 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Unexpected arene ligand exchange results in the oxidation of an organoruthenium anticancer agent: the first X-ray structure of a protein–Ru(carbene) adduct†
Matthew P.
Sullivan
 ab,
Michél K.
Nieuwoudt
ab,
Michél K.
Nieuwoudt
 ac,
Graham A.
Bowmaker
a,
Nelson Y. S.
Lam
ac,
Graham A.
Bowmaker
a,
Nelson Y. S.
Lam
 a,
Dianna
Truong
a,
Dianna
Truong
 a,
David C.
Goldstone
a,
David C.
Goldstone
 *b and
Christian G.
Hartinger
*b and
Christian G.
Hartinger
 *a
*a
aSchool of Chemical Sciences, University of Auckland, Private Bag 92019, Auckland 1142, New Zealand. E-mail: c.hartinger@auckland.ac.nz; Web: http://www.hartinger.auckland.ac.nz/
bSchool of Biological Sciences, University of Auckland, Private Bag 92019, Auckland 1142, New Zealand. E-mail: d.goldstone@auckland.ac.nz
cThe Photon Factory, University of Auckland, New Zealand
First published on 23rd April 2018
Abstract
The first X-ray structures of adducts formed between a RuII(N-heterocyclic carbene)(η6-p-cymene) compound and a protein are reported. Coordination to the protein induced the cleavage of the cymene ligand and EPR spectroscopy demonstrated the oxidation of the Ru centre.
Platinum-based anticancer drugs have been widely used in the treatment of tumourigenic diseases and RuIII and other metal complexes are considered a promising addition to the toolbox of anticancer chemotherapeutics.1–4 Sodium [trans-tetrachloridobis(indazole)ruthenate(III)] is currently undergoing clinical trials, showing promising activity and good tolerability,2,5 while organoruthenium(II) piano stool compounds are on track toward clinical development.6 The latter scaffold consists of a Ru centre with a π-bound arene and either ancillary ligands and/or leaving groups to complete the coordination sphere around the metal centre. Altering the scaffold allows for fine-tuning of the pharmacological properties of the compound type.3,6,7 RAPTA-C [RuII(cym)(1,3,5-triaza-7-phosphatricyclo-[3.3.1.1]decane)Cl2] (cym = η6-p-cymene) and RAED [RuII(arene)(1,2-diaminoethane)Cl]+ are the most notable lead structures to date, and have been shown to selectively target amino acids and DNA, respectively.8–11 More recently, we identified the plecstatin-targeting properties of orally-active pyridinecarbothioamide complexes.12 Interestingly, most compounds that feature a monodentate non-leaving ligand exhibit only limited in vitro anticancer activity, unless the ligand itself is bioactive, while bidentate ligands as simple as 1,2-ethylenediamine often induce cytotoxicity.2–4,6–10,12–14 This is based on stability of the bond(s) between the metal centre and the ancillary ligands and the lipophilicity of the ligand and complex.
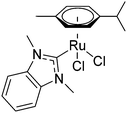
N-Heterocyclic carbenes (NHCs) have been widely studied as ancillary ligands in homogeneous catalysis and form stable complexes with metal centres but they have only recently been adopted for medicinal chemistry applications.15,16 Early Au(NHC) complexes showed promising cytotoxic activity, with inhibition of thioredoxin reductase (TrxR) being a possible mechanism.17 An ESI-MS study on a selenopeptide derived from TrxR suggested that the inhibition is caused by binding of the Au compound to the selenocysteine residue of TrxR.18 Recently structural data for an Au(NHC)–protein adduct has been reported. Merlino et al. found that Au(1-butyl-3-methyl-imidazole-2-ylidene)Cl formed six adducts with the model protein thaumatin.19 Ott et al. studied Ru(NHC) complexes, such as [dichlorido(dmb)(cym)ruthenium(II)] 1 (dmb = 1,3-dimethylbenzimidazol-2-ylidene; Fig. 1), for medicinal applications and showed that, like Au(NHC) complexes, they inhibited TrxR at μM concentrations.20,21 In order to add structural data to the discussion on the mode of action of this compound type, we report here the first crystal structure of a Ru(NHC)–protein adduct between 1 and the model protein hen egg white lysozyme (HEWL).
HEWL has a molecular weight of 14.3 kDa (129 amino acids) and is an ideal model for studies demonstrating protein modification with metal ions.22–31 Crystals of HEWL are readily obtained by hanging-drop vapour diffusion using a reservoir solution containing NaCl and NaOAc at pH 4.7. The crystals were formed when the reservoir solution was mixed with an equal volume of an aqueous solution of HEWL (100 mg ml−1). Crystals formed within 1 d and were soaked in a solution of NaNO3 and NaOAc containing 1 for a period of 3 d (PDB ID 6BO1) or 1 month (PDB ID 6BO2). The crystals were analysed at the Australian synchrotron MX1 beamline at a wavelength of 0.9537 Å. The phases were estimated by the molecular replacement method with a structure of HEWL (PDB ID 4NHI) as the search model. The Ru-containing fragments were identified in an anomalous difference map where they display significant anomalous signals compared with the proteinaceous light elements.32 Analysis of the data revealed that an initial adduct of the composition Ru(dmb)(OHx)Cl2–HEWL (x = 1, 2) was formed after a 3 d incubation with the protein acting as a bidentate ligand to the metal through the His15 and Arg14 residues (site I, Fig. 2). A second adduct was only present in a longer soak of one month, and formed at Lys33 where it was identified as Ru(dmb)(OHx)2Cl2–HEWL with the protein monodentately coordinated to the Ru centre (site II, Fig. 2). Notably, in both structures the cym ligand is absent, while the NHC remains coordinated to the metal centre. Upon compound placement, the structures were refined to Rwork/Rfree values of 0.20/0.23 and 0.18/0.23 for the structures of the adducts formed after 3 d and 1 month, respectively (ESI,† Table S2).
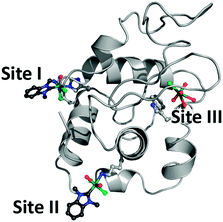 | ||
| Fig. 2 Sites of metalation on HEWL after reaction with 1 (PDB ID 6BO2) indicating formation of a bidentate Ru(dmb)(OHx)Cl2–HEWL adduct at site I and a monodentate Ru(dmb)(OHx)2Cl2–HEWL adduct at site II. Ru (teal), Cl (green), OHx (red), and NHC (blue and dark grey) are shown in ball and stick representation while the rest of the protein is presented as a light grey cartoon. | ||
A comparison of the ruthenated HEWL structures to native HEWL (PDB ID 4NHI) showed no major structural perturbations and only localised changes at sites I, II, and III in the side chain conformations to accommodate the organometallic fragment. This is supported by the small root-mean-square deviations (RMSD) of 0.164 and 0.289 Å (for overlays see Fig. S1, ESI†).
In the crystal of HEWL soaked for 3 d with 1, a peak was observed in the anomalous difference map with a peak height of 12.2σ. This was coupled with a large area of 2Fo − Fc electron density (magenta) adjacent to the Nε1 atom of His15 (Fig. S2a, ESI†). His15 is located on the surface of the protein near the C terminus. Histidine residues have been shown to be binding partners for Ru in previous protein structures.11,33 The anomalous density allowed for the placement of the Ru centre with a Ru–Nε1His15 bond length of 2.54 Å (site I). This is slightly longer than that observed in the structure of HEWL modified with a Ru(cym)Cl2 moiety (PDB ID: 5V4G).30 This interaction perturbs the side chain position of His15 resulting in a rotation of approximately 130° compared to apo-HEWL.
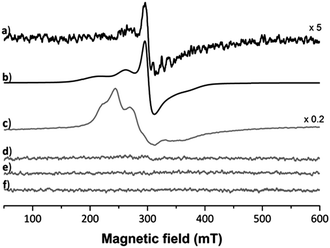
Five projections from the metal centre were observed in the 2Fo − Fc electron density map. In trans position to the coordinated His residue, a dagger-shaped region of density corresponding to dmb was observed. The distance between the Ru and carbene-C of 2.06 Å is in a similar range as found in Ru(NHC) complexes.34 During refinement it became evident that Arg14 was coordinated to the metal centre (Ru–Nη1Arg14 2.22 Å) and was displaced by 4.43 Å compared to a previously published structure (PDB ID: 5V4G, overlapped in Fig. S3, ESI†). In addition, density was evident for a further three spherical projections off the Ru centre. Two of these were modelled as chlorido ligands and the third as a coordinated water. The chlorido ligands are likely retained due to chloride present during crystal formation preventing ligand exchange reactions. The average Ru–Cl distance was determined as 2.44 Å. The Ru–Owater/hydroxido distance of 2.17 Å is consistent with a hydroxido/water ligand. This analysis demonstrates that the protein acts as a bidentate ligand, confirming an octahedral coordination geometry about the Ru centre. Moreover, no density was observed for the p-cymene ligand. This indicates that the p-cymene ligand was released during the binding event. The presence of the NHC ligand at the Ru centre may labilise the π-bound p-cymene, similar to the elongation effect NHC ligands have in complexes on bonds in trans position.34 Consequently, the bound metallofragment at site I was identified as a [Ru(dmb)(OHx)Cl2] fragment coordinated to the Nε1His15 and Nη1Arg14 atoms. The refined 2Fo − Fc electron density map about these ligands validated the atoms in our model and the positioning of these ligands was a good fit, with an occupancy of 0.65 (Fig. S2b; for a discussion on the occupancies see Table S2, ESI†). The addition of the NHC ligand seems to weaken the Ru–cym bond. This is in contrast to HEWL binding studies with other organoruthenium compounds, in which the cym ligand was retained in the protein structure.22,30 Notably, Ott et al. also showed that the dibenzyl derivative of 1 is significantly cytotoxic, demonstrating the need of the NHC ligand for the bioactivity to remain attached to the metal centre.21 As the arene ligand is known to stabilise the Ru centre in +2 oxidation state, we used EPR spectroscopy to investigate the impact of arene ligand exchange on the oxidation state of the Ru centre (Fig. 3). The 3 species obtained from simulated fitting of the HEWL–1 spectrum (Fig. 3b and Table S1, ESI†) are consistent with the mononuclear RuIII nature35 of HEWL–1 adducts, while neither HEWL, 1, nor the buffer are paramagnetic. While the cleavage of cym has been seen before in the binding of RAPTA-C to macromolecules,36,37 this is the first instance that the ruthenation was confirmed to result in the oxidation of the Ru centre. The spectrum for HEWL–1 KP1019 with 1.92 × 1018 spins, calculated using the area of the doubly integrated 1st-derivative EPR spectrum of a CuSO4 standard, is much stronger than that of HEWL–1 (5.94 × 1015 spins). This may be explained by only a small proportion of 1 undergoing ligand exchange with HEWL and oxidation to RuIII, while KP1019 with its RuIII centre is intrinsically paramagnetic.
To study the impact of longer soaking times on the adduct formation and protein structure, HEWL was treated with 1 for 1 month. The crystals analysed featured the identical adduct and interaction at site I but at higher occupancy (0.90 vs. 0.65 after 3 d; Fig. 4a and b). This indicates progressively complete metalation of the protein at site I. The anomalous density (Fig. 4a) disappeared at 14.2σ, and showed similar distances (Ru–Nε1His15 2.13 Å, Ru–Nη1Arg14 2.14 Å, Ru–CNHC 2.05 Å, Ru–O 2.00 Å, Ru–Cl 2.44 Å) and maps as for the 3 d incubation.
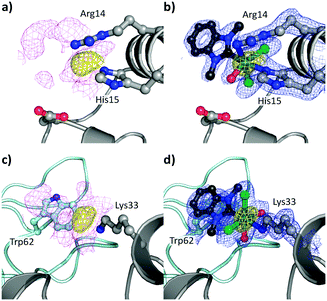 | ||
| Fig. 4 Details of the binding sites for 1 on HEWL after 1 month incubation (PDB ID 6BO2), binding site I on HEWL (a and b) and binding site II (c and d). The [Ru(dmb)(OHx)yCl2] (y = 1, 2) fragments are indicated in ball and stick representation with the Ru (teal). The electron density maps are contoured at 1σ (magenta and blue maps) while the anomalous difference maps are contoured at 4σ (yellow maps). (a) Unbiased electron density (magenta) and anomalous difference map showing binding site I, as well as (b) placement of the [Ru(dmb)(OHx)Cl2] fragment into the refined electron density map (blue) with the anomalous difference map. The interaction between HEWL and 1 at binding site II and the adjacent protein is shown as a light teal cartoon in the background with Trp62 highlighted. (c) Unbiased electron density and the anomalous difference map. (d) Placement of the Ru(dmb)(OHx)2Cl2 adduct into the refined electron density map with the anomalous difference map. | ||
In addition to the adduct at site I, the presence of anomalous density (yellow; Fig. 4c and d) was noted near Lys33 (site II) with a peak height of 12.3σ and was therefore identified as a Ru centre. The Ru centre was found with a large electron density observed in 2Fo − Fc maps (magenta) adjacent to the Nζ1Lys33 atom (Fig. 4c). The anomalous density allowed for the placement of the Ru centre with a Ru–Nζ1Lys33 bond length of 2.18 Å, which resulted in a shift in the side chain position of Lys33. Note that this binding site is different to site II identified for [Ru(cym)Cl2]2 on HEWL, which was found to form an adduct with Asp101 that acts as a bridging ligand for two Ru centres.30
Around the Ru centre, five distinct projections were observed emanating from the metal centre (Fig. S5 for the coordination modes at binding sites I and II, ESI†). Four projections appeared to adopt a planar coordination arrangement including the metal centre and these were initially modelled as chlorido ligands. However, for two of the projections negative peaks appeared in the Fo − Fc electron density maps, suggesting the presence of two OHx ligands at the metal centre. Thus, these were modelled with an average Ru–Cl distance of 2.45 Å and a Ru–Owater/OH distance of 2.18 Å. The last area of residual electron density was located trans to the Nζ1Lys33 atom, as at site I, dagger-shaped and accordingly modelled as dmb with a Ru–CNHC distance of 2.08 Å. The NHC forms a π-stacking interaction with the indole residue of Trp62, resulting in a twist of Trp62 by 21°, with the shortest distance between the aromatic systems being 3.44 Å. This interaction may have an additional stabilising effect on the binding of the organoruthenium fragment to the protein and also impacts its positioning on the protein. Therefore, the bound metallofragment was identified as Ru(dmb)(OHx)2Cl2 coordinated to Nζ1Lys33. The refined 2Fo − Fc electron density map about these ligands validated the atoms in our model and the positioning of these ligands were a good fit, with an occupancy of 0.8 (Fig. 4d). This indicates that, as at site I, the p-cymene ligand underwent a ligand exchange reaction. As the interaction occurs with the Nζ1 atom of Lys33 in a monodentate fashion, this further supports the hypothesis that the NHC ligand weakens the π-bond between the arene and the Ru centre.
A third Ru centre was identified in structural data collected from a crystal incubated for 1 month. It was found in the solvent channel with an occupancy of 0.5, however, with a coordination number of 6. We identified four OHx ligands coordinated to the Ru centre with an average Ru–Owater/hydroxido distance of 2.37 Å and one chlorido ligand with a Ru–Cl distance of 2.48 Å. This [RuCl(OHx)4] fragment was found to interact weakly with the peptide backbone through the carbonyl oxygen of Ala107 with a Ru–OAla107 distance of 2.76 Å (Fig. S4, ESI†).
Given the observed cleavage of the cym ligand from the Ru centre, we complemented the X-ray crystallographic studies with mass spectrometry experiments to understand the behaviour of 1 in presence of biomolecules in aqueous solution. Studies with His and HEWL in acetate buffer were conducted over a period of 6 d and 4 weeks, respectively, to mimic the conditions during soaking of HEWL. In the mass spectra recorded for the equimolar reaction mixture of 1 with His, the main peak was identified as [Ru(cym)(dmb) − H]+ followed by [Ru(cym)(dmb) + OAc]+ and a formate adduct formed after dilution of the sample with water/acetonitrile/formic acid (69/30/1). In addition minor peaks were detected that were assigned to the His adducts [Ru(cym)(dmb)(His) − H]+ (Fig. S8, ESI†) and [Ru(dmb)(His)Cl(CH3CN) + Na]2+, with the latter indicating the cleavage of the cym from the Ru centre. In reaction mixtures with a 5- or 10-fold excess of 1 over HEWL, the main adduct was identified as [HEWL + Ru(cym)(NHC)] while ions could be assigned to a species after cym cleavage. This indicates the influence of the protein microenvironment and soaking conditions on the type of adducts detectable by either method.
An analysis of all the published HEWL structures modified with Ru complexes shows that the primary binding site for most Ru complexes is at His15. However, the nature of the ancillary ligands at the Ru centres determines the modality of binding and potentially the presence and nature of further interactions (see ESI,† Table S3). Furthermore, in several of those reported structures, the original ancillary ligands, such as indazole, imidazole (and derivatives), DMSO, pyridine, and CO, at the metal centre underwent a ligand exchange reaction with most often only OHx or Cl ligands modelled. In contrast, π-bound arenes and, in this case, the NHC ligand remain tightly attached to the metal centre, indicating that the organoruthenium compounds retain their ligands to a larger extent than coordination compounds. We have also shown that the Ru(NHC) species are more selective in terms of binding sites compared to Au(NHC) complexes, despite the long soak times. However, we acknowledge that the crystallisation conditions may influence the species formed and that the intermolecular contacts required to maintain the crystal may limit the number of interactions observed. Nevertheless, these are promising results indicating that the NHC ligand is retained in a biological setting and allows for design of stable organoruthenium complexes for medicinal application.
We thank the University of Auckland (Frederick Douglas Brown Postgraduate Science Research Scholarship to M. P. S). D. G. is supported by a Rutherford Discovery Fellowship awarded by the NZ government and administered by the RSNZ. This research was, in part, undertaken on the crystallography beamlines MX1 at the Australian Synchrotron, Victoria, Australia with support from the NZ synchrotron group.
Conflicts of interest
There are no conflicts to declare.References
- W. A. Wani, S. Prashar, S. Shreaz and S. Gómez-Ruiz, Coord. Chem. Rev., 2016, 312, 67 CrossRef CAS.
- C. G. Hartinger and P. J. Dyson, Chem. Soc. Rev., 2009, 38, 391 RSC.
- M. A. Jakupec, M. Galanski, V. B. Arion, C. G. Hartinger and B. K. Keppler, Dalton Trans., 2008, 183 RSC.
- P. Zhang and P. J. Sadler, J. Organomet. Chem., 2017, 839, 5 CrossRef CAS.
- H. A. Burris, S. Bakewell, J. C. Bendell, J. Infante, S. F. Jones, D. R. Spigel, G. J. Weiss, R. K. Ramanathan, A. Ogden and D. Von Hoff, ESMO Open, 2016, 1, e000154 CrossRef PubMed.
- B. S. Murray, M. V. Babak, C. G. Hartinger and P. J. Dyson, Coord. Chem. Rev., 2016, 306, 86 CrossRef CAS.
- A. A. Nazarov, C. G. Hartinger and P. J. Dyson, J. Organomet. Chem., 2014, 751, 251 CrossRef CAS.
- R. E. Morris, R. E. Aird, S. Murdoch Pdel, H. Chen, J. Cummings, N. D. Hughes, S. Parsons, A. Parkin, G. Boyd, D. I. Jodrell and P. J. Sadler, J. Med. Chem., 2001, 44, 3616 CrossRef CAS PubMed.
- C. S. Allardyce, P. J. Dyson, D. J. Ellis and S. L. Heath, Chem. Commun., 2001, 1396 RSC.
- P. J. Dyson, Chimia, 2007, 61, 698 CrossRef CAS.
- Z. Adhireksan, G. E. Davey, P. Campomanes, M. Groessl, C. M. Clavel, H. Yu, A. A. Nazarov, C. H. Yeo, W. H. Ang, P. Droge, U. Rothlisberger, P. J. Dyson and C. A. Davey, Nat. Commun., 2014, 5, 3462 Search PubMed.
- S. M. Meier, D. Kreutz, L. Winter, M. H. M. Klose, K. Cseh, T. Weiss, A. Bileck, B. Alte, J. C. Mader, S. Jana, A. Chatterjee, A. Bhattacharyya, M. Hejl, M. A. Jakupec, P. Heffeter, W. Berger, C. G. Hartinger, B. K. Keppler, G. Wiche and C. Gerner, Angew. Chem., Int. Ed., 2017, 56, 8267 CrossRef CAS PubMed.
- M. Patra, T. Joshi, V. Pierroz, K. Ingram, M. Kaiser, S. Ferrari, B. Spingler, J. Keiser and G. Gasser, Chem. – Eur. J., 2013, 19, 14768 CrossRef CAS PubMed.
- B. Bernhard, Mini-Rev. Med. Chem., 2016, 16, 804 CrossRef.
- F. E. Hahn and M. C. Jahnke, Angew. Chem., Int. Ed. Engl., 2008, 47, 3122 CrossRef CAS PubMed.
- M. N. Hopkinson, C. Richter, M. Schedler and F. Glorius, Nature, 2014, 510, 485 CrossRef CAS PubMed.
- R. Rubbiani, E. Schuh, A. Meyer, J. Lemke, J. Wimberg, N. Metzler-Nolte, F. Meyer, F. Mohr and I. Ott, MedChemComm, 2013, 4, 942 RSC.
- A. Pratesi, C. Gabbiani, E. Michelucci, M. Ginanneschi, A. M. Papini, R. Rubbiani, I. Ott and L. Messori, J. Inorg. Biochem., 2014, 136, 161 CrossRef CAS PubMed.
- G. Ferraro, C. Gabbiani and A. Merlino, Bioconjugate Chem., 2016, 27, 1584 CrossRef CAS PubMed.
- A. Casini, C. Gabbiani, F. Sorrentino, M. P. Rigobello, A. Bindoli, T. J. Geldbach, A. Marrone, N. Re, C. G. Hartinger, P. J. Dyson and L. Messori, J. Med. Chem., 2008, 51, 6773 CrossRef CAS PubMed.
- L. Oehninger, M. Stefanopoulou, H. Alborzinia, J. Schur, S. Ludewig, K. Namikawa, A. Munoz-Castro, R. W. Koster, K. Baumann, S. Wolfl, W. S. Sheldrick and I. Ott, Dalton Trans., 2013, 42, 1657 RSC.
- I. W. McNae, K. Fishburne, A. Habtemariam, T. M. Hunter, M. Melchart, F. Wang, M. D. Walkinshaw and P. J. Sadler, Chem. Commun., 2004, 1786 RSC.
- A. Casini, G. Mastrobuoni, C. Temperini, C. Gabbiani, S. Francese, G. Moneti, C. T. Supuran, A. Scozzafava and L. Messori, Chem. Commun., 2007, 156 RSC.
- O. Pinato, C. Musetti, N. P. Farrell and C. Sissi, J. Inorg. Biochem., 2013, 122, 27 CrossRef CAS PubMed.
- C. Muegge, T. Marzo, L. Massai, J. Hildebrandt, G. Ferraro, P. Rivera-Fuentes, N. Metzler-Nolte, A. Merlino, L. Messori and W. Weigand, Inorg. Chem., 2015, 54, 8560 CrossRef CAS PubMed.
- Y. Gothe, T. Marzo, L. Messori and N. Metzler-Nolte, Chem. – Eur. J., 2016, 22, 12487 CrossRef CAS PubMed.
- M. Hanif, S. Moon, M. P. Sullivan, S. Movassaghi, M. Kubanik, D. C. Goldstone, T. Sohnel, S. M. F. Jamieson and C. G. Hartinger, J. Inorg. Biochem., 2016, 165, 100 CrossRef CAS PubMed.
- G. Ferraro, A. Pica, I. Russo Krauss, F. Pane, A. Amoresano and A. Merlino, J. Biol. Inorg. Chem., 2016, 21, 433 CrossRef CAS PubMed.
- L. Messori and A. Merlino, Coord. Chem. Rev., 2016, 315, 67 CrossRef CAS.
- M. P. Sullivan, M. Groessl, S. M. Meier, R. L. Kingston, D. C. Goldstone and C. G. Hartinger, Chem. Commun., 2017, 53, 4246 RSC.
- F. Zobi and B. Spingler, Inorg. Chem., 2012, 51, 1210 CrossRef CAS PubMed.
- G. Strahs and J. Kraut, J. Mol. Biol., 1968, 35, 503 CrossRef CAS PubMed.
- A. Bijelic, S. Theiner, B. K. Keppler and A. Rompel, J. Med. Chem., 2016, 59, 5894 CrossRef CAS PubMed.
- M. Hollering, M. Albrecht and F. E. Kühn, Organometallics, 2016, 35, 2980 CrossRef CAS.
- N. Cetinbas, M. I. Webb, J. A. Dubland and C. J. Walsby, J. Biol. Inorg. Chem., 2010, 15, 131 CrossRef CAS PubMed.
- B. M. Blunden, D. S. Thomas and M. H. Stenzel, Polym. Chem., 2012, 3, 2964 RSC.
- S. J. Dougan, M. Melchart, A. Habtemariam, S. Parsons and P. J. Sadler, Inorg. Chem., 2006, 45, 10882 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, literature analysis of HEWL–Ru adducts, additional mass spectrometry, X-ray crystallography, and EPR spectroscopy data. See DOI: 10.1039/c8cc02433b |
| This journal is © The Royal Society of Chemistry 2018 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: