Halogen bonding at the wet interfaces of an amyloid peptide structure†
Andrea
Pizzi
 a,
Nicola
Demitri
a,
Nicola
Demitri
 b,
Giancarlo
Terraneo
b,
Giancarlo
Terraneo
 *ac and
Pierangelo
Metrangolo
*ac and
Pierangelo
Metrangolo
 a
a
aLaboratory of Supramolecular and Bio-Nanomaterials (SBNLab), Department of Chemistry, Materials, and Chemical Engineering “Giulio Natta”, Politecnico di Milano, Via Luigi Mancinelli 7, Milano I-20131, Italy. E-mail: giancarlo.terraneo@polimi.it
bElettra – Sincrotrone Trieste, S.S. 14 Km 163.5 in Area Science Park, 34149 Basovizza – Trieste, Italy
cIstituto di Chimica del Riconoscimento Molecolare (ICRM), C.N.R., Via Luigi Mancinelli 7, Milano I-20131, Italy
First published on 17th August 2018
Abstract
Amyloid peptide hydrogels are a class of materials of great interest due to their structural simplicity, good performances and easy tuning of their properties by chemical modification. Among the possible modifications, halogenation has not yet been exploited extensively. Here, we report the single-crystal X-ray structure of two dihalogenated derivatives of the amyloidogenic sequence DFNKF. The obtained results show how halogenation is a promising tool to stabilize – through halogen bonds – the wet interface of amyloid structures, to determine an increase in the water uptake, hence the hydrogelation properties of the peptide sequence.
Hydrogels are unique materials, consisting of self-supporting, 3D, entangled networks that are capable of trapping huge amounts – up to a thousand-fold of their dry weight – of water or biological fluids. The mechanism of gelation deals with cross-linking phenomena – covalent or supramolecular – occurring between the chains of the gel matrix that produce complex entangled architectures where water molecules are trapped.1 This confinement drastically changes their mobility allowing the systems to gain different properties from those of classical Newtonian fluids. Thanks to these properties, hydrogels are commonly used in biomedical applications, such as tissue engineering, drug-delivery, wound dressing, and as biosensors.2–6
Hydrogels encompass a wide range of chemical structures, either small molecules or polymers. Notably, in recent years, amino acids and peptide-based hydrogels7–10 have grown exponentially due to the interest in their high biocompatibility and outstanding mechanical properties linked to intrinsic structural simplicity. A common structural motif of peptide hydrogels consists of β-sheets, which are a classical feature of naturally occurring amyloid sequences.11,12
Amyloid peptides usually possess alternating hydrophobic and hydrophilic amino acids, providing the peptide backbone an amphiphilic behaviour that directs the formation of β-sheets.13–15 This alternation of hydrophilic and hydrophobic residues, sometimes combined with an additional aliphatic chain to amplify the formation of high-aspect ratio nanostructures, has been used as a design guideline for new peptide-based hydrogels. Recently, alongside the de novo synthesis of amyloid sequences, chemical functionalization of natural peptide sequences was shown to be a powerful tool for controlling amyloid self-assembly and, thus, new hydrogel materials are obtained.16–19
In this context, we have recently demonstrated that, among the possible chemical modifications, halogenation is a very effective tool for conferring new properties and morphologies to amyloid materials.20–22 Specifically, we have shown that halogenation at the para-position of the phenylalanine (Phe) benzene rings improves the amyloidogenic propensity of the peptide sequences.20,21 The effects of halogenation on the self-assembly were found, in our case, to be sequence-independent. In fact, two different amyloid peptide fragments containing phenylalanine residues – KLVFF and DFNKF, core-sequences triggering the fibrillation of amyloid-beta (Aβ) and human calcitonin (h-CT) polypeptides, respectively – showed a very similar self-assembly behaviour.
Specifically, for the sequence DFNKF, the efficiency of hydrogel formation, at a 15 mM concentration, followed the order DF(I)NKF(I) ≫ DF(Cl)NKF(Cl) ≥ DFNKF(I), whereas DFNKF did not form a gel even after 30 days (Fig. 1). The oscillatory rheology analysis confirmed that the diiodinated peptide formed a very strong hydrogel with a G′ > 104 Pa while, under the same conditions, the DF(Cl)NKF(Cl) and DFNKF(I) derivatives formed much weaker hydrogels.
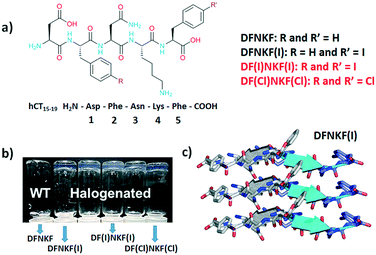 | ||
| Fig. 1 a) Chemical structures of the halogenated derivatives of the DFNKF sequence; in red, the peptides studied in this work, while in black, the peptides analysed in ref. 20. b) Photograph, adapted from ref. 20, of vials containing 15 mM peptide solutions left for 12 hours at r.t. After this time, the halogenated peptides formed clear hydrogels whereas the wild-type (WT) peptide did not. c) Representation of three parallel β-strands in one sheet of DFNKF(I), which are antiparallel to those in the adjacent sheet. β-Strands are depicted as cartoons (from ref. 23). | ||
In addition, single crystal X-ray studies on the DFNKF(I)23 segment showed that the halogen atom promoted the formation of an extended network of non-covalent interactions such as π⋯π, hydrophobic, and C–H⋯π interactions that contribute to the stabilization of the β-sheet and, thus, the overall aggregation of the sequence (Fig. 1). These findings have highlighted that halogenation of the DFNKF sequence amplified its ability to form hydrogels, with clear dependence on the nature, number, and position of the halogen atoms in the peptide sequence.
Herein, we report a structural study of DF(I)NKF(I) and DF(Cl)NKF(Cl) (Fig. 1), strong and weak gelators, respectively, in order to get more insights into the role of halogenation in determining hydrogel properties. Accurate structure solution with a resolution of ∼1.2 Å was only possible by using synchrotron radiation, due to the weakly diffracting and poorly ordered crystals obtained (see the ESI†). However, the diffraction experiments allowed a detailed structural description of the packing motifs for both the dihalogenated derivatives, revealing the classical features of the amyloid packing. Furthermore, these studies have highlighted a different water content in the structures of the two dihalogenated sequences suggesting that the different polarizability of the two involved halogen atoms, namely iodine and chlorine, may influence water uptake and stabilization of the wet interface of amyloid structures.
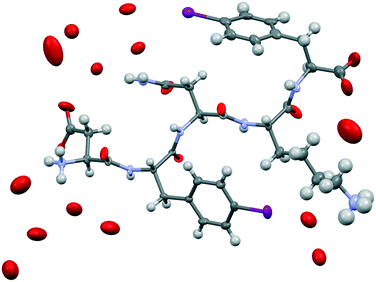
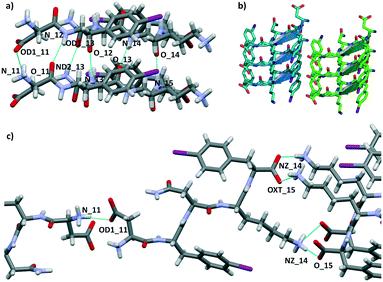
We attempted crystallization of the DF(I)NKF(I) and DF(Cl)NKF(Cl) sequences from water, i.e., the solvent used for gel formation. Single crystals of DF(I)NKF(I) suitable for synchrotron radiation analysis were obtained after two months by slow evaporation of the solvent at RT. DF(I)NKF(I) crystallized in the chiral monoclinic C2 space group and the asymmetric unit consists of a single molecule of the peptide solvated by 11.5 water molecules (Fig. 2). The peptide molecules adopt an extended conformation and are assembled in hydrogen-bonded strands, which pack in parallel β-sheets running along the crystallographic b-axis. Each DF(I)NKF(I) peptide forms six N–H⋯O hydrogen bonds (HBs) with neighbouring strands belonging to the same β-sheet (Fig. 3a and b) giving rise to the classical strand–strand separation of 4.8 Å.
The β-sheet arrangement is also stabilized by the presence of additional hydrogen bonds involving the charged group of the C and N-termini of paired peptide strands (Fig. 3). The geometrical parameters in the β-sheet, distances and angles, are the following: N11H⋯OD1_11 2.962 Å and 153.5°; N12H⋯O11 2.976 Å and 158.2°; ND2H⋯OD1_13 2.883 Å and 158.2°; N13H⋯O12 2.901 Å and 161.1°; N14H⋯O13 2.837 Å and 172.2°; N15H⋯O14 2.833 Å and 173.2°.
The lateral assembly of the peptides (Fig. 3c) is promoted by several electrostatic interactions and HBs involving the ionisable groups at both ends and water molecules. Specifically, charge-assisted HB occurs between the C terminus and lysine side chain of adjacent peptides (NZ_14H⋯O15: 2.798 Å and 175.2°; NZ_14H⋯OXT15 2.892 Å and 155.9°), and between the aspartate and the N terminus of the following peptide (N11H⋯OD1_11: 2.803 Å and 146.8°).
The features and values detected for the β-sheet formation by the DF(I)NKF(I) peptide are very similar to those reported for the analogous monoiodinated DFNKF(I) peptide23 and coherent with other reported non-halogenated amyloid sequences.24 This demonstrates, that in these structures the β-sheet is the dominating self-assembly motif.
The structure of DF(I)NKF(I) shares other similarities with that of DFNKF(I), such as the, so-called, Asn ladder and an extended network of aromatic–aromatic interactions (Fig. 4 top). It has been shown that aromatic–aromatic interactions (π⋯π interactions) are important stabilizing forces in DFNKF(I) and, more general, in amyloid structures. In the parallel β-sheets of DF(I)NKF(I), aromatic–aromatic interactions are detected either between the rings of Phe(I)2 and the rings of Phe(I)5 residues. All phenyl units adopt a parallel-displaced arrangement with the shortest distance between two carbon atoms of the interacting rings of around 3.4 Å for both Phe(I)2 and Phe(I)5 residues, respectively.
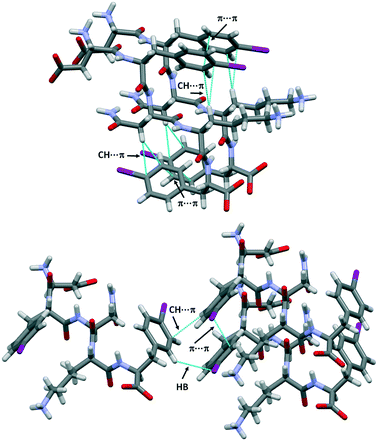 | ||
| Fig. 4 Partial crystal packing views of DF(I)NKF(I) where the aromatic–aromatic interactions and intramolecular CH⋯π interactions are shown as dotted blue lines (top). CH⋯π interactions and hydrogen⋯iodine contacts promote the T-shape antiparallel arrangement (edge-to-face) between two adjacent sheets (bottom). Colour and contact codes as those in Fig. 3. | ||
Notably, both the monoiodinated phenylalanine residues are also involved in weak intramolecular CH⋯π interactions occurring between the hydrogen atoms of the CH2 groups and the electron-rich π-surface of the aromatic rings. Specifically, the ring of Phe(I)2 interacts with the first CH2 unit of the Lys chain (shortest distance of the CH⋯Cring, 3.1 Å), while the ring of Phe(I)5 forms a short contact with the CH2 groups of Asn (shortest distance of the CH⋯Cring, 2.9 Å). These contacts contribute to stabilizing the “U-shape” conformation adopted by the Phe(I)2-Asn3-Lys4 and Asn3-Lys4-Phe(I)5 sub-segments favouring the orientation of the aromatic ring towards the formation of π⋯π interactions (Fig. 4, top).
Furthermore, CH⋯π interactions take place between adjacent β-sheets. Two adjacent iodinated phenyl moieties of different strands adopt a T-shape arrangement (edge-to-face) with the shortest distance CH⋯Cring of 2.9 Å. This edge-to-face motif is also stabilized by the presence of one weak hydrogen bond involving the iodine atom of the neighbouring iodinated ring. The HB-donor is a hydrogen atom located at the ortho position of the same aromatic ring involved in the T-shape arrangement and it further interacts with the electronegative belt of the iodine atom (H⋯I distance, 3.16 Å, Fig. 4 bottom). This antiparallel arrangement of the aromatic rings was also observed in the DFNKF(I) sequence.23
This evidence highlights two important structural features: (i) the key role of these CH⋯π interactions in the assembly of adjacent β-sheets and (ii) the iodine atom functions as an active moiety to further stabilize the lateral β-sheet aggregation by exploiting the anisotropic distribution of its electron density.
In DF(I)NKF(I), parallel β-sheets interact to form the typical cross-β spine structure,25,26 which is a peculiar structural feature of different amyloidogenic sequences. The “spine” is a pair of β-sheets running parallel to the fibril axis, which, by facing their side-chains, form a self-complementary dry interface commonly called “steric zipper”.27 Steric zippers are named “homosteric” when they are composed of a single sequence and can be classified considering three different features: (i) orientation of the β-strands in the same β-sheet; (ii) orientation of the paired β-sheets with respect to one another; and (iii) packing of the paired β-sheets. According to these criteria, the steric zipper in the crystal structure of DF(I)NKF(I) can be classified as up–up face-to-back class 2.
The steric zipper is characterized by alternating dry and wet interfaces. The wet interface of DF(I)NKF(I) contains several water molecules that are held in place by an extended network of hydrogen bonds involving the charged termini of the side chains of the peptide strands and the water molecules themselves. However, not only HBs are responsible in trapping the water molecules at the wet interface but also a halogen bond (XB)28,29 involving the iodine atom of Phe(I)5, which further stabilizes a water molecule in the wet interface. Specifically, the iodine atom at the para position on the aromatic ring functions as a good XB-donor, interacting, through its positive σ-hole, with the oxygen atom of one water molecule (the IZ_15⋯O_21 distance is 3.292 Å while the C–I⋯O angle is 170°, Fig. 5b and S4†). Although the observed XB is not particularly short, only a 6% reduction in the sum of the Van der Walls radii of the involved elements,30 its high directionality highlights its typical XB character where an iodine atom, with an anisotropically distributed electron density, accepts electron density from a Lewis base. In the context of electron density donor/acceptor ability, the amphoteric character of this iodine atom is also highlighted by the presence, as mentioned before, of a HB involving its negative belt and a hydrogen atom of the phenyl ring (Fig. 4, bottom).
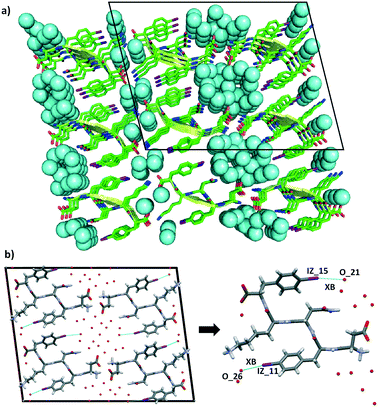 | ||
| Fig. 5 Representation of the steric zipper in DF(I)NKF(I). a): Partial DF(I)NKF(I) crystal packing viewed along the b-axis showing nine sheets. Pairing of the β-sheets into steric zippers creates the dry and wet interfaces. The two wet interfaces are filled in with water molecules depicted in blue in the space-filling model. b) View along the crystallographic b-axis of four facing β-sheets (left) formed by the steric zipper where the XB contacts between iodine atoms and water molecules are shown as blue dotted lines. Magnification of a single DF(I)NKF(I) peptide structure where the XBs are shown (right). Colour code as that in Fig. 3. | ||
The conformation adopted by the Phe(I)5 in DF(I)NKF(I) is very similar to that observed in DFNKF(I) where, as a result of the antiparallel edge-to-face organization of the aromatic rings, the iodine atoms point radially towards the wet interface. Although the iodine atom in the monoiodinated sequence does not exhibit any interaction with the water molecules, its presence in close proximity with the wet interface may suggest a possible involvement in some non-covalent contacts with the water molecules, phenomena that have been verified in the structure of DF(I)NKF(I).
The ability of the iodine atom on the phenylalanine ring to function as a XB-donor holds true also for the Phe(I)2 residue. In fact, also in this case, the iodine atom interacts with the oxygen atom of a different water molecule (the IZ_11⋯O_26 distance is 3.364 Å while the C–I⋯O angle is 164°, Fig. 5b). This XB takes place at an additional wet interface that is formed between the Lys residues of adjacent β-sheets and the Phe(I)2 moieties.
Although not very common, halogen–water XBs31,32 are not new in protein or DNA structures.33–37 In the studied peptide structures, the role of the iodine atoms is twofold. On the one hand, iodination contributes to stabilizing the β-sheet aggregation thanks to promoting multiple noncovalent interactions.23,38 On the other hand, the XBs work cooperatively with HBs,39–43 in holding in place the water molecules at the wet interface. The presence of two wet interfaces associated to the formation of two halogen bonds with water molecules drastically reduces the dry interface in the DF(I)NKF(I) steric zipper. Specifically, the only area of the packing where the water molecules are excluded is promoted by the edge-to-face (T-shape) arrangement of the two iodophenylalanine residues. Also in this case, the amphoteric character of the iodine atom plays a key role; in fact, the HB formed with its negative belt is one of the interactions that stabilize the arrangement of neighbouring iodobenzene moieties.
To determine whether the polarizability of the inserted halogen atom may play a role in the packing of amyloidogenic peptide sequences and in influencing the properties of the hydrogel thereof, we also attempted crystallization of the dichlorinated sequence DF(Cl)NKF(Cl) from water.
DF(Cl)NKF(Cl) crystallized, using the same experimental conditions adopted for the diiodinated fragment, in the chiral monoclinic C2 space group having one peptide molecule and 7 water molecules in the asymmetric unit. As expected, the crystal structure of DF(Cl)NKF(Cl) shares large structural similarities with that of DF(I)NKF(I). The DF(Cl)NKF(Cl) molecules are assembled through the supramolecular C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯NH synthon in an extended hydrogen-bonded network forming a parallel β-sheet architecture (Fig. 6a). The geometrical parameters of the β-sheet, distances and angles, are very similar to those observed in DF(I)NKF(I) (see Table S2 in the ESI† for details).
O⋯NH synthon in an extended hydrogen-bonded network forming a parallel β-sheet architecture (Fig. 6a). The geometrical parameters of the β-sheet, distances and angles, are very similar to those observed in DF(I)NKF(I) (see Table S2 in the ESI† for details).
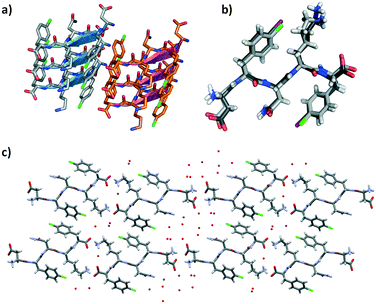 | ||
| Fig. 6 a) Geometrical arrangement of adjacent β-sheets formed by three strands each in DF(Cl)NKF(Cl). Hydrogen atoms have been omitted for clarity. β-Strands are depicted as cartoons. b) Superimposition of DF(Cl)NKF(Cl) and DF(I)NKF(I) conformations (light grey and dark grey sticks respectively). c) Partial representation of the steric zipper in DF(Cl)NKF(Cl) viewed along the b-axis showing eight sheets. Pairing of the β-sheets into the steric zippers creates the dry and wet interfaces. Water molecules are in red, while the colour code is as that in Fig. 3 and Cl in green. | ||
Also in this dichlorinated sequence, the amide groups of the Asn residues stack within a sheet forming the Asn ladder, and the halogenated aromatic rings adopt a parallel-displaced arrangement along the β-sheet. Similar to that of DF(I)NKF(I), the lateral self-assembly of DF(Cl)NKF(Cl) is driven by charged-assisted hydrogen bonds involving the terminal parts of the pentapeptide and an extended network of hydrogen bonds created by the water molecules. With the two halogenated peptides being structurally similar, with a root mean square deviation (RMSD) between the overlapped atoms of 0.74 Å (Fig. 6b), the steric zipper of the chlorinated peptide belongs to the same class (up–up face-to-back class 2), showing two well distinct wet interfaces and one dry interface. The dry interface is promoted by the edge-to-face (T-shape) arrangement of two aromatic rings of close phenylalanine residues; however, different from that in DF(I)NKF(I), this arrangement is held in place only by CH⋯π interactions occurring between the aromatic hydrogen atoms of one ring that point towards the electron rich π-cloud of the other phenyl ring (Fig. 6c).
The most remarkable difference between the structure of DF(Cl)NKF(Cl) and that of the diiodinated peptide appears at the wet interface. The overall water content is different; in the two wet interfaces of the dichlorinated peptide, there are 7 water molecules in total while in the DF(I)NKF(I) structure there were 11.5. More important, none of the chlorine atoms form any noncovalent interaction, neither XB nor HB, with the water molecules, although they both point towards the wet interfaces. This different behaviour may be related to the different polarizability, namely amphoteric nature, of the chlorine atom with respect to the iodine atom.28 This reduced polarizability makes the chlorine atom less prone to interact with the water molecules and thus be less effective in stabilizing the wet interface (Fig. S4†).
In conclusion, we have reported the single-crystal X-ray structures of two dihalogenated variants of DFNKF, DF(I)NKF(I) and DF(Cl)NKF(Cl). Our studies show that the β-sheet motif is structurally dominant in amyloid peptides and is always present regardless of the number and position of halogen atoms in the sequence. However, these studies further demonstrate that halogenation in the para position of the phenylalanine residues promotes the formation of weak CH⋯π or π⋯π interactions, which strengthen the construction of the steric zipper stabilizing the overall crystal packing. The strengthening of aromatic–aromatic interactions associated to the presence of the halogen atoms seems to be general and independent from the number and nature of the halogen atoms present in the amyloid sequence, as shown by the strong structural similarities observed between DF(I)NKF(I), DF(Cl)NKF(Cl), and DFNKF(I).
In addition, our structural studies point out, for the first time, that the presence of a halogen-bond donor, viz. the iodine atom, may contribute to stabilizing the wet interface of amyloid structures, determining an increase in the water uptake. Water acts as an in situ β-sheet extender by connecting the sheets with an extended network of hydrogen bonds. The occurrence of halogen bonding between iodine atoms and water molecules further increases these linking networks stabilizing the wet interfaces and thus the paring of adjacent β-sheets. Reducing the halogen atom polarizability from I to Cl, i.e., the strength of the halogen-bond donor, resulted in no halogen bonding with the water molecules, thus reducing the water content in the structure. The presence and the nature of the halogen atoms in the amyloid sequences can thus be exploited for modulating the water uptake and potentially the properties of the hydrogels formed thereof.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The European Research Council (ERC) is acknowledged for funding the project ‘FoldHalo’ (grant no. 307108).Notes and references
- J. Desvergne, Beilstein J. Org. Chem., 2010, 6, 846 CrossRef PubMed.
- K. Varaprasad, G. M. Raghavendra, T. Jayaramudu, M. M. Allapu and R. Sadiku, Mater. Sci. Eng., C, 2017, 79, 958 CrossRef PubMed.
- J. Tavakoli and Y. Tang, Polymer, 2017, 9, 364, DOI:10.3390/polym9080364.
- W. Wang, Y. Zhang and W. Liu, Prog. Polym. Sci., 2017, 71, 1 CrossRef.
- C. Guo, H. Kim, E. M. Ovadia, C. M. Mourafetis, M. Yang, W. Chen and A. M. Kloxin, Acta Biomater., 2017, 56, 80 CrossRef PubMed.
- H. Thérien-Aubin, Z. L. Wu, Z. Nie and E. Kumacheva, J. Am. Chem. Soc., 2013, 135, 4834 CrossRef PubMed.
- M. J. Webber, E. A. Appel, E. W. Meijer and R. Langer, Nat. Mater., 2016, 15, 13 CrossRef PubMed.
- A. M. Jonker, D. W. P. M. Löwik and J. C. M. van Hest, Chem. Mater., 2012, 24, 759 CrossRef.
- A. Dasgupta, J. Hassan Mondal and D. Das, RSC Adv., 2013, 3, 9117 RSC.
- W. Yang Seow and C. A. E. Hauser, Mater. Today, 2014, 17, 381 CrossRef.
- J. Greenwald and R. Riek, Structure, 2010, 18, 1244 CrossRef PubMed.
- O. S. Makin, E. Atkins, P. Sikorski, J. Johansson and L. C. Serpell, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 315 CrossRef PubMed.
- M. P. Hendricks, K. Sato, L. C. Palmer and S. I. Stupp, Acc. Chem. Res., 2017, 50, 2440 CrossRef PubMed.
- A. Dehsorkhi, V. Castelletto and I. W. Hamley, J. Pept. Sci., 2014, 20, 453 CrossRef PubMed.
- H. Cui, M. J. Webber and S. I. Stupp, Biopolymers, 2010, 94, 1 CrossRef PubMed.
- E. T. Pashuck, H. Cui and S. I. Stupp, J. Am. Chem. Soc., 2010, 132, 6041 CrossRef PubMed.
- J. D. Hartgerink, E. Beniash and S. I. Stupp, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5133 CrossRef PubMed.
- A. D. Martin, J. P. Wojciechowski, H. Warren, M. Panhuisb and P. Thordarson, Soft Matter, 2016, 12, 2700 RSC.
- A. D. Martin, A. B. Robinson, A. F. Mason, J. P. Wojciechowski and P. Thordarson, Chem. Commun., 2014, 50, 15541 RSC.
- A. Bertolani, L. Pirrie, N. Houbenov, J. Haataja, L. Stefan, L. Catalano, G. Terraneo, G. Giancane, L. Valli, R. Milani, O. Ikkala, G. Resnati and P. Metrangolo, Nat. Commun., 2015, 6(7574), 1 Search PubMed.
- A. Pizzi, C. Pigliacelli, A. Gori, Nonappa, O. Ikkala, N. Demitri, G. Terraneo, V. Castelletto, I. W. Hamley, F. Baldelli Bombelli and P. Metrangolo, Nanoscale, 2017, 9, 9805 RSC.
- A. Pizzi, L. Lascialfari, N. Demitri, A. Bertolani, D. Maiolo, E. Carretti and P. Metrangolo, CrystEngComm, 2017, 19, 1870 RSC.
- A. Bertolani, A. Pizzi, L. Pirrie, L. Gazzera, G. Morra, M. Meli, G. Colombo, A. Genoni, G. Cavallo, G. Terraneo and P. Metrangolo, Chem. – Eur. J., 2017, 23, 2051 CrossRef PubMed.
- J.-P. Colletier, A. Laganowsky, M. Landau, M. Zhao, A. B. Soriaga, L. Goldschmidt, D. Flot, D. Cascio, M. R. Sawaya and D. Eisenberg, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16938 CrossRef PubMed.
- M. R. Sawaya, S. Sambashivan, R. Nelson, M. I. Ivanova, S. A. Sievers, M. I. Apostol, M. J. Thompson, M. Balbirnie, J. J. W. Wiltzius, H. T. McFarlane, A. Ø. Madsen, C. Riekel and D. Eisenberg, Nature, 2007, 447, 453 CrossRef PubMed.
- R. Nelson, M. R. Sawaya, M. Balbirnie, A. Ø. Madsen, C. Riekel, R. Grothe and D. Eisenberg, Nature, 2005, 435, 773 CrossRef PubMed.
- J. C. Stroud, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2013, 69, 540 CrossRef PubMed.
- G. Cavallo, P. Metrangolo, R. Milani, T. Pilati, A. Priimägi, G. Resnati and G. Terraneo, Chem. Rev., 2016, 116, 2478 CrossRef PubMed.
- L. C. Gilday, S. W. Robinson, T. A. Barendt, M. J. Langton, B. R. Mullaney and P. D. Beer, Chem. Rev., 2015, 115, 7118 CrossRef PubMed.
- A. Bondi, J. Phys. Chem., 1964, 68(3), 441 CrossRef.
- Z. Xu, Z. Yang, Y. Liu, Y. Lu, K. Chen and W. Zhu, J. Chem. Inf. Model., 2014, 54, 69 CrossRef PubMed.
- P. Zhou, J. Lv, J. Zou, F. Tian and Z. Shang, J. Struct. Biol., 2010, 169, 172 CrossRef PubMed.
- P. Auffinger, F. A. Hays, E. Westhof and P. S. Ho, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16789 CrossRef PubMed.
- M. R. Scholfield, C. M. Vander Zanden, M. Carter and P. S. Ho, Protein Sci., 2013, 22, 139 CrossRef PubMed.
- F. A. Hays, J. M. Vargason and P. S. Ho, Biochemistry, 2003, 42, 9586 CrossRef PubMed.
- T. Eneqvist, E. Lundberg, A. Karlsson, S. Huang, C. R. A. Santos, D. M. Power and A. E. Sauer-Eriksson, J. Biol. Chem., 2004, 279, 26411 CrossRef PubMed.
- E. Parisini, P. Metrangolo, T. Pilati, G. Resnati and G. Terraneo, Chem. Soc. Rev., 2011, 40, 2267 RSC.
- A. Pizzi, V. Dichiarante, G. Terraneo and P. Metrangolo, Biopolymers, 2017, e23088, DOI:10.1002/bip.23088.
- A. R. Voth, P. Khuu, K. Oishi and P. S. Ho, Nat. Chem., 2009, 1, 74 CrossRef PubMed.
- C. B. Aakeroy, M. Fasulo, N. Schultheiss, J. Desper and C. Moore, J. Am. Chem. Soc., 2007, 129, 13772 CrossRef PubMed.
- V. Vasylyeva, S. K. Nayak, G. Terraneo, G. Cavallo, P. Metrangolo and G. Resnati, CrystEngComm, 2014, 16, 8102 RSC.
- A. M. S. Riel, D. A. Decato, J. Sun, C. J. Massena, M. J. Jessop and O. B. Berryman, Chem. Sci., 2018, 9, 5828 RSC.
- A.-C. C. Carlsson, M. R. Scholfield, R. K. Rowe, M. Coates Ford, A. T. Alexander, R. A. Mehl and P. S. Ho, Biochemistry, 2018, 57, 4135 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures and technical details, including the discussion of crystallographic data. CCDC 1574297 and 1574298 contain the supplementary crystallographic data for compounds DF(Cl)NKF(Cl) and DF(I)NKF(I). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ce01205a |
| This journal is © The Royal Society of Chemistry 2018 |