Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Lysine-based amino-functionalized lipids for gene transfection: the influence of the chain composition on 2D properties†
Stephanie
Tassler
a,
Dorota
Pawlowska
a,
Christopher
Janich
b,
Bodo
Dobner
b,
Christian
Wölk
 b and
Gerald
Brezesinski
b and
Gerald
Brezesinski
 *a
*a
aMax Planck Institute of Colloids and Interfaces, Science Park Potsdam-Golm, Am Mühlenberg 1, 14476 Potsdam, Germany. E-mail: brezesinski@mpikg.mpg.de
bMartin-Luther-University (MLU) Halle-Wittenberg, Institute of Pharmacy, Wolfgang-Langenbeck-Straße 4, 06120 Halle (Saale), Germany
First published on 7th February 2018
Abstract
The influence of the chain composition on the physical–chemical properties will be discussed for five transfection lipids containing the same lysine-based head group. For this purpose, the chain composition will be gradually varied from saturated tetradecyl (C14:0) and hexadecyl (C16:0) chains to longer but unsaturated oleyl (C18:1) chains with double bonds in the cis configuration. In this work, we investigated the lipids as Langmuir monolayers at the air–water-interface in the absence and presence of calf thymus DNA applying different techniques such as infrared reflection absorption spectroscopy (IRRAS) and grazing incidence X-ray diffraction (GIXD). The replacement of saturated tetradecyl (C14:0) and hexadecyl (C16:0) chains by unsaturated oleyl (C18:1) chains increases the fluidity of the lipid monolayer: TH10 < TT10 < OH10 < OT10 < OO10 resulting in a smaller packing density. TH10 forms the stiffest and OO10 the most fluid monolayer in this structure–property study. OO10 has a higher protonation degree compared to the saturated lipids TT10 and TH10 as well as to the hybrids OT10 and OH10 because of a better accessibility of the amine groups. Depending on the bulk pH, different scenarios of DNA coupling to the lipid monolayers have been proposed.
1. Introduction
Gene therapy is a promising tool to cure monogenetic diseases like cystic fibrosis and sickle-cell anemia.1,2 Gene therapy stands for the insertion of missing or the replacement of defect genes by healthy ones.2 Current definitions also include the application of siRNA (gene silencing) or the gene editing strategy (CRISPR-Cas9).3,4 These concepts require efficient delivery of polynucleotides into biological tissue or cells – the so called gene transfection.5 Due to nuclease degradation it is impossible for pure DNA/RNA to penetrate the cell membrane without physical methods like needle injection or gene gun, therefore, specific carriers are needed. Their main challenges are to transport the DNA through the bloodstream, to cross cell membranes and to efficiently release the genetic material near by the cell nucleus. Particularly, viral vector systems reach a higher efficiency than physical methods and non-viral transfer vehicles since they were evolved with the express purpose of infecting mammalian cells with foreign genetic materials. Despite their high transfection efficiency, viral systems have also disadvantages: retroviral systems have an oncogenic potential and all viral systems can cause immune responses to a greater or lesser extent.6 An alternative approach is the non-viral gene transfer, which is realised by non-viral vectors like cationic lipids. They are anyway easier and cheaper to produce and can be much better controlled.7Besides structural parameters such as the head group, the spacer and the backbone, the chain composition is crucial for the formation of liposomes and lipoplexes, and finally for the cellular uptake. The structure of the hydrophobic domain determines the phase transition temperature and the fluidity of the bilayer, further it influences the stability of liposomes, the DNA protection from nucleases, the endosomal escape, the DNA release from the lipoplex and the nuclear penetration. Especially the number and the length of aliphatic chains play a crucial role. While single-chain lipids tend to form micelles, they transfect poorly compared to their double- and triple-chain derivatives.8,9 Furthermore, the length of the aliphatic chain is important and determines, together with the size of the head group and the nature of the backbone and spacer, the overall molecule shape. There are many different opinions in the literature considering the minimum chain lengths, which is required for gene transfection (C10,10 C1211,12 or C1413). Aliphatic chains varying from C5 to C25 have been investigated, with the conclusion that the transfection efficiency is not a linear function of the length. A few groups are convinced that C14 is the best choice for efficient transfection, but obviously not for all lipid composites.12–14 For gemini lipids C18 turned out to be a suitable chain length.15,16 Further, the toxicity of lipids is influenced by the hydrophobic domain.17 Generally, it was found that the cytotoxicity increases with decreasing chain length. Shorter-chain lipids have lower phase transition temperatures, which prohibits them from developing stable liposomes.13,18 In contrast, lipids with longer aliphatic chains increase the rigidity of bilayers, which is necessary to form stable liposomes.11 In the liquid-crystalline state, longer saturated chains can be very flexible (gauche conformation) due to rotational freedom of each single bond.19 However, under physiological conditions they are usually in the gel state with chain segments in all-trans conformation. Except a few authors,20,21 most groups report that unsaturated C18:1 chains are frequently the best choice for good transfection.22–25 Oleyl chains can promote the endosomal escape due to the enhancement of membrane fluidity of the transfection complexes.10 Furthermore, they allow strong anchoring in the membrane and have a good miscibility with unsaturated helper lipids like DOPC and DOPE.26
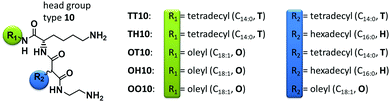
The effect of asymmetry is another variable influencing the transfection efficiency of cationic lipids. There are two possibilities to introduce asymmetry. One way is to choose a helper lipid with a chain length different from the cationic lipid or to introduce a mismatch in the cationic lipid itself by using different chains. For the first scenario it was found that a high level of asymmetry (C18:1 cationic lipid, C12:0 helper lipid) as well as high symmetry (C16:0 cationic lipid, C16:0 helper lipid) results in high transfection rates.27 In the case of the asymmetry being based on a mismatch within the cationic lipid itself, no clear trend was described in the literature. Some research groups claim that asymmetry enhances the transfection efficiency28–30 but a few groups report contrary findings.31,32 Our research goal is to correlate the structure of malonic acid diamide lipids with the physical–chemical properties and the interaction with DNA. Such data can be used for future correlations with the biological activity.33 To enable a systematic study, lipids with different chain patterns have been synthesized,34–36 namely TT (C14:0, C14:0), TH (C14:0, C16:0), OT (C18:1, C14:0), OH (C18:1, C16:0) and OO (C18:1, C18:1). In this paper, the physical–chemical properties of lipids with the head group type 10 (see Fig. 1) and the above described alkyl chain patterns have been examined in 2D model systems (Langmuir monolayers). The focus is set on the lipid self-assembling as well as the complex formation with DNA. Consequently, this article gives information on how to adjust physical–chemical parameters of transfection lipids by variation of the alkyl chain patterns to influence the coupling of DNA.
2. Materials
2.1. Experimental details
For the monolayer experiments, 1 mM stock solutions of TT10, TH10, OT10, OH10 and OO10 were prepared in chloroform:methanol (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v
2 v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) (CHCl3: J. T. Baker, Netherlands; stabilized with 0.75% of ethanol; CH3OH: Merck, Germany; purity >99.9%). The synthesis and the analytical data of TT10, TH10, OT10 and OH1035 as well as OO1037 have already been described.
v) (CHCl3: J. T. Baker, Netherlands; stabilized with 0.75% of ethanol; CH3OH: Merck, Germany; purity >99.9%). The synthesis and the analytical data of TT10, TH10, OT10 and OH1035 as well as OO1037 have already been described.
The lipids containing an oleyl chain have been synthesized from oleylamine with technical grade.38
Milli-Q Millipore water with a specific resistance of 18.2 MΩ cm was taken for all measurements and sample preparations. All chemicals used for the preparation of the bromide ion based buffers were purchased from Sigma Aldrich. The buffer had a constant concentration of 2 mM bromide anions. The pH 3 was adjusted by taking 1,4-diazabicyclo(2,2,2)octane (pKa1 = 4.2, pKa2 = 8.2), and pH 10 was obtained by using piperazine (pKa1 = 5.7, pKa2 = 9.8). The deoxyribonucleic acid sodium salt from calf thymus was purchased from Sigma Aldrich (Type 1, CAS: 73049-39-5). A stock solution of 1 mM ct-DNA was prepared freshly in 1 mM NaCl solution by gently stirring in the fridge (5 °C) overnight. The molar mass of DNA refers to a monomer containing one charge per phosphate moiety with 10% of hydration, therefore, M ∼ 370 g mol−1.39 The DNA solutions were diluted with water or the corresponding bromide ion based buffer to a concentration of 0.1 mM directly before the experiments.
2.2. Structural properties
TT10, TH10, OT10, OH10 and OO10 are lysine-based amino-functionalised peptide-mimicing lipids, which were synthesised as non-viral carriers for gene transfection (Fig. 1). The hydrophobic lipid domain changes gradually from short and saturated tetradecyl (T = C14:0) to longer saturated hexadecyl (H = C16:0) and finally to unsaturated oleyl (O = C18:1) chains.The oleyl chain was introduced for two reasons. First, because it is known for a high transfection efficiency, and second, it ensures a sufficient miscibility with the commonly used helper lipid DOPE.
The malonic acid/lysine backbone of TT10, TH10, OT10, OH10 and OO10 is biocompatible and ensures low cytotoxicity resulting in a low immune response. Due to this backbone, the lipids exhibit a peptide-like character. Head group 10 has a linear ethylene diamine spacer and two primary amine groups (Fig. 1). The pKa value increases from 5 to 5.6 for TH10 to OO10, respectively, indicating that their readiness to act as proton donors decreases with increasing chain fluidity.37 This can also influence the interaction with DNA.
3. Methods
3.1. Langmuir film balance
The lipid monolayers were examined on a computer-interfaced Langmuir trough equipped with a Wilhelmy balance to measure the surface tension with an accuracy of ±0.1 mN m−1. A connected thermostat was kept at 20 °C with a precision of ±0.1 °C. The lipid monolayers were compressed using a moveable barrier with a velocity of 5 Å2 (molecule min)−1 to a surface pressure below 45 mN m−1 to avoid the layer collapse. Afterwards, the lipid monolayers were expanded with the same velocity to record the hysteresis. Compression and expansion isotherms are very similar for all lipids investigated. The 1 mM lipid solution was spread carefully from a micro-syringe (Hamilton, Switzerland) onto the aqueous subphases. Before compression, 10 min were given for the evaporation of the organic solvent. In the case of DNA adsorbing to the lipid monolayer, 60 min were given. All isotherms were measured at least twice for reproducibility.3.2. Infrared reflection absorption spectroscopy
The infrared reflection absorption spectra were collected on a Vertex 70 FT-IR spectrometer (Bruker Optics, Ettlingen, Germany). The set-up includes a film balance (R&K, Potsdam, Germany) located inside an enclosed container (external air–water-reflection unit XA-511, Bruker). A sample trough with two movable barriers and a reference trough (only water or buffer) allow the fast recording of the sample and reference spectra using a shuttle technique. The infrared beam is focused on the liquid surface by a set of mirrors. The IR-beam angle of incidence (ϕ) normal to the surface has been adjusted to be 40° moveable arms. A KRS-5 wire grid polarizer is used to polarize the infrared radiation either in the parallel (p) or perpendicular (s) direction. After reflection from the surface, the beam is directed to a narrow band Mercury Cadmium Telluride (MCT) detector cooled with liquid nitrogen. The final reflectance–absorbance spectra were obtained using −log(R/R0), with R being the reflectance of the film covered surface and R0 being the reflectance of the pure subphase. For each single beam spectrum 200 scans (s-polarized light) or 400 scans (p-polarized light) were added with a scanning velocity of 20 kHz and a resolution of 8 cm−1, apodized using the Blackman–Harris 3-term function, and fast Fourier transformed after one level of zero filling.40–42 All spectra were corrected for atmospheric interference using the OPUS software and baseline corrected applying the spectra-subtraction software. The spectra are not smoothed.43 The Lorentzian fit maximum of the antisymmetric CH2-band (±0.2 cm−1) was taken to determine the lipid phase state, while the intensity of the antisymmetric PO2− band was used to estimate the amount of DNA coupled to the monolayers of TH10 and OO10.3.3. Grazing incidence X-ray diffraction
The grazing incidence X-ray diffraction patterns were recorded using the liquid surface diffractometer at the undulator beamline BW1 in the HASYLAB (DESY, Hamburg, Germany).44 A Langmuir film microbalance (R&K, Potsdam, Germany) with a glass block inside in order to dampen the mechanically excited surface wave was set on an anti-vibration table. The trough is in an air-tight container with a Kapton (polyimide film) window and under a helium atmosphere. A connected thermostat was kept at constant temperature during the measurements. To avoid radiation damage on the sample, the trough was laterally moved so that the strong X-ray beam (9 keV) hits different sample spots. A beryllium (002) crystal is used as a monochromator to obtain an X-ray beam with a wavelength of 1.304 Å. To gain maximal surface sensitivity the incident angle of the synchrotron beam is adjusted to 0.1° (85% of the critical angle αC) in order to reach total external reflection. The reflected beam travels as an evanescence wave along the interface and decays exponentially in the medium. The experimental details are described elsewhere.45–494. Results and discussion
The cationic lipids TT10, TH10, OT10, OH10 and OO10 have been investigated as lipid monolayers on different subphases in the absence and presence of calf thymus DNA at 20 °C.4.1. 2D phase behaviour of pure lipids
The π–A-isotherm of the TT10, TH10, OT10, OH10 and OO10 Langmuir monolayers on water at 20 °C are presented in Fig. 2A. Water saturated with CO2 has a pH value of approximately 5.8, therefore, the lipids are less charged than on the pH 3 buffer and exhibit weaker electrostatic repulsions. Hence, they have lower LE–LC transition pressures. The saturated lipids TT10 and TH10 are in the liquid-condensed phase state already at close to zero pressures (re-sublimation). Their π–A-isotherms are almost identical. The hybrid OH10 is in the LC phase as well, but the molecular area of OH10 is larger compared with TT10 and TH10. OT10 and OO10 are in the liquid-expanded phase state, but OT10 seems to be in a coexistence between LE and LC of 5 mN m−1 and 40 mN m−1. OO10 has the largest molecular area and is in the LE state.
On the pH 10 buffer (Fig. S1B, ESI†), the lipids are completely unprotonated. Only OO10 is in the LE phase state, while the saturated TH10 and the hybrid OH10 are in the LC phase state. Because of the saturated tetradecyl chain TH10 achieves a tighter packing and occupies a less area than the oleyl chain containing OH10.
4.2. 2D structure of pure lipids
Additionally, GIXD has been performed using TT10, TH10 and OH10 monolayers spread on water at 20 °C and 30 mN m−1 (Fig. 3A–C) and OT10 at 5 °C and 30 mN m−1 (Fig. 3D). 30 mN m−1 is the assumed lateral pressure in a cell membrane.50 Since OO10 is in the LE phase state with chains in the gauche conformation, the GIXD method was not applied. Fluid chains do not order and cause only a diffuse scattering, which results in a broad halo.For the lipid TT10, three Bragg peaks can be correlated with an oblique chain lattice, namely Qxy = 1.19 Å−1, Qxy = 1.34 Å−1 and Qxy = 1.53 Å−1. The reflections could be indexed as (1,0), (0,1) and (1,−1). Since their Qz values are in the ratio Q3z = Q2z + Q1z, an oblique unit cell with a distortion equal to 0.29 has been determined. Very similar Bragg peaks were found for TH10 and OH10 as well, namely Qxy = 1.2 Å−1, Qxy = 1.36 Å−1 and Qxy = 1.53 Å−1, and Qxy = 1.19 Å−1, Qxy = 1.39 Å−1 and Qxy = 1.53 Å−1, respectively. But since the signal intensity for OH10 is very low, it was not possible to determine the corresponding Qz values. We can only assume an oblique unit cell, because the three peaks found are at very similar positions compared with TT10 and TH10.
Compared to TT10, TH10 has two extra CH2 groups resulting in slightly stronger van der Waals interactions. As discussed before, TH10 forms a stiffer monolayer due to a tighter molecular packing. Therefore, the lattice parameters of TH10 (t = 35.3°, A0 = 20.6 Å2, Axy = 25.3 Å2, and distortion d = 0.28) are a bit smaller than for TT10 (t = 38.5°, A0 = 21 Å2, Axy = 25.7 Å2 and d = 0.29).
TT10 and TH10 have a high tilt angle, which can be explained by the large area requirement of the head group. In order to compensate the large head group areas the chains are strongly tilted. Subsequently, the cross-sectional areas per chain are a bit higher than the typical values for free rotating chains (A0 ∼ 20 Å2), because these strongly tilted chains are less tightly packed (see also the increased wavenumbers of CH2 stretching vibrations of TT10 (Fig. 2B)). The in-plane area Axy, which describes the real in-plane area occupied by one chain at the surface, is consistent with the area per molecule of the corresponding π–A-isotherms (Fig. 2A).
The GIXD pattern of OT10 reveals several Bragg peaks. The three Braggs peaks at Qxy = 1.19 Å−1, Qxy = 1.39 Å−1 and Qxy = 1.52 Å−1 can be associated with an oblique chain structure, but, as for OH10, the signal intensity was too low for the determination of the corresponding Bragg rods.
Besides the Bragg peaks assigned to the chain lattice, there are additional Bragg peaks in the diffraction pattern of TT10, TH10, OH10 and OT10 indicating further 2D alignments. The Bragg peak at Qxy = 1.61 Å−1 gives a real spacing distance of 3.9 Å, which is a typical value for the distance between the Cα carbons in a β-sheet structure. These transfection lipids contain a lysine moiety in the head group, which results in a peptide-mimic character. Therefore, this peak can be indexed as Cα–Cα spacing. The hypothesis is supported by the Bragg peaks found at Qxy = 1.31 Å−1, indicating intermolecular hydrogen bonds. Qxy = 1.31 Å−1 gives a real spacing distance of 4.8 Å, which is a typical value for strand separation in β-sheets due to their hydrogen bonds (strand-NH⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C-opposite strand). TT10, TH10, OH10 and OT10 have a high ability to form intra- and intermolecular hydrogen bonds between the primary amine and carbonyl groups. Since the intramolecular hydrogen bonds and the hydrogen bonds with water molecules from the subphase are not ordered, they do not cause Bragg peaks. Consequently, the observed Bragg peaks in the GIXD pattern are associated with intermolecular hydrogen bonds between different TT10, TH10, OH10 and OT10 molecules, respectively. The Bragg peak at Qxy = 0.59 Å−1 (d = 10.6 Å) indicates ordering of the head groups.
C-opposite strand). TT10, TH10, OH10 and OT10 have a high ability to form intra- and intermolecular hydrogen bonds between the primary amine and carbonyl groups. Since the intramolecular hydrogen bonds and the hydrogen bonds with water molecules from the subphase are not ordered, they do not cause Bragg peaks. Consequently, the observed Bragg peaks in the GIXD pattern are associated with intermolecular hydrogen bonds between different TT10, TH10, OH10 and OT10 molecules, respectively. The Bragg peak at Qxy = 0.59 Å−1 (d = 10.6 Å) indicates ordering of the head groups.
The same two Bragg peaks, namely Qxy = 1.31 Å−1 and Qxy = 1.61 Å−1, have been found for lipid 7, which is a representative of the first generation of our malonic acid amide lipids,51,52 but the low Qxy range was not investigated for lipid 7. Therefore, the lattices could be very similar.
Moreover, in the GIXD pattern of TT10 and OT10 two Bragg peaks at Qxy = 1.52 Å−1 (degenerated peak) and Qxy = 1.68 Å−1 (non-degenerated peak) have been observed. The Bragg rods have a FWHM of 0.17 Å−1. By using the Scherrer formula,  to calculate the length of the scattering unit, one obtains Lz equal to 32.5 Å, which is too large for a lipid molecule. It might be that the lipids TT10 and OT10 were not completely dissolved in the solution (chloroform/methanol 3
to calculate the length of the scattering unit, one obtains Lz equal to 32.5 Å, which is too large for a lipid molecule. It might be that the lipids TT10 and OT10 were not completely dissolved in the solution (chloroform/methanol 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). In this case, lipid microcrystals could be present at the surface. The crystals are most probably made of lipid layers and double layers. Since no Scherrer rings or any bending of contour plots are visible, the signals have to come from oriented crystals that give a spot-like signal rather than from a powder-like structure where the Scherrer rings are expected. Probably only the in-plane cross-section of the double layer can be seen so no 3D structures contribute to the signal. The presence of crystals is the most reasonable explanation. The two Bragg peaks are indicative of a rectangular unit cell. Both peaks are located at the horizon characterizing a lattice of non-tilted chains. Lz = 32.5 Å corresponds to interdigitated chains of two layers. The packing in such interdigitated layers is very tight (A0 ∼ 18.5 Å2) and corresponds to the herringbone chain packing mode.
1). In this case, lipid microcrystals could be present at the surface. The crystals are most probably made of lipid layers and double layers. Since no Scherrer rings or any bending of contour plots are visible, the signals have to come from oriented crystals that give a spot-like signal rather than from a powder-like structure where the Scherrer rings are expected. Probably only the in-plane cross-section of the double layer can be seen so no 3D structures contribute to the signal. The presence of crystals is the most reasonable explanation. The two Bragg peaks are indicative of a rectangular unit cell. Both peaks are located at the horizon characterizing a lattice of non-tilted chains. Lz = 32.5 Å corresponds to interdigitated chains of two layers. The packing in such interdigitated layers is very tight (A0 ∼ 18.5 Å2) and corresponds to the herringbone chain packing mode.
Besides the indexed Bragg peaks there are other peaks in the GIXD patterns of TH10, OH10 and OT10, which are marked in grey (Fig. 3B–D). Similar peaks were found for a highly ordered head group structure of a sugar-unit containing the phospholipid GPI-fragment, namely GlcNα1-6myoIno-1-phosphodistearylglycerol.53,54 The chains were strongly tilted (43°). In the present case of the transfection lipids, the observed Bragg peaks could also indicate a sub-gel phase,55–57 which is formed by the ordering of whole molecules instead of only chains. However, the signal intensity is very weak compared to the other Bragg peaks. Most likely, the van der Waals interactions cannot be optimized due to the large spacing between the head groups ordered by a hydrogen bonding network.
4.3. Adsorption of calf thymus DNA to TH10 and OO10 monolayers
Since the investigated lipids are designed for gene transfection, they were also examined on calf thymus DNA containing buffers using a Langmuir film balance combined with the IRRAS technique. The adsorption behaviour of DNA will be discussed depending on the pH value and the chain composition for the two extreme cases, namely TH10 and OO10. TH10 forms the stiffest and OO10 the most fluid monolayer of the investigated transfection lipids.The resulting increase in the surface pressure (Δπ = π60min − πinitial) due to the adsorption of ct-DNA after 60 min is given in Table 1. The initial pressure was in all cases ∼8 mN m−1.
| ct-DNA containing bromide ion based buffer | Δπ [mN m−1] of TH10 monolayer | Δπ [mN m−1] of OO10 monolayer |
|---|---|---|
| pH 3 | 0.4 | 0 |
| pH 7 | 11.3 | 12.2 |
| pH 10 | 8 | 6.4 |
The protonation state of DNA and the cationic lipids explain the strong increase of the surface pressure at pH 7. The negatively charged DNA is attracted by the weakly protonated cationic lipids due to electrostatic interactions between the phosphate diester backbone (PO2−) of DNA and the amine groups (R-NH3+) in the lipid head group. The adsorbed DNA requires a large area. Even at pH 10, at which the cationic lipids are unprotonated, a large amount of negatively charged DNA is attracted to the interface and occupies space, which results in an increasing surface pressure for both TH10 and OO10. Almost no increase in the surface pressure is detected for the TH10 and OO10 monolayers at pH 3 (Table 1). The monolayers of protonated TH10 and OO10 are not influenced by the presence of weakly charged DNA.
As expected, the unsaturated lipid OO10 is more fluid than the saturated derivative TH10. DNA has a fluidising effect on the saturated lipid TH10 at pH 3 and pH 7. At pH 10, the monolayer remains in the LC phase state with chains in all-trans conformation. Surprisingly, the wavenumbers even decrease in the presence of DNA. It seems like adsorbing and penetrating DNA compresses the TH10 monolayer leading to a tighter packing (Fig. 5C).
 | ||
| Fig. 5 Possible scenarios of DNA adsorption to the lipid monolayers at different pH values. (A) pH 3, (B) pH 7 and (C) pH 10. | ||
At pH 7, the wavenumbers are intermediate, which either means that some chain segments are in the gauche conformation whereas other chain segments remain in the all-trans conformation, or that there are patches of lipids in LE and LC states as suggested in Fig. 5B. The idea of coexisting LE and LC phases is supported by the strong increase in the surface pressure as shown in the adsorption isotherms (ΔπTH10 = 11.3 mN m−1 and ΔπOO10 = 12.2 mN m−1). Comparing the wavenumbers for OO10 between 20 and 25 mN m−1 (Fig. 4B), it seems that OO10 is more fluid without the DNA than with the DNA adsorbed. This implies that DNA penetrates the monolayer and compresses the lipids, which results in lower wavenumbers. Upon further compressing by the barriers, the penetrated DNA seems to be squeezed out at 27 mN m−1, because the wavenumbers increase by about 2 cm−1.
At pH 3, both monolayers are in the LE phase state at low surface pressures, but upon compression TH10 undergoes a transition to the LC state. The adsorbed DNA changes the wavenumbers only marginally. Since the increase in the surface pressure observed in the adsorption isotherms for TH10 and OO10 at pH 3 is also negligible, it might be that DNA does not penetrate and influence the lipid monolayers. More likely, the DNA is coordinated by the protonated head groups underneath the monolayer (Fig. 5A). In order to get deeper insights into the interaction between ct-DNA and the cationic lipids, the amount of attached DNA has been estimated.
The intensity plots of the νasym PO2− band versus the area per molecule of the corresponding lipids are given in Fig. 6 for TH10 and OO10. Here, the increase in chain unsaturation results in a higher amount of DNA attached to the lipid monolayer. The smaller packing density of the OO10 monolayer assists the adsorption of DNA at pH 3 and pH 7. On DNA containing pH 10 buffer, the performance of both lipids is very weak. The relative amount of attached DNA is negligible. Upon compression, the little amount of adsorbed DNA is squeezed out from the monolayer, indicating weak electrostatic interactions. Further, the shift of the νasym PO2− band to higher wavenumbers (1260 cm−1) indicates dehydration of the DNA phosphate diester groups and the absence of hydrogen bonds (see ESI,† Fig. S8). On DNA containing pH 3 and pH 7 buffers, the νasym PO2− band is at 1220 cm−1 which implies stronger hydration of the DNA phosphate groups and the presence of hydrogen bonds. Interestingly, the slope of the linear fit differs dramatically for TH10 and OO10 on the DNA containing pH 7 buffer. With further compression the intensity of the phosphate band for TH10 strongly increases, while it slightly decreases for OO10. Here it looks like DNA is squeezed out from the OO10 monolayer as already assumed in the discussion regarding the phase state. For TH10 and OO10, a small amount of attached DNA introduces further adsorption of DNA to the lipid monolayer (Manning condensation58). The negatively charged polyelectrolyte would only go to the interfaces, if the interaction with the interfaces is more attractive than the interactions between the polyelectrolyte and subphase. The adsorption of DNA to the lipid monolayer is a competition between an attractive surface potential and the entropic repulsion, which keeps the polyelectrolyte in the subphase. The electrostatic interactions depend on the charge density of DNA and the lipids,59 the salt concentration60 and the pH value.61 In case a DNA chain segment adsorbs at the charged monolayer, the system loses translation energy equal to the thermic energy (kB·T, where kB is the Boltzmann constant and T the temperature), but increases the entropy due to the released counter ions (Z·kB·T, where Z is the charge of the counter ion). As a result, the surface charge decreases and subsequently an equilibrium of adsorption and desorption will be established. For polyelectrolytes this phenomenon is described as the Manning condensation.58 A weakly charged monolayer at the interface will attract only a small amount of DNA inducing further attraction of DNA to the interface. Additionally, the adsorbed DNA affects the potential of the electrical double layer in the way that the surface pH changes in accordance with the Boltzmann equation. This leads to an increase in the protonation degree of the TH10 and OO10 monolayers and consequently to further binding of DNA chain segments.62 This explains why there is still a large amount of DNA attached to TH10 and OO10 at pH 7, where their protonation degrees are rather small.
Despite the higher protonation degree of OO10, the charge densities of TH10 and OO10 are comparable at 30 mN m−1on the bromide ion based buffer at pH 3.37 Both lipids are in the same phase state (liquid-expanded), but TH10 is closer to the phase transition to the liquid-condensed phase state (πtr = 35 mN m−1, ESI,† Fig. S1). Therefore, the TH10 monolayer is already better ordered than the OO10 monolayer. This order seems to disturb the interactions with DNA in the case of TH10, while the flexibility of the fluid OO10 monolayer benefits the interaction with DNA. More DNA is attached to the OO10 monolayer at pH 3 (Fig. 6).
5. Conclusion
In this work, the influence of the chain composition on the physical–chemical properties of five lipids, designed for non-viral gene transfection, was investigated in 2D model layers. These lipids possess the same head group. The replacement of saturated tetradecyl (C14:0) and hexadecyl (C16:0) chains by unsaturated oleyl (C18:1) chains increases the fluidity of the lipid monolayer resulting in a smaller packing density. The exchange of a short tetradecyl (C14:0) chain by a longer hexadecyl (C16:0) chain enhances the van der Waals interactions and leads to a more rigid monolayer. TH10 forms the stiffest and OO10 the most fluid monolayer in this structure–property study. The fluidity increases with shorter chains and double bonds TH10 < TT10 < OH10 < OT10 < OO10. Since the unsaturated OO10 occupies larger molecular areas, a better accessibility of the amine groups is given, resulting in a higher protonation degree compared to the saturated lipids TT10 and TH10 as well as to the hybrids OT10 and OH10. Different scenarios of DNA coupling to the lipid monolayers have been proposed for different bulk pH values. In a forthcoming study, the properties of these lipids as well as of lipoplexes will be examined in bulk and compared with transfection experiments.Abbreviations
| Arb. units | Arbitrary units |
| π–A-isotherm | Surface pressure–area-isotherm |
| ct-DNA | Calf thymus deoxyribonucleic acid |
| DOPE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine |
| GIXD | Grazing incidence X-ray diffraction |
| IRRAS | Infrared reflection absorption spectroscopy |
| LC | Liquid-condensed phase with chains in all-trans conformation |
| LE | Liquid-expanded phase state with chains in gauche conformation |
| OH10 | N-(2-Aminoethyl)-N′-{6-amino-1-[N-(9Z)-octadec-9-enylamino]-1-oxohexan-(2S)-2-yl}-2-hexadecylpropandiamide |
| OO10 | N-(2-Aminoethyl)-N′-{6-amino-1-[N-(9Z)-octadec-9-enylamino]-1-oxohexan-(2S)-2-yl}-2-[(9Z)-octadec-9-enyl]propandiamide |
| OT10 | N-(2-Aminoethyl)-N′-{6-amino-1-[N-(9Z)-octadec-9-enylamino]-1-oxohexan-(2S)-2-yl}-2-tetradecylpropandiamide |
| TH10 | N-(2-Aminoethyl)-N′-[6-amino-1-oxo-1-(N-tetradecylamino)hexan-(2S)-2-yl]-2-hexadecylpropandiamide |
| TT10 | N-(2-Aminoethyl)-N′-[6-amino-1-oxo-1-[(N-tetradecylamino)hexan-(2S)2-yl]]-2-tetradecylpropandiamid |
| TRXF | Total reflection X-ray fluorescence |
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Max Planck Society. We thank DESY for beamtime and support. Open Access funding provided by the Max Planck Society.Notes and references
- T. Friedman and R. Roblin, Science, 1972, 175, 949–955 Search PubMed.
- T. P. O'Connor and R. G. Crystal, Nat. Rev. Genet., 2006, 7, 261–276 CrossRef PubMed.
- B. L. Davidson and P. B. McCray (Jr), Nat. Rev. Genet., 2011, 12, 329–340 CrossRef CAS PubMed.
- M. L. Maeder and C. A. Gersbach, Mol. Ther., 2016, 24, 430–446 CrossRef CAS PubMed.
- R. Mulligan, Science, 1993, 260, 926–932 CAS.
- R. G. Crystal, Science, 1995, 270, 404–410 CAS.
- I. MacLachlan, P. Cullis and R. W. Graham, Curr. Opin. Mol. Ther., 1999, 1, 252–259 CAS.
- F. H. Cameron, M. J. Moghaddam, V. J. Bender, R. G. Whittaker, M. Mott and T. J. Lockett, Biochim. Biophys. Acta, Biomembr., 1999, 1417, 37–50 CrossRef CAS.
- P. Pinnaduwage, L. Schmitt and L. Huang, Biochim. Biophys. Acta, 1989, 985, 33–37 CrossRef CAS.
- Z. Hyvönen, M. Ruponen, S. Rönkkö, P. Suhonen and A. Urtti, Eur. J. Pharm. Sci., 2002, 15, 449–460 CrossRef.
- R. P. Balasubramaniam, M. J. Bennett, A. M. Aberle, J. G. Malone, M. H. Nantz and R. W. Malone, Gene Ther., 1996, 3, 163–172 CAS.
- A. E. Regelin, S. Fankhaenel, L. Gurtesch, C. Prinz, G. von Kiedrowski and U. Massing, Biochim. Biophys. Acta, Biomembr., 2000, 1464, 151–164 CrossRef CAS.
- G. V. Srilakshmi, J. Sen, A. Chaudhuri, Y. Ramadas and N. M. Rao, Biochim. Biophys. Acta, Biomembr., 2002, 1559, 87–95 CrossRef CAS.
- R. Koynova and B. Tenchov, Curr. Pharm. Biotechnol., 2014, 15, 806–813 CAS.
- C. McGregor, C. Perrin, M. Monck, P. Camilleri and A. J. Kirby, J. Am. Chem. Soc., 2001, 123, 6215–6220 CrossRef CAS PubMed.
- M. Damen, E. Cristóbal-Lecina, G. C. Sanmartí, S. F. M. van Dongen, C. L. G. Rodríguez, I. P. Dolbnya, R. J. M. Nolte and M. C. Feiters, Soft Matter, 2014, 10, 5702–5714 RSC.
- D. Niculescu-Duvaz, J. Heyes and C. J. Springer, Curr. Med. Chem., 2003, 10, 1233–1261 CrossRef CAS PubMed.
- G. Byk, C. Dubertret, V. Escriou, M. Frederic, G. Jaslin, R. Rangara, B. Pitard, J. Crouzet, P. Wils, B. Schwartz and D. Scherman, J. Med. Chem., 1998, 41, 224–235 CrossRef.
- M. J. Ruocco and G. G. Shipley, Biochim. Biophys. Acta, 1982, 691, 309–320 CrossRef CAS.
- A. Roosjen, J. Smisterova, C. Driessen, J. T. Anders, A. Wagenaar, D. Hoekstra, R. Hulst and J. Engberts, Eur. J. Org. Chem., 2002, 1271–1277 CrossRef CAS.
- M. Castro, D. Griffiths, A. Patel, N. Pattrick, C. Kitson and M. Ladlow, Org. Biomol. Chem., 2004, 2, 2814–2820 CAS.
- M. Dittrich, M. Heinze, C. Wölk, S. S. Funari, B. Dobner, H. Möhwald and G. Brezesinski, ChemPhysChem, 2011, 12, 2328–2337 CrossRef CAS PubMed.
- I. Van der Woude, A. Wagenaar, A. A. P. Meekel, M. B. A. TerBeest, M. H. J. Ruiters, J. Engberts and D. Hoekstra, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 1160–1165 CrossRef CAS.
- S. Loisel, V. Floch, C. Le Gall and C. Ferec, J. Liposome Res., 2001, 11, 127–138 CrossRef CAS PubMed.
- M. L. Fielden, C. Perrin, A. Kremer, M. Bergsma, M. C. Stuart, P. Camilleri and J. Engberts, Eur. J. Biochem., 2001, 268, 1269–1279 CrossRef CAS PubMed.
- A. Ahmad, H. M. Evans, K. Ewert, C. X. George, C. E. Samuel and C. R. Safinya, J. Gene Med., 2005, 7, 739–748 CrossRef CAS PubMed.
- M. E. Ferrari, D. Rusalov, J. Enas and C. J. Wheeler, Nucleic Acids Res., 2002, 30, 1808–1816 CrossRef CAS PubMed.
- J. A. Heyes, D. Niculescu-Duvaz, R. G. Cooper and C. J. Springer, J. Med. Chem., 2002, 45, 99–114 CrossRef CAS PubMed.
- M. H. Nantz, C. W. Dicus, B. Hilliard, S. Yellayi, S. Zou and J. G. Hecker, Mol. Pharmaceutics, 2010, 7, 786–794 CrossRef CAS PubMed.
- P. V. Dileep, A. Antony and S. Bhattacharya, FEBS Lett., 2001, 509, 327–331 CrossRef CAS PubMed.
- S. Obika, T. Uneda, T. Sugimoto, D. Nanbu, T. Minami, T. Doi and T. Imanishi, Bioorg. Med. Chem., 2001, 9, 1001–1011 CrossRef CAS PubMed.
- G. Byk, B. Wetzer, M. Frederic, C. Dubertret, B. Pitard, G. Jaslin and D. Scherman, J. Med. Chem., 2000, 43, 4377–4387 CrossRef CAS PubMed.
- C. Wölk, C. Janich, U. Bakowsky, A. Langner and G. Brezesinski, Adv. Colloid Interface Sci., 2017, 248, 20–34 CrossRef PubMed.
- C. Wölk, M. Heinze, P. Kreideweiss, M. Dittrich, G. Brezesinski, A. Langner and B. Dobner, Int. J. Pharm., 2011, 409, 46–56 CrossRef PubMed.
- C. Wölk, S. Drescher, A. Meister, A. Blume, A. Langner and B. Dobner, Chem.–Eur. J., 2013, 19, 12824–12838 CrossRef PubMed.
- C. Janich, C. Wölk, S. Taßler, S. Drescher, A. Meister, G. Brezesinski, B. Dobner and A. Langner, Eur. J. Lipid Sci. Technol., 2014, 116, 1184–1194 CrossRef CAS.
- S. Tassler, C. Wölk, C. Janich, B. Dobner and G. Brezesinski, Phys. Chem. Chem. Phys., 2017, 19, 20271–20280 RSC.
- M. Dittrich, M. Böttcher, J. S. L. Oliveira, B. Dobner, H. Möhwald and G. Brezesinski, Soft Matter, 2011, 7, 10162–10173 RSC.
- T. Schindler and E. Nordmeier, Macromol. Chem. Phys., 1997, 198, 1943–1972 CrossRef CAS.
- S. Gromelski and G. Brezesinski, Langmuir, 2006, 22, 6293–6301 CrossRef CAS PubMed.
- E. Maltseva and G. Brezesinski, ChemPhysChem, 2004, 5, 1185–1190 CrossRef CAS PubMed.
- A. H. Muenter, J. Hentschel, H. G. Börner and G. Brezesinski, Langmuir, 2008, 24, 3306–3316 CrossRef CAS PubMed.
- J. K. Kauppinen, D. J. Moffatt, H. H. Mantsch and D. G. Cameron, Appl. Opt., 1982, 21, 1866–1872 CrossRef CAS PubMed.
- R. Frahm, J. Weigelt, G. Meyer and G. Materlik, Rev. Sci. Instrum., 1995, 66, 1677–1680 CrossRef CAS.
- J. Als-Nielsen, D. Jacquemain, K. Kjaer, F. Leveiller, M. Lahav and L. Leiserowitz, Phys. Rep., 1994, 246, 251–313 CrossRef CAS.
- K. Kjaer, Phys. B, 1994, 198, 100–109 CrossRef CAS.
- T. R. Jensen and K. Kjaer, in Novel Methods to Study Interfacial Layers, ed. D. Möbius and R. Miller, Elsevier Science, 2001, vol. 11, pp. 205–254 Search PubMed.
- C. Stefaniu and G. Brezesinski, Adv. Colloid Interface Sci., 2014, 207C, 265–279 CrossRef PubMed.
- C. Stefaniu and G. Brezesinski, Curr. Opin. Colloid Interface Sci., 2014, 19, 216–227 CrossRef CAS.
- D. Marsh, Biochim. Biophys. Acta, 1996, 1286, 183–223 CrossRef CAS.
- M. Heinze, G. Brezesinski, B. Dobner and A. Langner, Bioconjugate Chem., 2010, 21, 696–708 CrossRef CAS PubMed.
- M. Dittrich, PhD thesis, University of Potsdam, Germany, 2011.
- C. Stefaniu, I. Vilotijevic, M. Santer, D. V. Silva, G. Brezesinski and P. H. Seeberger, Angew. Chem., Int. Ed., 2012, 51, 12874–12878 CrossRef CAS PubMed.
- C. Stefaniu and G. Brezesinski, Curr. Opin. Colloid Interface Sci., 2014, 19, 216–227 CrossRef CAS.
- D. Marsh, Chem. Phys. Lipids, 2012, 165, 59–76 CrossRef CAS PubMed.
- J. Katsaras, V. A. Raghunathan, E. J. Dufourc and J. Dufourcq, Biochemistry, 1995, 34, 4684–4688 CrossRef CAS PubMed.
- F.-G. Wu, H.-Y. Sun, Y. Zhou, R.-G. Wu and Z.-W. Yu, RSC Adv., 2014, 4, 51171–51179 RSC.
- G. S. Manning, J. Chem. Phys., 1969, 51, 924–933 CrossRef CAS.
- H. G. M. Van de Steeg, M. A. C. Stuart, A. Dekeizer and B. H. Bijsterbosch, Langmuir, 1992, 8, 2538–2546 CrossRef CAS.
- J. Papenhuijzen, H. A. Vanderschee and G. J. Fleer, J. Colloid Interface Sci., 1985, 104, 540–552 CrossRef CAS.
- G. J. Fleer, M. A. Cohen Stuart, J. M. H. M. Scheutjens, T. Cosgrove and B. Vincent, Polymers at Interfaces, Chapman & Hall, 1993 Search PubMed.
- M. N. Antipina, B. Dobner, O. V. Konovalov, V. L. Shapovalov and G. Brezesinski, J. Phys. Chem. B, 2007, 111, 13845–13850 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cp00047f |
| This journal is © the Owner Societies 2018 |