Open Access Article
Open Access ArticleSynthesis, isomerisation and biological properties of mononuclear ruthenium complexes containing the bis[4(4′-methyl-2,2′-bipyridyl)]-1,7-heptane ligand†
Biyun
Sun
a,
Hannah M.
Southam
b,
Jonathan A.
Butler‡
b,
Robert K.
Poole
b,
Alexandre
Burgun
c,
Andrew
Tarzia
c,
F. Richard
Keene
 *cd and
J. Grant
Collins
*cd and
J. Grant
Collins
 *a
*a
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au
bDepartment of Molecular Biology and Biotechnology, The University of Sheffield, Sheffield, S10 2TN, UK
cSchool of Physical Sciences, University of Adelaide, Adelaide, SA 5005, Australia. E-mail: richard.keene@adelaide.edu.au
dAustralian Institute of Tropical Health & Medicine/Centre for Biodiscovery & Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
First published on 30th January 2018
Abstract
A series of mononuclear ruthenium(II) complexes containing the tetradentate ligand bis[4(4′-methyl-2,2′-bipyridyl)]-1,7-heptane have been synthesised and their biological properties examined. In the synthesis of the [Ru(phen′)(bb7)]2+ complexes (where phen′ = 1,10-phenanthroline and its 5-nitro-, 4,7-dimethyl- and 3,4,7,8-tetramethyl- derivatives), both the symmetric cis-α and non-symmetric cis-β isomers were formed. However, upon standing for a number of days (or more quickly under harsh conditions) the cis-β isomer converted to the more thermodynamically stable cis-α isomer. The minimum inhibitory concentrations (MIC) and the minimum bactericidal concentrations (MBC) of the ruthenium(II) complexes were determined against six strains of bacteria: Gram-positive Staphylococcus aureus (S. aureus) and methicillin-resistant S. aureus (MRSA); and the Gram-negative Escherichia coli (E. coli) strains MG1655, APEC, UPEC and Pseudomonas aeruginosa (P. aeruginosa). The results showed that the [Ru(5-NO2phen)(bb7)]2+ complex had little or no activity against any of the bacterial strains. By contrast, for the other cis-α-[Ru(phen′)(bb7)]2+ complexes, the antimicrobial activity increased with the degree of methylation. In particular, the cis-α-[Ru(Me4phen)(bb7)]2+ complex showed excellent and uniform MIC activity against all bacteria. By contrast, the MBC values for the cis-α-[Ru(Me4phen)(bb7)]2+ complex varied considerably across the bacteria and even within S. aureus and E. coli strains. In order to gain an understanding of the relative antimicrobial activities, the DNA-binding affinity, cellular accumulation and water–octanol partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of the ruthenium complexes were determined. Interestingly, all the [Ru(phen′)(bb7)]2+ complexes exhibited stronger DNA binding affinity (Ka ≈ 1 × 107 M−1) than the well-known DNA-intercalating complex [Ru(phen)2(dppz)]2+ (where dppz = dipyrido[3,2-a:2′,3′-c]phenazine).
P) of the ruthenium complexes were determined. Interestingly, all the [Ru(phen′)(bb7)]2+ complexes exhibited stronger DNA binding affinity (Ka ≈ 1 × 107 M−1) than the well-known DNA-intercalating complex [Ru(phen)2(dppz)]2+ (where dppz = dipyrido[3,2-a:2′,3′-c]phenazine).
Introduction
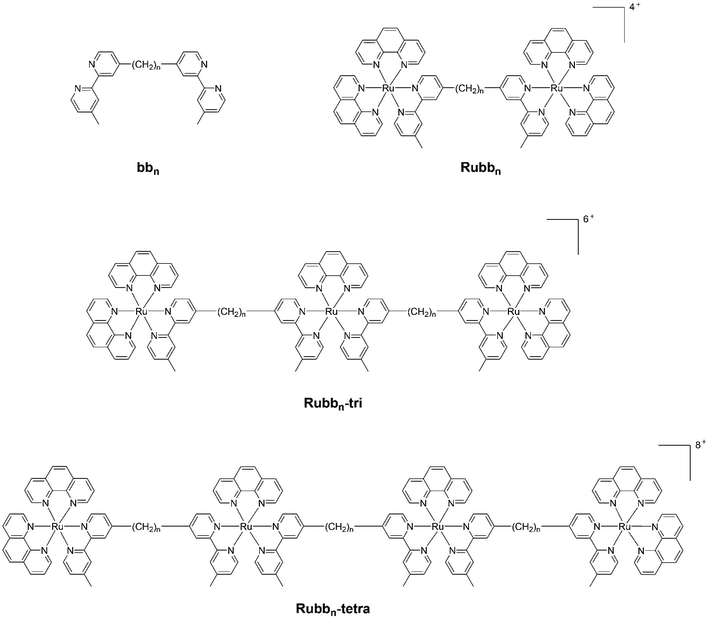
Due to a developing resistance to antimicrobial drugs, bacterial diseases are becoming a significantly greater threat to humans. Of note, the World Health Organisation has stated that antimicrobial resistance is one of the most important problems currently affecting global health, food security and development.1 As a consequence, there is considerable interest in the development of new classes of antimicrobial agents, and while the development of new drugs based upon organic compounds continues, transition metal-based antimicrobial agents in general have been attracting attention – and ruthenium(II) complexes in particular.2–14 A range of ruthenium(II) complexes have been studied, and generally have shown good activity against Gram-positive bacteria, but variable and often poor activity against Gram-negative species.10–12Dwyer and co-workers were the first to study the biological activity of mononuclear tris(bidentate) inert metal complexes with ligands such as 1,10-phenanthroline (phen) and its derivatives {e.g. 3,4,7,8-tetramethyl-1,10-phenanthroline (Me4phen) and 5-nitro-1,10-phenanthroline (5-NO2phen)}.9 More recently, a wide range of mononuclear {e.g. [Ru(2,9-Me2phen)2(dppz)]2+} (2,9-Me2phen = 2,9-dimethyl-1,10-phenanthroline; dppz = dipyrido[3,2-a:2′,3′-c]phenazine)10 and di- and oligo-nuclear polypyridylruthenium(II)12,14,15 complexes have been examined. In particular, earlier studies from our laboratories have examined the antimicrobial properties of di-, tri- and tetra-nuclear polypyridylruthenium(II) complexes in which the metal centres are linked by the bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane ligand (“bbn”; see Fig. 1).14 While these oligonuclear ruthenium complexes showed excellent activity against drug-sensitive Gram-positive bacterial strains, and maintained the activity against drug-resistant strains, they exhibited relatively poor activity against some Gram-negative strains.16 More recently, we synthesised mononuclear ruthenium complexes that contained the bbn moiety (for n = 10 and 12), but as a tetradentate ligand,17 rather than as a ligand linking metal centres. While the cis-α-[Ru(phen)(bb12)]2+ species showed better activity towards the clinically-important Gram-negative Pseudomonas aeruginosa, its activity against other bacteria was only equivalent to [Ru(Me4phen)3]2+,8 the most active of the ruthenium complexes prepared by Dwyer and co-workers sixty years ago. The cis-α-[Ru(phen)(bb10)]2+ complex was less active than cis-α-[Ru(phen)(bb12)]2+ and [Ru(Me4phen)3]2+ against most of the bacterial strains.17 The antimicrobial activity of the [Ru(phen)(bbn)]2+ complexes could be potentially improved through an increase of the lipophilicity, for example by utilising the bb16 ligand. However, an alternative approach is to decrease the lipophilicity of the bbn chain but increase the electron density at the ruthenium centre through the incorporation of electron-donating groups on the 1,10-phenanthroline ligand. It has been established that the antimicrobial activity of the metal complexes is significantly affected by the cellular uptake,15 which in turn is presumably sensitive to the charge on the metal centre as well as the overall lipophilicity. Consequently, the modulation of the electron density at the metal centre could provide higher uptake and greater ability to modulate the differential uptake between bacterial and eukaryotic cells.
Herein, we describe the synthesis of a series of complexes involving 1,10-phenanthroline and some of its derivatives – [Ru(phen′)(bb7)]2+ {where phen′ = phen; 4,7-dimethyl-1,10-phenanthroline (Me2phen); Me4phen; and 5-NO2phen} – and an examination of their DNA binding, cellular uptake and antimicrobial activity against a range of bacteria. Synthetically, the results indicate that although the cis-β-[Ru(phen′)(bb7)]2+ complexes are formed initially they are unstable and are converted to the cis-α isomer and, to a very small extent, the corresponding [Ru(phen′)(Me2bpy)2]2+ species. The cis-α-[Ru(Me4phen)(bb7)]2+ complex showed uniformly high activity against Gram-positive and Gram-negative bacteria. Furthermore, the results demonstrate the effect of the electron density at the ruthenium centre on the cellular uptake of the metal complexes.
Results
Synthesis of cis-α-[Ru(phen′)(bb7)]2+ complexes
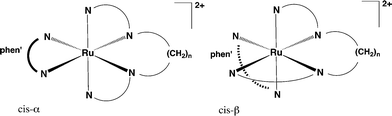
The synthesis of the cis-α and cis-β isomers of [Ru(phen)(bb12)]2+ was carried out as previously reported.17The synthesis of a series of mononuclear cis-α-[Ru(phen′)(bb7)]2+ complexes was achieved with good yields, as shown in Scheme 1, by heating cis,cis-[RuCl2(DMSO)2(phen′)]17 to 130–140 °C with the bb7 ligand in ethylene glycol: the 2+ complexes were isolated from higher-charged species using cation-exchange chromatography on a SP Sephadex C-25 column with aqueous sodium chloride as the eluent, and the resulting solid products recrystallised from acetonitrile/diethyl ether. Subsequent cation-exchange chromatography on a SP Sephadex C-25 column (1 metre) with sodium toluene-4-sulfonate as the eluent realised two bands – the cis-α isomer and corresponding [Ru(phen′)(Me2bpy)2]2+ species as a minor product. All the complexes were characterised by microanalysis, NMR spectroscopy and high-resolution electrospray ionisation (ESI) mass spectrometry. The cis-α-[Ru(Me4phen)(bb7)]2+ complex was also characterised by X-ray crystallography (see Table 1 and Fig. 2).
| Compound | cis-α-[Ru(Me4phen)(bb7)](PF6)2·3(CHCl3) (Fig. 2) | [Ru(Me4phen)(Me2bpy)](PF6)2·0.25(H2O) (Fig. S1; ESI) |
|---|---|---|
| Empirical formula | C48H51Cl9F12N6P2Ru | C40H40.5F12N6O0.25P2Ru |
| Formula weight | 1422.01 | 1000.29 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | P21/c | P21/c |
| a (Å) | 9.853(2) | 10.782(2) |
| b (Å) | 28.124(6) | 28.514(6) |
| c (Å) | 20.632(4) | 13.176(3) |
| α (°) | ||
| β (°) | 92.25(3) | 96.63(3) |
| γ (°) | ||
| Volume (Å3) | 5713(2) | 4023.7(14) |
| Z | 4 | 4 |
| Density (calc.) g m−3 | 1.653 | 1.651 |
| Absorption coefficient (mm−1) | 0.832 | 0.566 |
| F(000) | 2864 | 2026 |
| Crystal size (mm3) | 0.567 × 0.100 × 0.017 | 0.289 × 0.255 × 0.006 |
| θ range for collection (°) | 1.225 to 31.928 | 1.428 to 31.900 |
| Reflections collected | 105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 036 036 |
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249 249 |
| Observed reflections [R(int)] | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 330 [0.0463] 330 [0.0463] |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 496 [0.0794] 496 [0.0794] |
| Goodness-of-fit on F2 | 1.049 | 1.036 |
| R 1 [I > 2σ(I)] | 0.0488 | 0.0489 |
| wR2 (all data) | 0.1349 | 0.1272 |
| Largest diff. peak and hole (e Å−3) | 1.553, −2.055 | 0.535, −1.420 |
In our recent synthetic study17 of the analogous [Ru(phen)(bb10)]2+ and [Ru(phen)(bb12)]2+ complexes, in both cases the cis-α isomers and cis-β isomers (see Fig. 3) were isolated and individually characterised. In the present case, only the cis-α-[Ru(phen′)(bb7)]2+ isomer was isolated, and consequently the cis,cis-[RuCl2(DMSO)2(Me4phen)] reaction with the bb7 ligand was examined in detail. When samples were taken from the reaction mixture during the course of the synthesis and studied by 1H NMR, both the cis-α and cis-β isomers were unambiguously identified in COSY spectra of the reaction mixture. However, after either heating the reaction mixture to 200 °C (see Fig. 4) or allowing the mixture of the isolated isomers stand at room temperature for several weeks, only the cis-α isomer was observed. In addition, during the purification process for the cis-α isomer for all the [Ru(phen′)(bb7)]2+ cases, a small amount (<5% yield based upon starting material) of [Ru(phen′)(Me2bpy)2]2+ was observed: for phen′ = Me2phen and Me4phen, the products were isolated and the 1H NMR and TOF-MS (ESI+) characterisation data are provided in the ESI.† For [Ru(Me4phen)(Me2bpy)2]2+, the structure confirmed by X-ray crystallography (see Fig. S1: ESI†).
 | ||
| Fig. 4 Aromatic region of the 1H NMR spectra of cis-α and cis-β isomers of [Ru(Me4phen)(bb7)]2+ and reaction mixtures in CD3CN at different reaction conditions. (a) Isolated mixture of the cis-α and cis-β isomers (sample from reaction after 2 h at 130–140 °C). (b) Isolated products from reaction (Scheme 1) after 1 h at 130–140 °C. (c) Isolated products from reaction (Scheme 1) after 1 h at 130–140 °C and then a second hour at ∼200 °C. (d) cis-α-[Ru(Me4phen)(bb7)]2+ and (e) [Ru(Me4phen)(Me2bpy)]2+. | ||
The same observations were made for reactions conducted under normal laboratory light or in the dark.
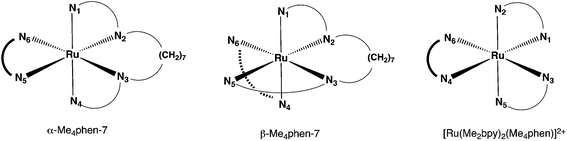
As the cis-β-[Ru(Me4phen)(bb7)]2+ complex appeared to be less stable than the corresponding cis-α isomer, it was of interest to compare the relative energies of the isomers through DFT calculations. In addition, the differences in energies between the cis-α and cis-β isomers for the bb7 complex were compared to those for the corresponding bb12 complex. As shown in Table 2, there was good agreement between the X-ray and DFT-optimised structures based upon the cis-α-[Ru(Me4phen)(bb7)]2+ N–Ru–N angles for the corresponding chelate rings. While there were only small differences in the N–Ru–N bond angles between the cis-α-[Ru(Me4phen)(bb7)]2+ (designated α-Me4phen-7) and the cis-α-[Ru(Me4phen)(bb12)]2+(designated α-Me4phen-12) complexes, significant differences were observed between the corresponding cis-β isomers (designated β-Me4phen-7 and β-Me4phen-12, respectively): see Table 3. Furthermore, the β-Me4phen-12 complex was determined to be 5.6 kJ mol−1 less stable than the α-Me4phen-12, whereas by contrast β-Me4phen-7 was 76.8 kJ mol−1 less stable than the α-Me4phen-7 isomer. It is observed that the calculated difference in the N1–Ru–N5 angle between the β-Me4phen-7 and β-Me4phen-12 species is significant (Table 3), and this may well be the origin of the instability of the β-Me4phen-7 species relative to both the α-Me4phen-7 isomer and the β-Me4phen-12 analogue. However, no significant differences in the calculated Ru–N bond lengths were observed between the α-Me4phen-7 and β-Me4phen-7 isomers.
| Angle | α-Me4phen-7 (crystal structure) | α-Me4phen-7 (DFT) | % deviation |
|---|---|---|---|
| N1–Ru–N2 | 78.9 | 77.5 | 1.77 |
| N1–Ru–N3 | 94.6 | 95.5 | 0.95 |
| N1–Ru–N5 | 97.1 | 97.3 | 0.21 |
| N1–Ru–N6 | 88.9 | 89.5 | 0.67 |
| N4–Ru–N2 | 94.6 | 95.6 | 1.06 |
| N4–Ru–N3 | 78.5 | 77.4 | 1.40 |
| N4–Ru–N5 | 89.4 | 89.9 | 0.56 |
| N4–Ru–N6 | 98.2 | 97.8 | 0.41 |
| N2–Ru–N6 | 100.6 | 99.5 | 1.09 |
| N2–Ru–N3 | 82.1 | 83.7 | 1.95 |
| N3–Ru–N5 | 98.4 | 99.1 | 0.71 |
| N3–Ru–N6 | 176.0 | 174.6 | 0.80 |
| Angle | α-Me4phen-7 | α-Me4phen-12 | β-Me4phen-7 | β-Me4phen-12 | [Ru(Me2bpy)2-(Me4phen)]2+ |
|---|---|---|---|---|---|
| N1–Ru–N2 | 77.5 | 77.3 | 76.9 | 77.4 | 77.2 |
| N1–Ru–N3 | 95.5 | 98.1 | 89.2 | 88.8 | 89.6 |
| N1–Ru–N5 | 97.3 | 96.5 | 103.9 | 99.1 | 97.5 |
| N1–Ru–N6 | 89.5 | 88.4 | 96.5 | 97.2 | 96.4 |
| N4–Ru–N2 | 95.6 | 98.0 | 96.2 | 96.7 | 97.3 |
| N4–Ru–N3 | 77.4 | 77.3 | 97.2 | 96.5 | 96.5 |
| N4–Ru–N5 | 89.9 | 88.4 | 84.2 | 87.3 | 88.4 |
| N4–Ru–N6 | 97.8 | 96.4 | 77.6 | 77.9 | 78.0 |
| N2–Ru–N6 | 99.5 | 97.6 | 94.2 | 89.3 | 88.1 |
| N2–Ru–N3 | 83.7 | 87.3 | 91.0 | 96.6 | 98.2 |
| N3–Ru–N5 | 99.1 | 97.7 | 75.8 | 77.3 | 77.2 |
| N3–Ru–N6 | 174.6 | 172.6 | 173.0 | 172.3 | 172.1 |
Antimicrobial activity
The minimum inhibitory concentrations (MIC) for the ruthenium complexes against six bacterial strains (methicillin-resistant Staphylococcus aureus (MRSA) and a methicillin-sensitive strain of S. aureus, both avian pathogenic (APEC) and uropathogenic (UPEC) strains of Escherichia coli as well as a laboratory strain (E. coli MG1655), and Pseudomonas aeruginosa PAO1) were determined and the results are summarised in Table 4. Interestingly, the [Ru(5-NO2phen)(bb7)]2+ complex (designated α-NO2phen-7) showed very little or no activity against any of the bacterial strains. By contrast, for the other cis-α-[Ru(phen′)(bb7)]2+ complexes, the antimicrobial activities increased with the degree of methylation. Most importantly, cis-α-[Ru(Me4phen)(bb7)]2+ displayed very similar activity against all bacterial strains: for ruthenium(II)-based complexes, the observed similarity in activity against Gram-positive and Gram-negative is rare. In absolute terms, the cis-α-[Ru(Me4phen)(bb7)]2+ complex exhibited comparable or better activities to the Gram-negative species compared to cis-α-[Ru(Me4phen)(bb12)]2+ and [Ru(Me4phen)3]2+ {designated (Me4phen)3}. The dinuclear complexes Rubb7 and Rubb12 (see Fig. 1) were also examined, and the results indicated that the cis-α-[Ru(Me4phen)(bb7)]2+ complex was more active than Rubb7, but less active than Rubb12.| Compound | Gram-positive | Gram-negative | ||||
|---|---|---|---|---|---|---|
| S. aureus | MRSA | E. coli MG1655 | E. coli APEC | E. coli UPEC | P. aeruginosa PAO1 | |
| α-phen-7 | 8 | 16 | >128 | >128 | >128 | >128 |
| α-Me2phen-7 | 8 | 8 | 32 | 64 | 128 | >128 |
| α-Me4phen-7 | 4 | 4 | 4 | 8 | 8 | 8 |
| α-NO2phen-7 | 128 | 128 | >128 | >128 | >128 | >128 |
| α-phen-12 | 1–2 | 2 | 4 | 8 | 8 | 16 |
| β-phen-12 | 2 | 2 | 8 | 16 | 16 | 32 |
| (Me4phen)3 | 0.5–1 | 0.5 | 4 | 4 | 8 | 32 |
| Rubb7 | 8 | 8 | 16 | 4 | 16 | 128 |
| Rubb12 | 2 | 2 | 2 | 2 | 2 | 16 |
| Gentamicin | 0.25 | 0.25 | 0.5 | 0.5 | 1 | 0.25 |
The minimum bactericidal concentrations (MBC) of the ruthenium complexes were also determined, and the results are summarised in Table 5. Consistent with previous results, the MBC values for Rubb12 and [Ru(Me4phen)3]2+ were generally ≤2 × MIC, indicating that the two ruthenium complexes are bactericidal. By contrast, cis-α-[Ru(phen)(bb12)]2+ is borderline bactericidal/bacteriostatic. Surprisingly, the MBC values for cis-α-[Ru(Me4phen)(bb7)]2+ show considerable variation, e.g. S. aureus compared to MRSA, or APEC to UPEC. Based upon the MBC/MIC ratios, cis-α-[Ru(Me4phen)(bb7)]2+ is clearly bactericidal against MRSA and APEC, but bacteriostatic against S. aureus and UPEC. These significant differences within both the Gram-positive and Gram-negative classes of bacteria indicate that cis-α-[Ru(Me4phen)(bb7)]2+ has significant specific toxicity to some bacteria and suggests that the mechanism of the activity is considerably different compared to [Ru(Me4phen)(bb12)]2+, [Ru(Me4phen)3]2+ and Rubb12.
| Compound | Gram-positive | Gram-negative | ||||
|---|---|---|---|---|---|---|
| S. aureus | MRSA | MG1655 | APEC | UPEC | PAO1 | |
| α-phen-7 | 64 | 16 | >128 | >128 | >128 | >128 |
| α-Me2phen-7 | 64 | 16 | ≥128 | ≥128 | >128 | >128 |
| α-Me4phen-7 | 64 | 4 | 32–64 | 16 | >128 | ≥128 |
| α-NO2phen-7 | 128 | ≥128 | >128 | >128 | >128 | >128 |
| α-phen-12 | 4 | 2 | 8–16 | 32 | 32 | ≥64 |
| β-phen-12 | 4–8 | 8 | 16–32 | 32 | 32 | 128 |
| (Me4phen)3 | 1 | 1 | 8 | 8 | 16–32 | 64 |
| Rubb7 | ≥32 | ≥16 | 32 | 32 | 64 | >128 |
| Rubb12 | 2 | 2 | 2 | 2 | 2–4 | 64 |
| Gentamicin | 2 | 1 | 2 | 1 | 4 | 1 |
Lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P)
P)
Lipophilicity is a factor that affects the biological activity of any metal complex because it is often correlated with the capacity of the drug to penetrate through the cell membrane. The standard octanol/water partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) were determined for the [Ru(phen′)(bbn)]2+ and [Ru(Me4phen)3]2+ complexes, and the results are summarised in Table 6. As expected, cis-α-[Ru(Me4phen)(bb7)]2+ was more lipophilic than the other cis-α-[Ru(phen′)(bb7)]2+ analogues. The introduction of the nitro-substituent on 1,10-phenanthroline ligand in cis-α-[Ru(5-NO2phen)(bb7)]2+ did not decrease the lipophilicity compared with the cis-α-[Ru(phen)(bb7)]2+ analogue. More significantly, the cis-α-[Ru(Me4phen)(bb7)]2+ isomer is more lipophilic than the Rubbn complexes, but of similar lipophilicity to [Ru(Me4phen)3]2+, and less lipophilic than their [Ru(phen)(bb12)]2+ analogues.
P) were determined for the [Ru(phen′)(bbn)]2+ and [Ru(Me4phen)3]2+ complexes, and the results are summarised in Table 6. As expected, cis-α-[Ru(Me4phen)(bb7)]2+ was more lipophilic than the other cis-α-[Ru(phen′)(bb7)]2+ analogues. The introduction of the nitro-substituent on 1,10-phenanthroline ligand in cis-α-[Ru(5-NO2phen)(bb7)]2+ did not decrease the lipophilicity compared with the cis-α-[Ru(phen)(bb7)]2+ analogue. More significantly, the cis-α-[Ru(Me4phen)(bb7)]2+ isomer is more lipophilic than the Rubbn complexes, but of similar lipophilicity to [Ru(Me4phen)3]2+, and less lipophilic than their [Ru(phen)(bb12)]2+ analogues.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) for the ruthenium complexes
P) for the ruthenium complexes
Cellular accumulation
The cellular accumulations of the cis-α-[Ru(phen′)(bbn)]2+ analogues (and Rubb12 and [Ru(Me4phen)3]2+) in MRSA, UPEC and PAO1 were determined by measuring the concentration of the complex remaining in the culture supernatant after removing the bacteria by centrifugation. The concentration of the ruthenium complex in the supernatant was calculated from a luminescence calibration curve obtained by adding known concentrations of the ruthenium complex to a blank supernatant. As the luminescence of the ruthenium complexes varied with the different broths and supernatants for each bacterial strain, a calibration curve was determined for each complex in the supernatant of each bacterial strain.Fig. 6 shows the cellular accumulation of the ruthenium complexes against MRSA, UPEC and PAO1 at various time points. The uptake of the ruthenium complexes was greater with the Gram-positive bacteria compared with the Gram-negative strains, with PAO1 showing the lowest accumulation – consistent with the observed MIC/MBC values. In addition, while the accumulation in Gram-positive bacteria gradually increased with time for all complexes, the same result was not observed for the Gram-negative bacteria strains. Interestingly, the cellular accumulation of the cis-α-[Ru(Me4phen)(bb7)]2+ and [Ru(Me4phen)3]2+ in PAO1 reached its highest within 15 minutes and then maintained this level; in contrast, the accumulation of Rubb12 and cis-α-[Ru(phen)(bb12)]2+ steadily increased over the two hours. Moreover, the uptake of the cis-α-[Ru(phen′)(bb7)]2+ family is consistent with the MIC/MBC values, with the degree of methylation of the phen ligand correlated with the accumulation. However, there was significantly less accumulation of cis-α-[Ru(Me4phen)(bb7)]2+ in PAO1 than with [Ru(Me4phen)3]2+, cis-α-[Ru(phen)(bb12)]2+ and Rubb12 despite the activity for cis-α-[Ru(Me4phen)(bb7)]2+ being two- to four-fold higher than for the other three complexes.
 | ||
| Fig. 6 Cellular accumulation of the cis-α-[Ru(phen′)(bb7)]2+, [Ru(Me4phen)3]2+ and Rubb12 complexes into bacteria after incubation for 15, 30, 60, 90 and 120 minutes. | ||
Interaction with DNA
As previous studies showed that the di-, tri- and tetra-nuclear ruthenium complexes linked by the bbn ligand preferentially localise with DNA and RNA in bacterial and eukaryotic cells,18,19 it was of interest to examine the DNA binding affinity of the [Ru(phen′)(bbn)]2+ complexes to DNA. The binding of the ruthenium complexes to calf-thymus (CT)-DNA was examined by UV/Vis spectroscopy and the intrinsic binding constants K to DNA calculated by a non-linear least-square method using eqn (1).20| (εa − εf)/(εb − εf) = (b − (b2 − 2K2Ct[DNA]/s)1/2)/2KCt | (1a) |
| b = 1 + KCt + K[DNA]/2s | (1b) |
All [Ru(phen′)(bbn)]2+ complexes showed strong binding to CT-DNA, with binding constants of approximately 1 × 107 M−1 (see Table 7). As the DNA binding affinities were surprisingly strong, the well-known high affinity metallointercalator complex [Ru(phen)2(dppz)]2+ (ref. 21) was examined as a control. The observed binding constant, 2 × 106 M−1, was comparable with previously reported values.20,21 The cis-α-[Ru(5-NO2phen)(bb7)]2+ complex exhibited the strongest binding affinity. However, no significant differences in the DNA binding affinities between the cis-α-[Ru(phen)(bb7)]2+ and cis-α-[Ru(Me4phen)(bb7)]2+ complexes were observed. In addition, the binding constants of cis-α-[Ru(phen′)(bb7)]2+ isomers are higher than for the [Ru(phen)(bb12)]2+ analogues, which is consistent with reported values for the DNA binding affinities for [Ru(phen)(bb10)]2+ and [Ru(phen)(bb12)]2+ analogues.17
| Complexes | K [M−1] × 107 |
|---|---|
| α-phen-7 | 1.73 ± 0.26 |
| α-Me2phen-7 | 0.42 ± 0.04 |
| α-Me4phen-7 | 1.46 ± 0.47 |
| α-NO2phen-7 | 4.22 ± 0.93 |
| α-phen-12 | 0.59 ± 0.05 |
| β-phen-12 | 0.20 ± 0.02 |
| (phen)2dppz | 0.21 ± 0.05 |
| (Me4phen)3 | 0.25 ± 0.01 |
Discussion
Due to the emergence of drug-resistant bacteria there is a need to develop new classes of antimicrobial agents. We have previously shown17 that ruthenium(II) complexes containing bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane (bbn; for n = 10 and 12) as a tetradentate ligand in a mononuclear complex (rather than as a bridging ligand in oligonuclear complexes) exhibit excellent antimicrobial activity in terms of MIC values, against both Gram-positive and Gram-negative bacteria. In this study we have extended the [Ru(phen)(bbn)]2+ family of complexes by altering both the bbn and phen ligands, and report their antimicrobial activities in terms of both MIC and MBC values.In the syntheses of the [Ru(phen′)(bb7)]2+ complexes, both the cis-α and cis-β isomers were initially observed. However, in contrast to the bb10 and bb12 homologues of [Ru(phen)bbn]2+, the cis-β isomer was not stable and converted predominantly to the cis-α isomer over time or in harsh conditions. It is concluded that the cis-β is a kinetic product but the cis-α is the thermodynamic product. The DFT calculations demonstrated that the cis-β complex is highly strained and considerably less stable than the corresponding cis-α isomer. As ruthenium(II) complexes with three bidentate chelating ligands groups (as is essentially the case in [Ru(phen′)(bb7)]2+ – but with an additional chelate ring involving the polymethylene chain in the tetradendate bb7) are considered to be “kinetically inert” the conversion from the cis-β to the cis-α is unusual, although isomerisation of ruthenium(II) complexes with monodentate ligands, e.g. pyridine, is well known.22 Examples of photoactivated isomerisation, ligand loss and degradation of tris(bidentate) complexes, e.g. [Ru(bpy)3]2+, are known23 and it has been suggested that ruthenium(II) complexes with distorted octahedral geometry photodecompose through ligand dissociation.24 The present DFT calculations have indicated that the cis-β-[Ru(Me4phen)(bb7)]2+ complex is unstable and the octahedral geometry is distorted: consequently, the cis-β and cis-α isomers can interconvert but demonstrably by a thermal mechanism in this case, as the presence of light does not affect the process.
The MIC/MBC values presented in this study demonstrate that apart from cis-α-[Ru(5-NO2phen)(bb7)]2+, the cis-α-[Ru(phen′)(bb7)]2+ complexes are active against Gram-positive bacteria but exhibit variable activity towards Gram-negative species. However, the cis-α-[Ru(Me4-phen)(bb7)]2+ complex shows good activity (MICs ≤ 8 μg mL−1) against both Gram-positive and Gram-negative bacteria, including the notoriously drug-resistant P. aeruginosa. Ruthenium(II) complexes,12,16 like most antimicrobial agents,25 have generally not shown good activity against P. aeruginosa. Recently we reported that the cis-α-[Ru(phen)(bb12)]2+ complex does readily accumulate in P. aeruginosa and exhibits good activity against this bacterial strain.17 The results of the current study indicate that cis-α-[Ru(Me4phen)(bb7)]2+ is slightly more active (in terms of MIC) against P. aeruginosa than cis-α-[Ru(phen)(bb12)]2+. However, a sixteen-fold difference between the MBC and MIC values for cis-α-[Ru(Me4phen)(bb7)]2+ against P. aeruginosa was observed, compared to the two-fold difference for [Ru(Me4phen)3]2+ and a four-fold difference for cis-α-[Ru(phen)(bb12)]2+. This suggests the cis-α-[Ru(Me4phen)(bb7)]2+ complex may exert its antimicrobial activity through a different mechanism. Furthermore, and of note, of all the mono-, di-, tri- and tetra-nuclear ruthenium complexes that incorporate the bbn ligand that we have examined,12–17 the cis-α-[Ru(Me4phen)(bb7)]2+ complex displays the most similar MIC values but the most varied MBC values across a range of Gram-positive and Gram-negative bacteria. This is also consistent with the cis-α-[Ru(Me4phen)(bb7)]2+ complex exerting its antimicrobial activity through a different mechanism. Alternatively, the highly variable MBC values observed for cis-α-[Ru(Me4phen)(bb7)]2+ could be due to different resistance mechanisms – e.g. the up-regulation of efflux pumps and plasmid-mediated resistance mechanisms – that exist between bacteria, even closely related bacteria such as the E. coli strains UPEC and APEC.
While we have previously reported the antimicrobial activities of mono- and oligo-nuclear ruthenium complexes that incorporate the bbn ligand, the present study also examined the effects of substituents on the 1,10-phenanthroline ligand. The antimicrobial activities were examined in relationship to the electron-withdrawing/-donating effect of substituents on the phenanthroline ligand through the analyses of their respective cellular accumulations and DNA binding. The results clearly demonstrate the importance of both cellular accumulation and DNA binding. The cis-α-[Ru(5-NO2phen)(bb7)]2+ complex bound DNA with the highest affinity but displayed no antimicrobial activity because it was not taken up by the bacterial cells. Given that cis-α-[Ru(5-NO2phen)(bb7)]2+ is slightly more lipophilic than cis-α-[Ru(phen)(bb7)]2+, the lack of uptake and antimicrobial activity of the nitro-substituted complex suggests that relatively minor charges of the charge density on the ruthenium centre play a critical role in the diffusion of the ruthenium complex across the bacterial membrane. The introduction of methyl substituents on the 1,10-phenanthroline progressively increased the cellular accumulation and antimicrobial activities of the cis-α-[Ru(phen′)(bb7)]2+ complexes, especially against Gram-negative bacteria.
All the ruthenium complexes showed better accumulation in the Gram-positive than the Gram-negative bacteria, suggesting a clear relationship between the cellular accumulation and MIC, but only to a point. The cis-α-[Ru(phen)(bb12)]2+ exhibited similar antimicrobial activity but a higher level of cellular accumulation than the cis-α-[Ru(phen)(bb7)]2+ across the bacteria, consistent with increasing lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.9 and −1.83 respectively). Furthermore, cis-α-[Ru(Me4phen)(bb7)]2+ has similar or even higher activity against Gram-negative bacteria than [Ru(Me4phen)3]2+ in terms of MIC, but cis-α-[Ru(Me4phen)(bb7)]2+ showed lower cellular accumulation in the Gram-negative bacteria compared to [Ru(Me4phen)3]2+. Notably, there was a different pattern of accumulation in P. aeruginosa between the cis-α-[Ru(Me4phen)(bb7)]2+ and [Ru(Me4phen)3]2+ complexes compared with the cis-α-[Ru(phen)(bb12)]2+ and Rubb12 complexes. The uptake of the latter two complexes steadily increased over the two-hour time period; whereas for the former complexes, maximum accumulation was achieved at the first time point (15 minutes), with the cellular accumulation then remaining constant up to two hours. These results potentially suggest that P. aeruginosa may have an effective efflux pump against the cis-α-[Ru(Me4phen)(bb7)]2+ and [Ru(Me4phen)3]2+ complexes, or that the ruthenium complexes incorporating the bb12 ligand can permeabilise the bacterial membrane.
P = −0.9 and −1.83 respectively). Furthermore, cis-α-[Ru(Me4phen)(bb7)]2+ has similar or even higher activity against Gram-negative bacteria than [Ru(Me4phen)3]2+ in terms of MIC, but cis-α-[Ru(Me4phen)(bb7)]2+ showed lower cellular accumulation in the Gram-negative bacteria compared to [Ru(Me4phen)3]2+. Notably, there was a different pattern of accumulation in P. aeruginosa between the cis-α-[Ru(Me4phen)(bb7)]2+ and [Ru(Me4phen)3]2+ complexes compared with the cis-α-[Ru(phen)(bb12)]2+ and Rubb12 complexes. The uptake of the latter two complexes steadily increased over the two-hour time period; whereas for the former complexes, maximum accumulation was achieved at the first time point (15 minutes), with the cellular accumulation then remaining constant up to two hours. These results potentially suggest that P. aeruginosa may have an effective efflux pump against the cis-α-[Ru(Me4phen)(bb7)]2+ and [Ru(Me4phen)3]2+ complexes, or that the ruthenium complexes incorporating the bb12 ligand can permeabilise the bacterial membrane.
The cis-α-[Ru(phen′)(bb7)]2+ complexes showed surprisingly high DNA binding affinity, even binding more strongly than the established high-affinity metallointercalator [Ru(phen)2(dppz)]2+. The high DNA binding affinity of the cis-α-[Ru(phen′)(bb7)]2+ complexes is likely to be due to an increased hydrophobic effect coupled with the decreased ability of the complexes to self-associate in water due to steric clashes between the methyl groups. The observations that cis-α-[Ru(Me4phen)(bb7)]2+ shows lower accumulation but exhibits the same or slightly better antimicrobial activities against the Gram-negative bacteria than the [Ru(Me4phen)3]2+ or cis-α-[Ru(phen)(bb12)]2+ complexes could suggest DNA (and RNA) binding is at least one aspect of the antimicrobial activity. Indeed, we have previously demonstrated that the dinuclear and oligonuclear ruthenium complexes target DNA and RNA in bacteria and eukaryotic cells.18,19 However, the mechanism of action for kinetically-inert polypyridylruthenium(II) complexes is still a matter of debate. In addition to nucleic acid binding, a variety of studies have proposed that these ruthenium complexes can permeabilise the bacterial membrane,26,27 inhibit the activity of important enzymes,28 or induce the production of reactive oxygen species (ROS) that could cause DNA damage and bacterial cell death.29 It is likely that for each ruthenium complex a number of these potential mechanisms are simultaneously active, with the balance between the various mechanisms modulated by the specific chemical structure.
Conclusions
The mononuclear cis-α-[Ru(Me4phen)(bb7)]2+ complex has significant potential as an antimicrobial agent. In this study, it has been shown that it is highly active against P. aeruginosa. Our results also suggest that the antimicrobial activity differences between the cis-α-[Ru(phen′)(bb7)]2+ complex and the mononuclear [Ru(phen)(bb12)]2+ complexes previously studied are significant in terms of the electron density imposed by substituents on the 1,10-phenanthroline ligand. As a consequence, and given that the structure can be readily modified, it is possible that ruthenium(II) complexes can be customised to particular bacteria. Our studies continue on this project: the toxicity on eukaryotic cells of [Ru(phen′)(bbn)]2+ species will be explored and a further probe of the working mechanisms of this class of ruthenium complexes is on the way.Experimental
Physical measurements
1H and 13C NMR spectra were recorded on a Varian Advance 400 MHz spectrometer at room temperature {CDCl3 (99.8%, CIL), CD3CN (>99.8%, Aldrich), or CD3OD (>99.8%, Aldrich)}. Luminescence was measured on a Cary Eclipse Fluorescence Spectrophotometer with λex = 488 nm: emission spectra were collected from λem = 500–800 nm. Absorbance (200–600 nm) was measured on a VARIAN CARY 50 Probe UV–visible spectrophotometer. Mass-spectroscopic analysis was performed by the RSC Mass-Spectrometry Facility (Research School of Chemistry, Australian National University, Canberra), and the Campbell Microanalytical Laboratory (Chemistry Department, University of Otago) performed the microanalyses.Materials and methods
Potassium hexafluorophosphate (KPF6), and 1,10-phenanthroline and its derivatives, were purchased from Aldrich and used as supplied. Amberlite IRA-402 (chloride form) anion-exchange resin and SP Sephadex C-25 cation exchanger were obtained from GE Health Care Bioscience. Cation-adjusted Mueller–Hinton broth (CAMHB), Mueller–Hinton Agar2 and the antibiotic gentamicin were purchased from Sigma-Aldrich (UK). LB broth was purchased from Formedium (UK).The syntheses of ligands bbn (n = 7, 12) and cis-[RuCl2(DMSO)4] were performed according to previously reported methods.30,31
Syntheses of metal complexes
[Ru(phen′)(bb7)](PF6)2
A solution of cis,cis-[RuCl2(DMSO)2(phen′)] {where phen′ = phen; Me2phen; Me4phen; and 5-NO2phen} (0.39 mmol) and the bb7 ligand (0.47 mmol) in N2-purged ethylene glycol (35 mL) was heated to 130–140 °C and stirred in an N2 atmosphere for 2 h. The reaction mixture turned from light green to bright orange during the course of the reaction. The reaction mixture was cooled to room temperature and water (10 mL) was added to the bright-orange solution, which was then loaded onto a SP Sephadex C-25 cation-exchange column (3 × 20 cm). The column was washed with water and the desired mononuclear complex was eluted with aqueous NaCl solution (0.3 M). Solid KPF6 (ca. 5 mg) was added to the eluate and the complex was extracted into DCM (2 × 30 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4 and evaporated to dryness to obtain the PF6− salt of the complex. The complex was further purified by recrystallisation from acetonitrile/diethyl ether to obtain a bright-red/orange solid of [Ru(phen′)(bbn)](PF6)2. Typical yields were approximately 20–25%. For cis-[RuCl2(DMSO)2(Me2phen)(bb7)](PF6)2 and cis [RuCl2(DMSO)2(Me4-phen)(bb7)](PF6)2, after recrystallisation, the symmetrical isomers were purified by the long SP Sephadex C-25 cation-exchange column (1.5 × 90 cm).Purification of cis-α isomers
[RuCl2(DMSO)2(Me2phen)(bb7)] or [RuCl2(DMSO)2(Me4phen)(bb7)](PF6)2 (35 mg) was converted into the chloride salt by stirring in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin for 1 h. After removal of the resin by filtration, the methanol filtrate was evaporated and the resultant chloride salt was dissolved in water (20 mL) and loaded onto a SP Sephadex C-25 cation-exchange column (1.5 × 90 cm). The cis-α isomers and impurities were eluted as separate bands with an aqueous solution of sodium toluene-4-sulfonate (0.125 M) as the eluent. Solid KPF6 was added to the eluents and the complexes were extracted into DCM (2 × 20 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain the PF6 salt of the complex.Density functional theory calculations
The geometries of five ruthenium metal complexes – cis-α- and cis-β-[Ru(Me4phen)(bbn)]2+ (n = 7 and 12) and [Ru(Me2bpy)2(Me4phen)]2+ – were optimised with density functional theory (DFT) using QCHEM 4.434 at the B3LYP/6-311G(d,p)35,36 level of theory with the LANL2DZ effective core potential37 used for ruthenium atoms. Calculations were carried out for both low- and high-spin electronic configurations of the complexes, with the low-spin state (multiplicity 1 and net charge 2) found to have the lower energy in all five cases, as expected for d6 octahedral ruthenium complexes. Initial structures for cis-α-[Ru(Me4phen)(bb7)]2+ and [Ru(Me2bpy)2(Me4phen)]2+ were obtained from crystal structures and were used to build the initial structures for cis-α-[Ru(Me4phen)(bb12)]2+, cis-β-[Ru(Me4phen)(bb7)]2+ and cis-β-[Ru(Me4phen)(bb12)]2+ using Materials Studio 5.0.38Single crystal X-ray diffraction
Crystals of compounds [Ru(Me4phen)(bb7)](PF6)2·3CHCl3 and [Ru(Me4phen)(Me2bpy)2](PF6)2 suitable for X-ray crystallographic studies were grown by slow diffusion of toluene into a chloroform solution of [Ru(Me4phen)(bb7)](PF6)2, and hexane into a DCM solution of [Ru(Me4phen)(Me2bpy)2](PF6)2, respectively. [Ru(Me4phen)(bb7)](PF6)2·3CHCl3 crystallised as orange rod-shaped crystals and [Ru(Me4phen)(Me2bpy)2](PF6)2 as orange plate-like crystals. Single crystals were selected and mounted on a nylon loop in paratone-N cryo-protectant. Single-crystal X-ray diffraction was performed at 100(2) K on the MX-1 beamline of the Australian Synchrotron (λ = 0.7107 Å).39 Data sets were corrected for absorption using a multi-scan method, and structures were solved by direct methods using SHELXS-2014 and refined by full-matrix least squares on F2 by SHELXL-2014,40 interfaced through the program X-Seed.41 In general, all non-hydrogen atoms were refined anisotropically and hydrogen atoms were included as invariants at geometrically estimated positions, unless specified otherwise.CIF data have been deposited with the Cambridge Crystallographic Data Centre, CCDC reference numbers 1559396 and 1558934† {where [Ru(Me4phen)(bb7)](PF6)2·3CHCl3 = 1559396 and [Ru(Me4phen)(Me2bpy)2](PF6)2 = 1558934}.
Special refinement details
Compound [Ru(Me4phen)(bb7)](PF6)2 crystallises with three molecules of chloroform in the unit cell, two of which are disordered over 2 positions. The disorder was modelled and both molecules were left to freely refine to give 70% and 76% occupancy for each main position.Bacterial strains
Two Staphylococcus aureus (Gram-positive) isolates {a wild type S. aureus strain (SH 1000) and a clinical multidrug-resistant MRSA strain (USA 300 LAC JE2)}, and three Gram-negative Escherichia coli isolates {MG 1655, NCTC 12241 (APEC) and ST 131 (UPEC)} and a Pseudomonas aeruginosa strain PAO1 (WT), were used for in vitro antimicrobial studies.MIC and MBC determination
The MIC tests were conducted by the broth micro-dilution method in duplicate as outlined in the CLSI guidelines.42 The MBC tests were performed in duplicate according to the standard microbiological techniques protocol.43 The bacteria were grown in LB media incubating at 37 °C for overnight. After washing and suspending in CAMHB, the bacteria were plated out on Mueller–Hinton agar, grown overnight and then suspended in growth medium CAMHB. Bacterial inocula were adjusted to a turbidity equivalent to that of a 0.5 McFarland standard and diluted to a final concentration of 4–8 × 105 cfu mL−1. Compounds tested were dissolved and serially diluted in CAMHB in sterile 96-well flat-bottom plates to a final volume of 100 μL in each well. An equal volume of inocula was added to each well, making a final concentration range of the compounds tested, including the control antibiotics gentamicin, of between 0.25 and 128 μg mL−1. MICs were recorded after 16–18 h of incubation at 37 °C. Colony counts of the inocula were performed for determination of the MBC. After MIC results were noted, 10 μL from each well was plated out on Mueller–Hinton agar. MBCs were recorded after overnight incubation at 37 °C, and the concentration of compounds that produced a 99.9% kill relative to the starting inoculum was recorded as the MBC.Cellular accumulation
The cellular accumulation of the ruthenium complexes was measured by monitoring the luminescence of the complexes remaining in the supernatant of the cultures after incubation for various periods. Bacterial inocula in log phase were adjusted to a cell concentration from 1–7 × 107 cfu mL−1. The cell culture (24 mL) was placed in a 250 mL conical flask and 75 μL of stock solution (2.56 mg mL−1) of the ruthenium complex was added to give a final concentration of 8 μg mL−1. Control flasks containing 50 mL of each bacterial suspension were set up as blank samples to obtain fluorescence calibration curves for each complex. Culture flasks and control flasks were incubated with shaking (Incu-shake TL6-5, SciQuip Ltd, Newtown, Wem, Shropshire, UK) at 200 rpm at 37 °C for 15, 30, 60, 90 or 120 min. At each time point, 3.3 mL of bacterial suspension was centrifuged (5500 rpm) at 4 °C for 10 min. Supernatants (3 mL) were carefully transferred to 5 mL tubes and the phosphorescence of the remaining ruthenium complex was measured on a Cary Eclipse Fluorescence Spectrophotometer with λex = 488 nm. The emission spectra were collected from λ = 500–800 nm. Volumes (21, 39, 57, 75 and 93 μL) of a stock solution (320 μg mL−1) of each complex were added to 3 mL aliquots of the supernatant from each control bacterial suspension (untreated with drug) to acquire a fluorescence-concentration linear correlation chart for calibration.Lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P) determination
P) determination
The partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) were measured using the shake-flask technique. Each ruthenium complex (0.1 mM) was dissolved in the water phase (Milli-Q water) and an equal volume of n-octanol was added. The two phases were mutually saturated by shaking overnight at ambient temperature and allowed to separate on standing. The concentration of the metal complex in each phase was determined spectrophotometrically at λ = 450 nm.
P) were measured using the shake-flask technique. Each ruthenium complex (0.1 mM) was dissolved in the water phase (Milli-Q water) and an equal volume of n-octanol was added. The two phases were mutually saturated by shaking overnight at ambient temperature and allowed to separate on standing. The concentration of the metal complex in each phase was determined spectrophotometrically at λ = 450 nm.
DNA-binding studies
Experiments were carried out in phosphate-buffered saline (PBS) at pH 7.4. The ratio of the UV absorbance of a solution of CT-DNA at λ = 260 and 280 nm was greater than 1.8, thus indicating that the CT-DNA was sufficiently free from protein. The DNA concentration of the stock solution (2.8 × 10−3 M) was determined by UV absorbance by using a molar absorption coefficient of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1 per base pair at λ = 260 nm. Absorption titration experiments were carried out by keeping the concentration of the ruthenium complex constant (2 × 10−5 M) and varying the CT-DNA concentration from 0 to fully binding. Absorbance values were recorded after each successive addition of the solution of CT-DNA.
300 M−1 cm−1 per base pair at λ = 260 nm. Absorption titration experiments were carried out by keeping the concentration of the ruthenium complex constant (2 × 10−5 M) and varying the CT-DNA concentration from 0 to fully binding. Absorbance values were recorded after each successive addition of the solution of CT-DNA.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
BS is grateful for a UNSW Canberra PhD scholarship and a Travel Grant to carry out the microbiology experiments at Sheffield University. A. T. thanks CSIRO Materials Science and Engineering for a Ph.D. top-up scholarship. RKP thanks the UK Biotechnology and Biological Sciences Research Council (BBSRC) for grant BB/M022579/1. Aspects of this research were undertaken on the MX1 beamline at the Australian Synchrotron, Victoria, Australia. This research was undertaken with the assistance of resources from the National Computational Infrastructure (NCI), which is supported by the Australian Government.References
- http://www.who.int/mediacentre/factsheets/antibiotic-resistance/en/ .
- F. Li, J. G. Collins and F. R. Keene, Chem. Soc. Rev., 2015, 44, 2529 RSC.
- H. M. Southam, J. A. Butler, J. A. Chapman and R. K. Poole, Adv. Microb. Physiol., 2017, 71, 1 CrossRef PubMed.
- X. Li, A. K. Gorle, M. K. Sundaraneedi, F. R. Keene and J. G. Collins, Coord. Chem. Rev., 2018 DOI:10.1016/jccr2017.11.011.
- A. D. Richards, A. Rodger, M. J. Hannon and A. Bolhuis, Int. J. Antimicrob. Agents, 2009, 33, 469 CrossRef CAS PubMed.
- N. S. Ng, P. Leverett, D. E. Hibbs, Q. Yang, J. C. Bulanadi, M. Jie Wu and J. R. Aldrich-Wright, Dalton Trans., 2013, 42, 3196 RSC.
- M. A. Neelakantan, M. Esakkiammal, S. S. Mariappan, J. Dharmaraja and T. Jeyakumar, Indian J. Pharm. Sci., 2010, 72, 216 CrossRef CAS PubMed.
- F. P. Dwyer, E. C. Gyarfas, W. P. Rogers and J. H. Koch, Nature, 1952, 170, 190 CrossRef CAS PubMed.
- F. P. Dwyer, I. K. Reid, A. Shulman, G. M. Laycock and S. Dixson, Aust. J. Exp. Biol. Med. Sci., 1969, 47, 203 CrossRef CAS PubMed.
- A. Bolhuis, L. Hand, J. E. Marshall, A. D. Richards, A. Rodger and J. Aldrich-Wright, Eur. J. Pharm. Sci., 2011, 42, 313 CrossRef CAS PubMed.
- C. Shobha Devi, D. Anil Kumar, S. S. Singh, N. Gabra, N. Deepika, Y. P. Kumar and S. Satyanarayana, Eur. J. Med. Chem., 2013, 64, 410 CrossRef CAS PubMed.
- F. Li, Y. Mulyana, M. Feterl, J. M. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC.
- M. Pandrala, F. Li, M. Feterl, Y. Mulyana, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 4686 RSC.
- A. K. Gorle, M. Feterl, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2014, 43, 16713 RSC.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686 CrossRef CAS PubMed.
- A. K. Gorle, X. Li, S. Primrose, F. Li, M. Feterl, R. T. Kinobe, K. Heimann, J. M. Warner, F. R. Keene and J. G. Collins, J. Antimicrob. Chemother., 2016, 71, 1547 CrossRef CAS PubMed.
- A. K. Gorle, M. Feterl, J. M. Warner, S. Primrose, C. C. Constantinoiu, F. R. Keene and J. G. Collins, Chem. – Eur. J., 2015, 21, 10472 CrossRef CAS PubMed.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. R. Keene and J. G. Collins, Chem. Sci., 2014, 5, 685 RSC.
- X. Li, A. K. Gorle, T. D. Ainsworth, K. Heimann, C. E. Woodward, J. G. Collins and F. Richard Keene, Dalton Trans., 2015, 44, 3594 RSC.
- R. B. Nair, E. S. Teng, S. L. Kirkland and C. J. Murphy, Inorg. Chem., 1998, 37, 139 CrossRef CAS PubMed.
- I. Haq, P. Lincoln, D. Suh, B. Norden, B. Z. Chowdhry and J. B. Chaires, J. Am. Chem. Soc., 1995, 117, 4788 CrossRef CAS.
- A. Breivogel, S. Wooh, J. Dietrich, T. Y. Kim, Y. S. Kang, K. Char and K. Heinze, Eur. J. Inorg. Chem., 2014, 2014, 2720 CrossRef CAS ; and references therein.
- G. Kalyuzhny, M. Buda, J. McNeill, P. Barbara and A. J. Bard, J. Am. Chem. Soc., 2003, 125, 6272 CrossRef CAS PubMed.
- B. S. Howerton, D. K. Heidary and E. C. Glazer, J. Am. Chem. Soc., 2012, 134, 8324 CrossRef CAS PubMed.
- T. Strateva and D. Yordanov, J. Med. Microbiol., 2009, 58, 1133 CrossRef CAS PubMed.
- F. Li, M. Feterl, J. M. Warner, F. R. Keene and J. G. Collins, J. Antimicrob. Chemother., 2013, 68, 2825 CrossRef CAS PubMed.
- S. V. Kumar, S. Ø. Scottwell, E. Waugh, C. J. McAdam, L. R. Hanton, H. J. L. Brooks and J. D. Crowley, Inorg. Chem., 2016, 55, 9767 CrossRef CAS PubMed.
- N. L. Kilah and E. Meggers, Aust. J. Chem., 2012, 65, 1325 CrossRef CAS.
- P.-L. Lam, G.-L. Lu, K.-M. Hon, K.-W. Lee, C.-L. Ho, X. Wang, J. C.-O. Tang, K.-H. Lam, R. S.-M. Wong, S. H.-L. Kok, Z.-X. Bian, H. Li, K. K.-H. Lee, R. Gambari, C.-H. Chui and W.-Y. Wong, Dalton Trans., 2014, 43, 3949 RSC.
- Y. Mulyana, D. K. Weber, D. P. Buck, C. A. Motti, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 1510 RSC.
- I. P. Evans, A. Spencer and G. Wilkinso, J. Chem. Soc., Dalton Trans., 1973, 204 RSC.
- H. A. Hudali, J. V. Kingston and H. A. Tayim, Inorg. Chem., 1979, 18, 1391 CrossRef CAS.
- R. C. Van der Drift, J. W. Sprengers, E. Bouwman, W. P. Mul, H. Kooijman, A. L. Spek and E. Drent, Eur. J. Inorg. Chem., 2002, 2147 CrossRef CAS.
- Y. Shao, et al. , Mol. Phys., 2015, 113, 184 CrossRef CAS ; full author list provided in ESI.†.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648 CrossRef CAS.
- R. Krishnan, J. S. Binkley, R. Seeger and J. A. Pople, J. Chem. Phys., 1980, 72, 650 CrossRef CAS.
- P. J. Hay and W. R. Wadt, J. Chem. Phys., 1985, 82, 299 CrossRef CAS.
- Materials Studio 5.0, Accelrys Software Inc., San Diego, 2009 Search PubMed.
- T. M. McPhillips, S. E. McPhillips, H. J. Chiu, A. E. Cohen, A. M. Deacon, P. J. Ellis, E. Garman, A. Gonzalez, N. K. Sauter, R. P. Phizackerley, S. M. Soltis and P. Kuhn, J. Synchrotron Radiat., 2002, 9, 401 CrossRef CAS PubMed.
- (a) G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed; (b) G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3 CrossRef PubMed.
- L. J. Barbour, J. Supramol. Chem., 2001, 1, 189 CrossRef CAS.
- Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100-S19, CLSI, Wayne, PA, USA, 2009 Search PubMed.
- K. D. M. Motyl, J. Barrett and R. Giacobbe, Current Protocols in Pharmacology, John Wiley & Sons, New York, 2005 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 1559396 and 1558934. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c7dt04595f |
| ‡ Current Address: School of Healthcare Science, Manchester Metropolitan University, Manchester M1, 5GD, UK. |
| This journal is © The Royal Society of Chemistry 2018 |