Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A sequence environment modulates the impact of methylation on the torsional rigidity of DNA†
Johanna
Hörberg
and
Anna
Reymer
 *
*
Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg 40530, Sweden. E-mail: anna.reymer@gu.se; Fax: +46 31 786 2599; Tel: +46 31 786 9117
First published on 28th September 2018
Abstract
We describe a potential molecular mechanism explaining how DNA methylation contributes to biological regulation. Using molecular dynamics together with a new torsional restraint, we identify the impact of methylation on DNA response to torsional stress. We observe that, depending on the sequence, DNA methylation hinders overwinding or underwinding molecular transitions.
DNA methylation is an important epigenetic modification, involved in the regulation of various biological processes, including development, synaptic plasticity, brain function, aging, and immune responses.1–3 In eukaryotes, DNA methylation is commonly found within a CpG dinucleotide, where a methyl group is covalently added at the 5th carbon of a cytosine base (denoted “M” in this article). Addition of a methyl group occurs in the DNA major groove. Structurally methylation does not affect the base-pair hydrogen bonds,4 but decreases the twist and increases the roll angles in CpG dinucleotides.5–7 The local structural modifications have implications for direct protein–DNA readout.8
Methylation affects the physical properties of DNA. Orozco and co-workers6 showed that DNA sequences containing MpG steps become very stiff and hard to bend, and display a lower propensity to circularize and form stable nucleosomes.9 Another study by groups of Schulten and Gaub revealed strong methylation dependence of DNA strand separation.10 These findings suggest a possible alternative mechanism for the regulatory effect of DNA methylation. The epigenetic mark may, by altering the topological environment of the genome, modify the organization of nucleosome arrays and hinder access to transcription starting sites for regulatory proteins.
The physical properties of DNA also depend on its sequence. We showed that when DNA is subjected to torsional stress, certain dinucleotides, termed “Twist Capacitors”, in specific sequence environments, are able to absorb most of the imposed torsional stress, leaving the rest of the stressed molecule in a close to relaxed conformation.11 One of the most effective twist capacitor dinucleotides is the CpG step in a Y-CpG-R environment (Y denotes pyrimidine and R denotes purine). This local heterogeneity affects the local torsional modulus of DNA, suggesting a molecular mechanism through which the topological environment can modulate interactions with proteins.
Here we can explain how DNA methylation may affect DNA response to torsional stress, and what role sequence-specific effects play in this process. We use all-atom molecular dynamics simulations together with a new torsional restraint,11 which controls the torsional state of DNA molecules. We observe that cytosine methylation makes DNA more torsionally rigid.6 But the impact of methylation depends on the sequence and could lead to an asymmetric response to torsional stress.
We study six DNA oligonucleotides that represent average-, most- and least-occurring tetranucleotide motifs in the human genome.12 The selected sequences are 20 base pairs long, containing repeating tetranucleotide motifs and their methylated counterparts: GG-(ACGT)4-GG, GG-(CCGC)4-GG, GG-(TCGA)4-GG, GG-ACGT-(AMGT)2-ACGT-GG, GG-CCGC-(CMGC)2-CCGC-GG, and GG-TCGA-(TMGA)2-TCGA-GG. We aim to consider the impact of methylation only on DNA twisting elasticity and exclude bending deformations, thus the torsional restraint is applied only to the central eight-base pair segments. The restraint is applied gradually starting from fully relaxed molecules, and then over and underwinding the selected segments by ±6° per base pair step with respect to the relaxed average twist. We use the umbrella sampling13 technique to obtain the potential of the mean force (PMF) profiles to describe the change in free energy as a function of average base pair twist. Each umbrella window consists of 300 ns of sampling allowing for sufficient equilibration. We use a force constant ktw of 0.06 kcal mol−1 degrees−2, the smallest value which enables achieving the desired torsional strain. Details of the simulation protocol are provided in the ESI.†
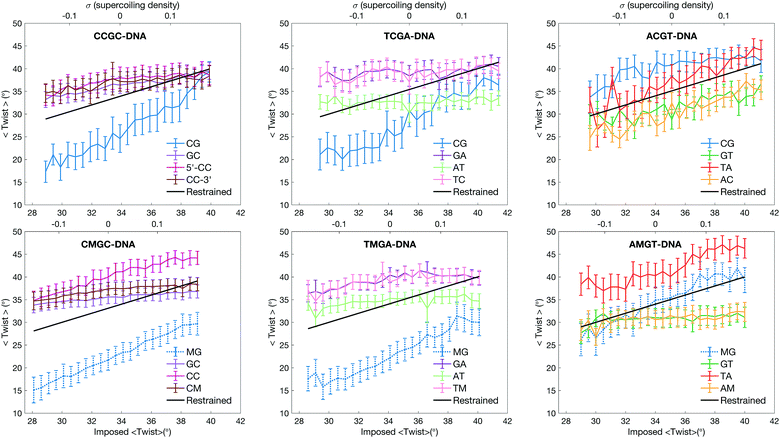
The PMF plots show that cytosine methylation in the CpG steps asymmetrically stiffens the twisting of DNA, in a sequence-specific manner (Fig. 1). To more easily compare the results, the PMF curves are plotted as a function of Δtwist/base pair – a change in average twist per base pair step with respect to the corresponding optimal average twist, derived from the unrestrained MD simulations. We subsequently derive the torsional force constants K by fitting the PMF curves with quadratic functions. We also derive the torsional force constants for overwinding (K+) and underwinding (K−) regimes, since the PMF profiles are asymmetrical about the minima. K values are also used to obtain the torsional moduli C, using the isotropic rod equation, T = KΔθ = CΔθ/L, where T is the torque resulting from a change in a base pair twist Δθ over a base pair step length L = 0.34 nm. Torsional moduli are then used to calculate the torsional persistence lengths, P = C/kBT (taking kBT at room temperature as 4.114 pN nm). The resulting order of torsional stiffness from the least to the most rigid is ACGT < AMGT ≈ CCGC ≈ TCGA < CMGC ≈ TMGA, validating that cytosine methylation makes DNA more torsionally rigid.6 When comparing the torsional constants for overwinding and underwinding independently, the effect of the sequence environment becomes more pronounced (Table 1).
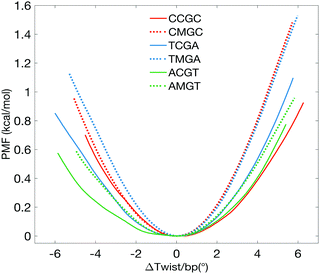 | ||
| Fig. 1 PMF plots showing the change in free energy with respect to the average change of twist per base pair (bp) step with respect to the corresponding value for the relaxed oligomers. | ||
| Oligomer | 〈Relaxed twist〉 (deg) | K (kcal mol−1 deg−2) | K + (kcal mol−1 deg−2) | K − (kcal mol−1 deg−2) | C (pN nm2) | P (nm) |
|---|---|---|---|---|---|---|
| CCGC | 33.6 | 0.055 | 0.051 | 0.058 | 427 | 108 |
| CMGC | 33.3 | 0.081 | 0.090 | 0.062 | 628 | 153 |
| TCGA | 35.5 | 0.058 | 0.065 | 0.050 | 450 | 109 |
| TMGA | 34.0 | 0.087 | 0.089 | 0.064 | 675 | 164 |
| ACGT | 35.5 | 0.042 | 0.052 | 0.030 | 326 | 79 |
| AMGT | 34.1 | 0.054 | 0.058 | 0.048 | 419 | 102 |
For the most abundant motif, CCGC-DNA, methylation of central CpG stiffens the molecule during overwinding, characterized by an additional energy cost of 0.47 kcal mol−1, when the segment is on average overwound by 4°. At the same time, the underwinding PMF profiles nearly overlap. For the averagely present ACGT-motif, the impact of methylation is much more pronounced in the underwinding regime. Underwinding by 4° per base pair costs an extra 0.18 kcal mol−1, while overwinding by 4° costs only 0.04 kcal mol−1. For the least abundant motif, TCGA, methylation stiffens the molecule in a more symmetric manner: underwinding by 4° per base pair will cost an extra 0.29 kcal mol−1, overwinding by 4° costs 0.21 kcal mol−1. We also observe that, in agreement with the earlier findings,6,7,10 cytosine methylation results in the overall unwinding of DNA molecules – for values of average base pair twist see Table 1 and Fig. S1 (ESI†).
The observed sequence-specific asymmetric impact of cytosine methylation on DNA response to torsional stress is coupled to the behaviour of individual base pairs. As expected, individual base pair steps do not respond equally to the imposed torsional stress, characterized by steep and flat twisting profiles (the data are presented in Fig. 2). Both in CCGC/CMGC- and in TCGA/TMGA-DNAs it is the CpG/MpG steps that play the role of twist capacitors; they show the most deviation in the average twist values. The CpG step ability to absorb torsional stress is due to its twist polymorphism.11,12,14 The unrestrained MD simulations show that the CpG step in the Y-CpG-Y/R environment can convert between low and high twist, with the two twist states separated by about 20° (see Fig. S2A and C, ESI† for twist distributions derived from unrestrained MD). However, the MpG step in the Y-MpG-Y/R environment prefers a low twist (Fig. S2B and D, ESI†), and can effectively absorb only the negative torsional stress during underwinding. During overwinding, however, it is the CpC step that takes over in CMGC-DNA, and to some extent – the TpM and GpA steps in TMGA-DNA. All three dinucleotide steps show a preference for a high twist during the unrestrained MD simulations.
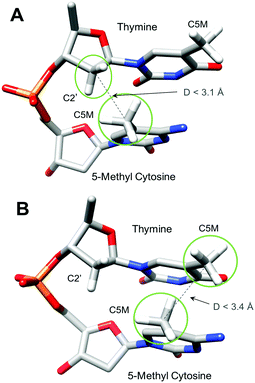
As demonstrated earlier,11,12,14 twist polymorphism of the CpG step is coupled to conformational transitions at the sugar phosphate backbone, which involve phosphate linkages on the 3′-side of the CpG step in both DNA strands. The high twist state is associated with the BI conformation (backbone torsional angles ε/ζ are in trans/gauche-) and the low twist state – with the BII conformation (ε/ζ in gauche-/trans). Our simulations show that cytosine methylation prevents the BI/BII flips, effectively locking the phosphodiester backbone in the BII conformation (Fig. S3A–D, ESI†). Reduced dynamics of the sugar phosphate backbone, as a result of cytosine methylation, in a similar sequence environment (T-MpG-C) was also detected by solid-state NMR.15 We notice that the bulky methyl group of cytosine creates a potential steric clash with the C2′ atom of the sugar phosphate backbone on the preceding 5′-nucleotide (see Fig. 3A, and Fig. S4A, B (ESI†) for information on the distances between C2′ and C5M atoms), and to avoid the clash the MpG dinucleotide has to increase the roll angle by additional 7° in comparison with the CpG step.5,6,8 The increased roll angle stabilizes the BII conformation, and thus the low twist state of the MpG dinucleotide. This steric clash results in an increased energy cost during overwinding of CMGC- and TMGA-DNA molecules. For TMGA-DNA, the methyl group of cytosine also creates a second steric clash with the methyl group of thymine during underwinding (Fig. 3B and Fig. S4C, ESI†). This explains the additional energy cost for TMGA-DNA underwinding.
In the case of ACGT-DNA, it is the TpA step that exhibits large twist changes (see Fig. 2) and absorbs major torsional stress.11 The TpA step exploits the same molecular mechanism when assuming low twist during underwinding: the phosphodiester linkages on 3′-flanks flip to the BII conformation (see Fig. S3E, ESI†). Cytosine methylation of ACGT-DNA blocks completely the TpA ability to convert to low twist (see Fig. 2 and Fig. S3F, ESI†). In our underwinding simulations the TpA step shows only high twist. We have considered several snapshots from several underwinding trajectories of the ACGT-sequence with TpA showing low twist, and manually mutated cytosine to 5-methylcytosine. Again we observe a steric clash between the methyl group of cytosine and the C2′-atom of the backbone on the 5′-flank (see Fig. S5, ESI†), which would prevent the BI/BII flip and block TpA from converting to low twist. Thus the negative torsional stress is absorbed by the MpG step, while the positive torsional stress is mostly absorbed by the TpA step. Interestingly, the CpG step in R-CpG-Y shows a relatively high twist of 35°,12,14 and in unrestrained MD simulations the phosphate linkages on the 3′-side of the step remain predominantly in the BI conformation.12 Cytosine methylation, as discussed above, induces an increased roll angle (see Fig. S6, ESI†), which in turn promotes the BI/BII backbone flips for the MpG step. The low twist state of the MpG step in the R-MpG-Y environment, however, remains less favourable, as phosphate linkages on the 3′-side of the MpG steps in both DNA strands appear in the BII conformation only in 35% of snapshots. This explains the additional energy cost during underwinding of AMGT-DNA.
Besides average base pair twist and roll, cytosine methylation also affects how other DNA helical parameters respond to torsional stress. Fig. S6 and S7 (ESI†) show translational and rotational parameters for torsionally active dinucleotide steps for the nonmethylated and methylated oligomers, as they undergo twisting deformations. For CCGC/CMGC- and TCGA/TMGA-DNAs, methylation blocks CpG shift-bimodality during overwinding – since similarly to twist-, shift-bimodality is coupled to the BI/BII backbone conformational flips.11 Methylation prevents increase of rise for the MpG step during overwinding. For CCGC/CMGC-DNAs methylation also removes shift-bimodality and decreases slide of the CpC step (GCCC/GCCM) during overwinding. For ACGT/AMGT-DNAs similar trends can be noted: the MpG step shows no shift-bimodality during overwinding while the CpG step does, MpG's slide in general shows a much more narrow distribution of values, and MpG's rise decreases during underwinding. For TpA, methylation brings significant changes in shift during overwinding, and increase in rise during underwinding. Finally, our data show no significant impact on the major and minor groove widths between nonmethylated and methylated DNA for the CCGC and TCGA sequences (Fig. S8, ESI†). For the ACGT-motif, methylation results in an increased minor groove width of CpG and GpT and a decreased major groove width of GpT.
Here we have demonstrated that cytosine methylation makes DNA more torsionally rigid. But the impact of methylation can be asymmetric: depending on the sequence, cytosine methylation can result in either additional energy cost for underwinding or for overwinding, which could explain the strong combinatorial effect of methylation and sequence context on the stability of DNA strand separation.10,16 The presence of a bulky methyl group creates several steric clashes that lead to changes in how the base pair steps react to torsional stress. In general, the MpG step prefers lower twist values than the CpG step. This is due to the increased roll angle of the dinucleotide that stabilizes the BII conformation of the backbone, allowing the MpG step to effectively reduce its twist and absorb negative torsional stress. Thus, in sequences where the CpG step plays a role of a twist capacitor, cytosine methylation would stiffen overwinding. Moreover, if the MpG step is flanked by thymine on the 5′-side, cytosine methylation would also hinder underwinding. In the ACGT-sequence where the CpG dinucleotide is not the torsionally active step, cytosine methylation would result in an additional energy cost for underwinding. Despite showing little preference for lower-twist values, dictated by the R-MpG-Y environment, MpG takes over the leading role in absorbing negative torsional stress.
In this study we illustrated what structural changes could be expected when methylated DNA is subjected to torsional stress and how heterogeneous these changes are, depending on the sequence environment. To complete the picture of the combined effect of the DNA sequence environment and methylation on how the duplex reacts to strong over or undertwisting, we plan to further study the effect by taking a more diverse set of DNA oligomers, both in sequence and in length. Our observations suggest a molecular mechanism through which methylation can contribute to biological regulation, by modulating the dynamics of DNA supercoiling transitions.17,18 Our data are also of significance for the future development of gene editing technology. Since it presents a method to predict, based on just DNA sequence, if cytosine methylation would contribute to molecular unwinding, which might have implications for chromatin packaging and stability. The demonstrated torsional stiffness asymmetry of methylated DNA could be exploited when designing sequence-specific inhibitors that could block the undesired DNA-motif.
This work was supported by the Swedish Research Council VR Grant [637–2014-437] and a Hasselblad Foundation Prize to A. R. The authors thank the Swedish National Infrastructure for Computing (SNIC) for the generous provision of computing resources.
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. Dulac, Nature, 2010, 465, 728–735 CrossRef CAS PubMed.
- M. Winnefeld and F. Lyko, Genome Biol., 2012, 13, 165 CrossRef PubMed.
- B. Suarez-Alvarez, R. M. Rodriguez, M. F. Fraga and C. López-Larrea, Trends Genet., 2012, 28, 506–514 CrossRef CAS PubMed.
- A. Jeltsch, ChemBioChem, 2002, 3, 274–293 CrossRef CAS PubMed.
- A. Lazarovici, T. Zhou, A. Shafer, A. C. Dantas Machado, T. R. Riley, R. Sandstrom, P. J. Sabo, Y. Lu, R. Rohs, J. A. Stamatoyannopoulos and H. J. Bussemaker, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6376–6381 CrossRef CAS PubMed.
- A. Pérez, C. L. Castellazzi, F. Battistini, K. Collinet, O. Flores, O. Deniz, M. L. Ruiz, D. Torrents, R. Eritja, M. Soler-López and M. Orozco, Biophys. J., 2012, 102, 2140–2148 CrossRef PubMed.
- A. T. P. Carvalho, L. Gouveia, C. R. Kanna, S. K. T. S. Wärmländer, J. A. Platts and S. C. L. Kamerlin, Epigenetics, 2014, 9, 1604–1612 CrossRef PubMed.
- A. C. D. Machado, T. Zhou, S. Rao, P. Goel, C. Rastogi, A. Lazarovici, H. J. Bussemaker and R. Rohs, Briefings Funct. Genomics, 2015, 14, 61–73 CrossRef PubMed.
- G. Portella, F. Battistini and M. Orozco, PLoS Comput. Biol., 2013, 9, e1003354 CrossRef PubMed.
- P. M. D. Severin, X. Zou, H. E. Gaub and K. Schulten, Nucleic Acids Res., 2011, 39, 8740–8751 CrossRef CAS PubMed.
- A. Reymer, K. Zakrzewska and R. Lavery, Nucleic Acids Res., 2018, 46, 1684–1694 CrossRef PubMed.
- P. D. Dans, I. Faustino, F. Battistini, K. Zakrzewska, R. Lavery and M. Orozco, Nucleic Acids Res., 2014, 42, 11304–11320 CrossRef CAS PubMed.
- G. M. Torrie and J. P. Valleau, J. Comput. Phys., 1977, 23, 187–199 CrossRef.
- M. Pasi, J. H. Maddocks, D. Beveridge, T. C. Bishop, D. A. Case, T. Cheatham, P. D. Dans, B. Jayaram, F. Lankas, C. Laughton, J. Mitchell, R. Osman, M. Orozco, A. Pérez, D. Petkevičiūt, N. Spackova, J. Sponer, K. Zakrzewska and R. Lavery, Nucleic Acids Res., 2014, 42, 12272–12283 CrossRef CAS PubMed.
- K. B. Geahigan, G. A. Meints, M. E. Hatcher, J. Orban and G. P. Drobny, Biochemistry, 2000, 39, 4939–4946 CrossRef CAS PubMed.
- A. Lefebvre, O. Mauffret, S. E. L. Antri, M. Monnot, E. Lescot and S. Fermandjian, Eur. J. Biochem., 1995, 229, 445–454 CrossRef CAS PubMed.
- S. Corless and N. Gilbert, Biophys. Rev., 2016, 8, 51–64 CrossRef CAS PubMed.
- F. Kouzine, A. Gupta, L. Baranello, D. Wojtowicz, K. Ben-Aissa, J. Liu, T. M. Przytycka and D. Levens, Nat. Struct. Mol. Biol., 2013, 20, 396–403 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details of simulation protocols and analysis, and additional figures. See DOI: 10.1039/c8cc06550k |
| This journal is © The Royal Society of Chemistry 2018 |