 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Field studies reveal functions of chemical mediators in plant interactions†
Meredith C.
Schuman
 * and
Ian T.
Baldwin
* and
Ian T.
Baldwin

Department of Molecular Ecology, Max Planck Institute for Chemical Ecology, Hans-Knöll-Straße 8, 07745 Jena, Germany. E-mail: mschuman@ice.mpg.de
First published on 17th May 2018
Abstract
Plants are at the trophic base of most ecosystems, embedded in a rich network of ecological interactions in which they evolved. While their limited range and speed of motion precludes animal-typical behavior, plants are accomplished chemists, producing thousands of specialized metabolites which may function to convey information, or even to manipulate the physiology of other organisms. Plants’ complex interactions and their underlying mechanisms are typically dissected within the controlled environments of growth chambers and glasshouses, but doing so introduces conditions alien to plants evolved in natural environments, such as being pot-bound, and receiving artificial light with a spectrum very different from sunlight. The mechanistic understanding gained from a reductionist approach provides the tools required to query and manipulate plant interactions in real-world settings. The few tests conducted in natural ecosystems and agricultural fields have highlighted the limitations of studying plant interactions only in artificial environments. Here, we focus on three examples of known or hypothesized chemical mediators of plants’ interactions: the volatile phytohormone ethylene (ET), more complex plant volatile blends, and as-yet-unknown mediators transferred by common mycorrhizal networks (CMNs). We highlight how mechanistic knowledge has advanced research in all three areas, and the critical importance of field work if we are to put our understanding of chemical ecology on rigorous experimental and theoretical footing, and demonstrate function.
1. Introduction: interactions are fundamental
To misquote Richard Feynman, “All [plants are] interaction.” (Feynman in fact said that “all mass is interaction”1 (p. 5), but plants are made of biomass and the same can be said for them). For much of their lives, most plants are rooted to the ground and have a limited range of motion. They have chosen a spot to germinate and must make the best of their neighborhood. Their zone of influence, and their success in life, will thus depend largely on their interactions. Plants in a community interact with each other when they come into direct contact or alter each other's environment in terms of light quality, water and nutrient availability, or chemistry: the focus of this review.1.1. Chemical mediators of plant interactions
Sessile plants can glean abundant information about their neighborhood by monitoring environmental cues. These include cues associated with neighbors, such as changes in light quality,2 emissions of varied volatile organic compounds including volatile hormones,3,4 root exudates containing amino acids, organic acids and carbohydrates as well as hormones,5 and even vibrations.6 Some of these cues, such as far-red light enrichment or ethylene (ET), are general indicators of neighboring plants.7 However, each of these cues comprises, or is directly affected by, specialized metabolites which represent signatures of plant identity and respond to specific environmental stimuli. Furthermore, many products of plant specialized metabolism are assimilated and metabolized by herbivores, pollinators, and their predators and parasitoids, as well as plant parasites, fungi and other microbes;8–10 and these products may also be transferred to other related or unrelated plants, even over long distances (Fig. 1). In this way, chemistry structures biological interactions: products of metabolism provide information and instructions to other organisms, and may be traceable through several nodes in an interaction network.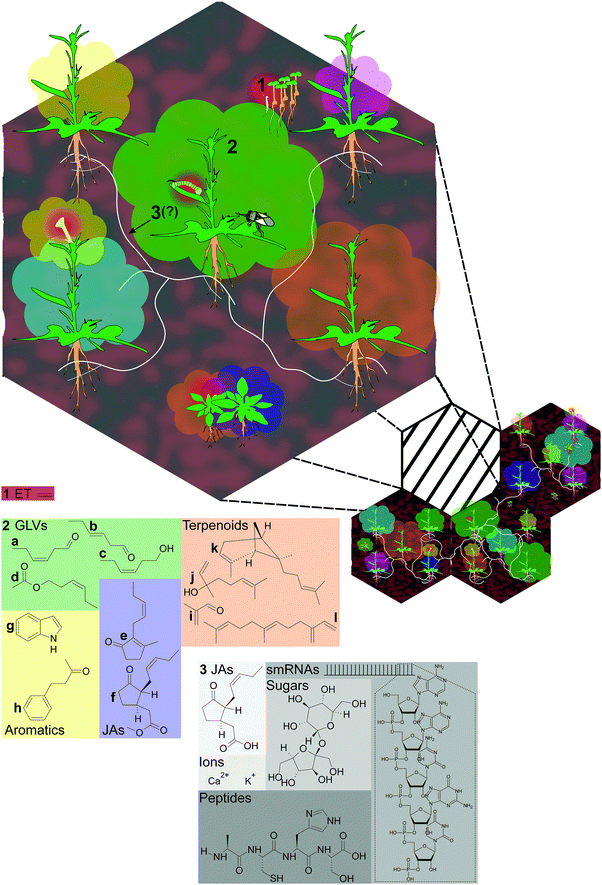 | ||
| Fig. 1 Placing chemical mediators in the context of natural communities. The numbered items in larger font in the illustration are given as examples in this review; see the text for a detailed description. (1) The volatile plant hormone ethylene (ET) (Section 2.1), shown in red, is involved in seedling emergence (upper right); emission from vegetative tissue is stimulated by herbivore feeding and alters plant defense responses (middle); and in flowers, ET emission precedes senescence and seed set after pollination (lower left). (2) Plant volatile blends (Section 2.2) are shown as clouds around plants, with some examples: the oxylipin green leaf volatiles (GLVs) (Z)-3-hexenal, (E)-2-hexenal, (Z)-3-hexenol, and (Z)-3-hexenyl acetate; and jasmonate volatiles (JAs) cis-jasmone, and methyl jasmonate (the (+)-7-iso isomer of methyl jasmonate is shown, corresponding to the active isomer of JA-Ile synthesized following demethylation22); the aromatic compounds indole and benzyl acetone; and several terpenoids: the sesquiterpenes (E)-α-bergamotene and (E)-β-farnesene, the monoterpenoid linalool, and the homoterpenoid methacrolein, derived from isoprene. (3) Common mycorrhizal networks (CMNs) (Section 2.3) link most established plants in communities, but infection takes time to establish and may not be detectable in younger plants (e.g., rosette-stage plants at bottom). Molecules hypothesized to be transferred by CMNs include ions (perhaps driving electrical potentials), hormones or hormone metabolites – the (+)-7-iso-isomer of jasmonic acid (JA) is shown, small RNAs, and peptides.34,35 | ||
Fig. 1 provides some examples of known or hypothesized chemical mediators of plants’ interactions. The structurally simple volatile phytohormone ET (C2H4) mediates diverse events in plants’ lives, from seedling emergence to seed set (Section 2.1). In nature, ET is likely more important as a plant-internal signal than as a between-plant cue, due to its high volatility and rapid dissipation in open systems, although ET from neighbors could reach active concentrations in very dense canopies.11 Structurally diverse plant volatile blends (Section 2.2), shown as clouds around plants accompanied by example structures in Fig. 1, are complex, genetically variable, tissue-specific, and plastic in their emission. This complexity and variation potentially conveys substantial information about plant identity and status.12 Plant volatiles may function both as within-plant signals,13 and as cues or signals for other organisms, including both herbivorous arthropods and their enemies,14–16 as well as neighboring plants.3 When damaged, most plants emit green leaf volatile (GLV) alcohols, aldehydes and esters, derivatives of α-linolenic and α-linoleic acid via the LOX/HPL pathway.17,18 Another group of α-linolenic acid-derived oxylipins, the jasmonates19,20 also include volatile products: cis-jasmone, reported to elicit a cytochrome P450 involved in the resistance of Arabidopsis thaliana to aphids,21 and methyl jasmonate, which can be demethylated and metabolized to the hormone jasmonoyl-L-isoleucine (JA-Ile).22,23 The aromatic compound indole, a precursor of tryptophan as well as indole acetic acid, IAA, is reported to be an important signaling compound in Zea mays (maize),24 and benzyl acetone is an essential component of pollinator-attracting floral volatiles in the wild tobacco Nicotiana attenuata.25
The most diverse class of plant volatiles are the terpenoids, products of two biosynthetic pathways: the plastidial 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate (MEP/DOXP) pathway (hemiterpenes, C5 and monoterpenes, C10) or the 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) pathway (sesquiterpenes, C15).26 Several terpenoids are shown in Fig. 1 which illustrate both the structural complexity, and the potential functional complexity of these compounds: the sesquiterpene (E)-α-bergamotene and the monoterpene alcohol linalool have been shown to attract carnivores in a mechanism termed indirect defense,14,27 but (E)-α-bergamotene can also both attract, and repel, herbivores in field studies,14,15,27 while the two enantiomers of linalool are reported to differently affect moth floral visitation and oviposition preferences in laboratory assays.28 The homoterpenoid methacrolein, derived from isoprene, can prime herbivore resistance in N. attenuata and may be one mechanism explaining reduced damage to N. attenuata growing near clipped Artemisia tridentata (big sagebrush) plants in field studies29,30 (Table S1, ESI†).
Also depicted in Fig. 1, common mycorrhizal networks (CMNs) (Section 2.3) link most established plants in communities, but infection takes time to establish and may not be detectable in younger plants. Priming of plant resistance via CMNs has been demonstrated in a handful of laboratory studies.31–33 The transfer of priming signals by CMNs has not been shown, but these are hypothesized to include ions (perhaps driving electrical potentials), hormones or hormone metabolites, small RNAs, and peptides;34,35 one laboratory study implicated CMNs in the transport of allelopathic thiophenes.36
Modern chemical and genetic tools enable the mapping, elucidation, and precise manipulation of chemical networks and are thereby enabling rigorous examinations of their functions. By identifying and (sometimes) synthesizing the molecules that mediate interactions among organisms – pheromones, defenses, allelochemicals, and so forth – chemical ecologists have developed the ability to manipulate these interactions in a real-world setting. The discipline of chemical ecology has long been one of the most experimental branches of ecology.37 Meanwhile, advances in the fields of molecular biology and plant physiology have provided indispensable means to increase the rigor of field studies by allowing researchers to manipulate the production, and the perception of these molecules in the organisms involved in the interactions. The ability to precisely manipulate a trait in a natural environment permits researchers to determine the natural function of that trait; in managed systems, this approach can test the utility of a trait for achieving management goals. The more precise the manipulation, the easier it is to interpret the data.
For example, consider whether a specific volatile odor compound, induced by feeding herbivores, is relevant for plants near to the attacked emitter. Of course, we can mimic the emitter using a dispenser of the compound in question (if we can synthesize or purify enough of it) – but in the natural scenario neighbors would usually be exposed to this compound as part of a blend of herbivore-induced plant volatiles. Surely the emission dynamics of our dispenser will also differ from the emission of a real plant. It would be better to remove the natural emission, generating “mute” plants in which everything else is intact, and see if this leads to a difference in neighbor responses. If we also have the pure compound, all the better – we can use it to complement the deficient headspace, which is an experimental gold standard.15,38–40 Genetic enhancement is also an option – the emission will still occur within a plant background and with kinetics at least partially determined by wild-type dynamics of substrate flux and other emission control mechanisms.41 And how do we know whether the neighbors are responding? We may have hypotheses about what such a response looks like, but our analysis would be more rigorous if we could identify the compound's receptor, and compare the phenotypes of neighbors in which the receptor has been blocked – “deaf” neighbors – to the wild-type.3 In the absence of a receptor, using “mute” neighbors is a good start,42 and these may also have altered sensitivity to the compound they can no longer produce, as is the case in ET signaling43,44 and jasmonate signaling.45,46
Yet such field trials remain rare15,38–40,47 (three of these studies employ native plants, the other two use maize), and thus we still know very little about how plants interact in nature. Specifically, we have a very poor understanding of how real plants integrate, respond to, and depend on the rich input they receive in complex real-world communities (Fig. 1), other than that “crosstalk” amongst the engaged signaling systems can dramatically change outcomes.48 Instead, we have some understanding of how plants in pots respond when suddenly removed from isolation and exposed to a few of the components of a real ecological community, usually one-by-one, in the artificial conditions of glasshouses and climate chambers. It is likely that the functional conclusions drawn from these studies are confounded by the artifacts that emerge from incoherent signal inputs in rarefied environments. To rectify this, we can use our growing mechanistic understanding, combined with inferences from natural history observations, to identify and manipulate key mediators in a complex community context. The aim of this review is to provide this real-world context and suggest rigorous, mechanistically informed approaches to elucidate complex, real chemical networks.
1.2. Studies of natural versus managed systems
Wherever possible, we focus this review on studies of the interactions of native plants in natural environments, and the mechanistic work enabling precise trait manipulations in such studies. Measures made in an environment which is evolutionarily novel to a plant, like a glasshouse or an agricultural field, may not reflect trait function, although they can reflect exaptation (the process by which traits acquire functions for which they were not previously selected). Furthermore, the traits of domesticated plants, which have either emerged from artificial selection by humans, or persisted in spite of it, cannot be assumed to any longer represent outcomes of natural selection and be functionally coherent. The study of domesticated plants in managed systems is thus doubly fraught when it comes to function: the plants express a set of artificially selected traits, and the environments are evolutionarily novel, e.g., uncommonly high-nutrient and low-diversity compared to the ancestral environments of most cultivated species. Thus, research into the chemical ecology of managed systems does not support inferences about the evolved function of plant traits.In contrast, the research on wild plants which is required to study trait function provides important insights that can improve the design of applied studies in domesticated systems. That is because the chemical ecology of domesticated systems is, however circuitously, derived from the chemical ecology of wild systems. Thus, we propose that hypotheses about domesticated systems which are grounded in principles learned from the study of wild ecosystems are more likely to bear fruit. In contrast, hypotheses based on human opinions about what would be convenient, or interesting, or logical, are more likely to be off-target and lead to dead ends in important lines of practical research. Some examples are discussed in Section 2. See also the Text boxes 1 and 2, which present issues relevant for the management of domesticated systems which can be resolved using studies of wild plants in natural environments.
Text box 1. The festering problem of functional redundancyGene functional redundancy describes the laboratory observation that many homologous genes seem to do the same thing. Similarly, partial functional redundancy describes observations of gene families, like the family of ET receptors (Section 2.1), in which the members clearly have signs of functional specification, yet seem to play redundant roles under laboratory conditions. These barriers to the understanding of gene function result from reliance on laboratory studies in the absence of field studies. Plants in complex natural environments experience a much broader spectrum of conditions than do plants in the laboratory, and these can reveal “hidden” functions of genes thought to be fully or partially redundant. The complexity of natural environmental conditions are the “known unknowns” in functional research. In contrast, artificial and alien aspects of laboratory cultivation are known artifacts. These include an unnaturally low light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) temperature ratio (photothermal ratio); patchy rather than continuous light spectra from which some wavelengths, most often UV light, are missing entirely; low growth densities in comparison to natural or agricultural fields; and growth of roots isolated from soil fungal networks and bound in pots which frequently experience temperatures above air temperatures – a situation which simply does not occur to field-grown roots.103,133 Unfortunately, there are also “unknown unknowns” in the laboratory. Many of these are likely to be laboratory artifacts of which experimenters are unaware.136 temperature ratio (photothermal ratio); patchy rather than continuous light spectra from which some wavelengths, most often UV light, are missing entirely; low growth densities in comparison to natural or agricultural fields; and growth of roots isolated from soil fungal networks and bound in pots which frequently experience temperatures above air temperatures – a situation which simply does not occur to field-grown roots.103,133 Unfortunately, there are also “unknown unknowns” in the laboratory. Many of these are likely to be laboratory artifacts of which experimenters are unaware.136 |
Text box 2. Examples of open questions about function in the field of plant–plant interactions research• How do plants integrate the diverse cues involved in sensing shade and neighbors, and what is the functional significance of specific cues in the context of a natural environment rich in cues?2,7• Does variation in neighbor responses correlated to relatedness indicate “kin recognition” in plants,137 or rather “phenotype matching”?138 Is the model of kin recognition helpful, or are models like phenotype matching, or neighbor response strategy, more powerful ways to understand these phenomena?139 • How does plant light perception influence the external metabolome; e.g., volatile emissions88–90 and root exudates, and what are the functional consequences for plant–plant interactions? • How, and to what extent, do plants detect and interpret variable volatile blends? Are self-volatiles distinguished from other-volatiles, and how? • Do common mycorrhizal networks transfer cues or signals among networked plants resulting in priming, in nature?34 |
One important goal of chemical ecology research on domesticated systems is to achieve sustainability. Understanding the mechanisms of wild ecologies is of great help for designing domesticated ecologies so that they “fit in”. As an example, so-called push–pull technology has been advocated as a sustainable, ecologically friendly agricultural solution derived from applied chemical ecological studies. These systems work by using discoveries from chemical ecology to make crops and livestock repellent to pest species, while additionally providing an attractive trap to lure pests away.49 However, the strategies developed so far depend on switching to genotypes or species having repellent or attractive traits, or using exposure to synthetic chemicals. These strategies are not as flexible or fine-tuned as the manipulation of individual traits, as discussed in the example of manipulating plant volatiles in Section 1.1. This may hinder adoption in modern high-tech, high-productivity agricultural systems,47 an essential goal for agrosystems if any natural biodiversity is to survive as human populations increase and adopt western diets.50
1.3. Function, communication, and chemistry
Functional measures, though challenging, are uniquely important because they provide a link between traits – which can be mechanistically elucidated and precisely manipulated – and the evolutionary context in which traits evolve: the unifying theory in biology. Traits which are dysfunctional are likely to be lost over evolutionary time, and neutral traits are free to vary until they are lost to drift or genome reduction, or evolve to be either functional or dysfunctional and are thus either conserved or eliminated. For traits which are not essential for growth and development – e.g., all specialized traits51 – function will be situational. In other words, most traits that are interesting for the study of biodiversity, specialization, and evolution are also context-dependent and can only be understood in the context in which they evolved, even if they can be exploited in other contexts – both evolution and biotechnology take advantage of the fact that this is often the case. While challenging in the laboratory, functional studies are easily done by planting out into a natural habitat and phenotyping plants as they grow and complete their life cycle by which individuals move their genomes forward in time to realize Darwian fitness (Fig. 2).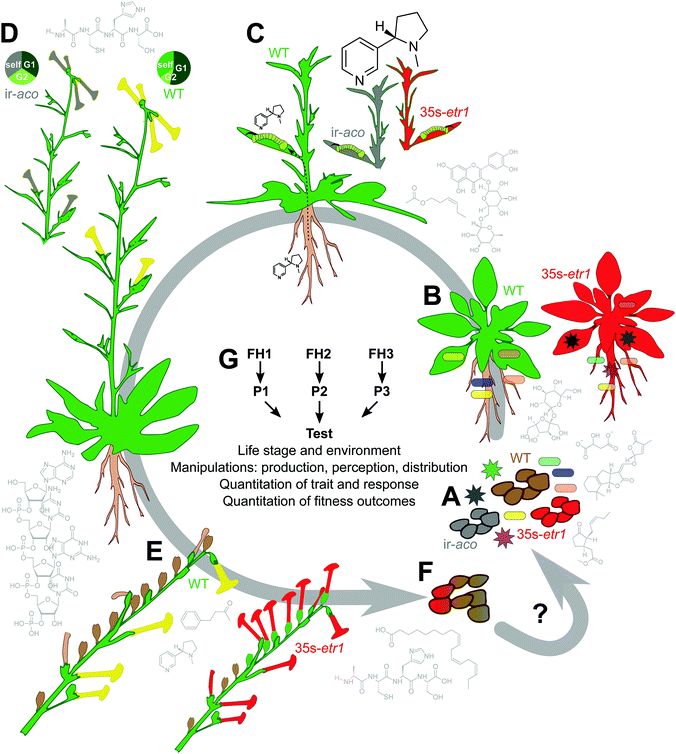 | ||
| Fig. 2 Rigorous functional tests measure how traits help an organism to move its genome forward in time. As an example, the use of plants both “deaf” and “mute” in ET signaling (35s-etr1, red and ir-aco, grey, respectively) permits rigorous elucidation of ET function. Examples of chemical mediators which may interact with, or mediate ET function are around the perimeter, from lower right (A): methyl jasmonate inhibits seed germination;140 strigolactones (here, 5-deoxystrigol), malate, and sugars (sucrose) are known components of root exudates;5 the flavonoid rutin is induced by UV exposure in leaves and is also in root exudates;5,141 the GLV (Z)-3-hexenyl acetate is one active component of plant volatile blends;142 nicotine is a genus-typical alkaloid neurotoxin in Nicotiana; protein and RNA factors contribute to floral mate selection and the regulation of nectar chemistry;78,143,144 nicotine and benzylacetone affect pollinator behavior;25,144 peptides like proteinase inhibitors may alter seed viability in seedbanks (I. T. Baldwin, personal observation), and lipids like α-linolenic acid may affect feeding preferences of seed predators.145 Parts (A–E) show hypothesized or demonstrated functions of ET (see text). (A) Germination and seedling emergence occur in a diverse soil microbial environment including both preferred endophytes (rod-shaped) and pathogens (starburst-shaped). The “triple response” shown in Fig. 3 likely promotes seedling emergence. (B) Microbiome recruitment and growth are altered in 35s-etr1 and ir-aco plants, which have a less diverse microbiome including some microbes not isolated from WT,74 symbolized here by a blue-green endophyte; for 35s-etr1, this affects growth and survival in the field.75 (C) Ethylene signaling limits the induction of root-synthesized nicotine when plants are elicited by the nicotine-tolerant specialist Manduca sexta.44 (D) Mate selection is abrogated in 35s-etr1 and ir-aco plants when flowers receive pollen from other genotypes;78 for ir-aco this has recently been demonstrated in field studies. Shown is a hypothetical example with self-pollen competing against a preferred genotype “G1” and a non-preferred genotype “G2”. (E) Floral advertisement and seed set depend on ET signaling.44 (F) The contribution to fitness of the phenotypic changes described in (A–E) are approximated by quantifying plant reproduction and survivorship, and could be determined by quantifying the contribution of 35s-etr1 and ir-aco plants to future generations, by combining seed production and viability data. Because seeds of N. attenuata may lay dormant for hundreds of years until a combination of stimulating cues from smoke and the absence of inhibitory cues from competing vegetation triggers germination, seedbank experiments are essential. (G) Rigorously testing functional hypotheses (FH) requires alternative hypotheses which generate different predictions (P) in order to design a falsification test. This test must consider the appropriate life stage and environment and apply rigorous manipulations and measures. | ||
In biology, communication is defined in the currency of Darwinian fitness as an adaptive exchange of a signal between a sender and a receiver, where a signal is a trait which “affects the behavior of other organisms,… evolved because of those effects,… and is effective because the response has evolved to be affected” by the trait.52 Dependence on the measurement of Darwinian fitness outcomes, or at least the best possible correlates, makes this a strict definition,53 but one which is difficult to discard in the absence of any other way to measure adaptive utility to senders and receivers (but see Bergstrom and Lachmann, 200454). Often, interactions between plants are loosely characterized as communication and a fitness benefit is presumed.55 We hypothesize that responding to neighbors generally confers a fitness advantage for plants, especially in the case of well conserved neighbor responses. However, this only half fulfills the criteria for communication and is perhaps better termed “eavesdropping”.3 Even eavesdropping, or the adaptive use of cues by a receiver,52 has rarely been rigorously demonstrated. Thus, we do not understand why plants demonstrate different types and magnitudes of responses to different neighbor-related cues, because we cannot connect the observed responses to fitness outcomes: our best proxy for intent.
With the exception of pheromones, for which fitness consequences are clear, there are vanishingly few cases in which biological communication via chemical mediators has been rigorously demonstrated. Here, we first seek to learn from a case in which there is a strong phenotype to screen, and where the molecular mechanisms are well elucidated: plant ET signaling. An easily screened phenotype, the seedling “triple response”, has led to the identification of insensitive or constitutively responding mutants, and thus to specific molecular receptors (Fig. 3). Yet although widely used in laboratory, climate chamber, and glasshouse studies, the relevant mutants and transgenic lines have seldom been deployed in field studies in natural environments. We discuss how artificial experimental conditions first led to a mis-classification of ET-mediated neighbor responses in plants. We then reflect on the greater gaps in our knowledge in two other areas of research which are hot topics, but much less well elucidated: plant–plant interactions mediated by more complex volatile blends, or by CMNs.
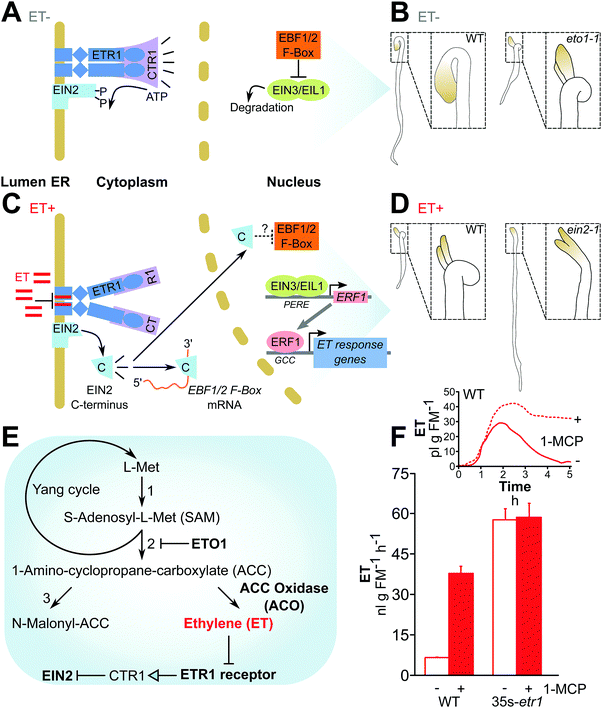 | ||
| Fig. 3 Ethylene signaling: an example in which understanding molecular mechanisms has led to the generation of “mute” and “deaf” plants. (A–D) The current model of ethylene (ET) signaling, adapted from Chang (2016)64 and phenotypes used to identify mis-regulation of ET production or perception in seedling screens drawn based on Guzmán and Ecker (1990).58 (A) In the absence of ET, the ETR1 receptor activates the CTR1 kinase, inhibiting EIN2 activity. The transcription factors EIN1/EIL1 are degraded. (B) Dark-grown WT seedlings are etiolated under low ET (left), whereas eto1 mutants show a “triple response” due to endogenous elevated ET production (see E): shortened and thickened hypocotyls and development of a pronounced apical hook (right). (C) ET inhibits ETR1 and thus CTR1 activity, permitting cleavage of EIN2 and activity of its C-terminal domain, which inhibits EBF1/2 translation and stabilizes the EIN1/EIL1 transcription factors, permitting the transcription of ET-responsive genes. (D) In response to ET, WT seedlings show the triple response (left), while ET-insensitive EIN2 mutants do not (right). (E) ET (highlighted in red) is synthesized from L-Met originating from the Yang cycle.146 The protein regulator ETO158,147 and biosynthetic enzymes like ACO44,148 have been manipulated to alter ET biosynthesis, while the ETR1 receptor149 and the downstream regulator of ET responses, EIN2,150 have been manipulated to alter ET perception. Numbers: 1, SAM synthetase; 2, ACC synthase (ACS); 3, ACC N-malonyl-transferase. (F) ET-insensitive plants expressing a mutant ETR1 receptor emit much larger amounts of ET,11,44 comparable to emission from WT plants in which ET perception has been transiently blocked by the application of 1-methylcyclopropane (1-MCP) application (drawn from data published by von Dahl and colleagues44). | ||
2. Functional analyses of chemical mediators in plant interactions
2.1. Example 1: ethylene signaling
The so-called triple response of etiolated seedlings exposed to ethylene (ET) comprises the shortening and thickening of the hypocotyl, shortening of the root, and a pronounced apical hook (Fig. 3), and is thought to be important for seedling emergence from the structural barriers imposed by soil.56–58 However, the functional significance of the triple response – a widely conserved seed plant phenotype critical for seedling genetic screens – has yet to be demonstrated in real seed banks in nature, and has only recently been suggested using sand-layered Petri dishes in the laboratory.56,59 That the triple response can faithfully be re-created in dark-grown seedlings on nutrient agar, with no physical barrier to seedling elongation, is likely a testimony to the importance of high ET concentrations as a self-generated signal allowing seedlings to anticipate, rather than succumbing to, frequent soil structural barriers in nature. A. thaliana mutants which either failed to show the triple response or did so constitutively in laboratory assays (in which such mutants survive their growth through agar and air) led to the discovery of known ET signaling components.ET signaling is characterized by dominant negative regulation. Plants have several ET receptors (4 in N. tabacum, 5 in A. thaliana, 6 in Solanum lycopersicum [cultivated tomato]) divided into two subfamilies, both of which have conserved His protein kinase (HPK) domains in common with 2-component kinase signaling systems first described in Escherichia coli and other bacteria.60–63 In fact, plants likely acquired ET receptors from the endosymbiotic cyanobacterium which became the chloroplast.64,65 The 5 ET receptors in A. thaliana are all located in the endoplasmic reticulum and constitutively suppress ET responses by activating the Raf-like Ser/Thr kinase constitutive triple response 1 (CTR1)62,66–68 (Fig. 3). Binding of ET to a receptor represses CTR1 activity: CTR1 no longer phosphorylates ET insensitive 2 (EIN2), and unphosphorylated EIN2 undergoes proteolysis to release its C-terminal domain, which migrates to the nucleus and activates a transcriptional cascade including EIN3/EIN3-like and ET response factor (ERF) transcription factors.69–71 Mutant studies indicate that the receptors have partially overlapping but distinct functions, but their functional differences are not well understood.72 The festering problem of partial functional redundancy, for which the study of ET receptors is only one example, is addressed in Text box 1.
The identification of ET biosynthetic enzymes and receptors, in turn, permitted functional studies with plants either “deaf” or “mute” in ET perception and biosynthesis.3,44 Because ET receptors work by suppressing signaling in the absence of ET, it is often sufficient to over-express the mutant ETR1 receptor from A. thaliana in order to abrogate ET sensitivity in other plants.44,73 ET- “deaf” (35s-etr1) or “mute” plants (ir-aco) have advanced the understanding of ET function throughout the life cycles of the ecological model wild plants N. attenuata (coyote tobacco) and Solanum nigrum (black nightshade) (Fig. 2). Microbiome recruitment and growth are both altered in 35s-etr1 and ir-aco plants, which have a less diverse microbiome including some microbes not isolated from wild-type (WT) plants.74 This unbalanced microbiome may make 35s-etr1 and ir-aco plants more susceptible to colonization by pathogens (M. Schuman and I. T. Baldwin, personal observation). Certain poor growth phenotypes of 35s-etr1, which are likely due to sulfur deficiency, can be rescued in nature by specific growth-promoting bacteria which colonize all three genotypes and release a volatile providing biologically available sulfur75,76 (Fig. 2B). Plastic defense responses are promoted by ethylene signaling, which limits the induction of root-synthesized nicotine when plants are elicited by the nicotine-tolerant specialist Manduca sexta. In contrast, both 35s-etr1 and ir-aco plants induce ca. 3 times as much nicotine in response to M. sexta elicitation,44 which may commit more nitrogen than necessary to resistance rather than tolerance, and interfere with more effective defenses against these specialists, such as predation77 (Fig. 2C). Mate selection is abrogated in 35s-etr1 and ir-aco plants when flowers are pollinated with pollen from other genotypes in controlled glasshouse trials,78 and similar results have recently been demonstrated in field studies with ir-aco (Fig. 2D). Floral advertisement and seed set depend on ET signaling. Flowers of 35s-etr1 plants remain turgid and non-senescent for days longer than flowers of WT plants and continue to advertise after pollination due to the lack of a post-pollination ET burst, or its perception44,78 (Fig. 2E).
In N. tabacum, “deaf” 35s-etr1 transgenic lines73 initially implicated ET in regulating the shade avoidance syndrome (SAS).11 The SAS is triggered by the presence of neighboring plants, which alters incident light due to filtration through, and reflectance off green tissues, reducing the ratio of red to far-red light (R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FR) and the fluence of blue and UV-B light, while enriching green wavelengths (reviewed by Pierik and de Wit, 20147). At high density, elevated ambient ET and possibly contact between leaf tips may also contribute to the SAS – at least under wind-free laboratory conditions.79 Over two decades ago, Schmitt and colleagues used transgenic lines of domesticated plants in which the plastic SAS is “always on,” due to manipulation of a phytochrome sensor of R
FR) and the fluence of blue and UV-B light, while enriching green wavelengths (reviewed by Pierik and de Wit, 20147). At high density, elevated ambient ET and possibly contact between leaf tips may also contribute to the SAS – at least under wind-free laboratory conditions.79 Over two decades ago, Schmitt and colleagues used transgenic lines of domesticated plants in which the plastic SAS is “always on,” due to manipulation of a phytochrome sensor of R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FR ratios, to show that a constitutive SAS reduces biomass and flower accumulation in sparse stands.80 More recently, López Pereira and colleagues demonstrated a 19–47% increased oil yield of sunflower crops in dense stands as a result of structural self-organization triggered by altered R
FR ratios, to show that a constitutive SAS reduces biomass and flower accumulation in sparse stands.80 More recently, López Pereira and colleagues demonstrated a 19–47% increased oil yield of sunflower crops in dense stands as a result of structural self-organization triggered by altered R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FR ratios.81 Thus, a plastic SAS in response to canopy density is likely adaptive, but there is a lack of data from wild plants and this hypothesis has not been directly tested.
FR ratios.81 Thus, a plastic SAS in response to canopy density is likely adaptive, but there is a lack of data from wild plants and this hypothesis has not been directly tested.
Pierik and colleagues showed that canopy ET levels from neighboring plants can themselves induce SAS, but sufficient concentrations were only obtained in dense experimental canopies, inside a glasshouse, composed of alternating WT plants with ETR1 transgenic plants which emit much more ET than WT.11 Initially, Pierik and colleagues showed that ET-insensitive 35s-etr1 plants had an attenuated SAS in response to reduced R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FR, and an abrogated response to low blue light fluence;11 later work with A. thaliana EIN mutants showed a failure to initiate SAS in response to reduced R
FR, and an abrogated response to low blue light fluence;11 later work with A. thaliana EIN mutants showed a failure to initiate SAS in response to reduced R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FR but a much milder attenuation in response to reduced blue light fluence.82 Pierik and colleagues also demonstrated increased ET emission from A. thaliana in response to reduced R
FR but a much milder attenuation in response to reduced blue light fluence.82 Pierik and colleagues also demonstrated increased ET emission from A. thaliana in response to reduced R![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FR.82 Thus, ET signaling is part of the initiation of a plastic SAS in response to altered light in dense canopies. The relative importance of specific wavelength changes is unclear and likely depends on overall light quality, which differs greatly by environment and in particular among growth chambers, glasshouses, and field conditions. It is also unclear whether plants in field conditions experience sufficiently high and sustained ET levels for ET to function as a neighbor cue, rather than as a self-signal (ca. 25–30 nL h−1 required to trigger an SAS response in N. tabacum).
FR.82 Thus, ET signaling is part of the initiation of a plastic SAS in response to altered light in dense canopies. The relative importance of specific wavelength changes is unclear and likely depends on overall light quality, which differs greatly by environment and in particular among growth chambers, glasshouses, and field conditions. It is also unclear whether plants in field conditions experience sufficiently high and sustained ET levels for ET to function as a neighbor cue, rather than as a self-signal (ca. 25–30 nL h−1 required to trigger an SAS response in N. tabacum).
Light perception in plants is a fascinating and complex topic on its own, and an important one, since plants “eat light”. Because it is tangential to our focus on chemical mediators, we address it in a ESI† (Text S1) and (Fig. S1).
2.2. Example 2: plant volatile blends
In 1983, Rhoades described evidence that feeding by Hyphantria cunea (fall webworm) larvae on Salix sitchensis (willow) trees induced resistance not only in the damaged trees, but in nearby undamaged trees, likely by airborne factors.83 In the same year, Baldwin and Schultz showed that potted, undamaged Populus x euroamericana (poplar) ramets and Acer saccharum (sugar maple) seedlings sharing only air contact in an enclosure with experimentally damaged conspecifics rapidly (within 36–52 h) increased concentrations and synthesis of foliar phenolics (P. euroamericana), and concentrations of phenolics and hydrolizable tannins (A. saccharum), to levels approaching or exceeding those in the damaged plants.84 These studies established that volatiles from damaged plants could induce resistance in neighboring undamaged plants, most likely in nature and not just in the laboratory.In the 34 years since, studies have identified other resistance-related responses to neighbor volatiles, and in many cases the specific plant volatile or group of volatiles responsible; for a detailed critical overview of specific plant volatiles other than ET, and their published activity in plant–plant interactions, see Table S1 (ESI†). Here, we focus on the activity of plant volatiles which are not volatile hormones, e.g., not ET, nitric oxide, or methylated forms of jasmonic and salicylic acid. It is not clear whether plants distinguish between external and internal sources of these hormones based on e.g. substrate-level feedback, but the activity spectrum of volatile hormones is well studied under laboratory conditions. However, it has been shown that non-hormonal volatiles from damaged plants, such as green leaf volatiles and terpenoids, can prime induced resistance: enhance the sensitivity and responsiveness of undamaged plants to future attack, resulting in greater resistance of plants when attacked.85,86 In other cases, these volatiles themselves induce, and do not just prime resistance (e.g. Baldwin and Schultz, 198384), whereas sometimes no effect on resistance to herbivores can be measured, including in well-designed experiments (e.g. Paschold et al., 200642). A few studies have found that light quality changes plant volatile emission, and Kegge and colleagues reported that volatiles from Hordeum vulgaris (barley) plants exposed to far red-rich light can alter carbon allocation in neighbors, which could be involved in tolerance or competitive growth.87–90
Recently, Karban and colleagues published a meta-analysis of 48 studies including at least two independent replicates of a volatile-exposure and control-exposure treatment, testing the effect on herbivores or herbivore damage, i.e., resistance measures.91 These studies span the 30 years from 1983 to 2013, and all of the published studies are included in Table S1 (ESI†). However, 11 of the 48 studies in the meta-analysis were unpublished studies from the authors, and it is unclear why the editors allowed these to be included in the meta-analysis, as none of the data from these unpublished studies have been deposited in public repositories.
Of the 48 studies, 39 (including 8 unpublished) indicated induction of resistance by neighbor volatiles, while 8 (including 3 unpublished) indicated induced susceptibility, and 1 indicated no measurable effect. The studies included 8 domesticated and 28 wild plant species from 14 different families. A funnel plot analysis indicated that there was no effect of publication bias on the authors’ identified effect size.91 It is questionable to draw this conclusion from a meta-analysis comprising nearly 25% unpublished studies, and it seems that publication bias against negative data may be a factor in a field of study for which response variables are often controversial, and there is no clearly described mechanism behind most of the published phenomena.
Univariate tests conducted by Karban and colleagues as part of their meta-analysis91 indicated that studies were more likely to demonstrate induced resistance when employing insect damage rather than experimental (controlled) damage; when conducted in the laboratory versus the field; and when using agricultural cultivars rather than wild plants. The authors suggest that these trends are due to increased power with reduced sources of variation, but that explanation is inconsistent with the fact that herbivore damage – which is much more spatiotemporally variable and more variable in intensity than experimental wounding – was more likely to produce a resistance effect.91 Alternative explanations are that many instances of resistance induction demonstrated in laboratory studies cannot be reproduced in the field, and that studies on cultivated plants are not representative of studies on wild plants, as discussed above. In a recent example, Triticum aestivum cv. Cadenza (allohexaploid common wheat) was engineered to release (E)-beta-farnesene, which has been described in laboratory studies as a common aphid alarm pheromone to which aphids are also able to habituate.92 Although volatiles from the transformed T. aestivum repelled three different aphid species in laboratory olfactometer studies, and tended to reduce settlement time of aphids in the laboratory, transformed T. aestivum did not have reduced aphid populations in 3 consecutive field studies over 2 years.93
Recently, in a study including a field trial, Sugimoto and colleagues showed that S. lycopersicum plants can take up and glycosylate the green leaf volatile (Z)-3-hexenol, thus increasing resistance to Spodoptera litura (common cutworm).94 They furthermore showed that A. thaliana and 22 domesticated species from 10 families, in addition to S. lycopersicum, produce (Z)-3-hexenyl glycosides when exposed to aerial (Z)-3-hexenol, indicating that the phenomenon is widespread. Sugimoto and colleagues demonstrated accumulation of the S. lycopersicum glycoside, (Z)-3-hexenyl vicianoside, in a field trial of undamaged potted plants grown for 5 d within ca. 22 cm of damaged, potted plants, suggesting that the uptake and decoration of aerial (Z)-3-hexenol can occur in dense stands under natural conditions.94 Aside from this study, progress in understanding the perception of non-hormonal plant volatiles has been limited to demonstrating the involvement of calcium signaling and perhaps membrane potential changes.95–97 Perception is generally defined as the ability to sense and respond to environmental stimuli. (Z)-3-Hexenol must be detected as a substrate in order for glycosylation to occur, but this is a much weaker example of perception than is the interaction of a ligand with a receptor protein, and it is not clear whether the resistance effect of the resulting glycoside is its primary function, or incidental – in part because the phenomenon is so far best studied in a domesticated plant species.94 The generation of mutants unable to glycosylate (Z)-3-hexenol would allow determination of whether this glycosylation is primarily a detoxification response. However, the power of such mutants to dissect responses to (Z)-3-hexenol would be severely limited and is not comparable to the power of e.g. ET biosynthesis and perception mutants to demonstrate ET functions (Fig. 2 and 3).
In the absence of volatile-“deaf” plants (with the exception of ET), the use of “mutes” unable to synthesize and emit specific classes of volatiles has brought the potential for mechanistic clarity to the field. In an elegant laboratory study, Paschold and colleagues demonstrated that N. attenuata plants subjected to neighbor volatiles in an open-flow design changed their transcriptional response when either the GLV or induced sesquiterpene components were absent from the WT volatile blend, and these transcriptional changes could largely be recovered by supplementing the blends from “mute” plants with pure standards of the missing volatiles.42 Interestingly, the transcriptional responses to green leaf volatiles described by Paschold and colleagues indicated that green leaf volatiles repress the transcription of many genes, rather than activating a response.3,42 More recently, Erb and colleagues used lines of the crop plant Zea mays (maize) deficient in the production of indole to show the importance of this auxin-related compound in priming defense responses.24 The combination of “mute” plants and supplementation by synthetic standards in field studies could provide an especially powerful approach, as has been shown in a few cases for studies of plant indirect defense and its potential agricultural application.15,38,39
Despite the fact that this field of study began with field observations of trees,83 there has been pervasive skepticism as to what extent these phenomena occur and are relevant in nature, which is justified by a paucity of field studies having mechanistic rigor (Table S1 (ESI†) and Fig. 2). There are a few exceptions: in 2000, Karban and Baldwin demonstrated that damaged A. tridentata neighbors could enhance resistance to herbivory in natural populations of N. attenuata, and hypothesized that the phenomenon was due to the hormone methyl jasmonate released by clipped sagebrush.29 However, in 2006, Kessler and colleagues reproduced this phenomenon and demonstrated that it could be due to resistance priming by methacrolein and (E)-2-hexenal.30 Similarly, Karban and colleagues have demonstrated increased resistance in field-grown or naturally occurring A. tridentata exposed to the headspace of damaged conspecifics and that chemotypes of neighbors and emitters affect the strength of the response.98,99 Using naturally growing Phaseolus lunatus (lima bean) plants in field studies, Heil and Kost demonstrated in 2006 that specific herbivore-induced volatiles induced or primed extrafloral nectar production in neighbors, and in 2007, Heil and Silva Bueno demonstrated that damaged self-volatiles elicit and prime defense in remote branches within a plant.13,100,101 Each of these represent case studies in particular plant species, and individual case studies may suffer from unclarities. For example, the studies by Karban, Heil, and colleagues frequently relied on enclosing branches in plastic bags; appropriate controls were employed, and in the case of Heil and Silva Bueno airflow was maintained in bags,13 but enclosure of focal plant tissues threatens to generate artifacts, including artificially high concentrations of ET.3
Many laboratory studies include artifacts which obfuscate the mechanism and phenotypic precision of measured responses, and field studies make it easier to avoid these artifacts as well as to establish ecological realism and demonstrate utility (Fig. 2). The unintentional accumulation of ET in closed exposure setups is a pervasive problem, so that this important hormone regulating plant neighbor responses also becomes an invisible force threatening reproducibility and mechanistic understanding of these phenomena.3 Similarly, light quality affects not only plant growth and neighbor growth responses, but also plant volatile emission,88 and thus artificial light conditions may lead to laboratory results which cannot be reproduced under natural light. Of general concern is the number of laboratory-demonstrated phenomena which have not been field-proven (see Table S1, ESI†).
2.3. Example 3: exchange through common mycorrhizal networks
Research on plant–mycorrhizal interactions offers sobering insight into the importance of field studies. In nature, about 83% of flowering plant species associate with mycorrhizal fungi; of the remaining 16%, around half belong to families with both mycorrhizal and non-mycorrhizal members and are found in habitats inhospitable to mycorrhizae.102 Thus, most flowering plants in nature are mycorrhized. The vast majority (74% of angiosperms) associate with arbuscular mycorrhizal fungi, as opposed to other forms of mycorrhizae.102 In contrast, ca. 100% of flowering plants in laboratories and glasshouses are not mycorrhized unless inoculated. A mycorrhized plant in a pot has little in common with a mycorrhized plant in nature: the plant in nature is likely to participate in a widespread fungal network which not only extends the reach of its root system, with hyphae finer than any fine roots,103 but also connects it to other plants in a common mycorrhizal network (CMN).31,34A handful of studies have indicated that putative signals transferred from attacked to unattacked plants through CMNs can prime or elicit resistance to disease (Alternaria solani) and herbivory (Spodoptera litura, oriental leafworm moth) in the crop plant S. lycopersicum,32,104 and render the crop Vicia fabae (common bean) less attractive to the aphid Acyrthosiphon pisum and more attractive to its parasitoid, Aphidius ervi.31 Barto and colleagues also demonstrated that CMNs can increase transfer rates of the herbicide imazamox from inoculated to uninoculated Z. mays, or of two phytotoxic thiophenes produced by Tagetes tenuifolia (Signet marigold) through soil.36
These studies, which each provide a nice proof of principle, have been conducted in climate chambers or glasshouses, and exclusively with domesticated plants. The common approach is to put plants in screen-divided pots which allow them to share soil and mycelial, but not root contact; half of replicates are inoculated with a convenient arbuscular mycorrhizal fungus mixture (locally available Glomus mosseae104 or Funelliformis mossae,32 commercially available Glomus intraradices31 or unknown inoculum from a local field36) and allowed to establish a CMN, and then half of CMNs are disrupted by regularly rotating screens. The published results generally lack temporal resolution and thus it is not clear whether the effects described are transient, or change in magnitude over time. None include an assessment of reproduction or other fitness-related outcomes for the sender or the receiver of the hypothesized signals carried by AMF (Fig. 1), and as discussed above, such data from domesticated plants grown in artificial environments would be difficult to place in an evolutionary context. The clearest case from this perspective is the study from Barto and colleagues, where it can be argued that the transfer of allelochemicals could represent coercion by the emitter as successful transfer increases receiver mortality.36,52 The more complex interactions described by Song, Babikova and colleagues are fascinating, but there is as yet no credible evidence that they represent real interactions which matter in nature.
The screen rotation technique used in these laboratory studies does not translate to field studies, where roots are not pot-bound; and approaches to manipulate AMF in natural communities have relied on fungicides,105–107 which has limitations as described e.g. by O’Connor and colleagues;108 or, in S. lycopersicum, mutants in which colonization is reduced, but not eliminated.109–111 However, it has been demonstrated in several legume species, and in Oryza sativa (rice), that plants deficient in a highly conserved calcium calmodulin protein kinase (CCaMK112), which is a key component of the symbiotic pathway,113 do not form symbioses with rhizobia or AMF.112,114,115 Recently, Groten and colleagues described a field screening of transgenic lines of the wild plant N. attenuata deficient in CCaMK, and demonstrated that these plants are not infected by native AMF communities, but have an otherwise similar fungal and bacterial community to EV controls and do not differ in resistance-related traits.116 Lines deficient in CCaMK may be a useful tool to put studies of AMF-mediated interaction among plants on a more solid ecological footing, and one which could work in any mycorrhizal flowering plant species amenable to transformation or gene editing.
3. Testing functional hypotheses about chemical mediators in the real world
There is no substitute for field studies when it comes to identifying functions of chemical mediators, or determining their utility for real-world applications. To determine whether a mediator can be applied in agriculture, it must first be demonstrated reproducibly in representative agricultural fields. If this fails, we do not understand the phenomenon well enough to apply it; if it succeeds, we will immediately want a cost-benefit analysis under field conditions to determine whether, and how, to develop it further. Similarly, to identify the function of a trait, we must first be able to manipulate it precisely and reliably in an environment representative of the one in which it evolved. If we cannot do that, we will not be able to investigate the evolved function. However, if we achieve a clean and reproducible manipulation of the trait in nature, we will immediately want to quantify the best possible measure of its Darwinian fitness effect (Fig. 2). Ideally, we would count how many grandchildren of manipulated organisms versus controls survive to reproduce, but in practice we often must settle for the best suitable proxy. Often, these proxies provide a reasonable estimate.For example, the green leaf volatiles (GLVs) – C6 aldehydes, alcohols, and esters produced by most plants and released upon wounding, the typical “cut grass” smell – are thought to have many functions including in plant–plant interactions. GLVs are also thought to function as so-called indirect defenses, by attracting predators and parasitoids of herbivores which then disable or remove the herbivores; in other words, by manipulating tri-trophic interactions to the plant's benefit.117,118 If the defensive function of GLVs against herbivores is indirect, then GLV-emitting plants should have a relative fitness advantage in the presence of responsive predators or parasitoids. This fitness benefit should be explained by a reduction in herbivore load due to predator or parasitoid activity: in the absence of the third trophic level, GLV-emitting and GLV-“mute” plants should experience similar herbivore loads and damage and not have large differences in fitness correlates. If there are differences in fitness correlates with GLV emission which cannot be explained by tri-trophic interactions, that is evidence of alternative functions (Fig. 2). GLVs had been shown to attract predators and parasitoids of herbivores, but also to attract some herbivores, and repel others, both in laboratory and field bioassays.14,15,119–123 Thus, it was unclear whether GLVs functioned as indirect defenses in nature. However, if they did, they could be a powerful tool to employ in biocontrol, given their ubiquity in plants.
Schuman and colleagues tested the indirect defense function of GLVs by quantifying herbivore damage, growth, and reproductive output of transgenic lines of N. attenuata which were identical except in their GLV emission, both in the absence and in the presence of a natural population of predators (no larval parasitoids have yet been observed in this system), in two consecutive years of field studies in the plant's native habitat.38 The GLV-emitting and GLV-“mute” lines were first monitored to show that their growth, reproduction, and herbivore damage rates did not differ in the absence of a predator population, and these plants were also subjected to damage by experimentally applied specialist Manduca sexta (tobacco hornworm) larvae, which, in the absence of predation, did not produce any differences in growth or reproduction of the different transgenic lines. In a second year, when native predators were again abundant, all plants were first baited with M. sexta larvae and supplemented with synthetic GLVs placed on cotton swabs placed adjacent to the focal plants, so that Geocoris spp. (big-eyed bugs) – the most abundant predator in this system, responsible for the great majority of M. sexta mortality to predation in most years124 – had the opportunity to locate prey and associate GLVs with prey on all plants, including the GLV-“mute” plants. This resulted in equal predation rates on all plants. GLV supplementation was then removed, and predation rates on GLV-“mute” plants dropped to ca. half the rates on GLV-emitting plants. A new set of wild Manduca spp. larvae (M. sexta and M. quinquemaculata, randomly distributed in the proportions found in wild ovipositions at the time) were then applied to plants matched for prior growth, reproduction, and herbivore damage. On average, larvae enjoyed higher survivorship and longer residence time on GLV-mute plants, corresponding to a decrease in reproduction of these plants. Because transgenic lines are not permitted by the regulatory agency (APHIS) to distribute mature seed when released in nature, Schuman and colleagues counted buds, flowers, and unripe seed capsules as proximate fitness measures. They concluded that GLVs likely function as indirect defenses for N. attenuata plants. An additional set of experiments in the study demonstrated a potential synergistic role of antidigestive direct defense compounds, trypsin protease inhibitors (TPIs), by showing that Manduca spp. larvae engaged in fewer defensive behaviors against attempted predation on plants producing TPIs in nature.
As shown by this example, demonstrating function for traits in complex natural interactions requires an understanding of the organism's natural history, careful design of field trials to eliminate confounding variables but permit natural variation (ecological realism), precise manipulation of only the traits under investigation; and quantification of function in terms of reproductive output, survival to reproductive maturity, or other close fitness correlates (Fig. 2). This approach can be improved by a gold standard of experimental manipulation, having both “deaf” and “mute” organisms, such as is the case for ET signaling (Fig. 3). It should be noted that field trials of all sorts are subject to year-to-year variability in conditions. In the case of GLV emission, several years of field trials in N. attenuata have demonstrated the importance of GLVs for increasing predation of herbivores, supporting the assertion that GLVs function as indirect defenses in nature,14,15,123,125 but it is possible that continued research with GLV-deficient plants in other years would reveal different functions of GLVs. For example, the GLV aldehyde (Z)-3-hexenal accumulates in large amounts in resting leaf tissue and is modified and released upon damage.126 This leaf-internal pool may have functions in defense against pathogens or the control of damage due to the oxidation of lipids as a result of photosynthesis, for example, which were not apparent under the conditions studied so far.18
Another limitation of the study described above is that it did not address the effects of volatiles on neighbor plants which, as discussed in Section 2.2, are relevant in the real world. In addition to possible direct effects, the emission of volatiles changes the neighborhood of ecological interactions and thus may have indirect consequences for neighbors.127 A subsequent field study manipulated the frequency of plants with genetically enhanced emission of the sesquiterpene volatiles (E)-alpha-bergamotene and (E)-beta-farnesene, in small experimental populations of N. attenuata.47 These experimental populations comprised plants having either abrogated, or intact indirect and direct defense responses, and reflected the chemical diversity and patchy clustering of plants in wild populations,27,47 although the half of populations having abrogated defenses may be more representative of domesticated than of wild plant phenotypes.128,129 These populations with abrogated defense were included in order to reveal effects of (E)-alpha-bergamotene and (E)-beta-farnesene independently of the wild-type defense and volatile emission profile. Schuman and colleagues found that the presence of a single plant having genetically enhanced (E)-alpha-bergamotene and (E)-beta-farnesene emission (ca. 5-fold WT emission), planted in the middle of a 5-plant cluster, altered several important life history measures in the 4 neighboring plants, such as predation rates of herbivores, infestation rates by a stem-boring weevil, and average survivorship to reproductive maturity. The effect size and direction depended on the defensive capacity of the neighboring plants and their own volatile emission.47
These two studies from Schuman and colleagues are examples employing precise molecular chemical tools, combined with an understanding of organisms’ natural history, to dissect complex biological phenomena in ecologically realistic scenarios. Controlled laboratory studies, such as those reviewed in the following recommended references on the chemical ecology of plant interactions, are essential for generating the tools which enable this sort of ecological research, and thus laboratory studies and field studies are synergistic and complementary endeavors.2,7,16,130,131 The ecological resolution provided by well-conducted field studies is no more dispensable than is the mechanistic resolution provided by controlled studies in the laboratory. Biological research moves forward most effectively by correctly deciding when to employ which approach.
This is because mechanistic studies in controlled environments provide a fundamentally different kind of information than do field studies, in which the manipulation is controlled, but the environment is not. Precise manipulations performed in a highly controlled environment can reveal cause and effect, or causal flow: the effect of a change in A on B when all other factors are held constant.132 This approach is essential for revealing genetic regulation, biosynthetic pathways, and other fundamentals of biological mechanism. However, in the real world, environmental factors greatly affect the probability of events. The use of precise manipulations in a realistic, but uncontrolled environment is therefore essential in order to determine conditional interventional probability: the effect of a change in A on B when other factors may vary.132 Field studies in realistic environments thus make it easier to predict and understand biological phenomena in the real world. This is the reason that several stages of clinical trials are required to take a candidate drug from biologically active substance to medication. Field trials of all kinds provide information required for predicting real-world outcomes, and do this most effectively when employing precise manipulations.
4. Conclusions: moving forwards by looking backwards
The literature on plant interactions often treats phenomena like neighbor detection and priming as though these were deviations from the control condition of an isolated plant. That is of course the inversion of the natural scenario. Plants in nature usually occur in populations and communities; are inoculated by a microbial community which, despite recent advances, remains poorly characterized; and in ca. 80% of cases, are connected to a mycorrhizal fungal network102,133 (Fig. 1). Thus the typical laboratory plant, in a pot which typically attains temperatures above air temperature (which rarely occurs to roots in nature except under dire conditions), under gnotobiotic conditions or having an artificial and likely depauperate microbiome, is an oddity.133 It is misleading to characterize the responses of plants exposed to neighbors and networks as “primed”. It would be more accurate to study these as manifestations of normal plant physiology, and ask what happens when these natural components are removed, in order to understand how real plants coordinate with their ecological communities in nature.Such a perspective might also be beneficial when tackling urgent practical problems, such as sustainability in modern agro-ecosystems, which, we would argue, have too long been approached based on the reductionist understanding of the potted plant. In agricultural fields, where plants are grown at high density (a more common natural scenario), manipulation of the shade avoidance syndrome (SAS) can be a powerful tool for forcing ground cover and bushy crops to invest in yield and foliage rather than overtopping,134 but a plastic response can increase yield in apically dominant crops like sunflowers by promoting efficient access to light in dense stands.81 In the realm of crop protection, the use of repellent and attractive stimuli in the so-called “push–pull” strategy depends on understanding dynamics in populations and communities, and knowledge of the relevant traits in wild plants might facilitate the employment of this strategy in a way that is more robust to pest adaptation.47,49,93 Recent work indicates that simultaneously manipulating jasmonate perception via JAZ proteins, and phytochrome B, permits the generation of well-defended plants which also invest in yield, eliminating the stereotypical yet elusive growth-defense trade-off.135
Understanding chemical mediators and signaling systems permits precise manipulation in a real-world context, when coupled with knowledge of natural history (Section 3). In this context, removal experiments are challenging but indispensable: akin to studying biodiversity effects by removal of single components from complex communities, versus assembly of artificial communities from standardized components. Except molecular chemical “removal experiments” are much more precise: rather than removing entire network nodes, one tweaks the individual network connections linking them (Fig. 1). This commonly requires the use of genetic and biotechnological tools (Fig. 2 and 3) for which field releases are not trivial and usually involve regulatory approval and oversight. Newer genome editing methods may change this situation. Regardless, it is only by employing such approaches that we will be able to address the open questions about the mechanisms and the course of plant–plant interactions in the real world, and gain the understanding required to take advantage of these traits in agriculture and other managed ecosystems.81
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank the Max Planck Society, and the Collaborative Research Centre “Chemical Mediators in Complex Biosystems – ChemBioSys” (SFB 1127) for funding, S.-G. Kim for providing the plant sketches used for the life cycle in Fig. 2 and for plant forms in Fig. 1, and A. Steppke for advice and assistance to improve clarity and aesthetics of figures. The phrase “looking forwards by moving backwards” is used in memory of T. Eisner and J. Meinwald. Open Access funding provided by the Max Planck Society.References
- J. Gleick, Genius: The Life and Science of Richard Feynman, Pantheon Books, New York, NY, USA, 1st edn, 1992 Search PubMed.
- C. L. Ballaré and R. Pierik, Plant, Cell Environ., 2017, 40, 2530–2543 CrossRef PubMed.
- I. T. Baldwin, R. Halitschke, A. Paschold, C. C. Von Dahl and C. A. Preston, Science, 2006, 311, 812–815 CrossRef PubMed.
- M. Heil and R. Karban, Trends Ecol. Evol., 2010, 25, 137–144 CrossRef PubMed.
- D. V. Badri and J. M. Vivanco, Plant, Cell Environ., 2009, 32, 666–681 CrossRef.
- M. Gagliano, S. Mancuso and D. Robert, Trends Plant Sci., 2012, 17, 323–325 CrossRef PubMed.
- R. Pierik and M. de Wit, J. Exp. Bot., 2014, 65, 2815–2824 CrossRef PubMed.
- M. D. Madritch and R. L. Lindroth, New Phytol., 2015, 208, 410–420 CrossRef PubMed.
- M. C. Schuman, N. M. Van Dam, F. Beran and W. S. Harpole, Curr. Opin. Insect. Sci., 2016, 14, 46–55 CrossRef PubMed.
- H. J. Subrahmaniam, C. Libourel, E.-P. Journet, J.-B. Morel, S. Muños, A. Niebel, S. Raffaele and F. Roux, Plant J., 2017, 12, 3218–3221 Search PubMed.
- R. Pierik, G. C. Whitelam, L. A. C. J. Voesenek, H. de Kroon and E. J. W. Visser, Plant J., 2004, 38, 310–319 CrossRef PubMed.
- M. Dicke, J. Indian Inst. Sci., 2015, 95, 35–42 Search PubMed.
- M. Heil and J. C. Silva Bueno, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 5467–5472 CrossRef PubMed.
- A. Kessler and I. T. Baldwin, Science, 2001, 291, 2141–2144 CrossRef PubMed.
- R. Halitschke, J. A. Stenberg, D. Kessler, A. Kessler and I. T. Baldwin, Ecol. Lett., 2008, 11, 24–34 Search PubMed.
- M. Dicke and I. T. Baldwin, Trends Plant Sci., 2010, 15, 167–175 CrossRef PubMed.
- K. Matsui, Curr. Opin. Plant Biol., 2006, 9, 274–280 CrossRef PubMed.
- M. C. Schuman, H. A. Valim and Y. Joo, in Deciphering Chemical Language of Plant Communication, ed. J. D. Blande and R. Glinwood, Springer, Switzerland, 2016, pp. 3–34 Search PubMed.
- S. Allmann, R. Halitschke, R. C. Schuurink and I. T. Baldwin, Plant, Cell Environ., 2010, 33, 2028–2040 CrossRef PubMed.
- C. Wasternack and B. Hause, Ann. Bot., 2013, 111, 1021–1058 CrossRef PubMed.
- T. J. Bruce, M. C. Matthes, K. Chamberlain, C. M. Woodcock, A. Mohib, B. Webster, L. E. Smart, M. A. Birkett, J. A. Pickett and J. A. Napier, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4553–4558 CrossRef PubMed.
- J. Wu, L. Wang and I. T. Baldwin, Planta, 2008, 227, 1161–1168 CrossRef PubMed.
- S. Fonseca, A. Chini, M. Hamberg, B. Adie, A. Porzel, R. Kramell, O. Miersch, C. Wasternack and R. Solano, Nat. Chem. Biol., 2009, 5, 344–350 CrossRef PubMed.
- M. Erb, N. Veyrat, C. A. M. Robert, H. Xu, M. Frey, J. Ton and T. C. J. Turlings, Nat. Commun., 2015, 6, 6273 CrossRef PubMed.
- D. Kessler, K. Gase and I. T. Baldwin, Science, 2008, 321, 1200–1202 CrossRef PubMed.
- D. A. Nagegowda, FEBS Lett., 2010, 584, 2965–2973 CrossRef PubMed.
- M. C. Schuman, N. Heinzel, E. Gaquerel, A. Svatos and I. T. Baldwin, New Phytol., 2009, 183, 1134–1148 CrossRef PubMed.
- C. E. Reisenman, J. A. Riffell, E. A. Bernays and J. G. Hildebrand, Proc.: Biol. Sci., 2010, 277, 2371–2379 CrossRef PubMed.
- R. Karban, I. T. Baldwin, K. J. Baxter, G. Laue and G. W. Felton, Oecologia, 2000, 125, 66–71 CrossRef PubMed.
- A. Kessler, R. Halitschke, C. Diezel and I. T. Baldwin, Oecologia, 2006, 148, 280–292 CrossRef PubMed.
- Z. Babikova, L. Gilbert, T. J. A. Bruce, M. Birkett, J. C. Caulfield, C. Woodcock, J. A. Pickett and D. Johnson, Ecol. Lett., 2013, 16, 835–843 CrossRef PubMed.
- Y. Y. Song, M. Ye, C. Li, X. He, K. Zhu-Salzman, R. L. Wang, Y. J. Su, S. M. Luo and R. Sen Zeng, Sci. Rep., 2013, 4, 3915 CrossRef PubMed.
- Y. Song, D. Chen, K. Lu, Z. Sun and R. Zeng, Front. Plant Sci., 2015, 6, 786 Search PubMed.
- D. Johnson and L. Gilbert, New Phytol., 2015, 205, 1448–1453 CrossRef PubMed.
- C. Hettenhausen, J. Li, H. Zhuang, H. Sun, Y. Xu, J. Qi, J. Zhang, Y. Lei, Y. Qin, G. Sun, L. Wang, I. T. Baldwin and J. Wu, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E6703–E6709 CrossRef PubMed.
- E. K. Barto, M. Hilker, F. Mueller, B. K. Mohney, J. D. Weidenhamer and M. C. Rillig, PLoS One, 2011, 6, e27195 Search PubMed.
- T. Hartmann, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4541–4546 CrossRef PubMed.
- M. C. Schuman, K. Barthel and I. T. Baldwin, eLife, 2012, e00007 Search PubMed.
- S. Rasmann, T. G. Köllner, J. Degenhardt, I. Hiltpold, S. Toepfer, U. Kuhlmann, J. Gershenzon and T. C. J. Turlings, Nature, 2005, 434, 732–737 CrossRef PubMed.
- J. Degenhardt, I. Hiltpold, T. G. Köllner, M. Frey, A. Gierl, J. Gershenzon, B. E. Hibbard, M. R. Ellersieck and T. C. J. Turlings, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17606 CrossRef PubMed.
- M. C. Schuman, E. P. C. Palmer-Young, A. Schmidt, J. Gershenzon and I. T. Baldwin, Plant Physiol., 2014, 166, 779–797 CrossRef PubMed.
- A. Paschold, R. Halitschke and I. T. Baldwin, Plant J., 2006, 45, 275–291 CrossRef PubMed.
- Inderjit, C. C. von Dahl and I. T. Baldwin, Planta, 2009, 229, 569–575 CrossRef PubMed.
- C. C. von Dahl, R. A. Winz, R. Halitschke, F. Kühnemann, K. Gase and I. T. Baldwin, Plant J., 2007, 51, 293–307 CrossRef PubMed.
- A. Paschold, R. Halitschke and I. T. Baldwin, Plant J., 2007, 51, 79–91 CrossRef PubMed.
- D. Li, I. T. Baldwin and E. Gaquerel, Plants, 2016, 5, 14 CrossRef PubMed.
- M. C. Schuman, S. Allmann and I. T. Baldwin, eLife, 2015, 4, e04490 Search PubMed.
- A. Robert-Seilaniantz, M. Grant and J. D. G. Jones, Annu. Rev. Phytopathol., 2011, 49, 317–343 CrossRef PubMed.
- S. M. Cook, Z. R. Khan and J. A. Pickett, Annu. Rev. Entomol., 2007, 52, 375–400 CrossRef PubMed.
- I. T. Baldwin, eLife, 2014, 3, e02394 CrossRef PubMed.
- G. S. Fraenkel, Science, 1959, 129, 1466–1470 Search PubMed.
- T. C. Scott-Phillips, J. Evol. Biol., 2008, 21, 387–395 CrossRef PubMed.
- R. Karban, Ecol. Lett., 2008, 11, 727–739 CrossRef PubMed.
- C. T. Bergstrom and M. Lachmann, IEEE Inf. Theory Work, 2004, 50–54 Search PubMed.
- R. Karban, Plant Sensing and Communication, University of Chicago Press, Chicago, IL, USA, 2015 Search PubMed.
- W. R. Briggs, Curr. Biol., 2016, 26, R68–R70 CrossRef PubMed.
- H. Shi, R. Liu, C. Xue, X. Shen, N. Wei, X. W. Deng and S. Zhong, Curr. Biol., 2016, 26, 139–149 CrossRef PubMed.
- P. Guzmán and J. R. Ecker, Plant Cell, 1990, 2, 513–523 CrossRef PubMed.
- S. Zhong, H. Shi, C. Xue, N. Wei, H. Guo and X. W. Deng, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3913–3920 CrossRef PubMed.
- A. J. Ninfa and B. Magasanik, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 5909–5913 CrossRef.
- B. T. Nixon, C. W. Ronson and F. M. Ausubel, Proc. Natl. Acad. Sci. U. S. A., 1986, 83, 7850–7854 CrossRef.
- D. R. Gallie, F1000Prime Rep., 2015, 7, 39 Search PubMed.
- K. M. Light, J. A. Wisniewski, W. A. Vinyard and M. T. Kieber-Emmons, J. Biol. Inorg. Chem., 2016, 21, 715–728 CrossRef PubMed.
- C. Chang, BMC Biol., 2016, 14, 7 CrossRef PubMed.
- B. M. Binder, C. Chang and G. E. Schaller, in Annual Plant Reviews: The Plant Hormone Ethylene, ed. M. T. McManus, Wiley-Blackwell, Oxford, England, UK, 2012, vol. 44 Search PubMed.
- J. Hua, H. Sakai, S. Nourizadeh, Q. G. Chen, A. B. Bleecker, J. R. Ecker and E. M. Meyerowitz, Plant Cell, 1998, 10, 1321–1332 CrossRef.
- K. L. Clark, P. B. Larsen, X. Wang and C. Chang, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 5401–5406 CrossRef.
- J. J. Kieber, M. Rothenberg, G. Roman, K. A. Feldmann and J. R. Ecker, Cell, 1993, 72, 427–441 CrossRef PubMed.
- Q. Liu and C.-K. Wen, Plant Physiol., 2012, 158, 1193–1207 CrossRef PubMed.
- C. Ju, G. M. Yoon, J. M. Shemansky, D. Y. Lin, Z. I. Ying, J. Chang, W. M. Garrett, M. Kessenbrock, G. Groth, M. L. Tucker, B. Cooper, J. J. Kieber and C. Chang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19486–19491 CrossRef PubMed.
- H. Qiao, Z. Shen, S. C. Huang, R. J. Schmitz, M. A. Urich, S. P. Briggs and J. R. Ecker, Science, 2012, 338, 390–393 CrossRef PubMed.
- S. N. Shakeel, X. Wang, B. M. Binder and G. E. Schaller, AoB Plants, 2013, 5, plt010 CrossRef PubMed.
- M. Knoester, L. C. van Loon, J. van den Heuvel, J. Hennig, J. F. Bol and H. J. Linthorst, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 1933–1937 CrossRef.
- H. H. Long, D. G. Sonntag, D. D. Schmidt and I. T. Baldwin, New Phytol., 2010, 185, 554–567 CrossRef PubMed.
- D. G. Meldau, S. Meldau, L. H. Hoang, S. Underberg, H. Wünsche and I. T. Baldwin, Plant Cell, 2013, 25, 2731–2747 CrossRef PubMed.
- D. G. Meldau, H. H. Long and I. T. Baldwin, Front. Plant Sci., 2012, 3, 112 Search PubMed.
- P. Kumar, S. S. Pandit, A. Steppuhn and I. T. Baldwin, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1245–1252 CrossRef PubMed.
- S. Bhattacharya and I. T. Baldwin, Plant J., 2012, 71, 587–601 CrossRef PubMed.
- M. de Wit, W. Kegge, J. B. Evers, M. H. Vergeer-van Eijk, P. Gankema, L. A. C. J. Voesenek and R. Pierik, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14705–14710 CrossRef PubMed.
- J. Schmitt, A. C. Mccormac and S. Harry, Am. Nat., 1995, 146, 937–953 CrossRef.
- M. López Pereira, V. O. Sadras, W. Batista, J. J. Casal and A. J. Hall, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 7975–7980 CrossRef PubMed.
- R. Pierik, T. Djakovic-Petrovic, D. H. Keuskamp, M. de Wit and L. A. C. J. Voesenek, Plant Physiol., 2009, 149, 1701–1712 CrossRef PubMed.
- D. F. Rhoades, in Plant Resistance to Insects, ed. P. Hedin, American Chemical Society, Washington, DC, USA, 1983, pp. 55–68 Search PubMed.
- I. T. Baldwin and J. C. Schultz, Science, 1983, 221, 277–279 Search PubMed.
- B. Mauch-Mani, I. Baccelli, E. Luna and V. Flors, Annu. Rev. Plant Biol., 2017, 68, 485–512 CrossRef PubMed.
- C. J. Frost, M. C. Mescher, J. E. Carlson and C. M. De Moraes, Plant Physiol., 2008, 146, 818–824 CrossRef PubMed.
- J. Schwachtje, P. E. H. Minchin, S. Jahnke, J. T. Van Dongen, U. Schittko and I. T. Baldwin, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12935–12940 CrossRef PubMed.
- W. Kegge, V. Ninkovic, R. Glinwood, R. A. M. Welschen, L. A. C. J. Voesenek and R. Pierik, Ann. Bot., 2015, 115, 961–970 CrossRef PubMed.
- W. Kegge and R. Pierik, Trends Plant Sci., 2009, 15, 126–132 CrossRef PubMed.
- W. Kegge, B. T. Weldegergis, R. Soler, M. V.-V. Eijk, M. Dicke, L. A. C. J. Voesenek and R. Pierik, New Phytol., 2013, 200, 861–874 CrossRef PubMed.
- R. Karban, L. H. Yang and K. F. Edwards, Ecol. Lett., 2014, 17, 44–52 CrossRef PubMed.
- M. de Vos, W. Y. Cheng, H. E. Summers, R. A. Raguso and G. Jander, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14673–14678 CrossRef PubMed.
- T. J. Bruce, G. I. Aradottir, L. E. Smart, J. L. Martin, J. C. Caulfield, A. Doherty, C. A. Sparks, C. M. Woodcock, M. A. Birkett, J. A. Napier, H. D. Jones and J. A. Pickett, Sci. Rep., 2015, 5, 11183 CrossRef PubMed.
- K. Sugimoto, K. Matsui, Y. Iijima, Y. Akakabe, S. Muramoto, R. Ozawa, M. Uefune, R. Sasaki, K. M. Alamgir, S. Akitake, T. Nobuke, I. Galis, K. Aoki, D. Shibata and J. Takabayashi, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7144–7149 CrossRef PubMed.
- S. A. Zebelo, K. Matsui, R. Ozawa and M. E. Maffei, Plant Sci., 2012, 196, 93–100 CrossRef PubMed.
- N. Asai, T. Nishioka, J. Takabayashi and T. Furuichi, Plant Signaling Behav., 2009, 4, 294–300 CrossRef.
- G. Arimura, R. Ozawa, T. Shimoda, T. Nishioka, W. Boland and J. Takabayashi, Nature, 2000, 406, 512–515 CrossRef PubMed.
- R. Karban, K. Shiojiri, M. Huntzinger and A. C. McCall, Ecology, 2006, 87, 922–930 CrossRef PubMed.
- R. Karban, W. C. Wetzel, K. Shiojiri, S. Ishizaki, S. R. Ramirez and J. D. Blande, New Phytol., 2014, 204, 380–385 CrossRef PubMed.
- M. Heil and C. Kost, Ecol. Lett., 2006, 9, 813–817 CrossRef PubMed.
- C. Kost and M. Heil, J. Ecol., 2006, 94, 619–628 CrossRef.
- M. C. Brundrett, Plant Soil, 2009, 320, 37–77 CrossRef.
- Y. Lekberg and R. T. Koide, Can. J. Bot., 2014, 251, 241–251 CrossRef.
- Y. Y. Song, R. Sen Zeng, J. F. Xu, J. Li, X. Shen and G. Yihdego, PLoS One, 2010, 5, e13324 Search PubMed.
- J. F. J. Cahill, E. Elle, G. R. Smith and B. H. Shore, Ecology, 2008, 89, 1791–1801 CrossRef PubMed.
- G. W. T. Wilson and M. W. Williamson, Mycologia, 2008, 100, 548–554 CrossRef PubMed.
- G. Yang, N. Liu, W. Lu, S. Wang, H. Kan, Y. Zhang and Y. Chen, J. Ecol., 2014, 102, 1072–1082 CrossRef.
- P. O’Connor, M. Manjarrez and S. E. Smith, Can. J. Microbiol., 2009, 55, 901–904 CrossRef PubMed.
- T. R. Cavagnaro, F. A. Smith, G. Hay, V. L. Carne-Cavagnaro and S. E. Smith, New Phytol., 2004, 161, 485–494 CrossRef.
- T. R. Cavagnaro, A. J. Langley, L. E. Jackson, S. M. Smukler and G. W. Koch, Funct. Plant Biol., 2008, 35, 228–235 CrossRef.
- S. J. Watts-Williams and T. R. Cavagnaro, Biol. Fertil. Soils, 2012, 48, 285–294 CrossRef.
- C. Chen, M. Gao, J. Liu and H. Zhu, Plant Physiol., 2007, 145, 1619–1628 CrossRef PubMed.
- S. Singh and M. Parniske, Curr. Opin. Plant Biol., 2012, 15, 444–453 CrossRef PubMed.
- J. Levy, C. Bres, R. Geurts, B. Chalhoub, O. Kulikova, G. Duc and F. Debelle, Science, 2004, 303, 1361–1364 CrossRef PubMed.
- R. M. Mitra, C. A. Gleason, A. Edwards, J. Hadfield, J. A. Downie, G. E. D. Oldroyd and S. R. Long, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4701–4705 CrossRef PubMed.
- K. Groten, A. Nawaz, N. H. T. Nguyen, R. Santhanam and I. T. Baldwin, Plant, Cell Environ., 2015, 38, 2398–2416 CrossRef PubMed.
- P. W. Price, C. E. Bouton, P. Gross, B. A. McPheron, J. N. Thompson and A. E. Weis, Annu. Rev. Ecol. Syst., 1980, 11, 41–65 CrossRef.
- M. C. Schuman and I. T. Baldwin, in The Ecology of Plant Secondary Metaboites, ed. G. R. Iason, M. Dicke and S. E. Hartley, Cambridge University Press, Cambridge, UK, 1st edn, 2012, pp. 287–307 Search PubMed.
- R. Halitschke, J. Ziegler, M. Keinänen and I. T. Baldwin, Plant J., 2004, 40, 35–46 CrossRef PubMed.
- R. Halitschke and I. T. Baldwin, Plant J., 2003, 36, 794–807 CrossRef PubMed.
- K. Shiojiri, K. Kishimoto, R. Ozawa, S. Kugimiya, S. Urashimo and G. Arimura, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 16672–16676 CrossRef PubMed.
- D. Kessler and I. T. Baldwin, Plant J., 2006, 49, 840–854 CrossRef PubMed.
- S. Allmann and I. T. Baldwin, Science, 2010, 329, 1075–1078 CrossRef PubMed.
- M. C. Schuman, D. Kessler and I. T. Baldwin, Psyche, 2013, 2013, 465108 Search PubMed.
- Y. Joo, M. C. Schuman, J. K. Goldberg, S.-G. Kim, F. Yon, C. Brütting and I. T. Baldwin, Funct. Ecol., 2018, 32, 136–149 CrossRef.
- K. Matsui, K. Sugimoto, J. Mano, R. Ozawa and J. Takabayashi, PLoS One, 2012, 7, e36433 Search PubMed.
- P. Barbosa, J. Hines, I. Kaplan, H. Martinson, A. Szczepaniec and Z. Szendrei, Annu. Rev. Ecol. Evol. Syst., 2009, 40, 1–20 CrossRef.
- E. Rowen, I. Kaplan and E. Rowen, New Phytol., 2016, 210, 284–294 CrossRef PubMed.
- M. Kallenbach, G. Bonaventure, P. A. Gilardoni, A. Wissgott and I. T. Baldwin, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E1548–E1557 CrossRef PubMed.
- G. Howe and G. Jander, Annu. Rev. Plant Biol., 2008, 59, 41–66 CrossRef PubMed.
- M. C. Schuman and I. T. Baldwin, Annu. Rev. Entomol., 2016, 61, 373–394 CrossRef PubMed.
- P. C. W. Davies and S. I. Walker, Rep. Prog. Phys., 2016, 79, 102601 CrossRef PubMed.
- H. Poorter, F. Fiorani, R. Pieruschka, W. H. Van Der Putten, M. Kleyer and U. Schurr, New Phytol., 2016, 212, 838–855 CrossRef PubMed.
- M. Ganesan, H. Lee, J. Kim and P. Song, Plant, Cell Environ., 2017, 40, 2469–2486 CrossRef PubMed.
- M. L. Campos, Y. Yoshida, I. T. Major, D. de Oliveira Ferreira, S. M. Weraduwage, J. E. Froehlich, B. F. Johnson, D. M. Kramer, G. Jander, T. D. Sharkey and G. A. Howe, Nat. Commun., 2016, 7, 12570 CrossRef PubMed.
- J. Alfred and I. T. Baldwin, eLife, 2015, 4, e06956 Search PubMed.
- M. A. Crepy and J. J. Casal, New Phytol., 2015, 205, 329–338 CrossRef PubMed.
- R. M. Callaway, S. C. Pennings and C. L. Richards, Ecology, 2003, 84, 1115–1128 CrossRef.
- B. Allen, M. A. Nowak and E. O. Wilson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 20135–20139 CrossRef PubMed.
- C. A. Preston, H. Betts and I. T. Baldwin, J. Chem. Ecol., 2002, 28, 2343–2369 CrossRef PubMed.
- S. N. T. T. Dinh, I. Gális and I. T. Baldwin, Plant, Cell Environ., 2013, 36, 590–606 CrossRef PubMed.
- S. Allmann, A. Späthe, S. Bisch-Knaden, M. Kallenbach, A. Reinecke, S. Sachse, I. T. Baldwin and B. S. Hansson, eLife, 2013, 2, e00421 Search PubMed.
- S. Bezzi, D. Kessler, C. Diezel, A. Muck, S. Anssour and I. T. Baldwin, Plant Physiol., 2010, 152, 2232–2242 CrossRef PubMed.
- D. Kessler, S. Bhatacharya, C. Diezel, E. Rothe, K. Gase, M. Schoettner, I. T. Baldwin, S. Bhattacharya, C. Diezel, E. Rothe, K. Gase, M. Schoettner, I. T. Baldwin, M. Schöttner and I. T. Baldwin, Plant J., 2012, 71, 529–538 CrossRef PubMed.
- M. A. Stanton, J. Pressler, C. Paetz, W. Boland and I. T. Baldwin, New Phytol., 2016, 211, 113–125 CrossRef PubMed.
- S. F. Yang and N. E. Hoffman, Annu. Rev. Plant Physiol. Plant Mol. Biol., 1984, 35, 155–189 CrossRef.
- K. C.-L. Wang, H. Yoshida, C. Lurin and J. R. Ecker, Nature, 2004, 428, 945–950 CrossRef PubMed.
- P. John, Physiol. Plant., 1997, 100, 583–592 CrossRef.
- J. Hua and E. M. Meyerowitz, Cell, 1998, 94, 261–271 CrossRef PubMed.
- J. M. Alonso, T. Hirayama, G. Roman, S. Nourizadeh and J. R. Ecker, Science, 1999, 284, 2148–2152 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cs00749c |
| This journal is © The Royal Society of Chemistry 2018 |


