Open Access Article
Open Access ArticleIdentification of the binding site between bovine serum albumin and ultrasmall SiC fluorescent biomarkers
Gabriella
Dravecz
a,
Tibor Z.
Jánosi
bc,
Dávid
Beke
 *ad,
Dániel Á.
Major
d,
Gyula
Károlyházy
ad,
János
Erostyák
ce,
Katalin
Kamarás
*ad,
Dániel Á.
Major
d,
Gyula
Károlyházy
ad,
János
Erostyák
ce,
Katalin
Kamarás
 a and
Ádám
Gali
a and
Ádám
Gali
 af
af
aInstitute for Solid State Physics and Optics, Wigner Research Centre for Physics, Hungarian Academy of Sciences, P.O. Box 49, H-1525 Budapest, Hungary. E-mail: beke.david@wigner.mta.hu
bMTA-PTE High-Field Terahertz Research Group, University of Pécs, Ifjúság u. 6., H-7624 Pécs, Hungary
cUniversity of Pécs, Szentágothai Research Centre, Spectroscopy Research Group, Ifjúság u. 20., H-7624 Pécs, Hungary
dFaculty of Chemical Technology and Biotechnology, Budapest University of Technology and Economics, Műegyetem rakpart 3., H-1111 Budapest, Hungary
eUniversity of Pécs, Faculty of Sciences, Ifjúság u. 6., H-7624 Pécs, Hungary
fDepartment of Atomic Physics, Budapest University of Technology and Economics, Budafoki út 8., H-1111 Budapest, Hungary
First published on 23rd April 2018
Abstract
Ultrasmall silicon carbide nanoparticles (SiC USNPs) are very promising biomarkers for developing new applications in diagnostics, cell monitoring or drug delivery, even though their interaction with biological molecules such as different proteins has not yet been investigated in detail. In this study, the biological behaviour of SiC USNPs in a medium modeling a living organism was investigated in detail through the dependence of the fluorescence on interactions between bovine serum albumin (BSA) and SiC USNPs. The interaction shows transient nanoparticle–protein associations due to the restricted diffusion behaviour of the nanoparticles in the vicinity of a protein. The transient association manifests in a complex fluorescence quenching mechanism where the dynamic component was dominated by Förster resonance energy transfer. By studying SiC nanoparticles of different sizes, it can be concluded that the transient effect is an ultrasmall nanoparticle behaviour.
Introduction
Nanoparticles (NPs) have many diverse applications in life- and physical sciences, for example, NP probes have great potential for bioimaging,1,2 diagnostics,3 and therapeutic purposes.4,5 There is tremendous interest in understanding the impact of NP systems on living organisms. Previous studies might imply6,7 that each individual collection of NPs should be investigated in detail before its application in biology. The interaction of NPs with proteins is the basis of NP research for biological application. Proteins are able to adsorb onto the surface of the NPs forming a dense protein coating known as protein corona (PC). The interaction determines the physiological response and effect of the NPs on living systems, including cellular uptake, circulation lifetime, signalling, biodistribution, therapeutic effects, and toxicity.8 It depends on many factors including the morphology of the NPs that is related to their size and shape.9 Compared to a nanoparticle with a size of several tens of nanometers, even a large protein is small, and the protein will experience a surface almost indistinguishable from a two-dimensional one; the protein can adsorb and be denatured, depending on the surface characteristics and the protein–surface interaction. If the sizes of the nanoparticle and the protein are comparable, then they will interact like two proteins or like two nanoparticles. For very small nanoparticles and large proteins, we can predict a specific interaction where a nanoparticle can interact with a specific surface epitope or binding pocket of a protein.10Systems consisting of NPs of sizes below 3 nm are often referred to as ultrasmall nanoparticles (USNPs).11 Considering that most plasma proteins present a hydrodynamic diameter of about 3–15 nm, the behaviour of NPs < 3 nm in biological systems could be dramatically different from that of larger particles12 described by the classical model. To name just a few examples: dispersions of USNPs lie in between complete molecular dispersions and colloid sols of larger-sized NPs and consequently exhibit intermediate structural, optical, electrical, catalytic, and magnetic properties11,13 representing an additional type of system having potential use in biology. The paramagnetic properties of iron oxide USNPs were adopted toward the development of non-toxic MRI contrast agents.14 The luminescent properties of metal USNPs have been applied to biosensing materials.15–18 Moreover, it has been shown that small quantum dots can be excreted with highly efficient renal clearance.19–21 The in vivo toxicity of nanoparticles mainly arises from their accumulation in the liver and spleen. Specifically, 10–200 nm nanoparticles show very high abundance in these organs due to reticuloendothelial system absorption.22 However, being small is not a sufficient condition for clearance, as nanoparticles can still react with serum proteins and form larger complexes. Despite many promising applications in biological research, studies of the interaction between biomolecules and USNPs are still rare in the literature and often controversial.23 Recently, some studies on the interactions between proteins and USNPs, such as Ag NPs,17 Au NPs,9,24,25 carbon-NPs,26,27 quantum dots,28 Si NPs29 and oxide NPs30 have been reported. They either assumed that the proteins and nanoparticles formed complexes or that they generated different protein coronas, which reflected the strength of the affinity of binding between the constituents. Boselli et al. studied the effect of sizes between 2 and 5 nm on differently PEGylated Au NPs in human plasma using gel assays.31 They observed that for 5 nm and 3 nm Au NPs, protein interactions led to significant changes in mobility. In contrast, 2 nm NPs exhibited few signs of protein interaction. In many cases, a more complex interaction was reported, where a simple model alone was not applicable.
Here we report the interaction of bovine serum albumin and ultrasmall silicon carbide (SiC) NPs which has not been presented before, in order to broaden the scope of USNP–protein interactions. We propose that restricted diffusion behaviour of the USNPs in the vicinity of the proteins should be considered when USNPs interact with proteins. The transient particle–protein associations can manifest themselves in physical complex formation that determines the characteristics of the system.
SiC NPs are promising candidates for in vivo medical applications due to their extremely high degree of biocompatibility, water solubility and chemical resistance.32–34 Furthermore, SiC can host magneto-optically active centres35–38 similar to the nitrogen-vacancy centres in nanodiamond.39 Room-temperature magnetically and optically active centres in solids are the most prominent objects for applications in new generations of biosensors, supersensitive magnetometers and thermometers.40–42 SiC NPs of sizes below 5 nm can be synthesized using electrochemical43 or stain etching44 of bulk SiC. The nanoparticle surface contains organic functional groups such as carboxyl and hydroxyl, stabilising the nanoparticles in polar solvents, including biologically relevant media, without the need for surfactants or capping layers. Thus, the hydrodynamic size of the ultrasmall SiC NPs is still below 5 nm. While many investigations focus primarily on a particular biological application of SiC NPs, the general knowledge of their behaviour in a complex biological environment is still rather limited.
Serum albumins are the most abundant proteins in the blood, and their major physiological role is to carry various ligands to their respective target organs.45 BSA is also the most studied protein, which has a single polypeptide chain with a molecular weight of 66 kDa and consists of 583 amino acid residues. BSA contains 76% sequence homology with human serum albumin (HSA). Being one of the most abundant blood plasma proteins, serum albumin forms the first layer of the corona on the NPs surface. To our knowledge, no study has been reported so far on the effect of SiC NPs on the structure and stability of a protein. Therefore, we studied the interaction between SiC USNPs as well as SiC NPs, a slightly larger series of particles, and BSA with various spectroscopic methods. By studying covalent USNPs and the effect of particle size, our results may help in the interpretation of the complex interactions between USNPs and proteins.
Experimental
Pure phosphate buffered saline (PBS) solution was prepared by dissolving PBS powder, pH = 7.4 from Sigma Aldrich, no weigh format, in 1 liter high purity MilliQ (Millipore, Bedford MA, USA) ion-exchanged, deionized water with a resistivity of 18.2 MΩ.BSA lyophilized powder ≥96% (agarose gel electrophoresis, from Sigma-Aldrich) was dissolved in the PBS solution described above for the measurements.
Synthesis of SiC USNPs and NPs
SiC USNPs were prepared by stain etching44 and filtered through a 30 kDa Pall Macrosep filter to reduce the size to below 4 nm.46 SiC NPs with a particle diameter between 4 and 15 nm were prepared by a modified stain etching method.46 We repeatedly etched and sonicated 10 grams of SiC powder four times. The size distribution of the NPs widens with each etching step. SiC NPs from the fourth etch were filtered through a 0.02 μm syringe filter (GE Whatman Anotop 10, Sigma Aldrich). This solution was further separated by using a 30 kDa Pall Macrosep centrifuge filter, but here, the retentate was used and is subsequently referred to as SiC NPs.The size and the crystallinity of SiC USNPs and SiC NPs were determined by high-resolution transmission electron microscopy (HRTEM – JEOL JEM-3010, JEOL Ltd), and atomic force microscopy (AFM – a NeaSNOM AFM instrument, NeaSpec GmbH). The size distribution was assessed by measuring the altitudes of at least 300 nanoparticles with AFM for all samples. The mean size of the SiC USNPs is 1.4 nm, the median is 1.5 nm and the mode is 1.5 nm. The SiC NPs have a mean size of 9.1 nm, the median is 8.6 nm, and the mode is 7.8 nm. Fig. 1a and b show the results of the AFM measurements as well as the HRTEM images of the SiC USNPs and SiC NPs, respectively.
 | ||
| Fig. 1 TEM images and size distributions of (a) SiC USNPs and (b) SiC NPs. (c) ATR-IR spectra of the samples, (d) titration curve of SiC USNPs. | ||
The concentration of SiC in the produced water dispersed samples was determined by high-resolution continuum source graphite furnace atomic absorption spectroscopy (HR-CS-GFAAS) using a ContrAA-700 tandem spectrometer (Analytik Jena AG, Jena, Germany) equipped with a transversally heated graphite tube atomizer (THGA).47 The Si content of the Si USNPs’ stock solution was found to be 0.85 mg ml−1. Assuming that all the Si atoms in the sample are in the form of SiC and from the average diameter of the SiC USNPs, the SiC concentration is 9.7 10−5 mol l−1. The Si content of the Si NPs’ stock solution was 1.05 mg ml−1. The SiC concentration is 4.6 10−6 mol l−1.
The surface termination of the SiC NPs was studied with ATR-IR (attenuated total reflectance infrared) spectroscopy (Bruker Tensor 37, Bruker Co.) with a 1 cm−1 resolution and a DTGS detector. Spectra were recorded in the 700–4000 cm−1 range using a ZnSe multiple reflection ATR crystal. (For measurement and evaluation details, see below). The spectra show that the surface is mostly hydroxyl terminated, with the presence of carboxyl, carbonyl, and ether moieties. There are no differences in the surface termination of the differently sized nanoparticles (Fig. 1c).
Different oxidation states of the surface groups can also be observed by recording the titration curve of the pure solution of the nanocrystal. Comparing the measured and calculated pKa values of the different functional groups in the literature48,49 we conclude that the first inflection point at about pH = 4 corresponds to the deprotonation of the SiC–COOH group, while the second at around pH = 9 corresponds to the deprotonation of the SiC–OH group50 (Fig. 1d).
The optimal pH value chosen for investigating the interaction between SiC and BSA was around 7 where the BSA molecule has the normal conformation45 and the –OH groups on the surface of SiC are in the deprotonated form.
The pH measurements were performed with a Consort C6030 electrochemical analyser.
ATR-IR measurements of the protein systems
ATR-IR spectra of the BSA and BSA/SiC samples were recorded using the setup that was mentioned previously. Aqueous solutions of BSA (10 μM), or a solution of 10 μM BSA and 10 μM SiC, were prepared and 12 ml batches of the solutions were slowly evaporated on one side of the ATR crystal under a stream of nitrogen.51 The formed gel was measured with a 1 cm−1 resolution in the 700–400 cm−1 range in a nitrogen atmosphere at a temperature of 18 ± 3 °C. The absorbance was calculated from the obtained single-beam signals using the spectrum of the blank ATR crystal as reported in ref. 48.One of the main difficulties of studying proteins by infrared spectroscopy is the strong IR absorbance of water with three prominent bands at around 3400 (the O–H stretching), 2125 (water association), and 1645 cm−1 (the H–O–H bending). The amide I vibration of the proteins overlaps directly with the H2O bending vibrational band at 1645 cm−1. In order to get a successful subtraction of absorption bands due to liquid water and water vapor in the atmosphere, a water spectrum was recorded with the same ATR crystal used for protein measurements. Subsequently, the absorbance calculated from the ATR spectrum of the water is subtracted from the BSA or the BSA/SiC spectra by adjusting the subtraction factor in such a way that it gives a straight line between 1750 and 2500 cm−1.52 When the BSA/SiC spectra were studied, the pure SiC spectra were subtracted as well to get the BSA spectra. All spectra were baseline corrected before subtraction.
In the case of quantitative measurements, precautions were taken to avoid known artefacts when recording ATR spectra. These artefacts are caused because the layer adsorbed on the surface of the ATR crystal interacts differently with the incident electromagnetic radiation than the bulk, due to possible symmetry changes and surface selection rules.52 In order to determine the minimum sample quantity where the contribution of the adsorbed layer becomes negligible, we performed the following procedure:
First, the concentration of BSA in the dropped solution was increased from 2 μm to 10 μM; second, sample quantity was increased from 3 ml to 12 ml while the concentration was kept at 10 μM. These measurements were repeated 5 times. Between 6 and 10 μM, the increase of the absorption bands was proportional to the increase of the concentration, showing that the signal of the protein adsorbed onto the ATR crystal surface is negligible compared to the concentration of proteins in the gel.52 Over 8 ml, there were no differences in the amide I/amide II ratios between the measurements, showing that neither surface adsorption nor the orientation influence the measurements.
Ultraviolet-visible (UV-VIS) absorption measurement
A two-beam differential spectrophotometer (Thermo Fisher Scientific, Evo600) was applied for the absorption measurements in the UV and the VIS spectral regions. All the absorption and fluorescence measurements were carried out in a 10 mm quartz cuvette (Hellma 110-QS). The absorbance spectrum of BSA shows two characteristic bands, at around 220 nm and 280 nm, due to the α-helix structure of the protein and the aromatic amino acid residues, respectively. Usually, an alteration in the 220 nm absorbance peak is associated with the perturbation of the secondary structure of the protein, while the changes observed in the 280 nm band indicate that the proximity of the aromatic amino acid residues is altered.30Fluorescence spectroscopy
A HORIBA Jobin-Yvon, NanoLog FL3-2iHR spectrofluorometer with a 450W xenon lamp was used to observe the fluorescence spectra. The time-correlated single photon counting (TCSPC) method was applied to obtain the fluorescence decay curves in the time-resolved measurements. TCSPC measurements were performed with the same NanoLog equipment, but pulsed excitation was achieved by a NanoLED295 laser source with ∼1.0 ns pulse duration at a wavelength of 295 nm. Lifetimes were derived from the fluorescence decay curves by using the least squares fitting deconvolution technique with a two-exponential decay model. The time resolution for the fluorescence lifetime measurements was 55 ps. We applied a Thermo Scientific circulating bath (AC200-A25) to keep the sample at a constant temperature of 15 °C.The fluorescence spectra were corrected to eliminate the decrease of excitation intensity through the sample in the cuvette due to the inner filter effect and to the absorption of the quencher at the excitation wavelength.53 It is well known that the intrinsic fluorescence of BSA is mainly due to the presence of two Trp residues at positions 134 and 212 in domains IB and IIA, respectively. Any changes in the fluorescence quantum yield or the peak position directly indicate changes in the polarity surrounding these two Trp residues due to unfolding of the native conformation. Quenching of fluorescence can occur by non-molecular, intramolecular or intermolecular (dynamic or static quenching) mechanisms54 and many different models already exist for describing and “visualizing” these mechanisms.
Dynamic quenching occurs when the excited fluorophore returns to the ground state due to some quencher related reactions without photon emission. This process appears both in the fluorescence intensity and lifetime. When the complex formation leads to static quenching, the created complex is unable to fluoresce, so it does not affect the fluorescence lifetime.
Results
Absorption measurements
Steady-state absorption measurements were carried out for aqueous solutions of BSA in the presence of SiC USNPs. As SiC USNPs and BSA both show absorption in the 200–300 nm region, their interaction was verified by subtracting the USNP absorption spectrum from the BSA/USNP mixture absorption spectra at the same nanoparticle concentration (Fig. 2a). The BSA absorption band calculated in this way did not show significant changes with a USNP concentration of around 280 nm. Fig. 2b presents the differential spectra of the measured and calculated BSA bands. There are differences, but no significant change or trend can be seen except the increasing discrepancy due to the diffraction of the NPs, indicating that changes in the environment near the tryptophan residue were not clearly observed.Steady-state fluorescence
The characteristic luminescence band of BSA in the samples containing different amounts of SiC and 15 μM BSA was recorded at around 340 nm (Fig. 2c). The characteristic peak due to Trp residue emission at 340 nm55 can be clearly seen. The wavelength of the peak maximum remains fundamentally constant, but the fluorescence intensity decreases with increasing SiC USNP content of the samples, which indicates fluorescence quenching, i.e. USNPs–protein interactions. The lack of spectral shift in the presence of SiC-USNPs indicates negligible changes in the protein secondary structure.Fluorescence lifetime
Samples created for the steady-state measurements were characterized by time-correlated emission experiments as well. A 295 nm excitation wavelength was chosen, and the emission was recorded at 340 nm (Fig. 2d) in order to keep the optical density of the measured samples as low as possible. A decrease of the decay time can be observed with increasing concentration of SiC NPs. The decreasing decay time indicates a dynamic quenching process.Stern–Volmer representation of the quenching process
Both the intensity and the lifetime changes were studied in the Stern–Volmer representation (Fig. 2e). The Stern–Volmer plot of the luminescence quenching constructed from the luminescence intensity shows complex or mixed interactions; if the interaction is either poorly dynamic or static, the Stern–Volmer plot of the intensity gives linear coherence between the quencher concentration and the intensity. However, even if a mixed interaction is the case for quenching, the Stern–Volmer representation of the change in the lifetime still gives linear coherence between the decay times and the quencher concentrations. For SiC USNPs, there is a deviation from the linearity for the lifetime, too.Temperature dependence of the quenching process
The luminescence spectra of the BSA samples containing different amounts of SiC were recorded at different temperatures as well, and the Stern–Volmer plot was constructed to monitor the temperature dependence of the quenching process (Fig. 2f). It can be seen that the shapes of the spectra did not change with temperature, however, there is a decrease in the gradient which is usually explained as a consequence of complex formation and static quenching.56IR measurements
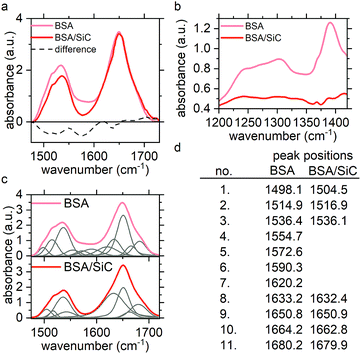
The infrared spectrum of the protein exhibits 5 characteristic bands, two of which are known as amide bands and used to elucidate the secondary structure of a protein. These are most commonly used in infrared protein studies. The amide I band is found between 1600 and 1700 cm−1. It is primarily caused by the stretching vibrations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O coupled weakly with the C–N stretching and N–H bending. The exact band position is determined by the backbone conformation and the hydrogen bonding pattern within the protein molecule. The amide II band occurs at 1500–1600 cm−1 and is mainly derived from the C–N stretching along with the N–H in-plane bending.45
O coupled weakly with the C–N stretching and N–H bending. The exact band position is determined by the backbone conformation and the hydrogen bonding pattern within the protein molecule. The amide II band occurs at 1500–1600 cm−1 and is mainly derived from the C–N stretching along with the N–H in-plane bending.45
The ATR-IR spectra of the BSA film and the SiC-BSA film in the amide I and amide II regions are plotted in Fig. 3a. One can see that that there is no shift in the amide I and amide II peak positions, while peak intensities are partially reduced in the presence of SiC USNPs. Another recognizable difference occurs between 1550 and 1590 cm−1, where the absorption was reduced further in the presence of SiC USNPs. This part of the FTIR spectra can be connected to the tyrosine (Tyr) amino acid as well as amide II. Tyr is one of the strong absorbing amino acids recognizable in the infrared spectrum57 with absorption bands at around 1520 cm−1, 1450 cm−1 and a broad band at around 1250 cm−1. The latter band in the BSA/SiC system experienced a similar decrease, and here the difference is more visible (Fig. 3b). Trp also has a weak band at around 1550 cm−1. However, this band has not been observed for the protein structure.57 These bands are also sensitive to environmental changes. Protein compression also decreases amide II intensity while the amide I band is basically unaltered.58 The amide I and amide II regions in the IR spectrum were fitted with Gaussian/Lorentzian functions (Voigt) to study the differences (Fig. 3c). Eleven peaks were needed to fit the IR spectrum of BSA and 8 for that of BSA/SiC. These can be connected to 8 peaks from the BSA spectrum with similar peak positions but different intensities in some cases, while there are missing peaks between 1550–1600 cm−1. Deconvolution should be considered with extra care because of the previously described manipulation of the spectra, nevertheless, the IR spectra show slight changes in the protein structure that are either stress-related or caused by environmental changes around the Tyr or Trp residues.
Discussion
We found that the results of the various analyses are rather contradictory. Neither the UV-VIS absorbance spectra nor the luminescence measurements imply strong complex formation. The results of IR measurements show little changes in the amide II band, which is sensitive to the side chain vibrations. However, the fluorescence quenching process cannot be described by simple collision quenching. The curve of I0/I shows higher values than τ0/τ (Fig. 2e) which is often explained by both static and dynamic quenching occurring during this process.59 Additionally, the temperature dependence of the quenching is typical of the complexes.To understand the interaction between BSA and SiC USNPs, we first examined the changes in the fluorescence lifetime. The Stern–Volmer plot of the lifetime as a function of SiC concentration shows positive deviation from linearity. If Stern–Volmer and bimolecular quenching constants were calculated from the equation of the collisional quenching, the results would be 0.10 μM−1 and 1.6 1013 M−1 s−1. This bimolecular quenching constant largely exceeds the limit of the diffusion-controlled bimolecular rate constant value (1010 M−1 s−1),60–64 which excludes that a diffusion-mediated collisional quenching causes our dynamic quenching. When both static and dynamic quenching occur, a modified Stern–Volmer equation is suggested,56 and I0/I = (1 + Ks[Q])(1 + KD[Q]) where Ks and KD stand for the static and dynamic quenching constants, respectively, and [Q] is the quencher concentration. By fitting with the second order S–V equation, using correlation coefficients close to unity (R2 = 0.999), the calculated KS and KD values are found to be negative and the mechanism of a ground-state complex is rejected as being responsible for the fluorescence quenching of BSA.
Not only collisional quenching is a dynamic process, however. Quenching can be caused by different mechanisms such as energy transfer and charge transfer.56 As there is a large spectral overlap between BSA emission and SiC NPs absorption (inset in Fig. 4a), the possibility of Förster resonance energy transfer (FRET) quenching should be examined. FRET occurs when the donor fluorophore absorbs a photon, and the excitation energy is transferred to an acceptor molecule close to the donor. FRET efficiency highly depends on the distance between the donor and acceptor molecules. From Förster's theory, the Förster radius (R0) can be calculated. The applied parameter values were κ2 = 2/3, n = 1.34 and ΦD = 0.15.60,65,66 The obtained Förster radius was R0 = 2.83 nm, which is in the interpretable range for the FRET mechanism. However, FRET is usually studied in a system where the donor–acceptor distance is fixed in some way (i.e., by labelling biomolecules). The donor–acceptor distances in an NP–protein solution are constantly changing in time and can only be described by an average distance67,68 which depends on the concentration of the NPs. We assume that – in the presence of acceptors – the ratio of the donors in the donor–acceptor pairs is x(cA) = cDA/cD0, where cDA and cD0 are the concentration of the donor–acceptor pairs and the initial concentration of the donors without any acceptors, respectively, and cA is the acceptor concentration. We further assume that the rest of the donors emit unaltered fluorescence. The efficiency of energy transfer in a single donor–acceptor pair is denoted by E. In this case, the decrease of the fluorescence lifetime of the entire sample can be described by the following equation:
 | (1) |
 | (2) |
We inspected our hypothesis by studying the luminescence intensity of the SiC USNPs as a function of BSA concentration. SiC USNPs have a low emission efficiency at 290 nm excitation. The addition of BSA should increase the luminescence intensity of the SiC USNPs as a result of energy transfer. What we found is that the emission intensity of SiC NPs does not increase with an increase of BSA concentration in the 0.015–3.75 μM BSA concentration range. A further increase in the BSA concentration obscures the SiC peak and deconvolution is needed to extract the emission intensity of the nanoparticles. At higher concentrations, a small increase can be deduced from the deconvolutions, however, the statistical analysis of five replicates gives us only a trend instead of significant correlation, weakening the possibility of energy transfer (not shown).
Conceding FRET, we still have to assume that other processes affect the interaction of the BSA-SiC USNPs. Only luminescence measurements (such as temperature-dependent quenching, which shows a decrease in the quenching efficiency with temperature) are usually connected to complex formation. Here, values of I0/I higher than τ0/τ denote a static quenching process. On the other hand, IR, UV-VIS and the peak position of the Trp emission peak imply the possibility of complex formation. The IR spectrum and the titration curve of the SiC USNPs show hydroxyl moieties on the surface with high concentration. Hydroxyl groups can lead to complex formation where hydrogen bonds are the main interaction force.70 Therefore, we tested the possibility of hydrogen bond-mediated complex formation by using sucrose as a competitive quencher that can protect BSA.71Fig. 4b shows that the addition of sucrose further increases the quenching which proves that SiC USNPs do not interact with BSA via hydrogen bonds.
Because of the lack of detectable binding between BSA and SiC USNPs, we instead consider the transient effect which is a pseudo-static effect in the quenching sphere of action.72 Indeed, deviations from linearity in the Stern–Volmer plots in both the steady and transient states are mostly referred to as a consequence of the transient effects in the Smoluchowski formulation,73 which is usually described by the quenching sphere of action model. Such a description of the fluorescence quenching considers that the quenching reaction itself occurs on a timescale shorter than the one necessary for significant diffusion. The time resolution of the fluorescence lifetime apparatus is not sufficient to detect what is occurring on a certain timescale, in the sub 55 ps range, for instance; then, the phenomena occurring within a distance smaller than the length that results from a random walk of the interacting molecules can be regarded as instantaneous. The whole quenching process is dynamic, but the instrumental limitations enable a separation of static phenomena occurring on timescales below the instrumental resolution and dynamic ones occurring on timescales above the instrumental resolution. Fluorescence quenching with exponential distance dependence is reported when diffusion is hindered.68,74 The modified S–V equation can be written as follows:
 | (3) |
The diffusion-limited association of suspended spherical particles is often described at the mass-action law level, according to Smoluchowski, where the diffusion coefficient is identified with that predicted from the Stokes–Einstein approximation. This expression, however, is valid only for single particles far from interfaces. During contact between particles or with an interface the diffusion coefficient is smaller than that calculated from the Stokes–Einstein equation.75 Moreover, proteins are macromolecules with numerous binding sites, cavities and pockets with sizes as large76 as 102–103 Å3. Molecules are much smaller, so a molecular quencher can diffuse even inside the smallest pocket or pore of the protein and this close proximity to the binding sites facilitates complex formation. Ultrasmall nanoparticles (USNPs) are larger than the molecules but the diffusion of USNPs close to the protein can be influenced by these pockets.57,77,78 Within the protein structure, protein flexibility is an essential factor concerning reactivity. The fluctuation in the protein structure allows diffusion of small molecules or clusters even inside the closed binding sites.79,80 Such a mechanism is able to temporarily trap the molecules and clusters. This mechanism can be described as physical complex formation where the binding force is steric hindrance.81
The energy transfer, however, is also temperature dependent, as R0 and E decrease with increasing temperature.67,68,82 Sucrose increases the viscosity of the medium and the stability of the protein, but changes its conformation making tryptophans more accessible. Sucrose also decreases the polarity of the medium83 and we have to consider sucrose–SiC interactions as well, which makes it difficult to understand the final effect of sucrose on SiC. Nevertheless, the increased viscosity decreases diffusion and increases the efficiency of energy transfer.
BSA contains two Trp around its binding site. Trp134 is outside of the pocket, surrounded by the polar environment; Trp212 is inside the hydrophobic pocket. Even if Trp212 is accessible for SiC USNPs, the hydrophobic pocket temporarily traps the particles causing static-like quenching. Increasing temperature increases the flexibility of the protein and reduces the interaction forces resulting in faster diffusion and a shorter detention time in the vicinity of Trp212. Table 1 presents the lifetimes obtained from the fits of the decay times with three exponentials, where An is the amplitude and τn is the decay time of the components (n is 1, 2, or 3). Even collisional quenching results in non-exponential decays, changing the overall intensity decay rate due to transient effects in quenching. These effects are caused by the rapid quenching of closely spaced donor–acceptor pairs, followed by a slower quenching rate due to quencher diffusion. As a result, the presence of transient effects can manifest itself in additional nanosecond decay time components.56 The fitting procedure yields a decrease in all lifetime components, suggesting the influence of the SiC USNPs on either tryptophan. The shorter decay component of the BSA increases its amplitude with the increase of SiC USNPs concentration, while the decay time slowly decreases. The second BSA decay component behaves in the opposite way. Its amplitude decreases relatively faster than its lifetime. A decrease in the amplitude means less contribution to the luminescence that is caused by static-like quenching, while a decrease in the lifetime is the result of a dynamic type quenching process. It can be seen that both processes affect both components but to different extents. The amplitudes and lifetimes of the third component, that are the result of the transient effect, show small variations with the addition of SiC USNPs to BSA.
| 3 exponential fit | ||||||
|---|---|---|---|---|---|---|
| [SiC] | A 1 | τ 1 | A 2 | τ 2 | A 3 | τ 3 |
| 0 | 1.7 | 4.2 | 3.1 | 7.3 | — | — |
| 0.625 | 0.5 | 1.4 | 3.0 | 5.6 | 1.4 | 8.1 |
| 1.25 | 0.6 | 1.3 | 2.3 | 5.0 | 2.1 | 7.5 |
| 1.875 | 0.8 | 1.0 | 2.6 | 5.2 | 1.8 | 7.8 |
| 2.5 | 0.9 | 1.3 | 2.4 | 5.2 | 1.9 | 7.7 |
| 3.125 | 1.1 | 1.1 | 2.3 | 4.9 | 2.0 | 7.6 |
| 3.75 | 1.3 | 1.1 | 2.0 | 4.6 | 2.2 | 7.4 |
| 5 | 1.7 | 1.0 | 2.1 | 4.6 | 1.9 | 7.4 |
| 6.25 | 2.1 | 1.0 | 2.0 | 4.2 | 1.9 | 7.3 |
| 7.5 | 2.5 | 1.0 | 2.1 | 4.2 | 1.6 | 7.3 |
| 10 | 3.2 | 0.8 | 2.0 | 3.3 | 1.6 | 6.7 |
Our results suggest that SiC USNPs and BSA interact in an inhibited diffusion manner and such a mechanism results in a complex quenching reaction, which shows second-order dynamic and static quenching without perceptible changes in the protein structure. This picture is in agreement with the binding kinetics results for negatively charged ultrasmall gold nanoparticles.84 Indeed, nanoparticles between 1 and 2 nm show transient particle–protein associations with lifetimes shorter than gel band resolution times in gel assays as a consequence of repetitive binding and unbinding processes.31
As Boselli et al. pointed out,31 this transient effect is extremly size dependent. NPs with sizes comparable to or larger than the protein cannot get inside the pockets and large NPs are adsorbed by proteins on the surface forming a dense protein corona (PC). We, therefore, measured the quenching characteristics of SiC NPs with average size of about 9 nm. Here, the particle is comparable but not larger than the size of the BSA. The result is shown in Fig. 5a and b. It can be seen that SiC NPs show a simpler interaction that can be described with a linear Stern–Volmer plot. The slopes of the intensity and the lifetime are almost the same suggesting pure dynamic quenching processes.
 | ||
| Fig. 5 (a) Fluorescence of BSA in the presence of SiC NPs with various concentrations representing the steady state quenching process. (b) Stern–Volmer representation of the quenching processes. | ||
Identification of the binding site
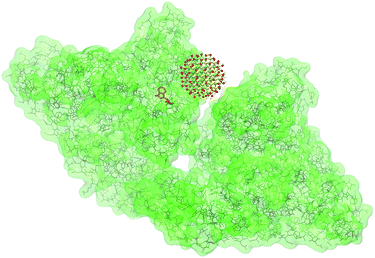
Our experimental results were tested using theoretical calculations in order to see if the modelled NPs–protein interaction gives a consistent solution to the models deduced from the experimental data. Auto Dock Vina85,86 was applied to find the optimal binding site of the SiC nanocrystal to BSA based on the optimization of free energy (calculated from molecular mechanics force fields). Python Molecule Viewer87 was used for the visualization of the result. The calculation was performed on the surface of the three dimensional model of the BSA molecule (3v03) downloaded from the Protein Data Bank. The model of a 1.4 nm SiC nanocrystal was constructed from our ab initio calculations. The binding affinity provided by the optimization process was ΔE = −35.7 kJ M−1. From this, the dissociation constant can be calculated as KDis = 0.33 μM at 288 K (since KDis = eΔE/(RT)). The distance between the tryptophan group of BSA and SiC was found to be approximately 2 nm. This result agrees well with our results obtained from the FRET interpretation. Fig. 6 depicts the constructed BSA–SiC complex.Conclusions
We studied the interaction of 1.5 nm and 9 nm SiC particles, a new candidate in bioimaging and nanoscale sensing, with BSA in aqueous solution by various spectroscopic techniques. Our results strengthen the observation of other researchers for USNPs of a transient effect in the ultrasmall nanoparticle–protein interaction. We explain such an interaction by a disturbance in the free particle diffusion of adjacent particles (ultrasmall nanoparticles and proteins). Otherwise, the mostly hydroxyl terminated SiC particles show no specific interaction with BSA regardless of the size. Only collisional quenching was found for 9 nm SiC NPs, while the best explanation of the 1.5 nm SiC USNPs quenching action, based on our experimental and theoretical results, is FRET.The biocompatibility and stability of SiC with its interesting magneto-optically active centres in the visible and near infrared range make SiC a very promising host material in nanobiotechnology. The lack of evidence for BSA denaturation in the presence of SiC, and the absence of a strong complex or protein corona formation imply in vivo biocompatibility for SiC below 10 nm as well. A functionalized nanoparticle will keep its designed properties in complex biological media with the additional capacity for purposeful engineering of the particle corona and formation of transient complex interactions that lead to a rich range of receptor target interactions, qualitatively different from molecules or larger particles.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the GINOP 2.3.2-15-2016-00022 grant, National Research Development and Innovation Office of Hungary (NKFIH) Grant No. NN118161 (V4-Japan Joint Research Project on Advanced Materials, NamSeN) and Grant No. NN127902 (QuantERA Project Nanospin), Grant No. NVKP_16-1-2016-0152958, and Quantum Technology National Excellence Program (Project No. 2017-1.2.1-NKP-2017-00001). The present scientific contribution is dedicated to the 650th anniversary of the foundation of the University of Pécs, Hungary.References
- T. Terai and T. Nagano, Fluorescent probes for bioimaging applications, Curr. Opin. Chem. Biol., 2008, 12, 515–521 CrossRef CAS PubMed.
- W. C. W. Chan and S. Nie, Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection, Science, 1998, 281, 2016–2018 CrossRef CAS PubMed.
- X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir and S. Weiss, Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics, Science, 2005, 307, 538–544 CrossRef CAS PubMed.
- P. Zrazhevskiy, M. Sena and X. Gao, Designing multifunctional quantum dots for bioimaging, detection, and drug delivery, Chem. Soc. Rev., 2010, 39, 4326–4354 RSC.
- J. Wang, Z. Hu, J. Xu and Y. Zhao, Therapeutic applications of low-toxicity spherical nanocarbon materials, NPG Asia Mater., 2014, 6, e84 CrossRef CAS.
- W. Jiang, X. Wang, O. J. Osborne, Y. Du, C. H. Chang, Y.-P. Liao, B. Sun, J. Jiang, Z. Ji, R. Li, X. Liu, J. Lu, S. Lin, H. Meng, T. Xia and A. E. Nel, Pro-Inflammatory and Pro-Fibrogenic Effects of Ionic and Particulate Arsenide and Indium-Containing Semiconductor Materials in the Murine Lung, ACS Nano, 2017, 11, 1869–1883 CrossRef CAS PubMed.
- M. Auffan, J. Rose, J.-Y. Bottero, G. V. Lowry, J.-P. Jolivet and M. R. Wiesner, Towards a definition of inorganic nanoparticles from an environmental, health and safety perspective, Nat. Nanotechnol., 2009, 4, 634–641 CrossRef CAS PubMed.
- J. C. Y. Kah, Stability and Aggregation Assays of Nanoparticles in Biological Media, Methods Mol. Biol., 2013, 1025, 119–126 CAS.
- J. Piella, N. G. Bastús and V. Puntes, Size-Dependent Protein–Nanoparticle Interactions in Citrate-Stabilized Gold Nanoparticles: The Emergence of the Protein Corona, Bioconjugate Chem., 2017, 28, 88–97 CrossRef CAS PubMed.
- M. Kopp, S. Kollenda and M. Epple, Nanoparticle–Protein Interactions: Therapeutic Approaches and Supramolecular Chemistry, Acc. Chem. Res., 2017, 50, 1383–1390 CrossRef CAS PubMed.
- B. H. Kim, M. J. Hackett, J. Park and T. Hyeon, Synthesis, characterization, and application of ultrasmall nanoparticles, Chem. Mater., 2014, 26, 59–71 CrossRef CAS.
- K. Zarschler, L. Rocks, N. Licciardello, L. Boselli, E. Polo, K. P. Garcia, L. De Cola, H. Stephan and K. A. Dawson, Ultrasmall inorganic nanoparticles: state-of-the-art and perspectives for biomedical applications, Nanomedicine Nanotechnology, Biol. Med., 2016, 12, 1663–1701 CAS.
- I. I. Vlasov, A. A. Shiryaev, T. Rendler, S. Steinert, S.-Y. Lee, D. Antonov, M. Vörös, F. Jelezko, A. V. Fisenko, L. F. Semjonova, J. Biskupek, U. Kaiser, O. I. Lebedev, I. Sildos, P. R. Hemmer, V. I. Konov, A. Gali and J. Wrachtrup, Molecular-sized fluorescent nanodiamonds, Nat. Nanotechnol., 2014, 9, 54–58 CrossRef CAS PubMed.
- K. Araki, M. Uchiyama, S. Toma, S. Rodrigues, A. Shimada, R. Loiola, H. Rodriguez, P. Oliveira, M. Luz, S. Rabbani, H. Toma and S. H. P. Farsky, Ultrasmall cationic superparamagnetic iron oxide nanoparticles as nontoxic and efficient MRI contrast agent and magnetic-targeting tool, Int. J. Nanomed., 2015, 4731 CrossRef PubMed.
- J. Gao, K. Chen, R. Xie, J. Xie, S. Lee, Z. Cheng, X. Peng and X. Chen, Ultrasmall near-infrared non-cadmium quantum dots for in vivo tumor imaging, Small, 2010, 6, 256–261 CrossRef CAS PubMed.
- L. Shang and G. U. Nienhaus, Small fluorescent nanoparticles at the nano-bio interface, Mater. Today, 2013, 16, 58–66 CrossRef CAS.
- L. Shang, R. M. Dörlich, V. Trouillet, M. Bruns and G. U. Nienhaus, Ultrasmall fluorescent silver nanoclusters: protein adsorption and its effects on cellular responses, Nano Res., 2012, 5, 531–542 CrossRef CAS.
- L. Shang, S. Brandholt, F. Stockmar, V. Trouillet, M. Bruns and G. U. Nienhaus, Effect of protein adsorption on the fluorescence of ultrasmall gold nanoclusters, Small, 2012, 8, 661–665 CrossRef CAS PubMed.
- H. C. Fischer, L. Liu, K. S. Pang and W. C. W. Chan, Pharmacokinetics of Nanoscale Quantum Dots: In Vivo Distribution, Sequestration, and Clearance in the Rat, Adv. Funct. Mater., 2006, 16, 1299–1305 CrossRef CAS.
- H. S. Choi, W. Liu, P. Misra, E. Tanaka, J. P. Zimmer, B. Itty Ipe, M. G. Bawendi and J. V. Frangioni, Renal clearance of quantum dots, Nat. Biotechnol., 2007, 25, 1165–1170 CrossRef CAS PubMed.
- X. D. Zhang, J. Yang, S. S. Song, W. Long, J. Chen, X. Shen, H. Wang, Y. M. Sun, P. X. Liu and S. Fan, Passing through the renal clearance barrier: toward ultrasmall sizes with stable ligands for potential clinical applications, Int. J. Nanomed., 2014, 9, 2069–2072 CrossRef PubMed.
- G. Zhang, Z. Yang, W. Lu, R. Zhang, Q. Huang, M. Tian, L. Li, D. Liang and C. Li, Influence of anchoring ligands and particle size on the colloidal stability and in vivo biodistribution of polyethylene glycol-coated gold nanoparticles in tumor-xenografted mice, Biomaterials, 2009, 30, 1928–1936 CrossRef CAS PubMed.
- N. Goswami, K. Zheng and J. Xie, Bio-NCs – the marriage of ultrasmall metal nanoclusters with biomolecules, Nanoscale, 2014, 6, 13328–13347 RSC.
- M. M. Yin, P. Dong, W. Q. Chen, S. P. Xu, L. Y. Yang, F. L. Jiang and Y. Liu, Thermodynamics and Mechanisms of the Interactions between Ultrasmall Fluorescent Gold Nanoclusters and Human Serum Albumin, γ-Globulins, and Transferrin: A Spectroscopic Approach, Langmuir, 2017, 33, 5108–5116 CrossRef CAS PubMed.
- J. Hou, D. M. Szaflarski and J. D. Simon, Quantifying the association constant and stoichiometry of the complexation between colloidal polyacrylate-coated gold nanoparticles and chymotrypsin, J. Phys. Chem. B, 2013, 117, 4587–4593 CrossRef CAS PubMed.
- Z. Q. Xu, Q. Q. Yang, J. Y. Lan, J. Q. Zhang, W. Peng, J. C. Jin, F. L. Jiang and Y. Liu, Interactions between carbon nanodots with human serum albumin and γ-globulins: the effects on the transportation function, J. Hazard. Mater., 2016, 301, 242–249 CrossRef CAS PubMed.
- J. Zhang, M. Zheng and Z. Xie, Co-assembled hybrids of proteins and carbon dots for intracellular protein delivery, J. Mater. Chem. B, 2016, 4, 5659–5663 RSC.
- V. Poderys, M. Matulionyte, A. Selskis and R. Rotomskis, Interaction of Water-Soluble CdTe Quantum Dots with Bovine Serum Albumin, Nanoscale Res. Lett., 2010, 6, 9 Search PubMed.
- S. Chatterjee and T. K. Mukherjee, Spectroscopic investigation of interaction between bovine serum albumin and amine-functionalized silicon quantum dots, Phys. Chem. Chem. Phys., 2014, 16, 8400 RSC.
- D. M. Togashi, A. G. Ryder, D. M. Mahon, P. Dunne, J. E. D. S. D. McManus and M. Fitzmaurice, Fluorescence study of Bovine Serum Albumin and Ti and Sn Oxide Nanoparticles Interactions, Diagn. Opt. Spectrosc. Biomed. IV, 2007, 6628, 6628_61 Search PubMed.
- L. Boselli, E. Polo, V. Castagnola and K. A. Dawson, Regimes of Biomolecular Ultrasmall Nanoparticle Interactions, Angew. Chem., Int. Ed., 2017, 56, 4215–4218 CrossRef CAS PubMed.
- C. Torres-Raya, D. Hernandez-Maldonado, J. Ramirez-Rico, C. Garcia-Gañan, A. R. de Arellano-Lopez and J. Martinez-Fernandez, Fabrication, chemical etching, and compressive strength of porous biomimetic SiC for medical implants, J. Mater. Res., 2008, 23, 3247–3254 CrossRef CAS.
- J. Botsoa, V. Lysenko, A. Géloën, O. Marty, J. M. Bluet and G. Guillot, Application of 3C-SiC quantum dots for living cell imaging, Appl. Phys. Lett., 2008, 92, 173902 CrossRef.
- J. Fan, H. Li, J. Jiang, L. K. Y. So, Y. W. Lam and P. K. Chu, 3C-SiC nanocrystals as fluorescent biological labels, Small, 2008, 4, 1058–1062 CrossRef CAS PubMed.
- S. Castelletto, B. Johnson, C. Zachreson, D. Beke, I. Balogh, T. Ohshima, I. Aharonovich and A. Gali, Room Temperature Quantum Emission from Cubic Silicon Carbide Nanoparticles, ACS Nano, 2014, 8, 7938–7947 CrossRef CAS PubMed.
- S. Castelletto, B. C. Johnson, V. Ivády, N. Stavrias, T. Umeda, A. Gali and T. Ohshima, A silicon carbide room-temperature single-photon source, Nat. Mater., 2013, 13, 151–156 CrossRef PubMed.
- D. J. Christle, A. L. Falk, P. Andrich, P. V. Klimov, J. U. Hassan, N. T. Son, E. Janzén, T. Ohshima and D. D. Awschalom, Isolated electron spins in silicon carbide with millisecond coherence times, Nat. Mater., 2015, 14, 160–163 CrossRef CAS PubMed.
- M. Widmann, S.-Y. Lee, T. Rendler, N. T. Son, H. Fedder, S. Paik, L.-P. Yang, N. Zhao, S. Yang, I. Booker, A. Denisenko, M. Jamali, S. A. Momenzadeh, I. Gerhardt, T. Ohshima, A. Gali, E. Janzén and J. Wrachtrup, Coherent control of single spins in silicon carbide at room temperature, Nat. Mater., 2015, 14, 164–168 CrossRef CAS PubMed.
- P. Deák, B. Aradi, M. Kaviani, T. Frauenheim and A. Gali, Formation of NV centers in diamond: a theoretical study based on calculated transitions and migration of nitrogen and vacancy related defects, Phys. Rev. B: Condens. Matter Mater. Phys., 2014, 89, 75203 CrossRef.
- A. Muzha, F. Fuchs, N. V. Tarakina, D. Simin, M. Trupke, V. A. Soltamov, E. N. Mokhov, P. G. Baranov, V. Dyakonov, A. Krueger and G. V. Astakhov, Room-temperature near-infrared silicon carbide nanocrystalline emitters based on optically aligned spin defects, Appl. Phys. Lett., 2014, 105, 243112 CrossRef.
- B. Somogyi and A. Gali, Computational design of in vivo biomarkers, J. Phys.: Condens. Matter, 2014, 26, 143202 CrossRef PubMed.
- B. Somogyi, V. Zólyomi and A. Gali, Near-infrared luminescent cubic silicon carbide nanocrystals for in vivo biomarker applications: an ab initio study, Nanoscale, 2012, 4, 7720–7726 RSC.
- X. L. Wu, J. Y. Fan, T. Qiu, X. Yang, G. G. Siu and P. K. Chu, Experimental evidence for the quantum confinement effect in 3C-SiC nanocrystallites, Phys. Rev. Lett., 2005, 94, 26102 CrossRef CAS PubMed.
- D. Beke, Z. Szekrényes, I. Balogh, Z. Czigány, K. Kamarás and A. Gali, Preparation of small silicon carbide quantum dots by wet chemical etching, J. Mater. Res., 2013, 28, 44–49 CrossRef CAS.
- T. Peters, All about albumin: biochemistry, genetics, and medical applications, Academic Press, 1996 Search PubMed.
- D. Beke, Z. Szekrényes, Z. Czigány, K. Kamarás and Á. Gali, Dominant luminescence is not due to quantum confinement in molecular-sized silicon carbide nanocrystals, Nanoscale, 2015, 7, 10982–10988 RSC.
- G. Dravecz, L. Bencs, D. Beke and A. Gali, Determination of silicon and aluminum in silicon carbide nanocrystals by high-resolution continuum source graphite furnace atomic absorption spectrometry, Talanta, 2016, 147, 271–275 CrossRef CAS PubMed.
- D. D. Perrin, B. Dempsey and E. P. Serjeant, PKa prediction for organic acids and bases, Springer, 1981 Search PubMed.
- K. Leung, I. M. B. Nielsen and L. J. Criscenti, Elucidating Acid-base Behavior of Silica with ab initio Molecular Dynamics, Nanosci. Nanotechnol., 1992, 56–57 Search PubMed.
- D. Beke, T. Z. Jánosi, B. Somogyi, D. Á. Major, Z. Szekrényes, J. Erostyák, K. Kamarás and A. Gali, Identification of Luminescence Centers in Molecular-Sized Silicon Carbide Nanocrystals, J. Phys. Chem. C, 2016, 120, 685–691 CAS.
- E. Goormaghtigh, R. Gasper, A. Bénard, A. Goldsztein and V. Raussens, Protein secondary structure content in solution, films and tissues: redundancy and complementarity of the information content in circular dichroism, transmission and ATR FTIR spectra, Biochim. Biophys. Acta, Proteins Proteomics, 1794, 2009, 1332–1343 Search PubMed.
- M. E. Goldberg and A. F. Chaffotte, Undistorted structural analysis of soluble proteins by attenuated total reflectance infrared spectroscopy, Protein Sci., 2005, 14, 2781–2792 CrossRef CAS PubMed.
- T. Z. Janosi, J. Korppi-Tommola, Z. Csok, L. Kollar, P. Myllyperkio and J. Erostyak, Anthracene Fluorescence Quenching by a Tetrakis (Ketocarboxamide) Cavitand, J. Spectrosc., 2014, 2014, 1–8 CrossRef.
- V. Biju, Y. Makita, A. Sonoda, H. Yokoyama, Y. Baba and M. Ishikawa, Temperature-sensitive photoluminescence of CdSe quantum dot clusters, J. Phys. Chem. B, 2005, 109, 13899–13905 CrossRef CAS PubMed.
- N. Tayeh, T. Rungassamy and J. R. Albani, Fluorescence spectral resolution of tryptophan residues in bovine and human serum albumins, J. Pharm. Biomed. Anal., 2009, 50, 107–116 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer US, Boston, MA, 2006 Search PubMed.
- A. Barth, The infrared absorption of amino acid side chains, Prog. Biophys. Mol. Biol., 2000, 74, 141–173 CrossRef CAS PubMed.
- R. I. Litvinov, D. A. Faizullin, Y. F. Zuev and J. W. Weisel, The α-helix to β-sheet transition in stretched and compressed hydrated fibrin clots, Biophys. J., 2012, 103, 1020–1027 CrossRef CAS PubMed.
- J. Keizer, Nonlinear fluorescence quenching and the origin of positive curvature in Stern-Volmer plots, J. Am. Chem. Soc., 1983, 105, 1494–1498 CrossRef CAS.
- Y.-Q. Wang, H.-M. Zhang, G.-C. Zhang, W.-H. Tao, Z.-H. Fei and Z.-T. Liu, Spectroscopic studies on the interaction between silicotungstic acid and bovine serum albumin, J. Pharm. Biomed. Anal., 2007, 43, 1869–1875 CrossRef CAS PubMed.
- A. Papadopoulou, R. J. Green and R. Frazier, Interaction of Flavonoids with Bovine Serum Albumin: A Fluorescence Quenching Study, J. Agric. Food Chem., 2005, 53, 158–163 CrossRef CAS PubMed.
- D. Wu, Z. Chen and X. Liu, Study of the interaction between bovine serum albumin and ZnS quantum dots with spectroscopic techniques, Spectrochim. Acta, Part A, 2011, 84, 178–183 CrossRef CAS PubMed.
- X. Shi, D. Li, J. Xie, S. Wang, Z. Wu and H. Chen, Spectroscopic investigation of the interactions between gold nanoparticles and bovine serum albumin, Chin. Sci. Bull., 2012, 57, 1109–1115 CrossRef CAS.
- D. Li, T. Zhang, C. Xu and B. Ji, Effect of pH on the interaction of vitamin B12 with bovine serum albumin by spectroscopic approaches, Spectrochim. Acta, Part A, 2011, 83, 598–608 CrossRef CAS PubMed.
- T. Sen, K. K. Haldar and A. Patra, Au nanoparticle-based surface energy transfer probe for conformational changes of BSA protein, J. Phys. Chem. C, 2008, 112, 17945–17951 CAS.
- A. Satheshkumar and K. P. Elango, Spectroscopic and molecular docking studies on the charge transfer complex of bovine serum albumin with quinone in aqueous medium and its influence on the ligand binding property of the protein, Spectrochim. Acta, Part A, 2014, 130, 337–343 CrossRef CAS PubMed.
- B. Wallace and P. J. Atzberger, Förster resonance energy transfer: role of diffusion of fluorophore orientation and separation in observed shifts of FRET efficiency, PLoS One, 2017, 12, 1–22 Search PubMed.
- M. Rae, A. Fedorov and M. N. Berberan-Santos, Fluorescence quenching with exponential distance dependence: Application to the external heavy-atom effect, J. Chem. Phys., 2003, 119, 2223–2231 CrossRef CAS.
- M. Lunz, A. L. Bradley, V. A. Gerard, S. J. Byrne, Y. K. Gun’Ko, V. Lesnyak and N. Gaponik, Concentration dependence of Förster resonant energy transfer between donor and acceptor nanocrystal quantum dot layers: effect of donor–donor interactions, Phys. Rev. B: Condens. Matter Mater. Phys., 2011, 83, 1–10 CrossRef.
- C. Dufour and O. Dangles, Comparative Binding Affinities of Flavonoid Phytochemicals with Bovine Serum Albumin, Iran. J. Pharm. Res., 2014, 13, 1019 Search PubMed.
- J.-H. Shi, Y.-Y. Lou, K.-L. Zhou and D.-Q. Pan, Elucidation of intermolecular interaction of bovine serum albumin with Fenhexamid: a biophysical prospect, J. Photochem. Photobiol., B, 2018, 180, 125–133 CrossRef CAS PubMed.
- H. Zeng and D. Gilles, Analysis of fluorescence quenching in some antioxidants from non-linear Stern-Volmer plots, J. Lumin., 1995, 63, 75–84 CrossRef CAS.
- M. A. R. Castanho and M. J. Prieto, Fluorescence quenching data interpretation in biological systems, Biochim. Biophys. Acta, Biomembr., 1998, 1373, 1–16 CrossRef CAS.
- M. H. Gehlen, Spectral analysis of the fluorescence quenching kinetics in micelles with probe migration, Chem. Phys., 1997, 224, 275–279 CrossRef CAS.
- V. P. Zhdanov, Note: The effect of viscosity on the rate of diffusion-limited association of nanoparticles, J. Chem. Phys., 2015, 143, 63–65 CrossRef PubMed.
- J. Liang, H. Edelsbrunner and C. Woodward, Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design, Protein Sci., 1998, 7, 1884–1897 CrossRef CAS PubMed.
- Y. Wang, C. Li and G. J. Pielak, Effects of Proteins on Protein Diffusion, J. Am. Chem. Soc., 2010, 132, 9392–9397 CrossRef CAS PubMed.
- I. Kohli and A. Mukhopadhyay, Diffusion of Nanoparticles in Semidilute Polymer Solutions: Effect of Different Length Scales, Macromolecules, 2012, 45, 6143–6149 CrossRef CAS.
- C.-H. Tang and L. Shen, Role of Conformational Flexibility in the Emulsifying Properties of Bovine Serum Albumin, J. Agric. Food Chem., 2013, 61, 3097–3110 CrossRef CAS PubMed.
- J. Mukherjee and M. N. Gupta, Increasing importance of protein flexibility in designing biocatalytic processes, Biotechnol. Rep., 2015, 6, 119–123 CrossRef PubMed.
- R. Elber, Ligand diffusion in globins: simulations versus experiment, Curr. Opin. Struct. Biol., 2010, 20, 162–167 CrossRef CAS PubMed.
- L. Momeni, S. Mahmodian, B. Shareghi, A. A. Saboury and S. Farhadian, The functional and structural stabilization of trypsin by sucrose, Int. J. Biol. Macromol., 2017, 99, 343–349 CrossRef CAS PubMed.
- C. G. Malmberg and A. A. Maryott, Dielectric constants of aqueous solutions of dextrose and sucrose, J. Res. Natl. Bur. Stand., 1950, 45, 299 CrossRef CAS.
- A. L. Lira, R. S. Ferreira, R. J. S. Torquato, H. Zhao, M. L. V. Oliva, S. A. Hassan, P. Schuck and A. A. Sousa, Binding kinetics of ultrasmall gold nanoparticles with proteins, Nanoscale, 2018, 10, 3235–3244 RSC.
- D. S. Goodsell and A. J. Olson, Automated docking of substrates to proteins by simulated annealing, Proteins: Struct., Funct., Genet., 1990, 8, 195–202 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31, 455–461 CAS.
- M. F. Sanner, Python: a programming language for software integration and development, J. Mol. Graphics Modell., 1999, 17, 57–61 CAS.
| This journal is © the Owner Societies 2018 |