. J. Bard and L. R. Faulkner, Electrochemical Methods–Fundamentals and Applications, John Wiley & Sons, New York, 2000 Search PubMed.
- K. Juvale, V. F. S. Pape and M. Wiese, Bioorg. Med. Chem., 2012, 20, 346–355 CrossRef CAS PubMed.
- H. Mueller, M. U. Kassack and M. Wiese, J. Biomol. Screening, 2004, 9, 506–515 CrossRef CAS PubMed.
- I. GraphPad Software, GraphPad Prism, GraphPad Software, Inc., 2007 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Crystal and UV-Vis spectral data. EPR parameters for the Cu(II) complexes, UV-Vis and EPR spectra, distribution diagrams and cyclic voltammograms for the studied complexes. NMR spectra for Q-4. CCDC 1575107–1575111. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8dt03088j |
| ‡ Present address: Department of Physiology, Semmelweis University, Faculty of Medicine, Tűzoltó utca 37-47, H-1094 Budapest, Hungary. |
| § Present address: Department of Dermatology and Cutaneous Biology, Sidney Kimmel Medical College, Thomas Jefferson University, 233 S. 10th Street, 19107 Philadelphia (PA), USA. |
| This journal is © The Royal Society of Chemistry 2018 |