Open Access Article
Open Access ArticleOptochemical control of gene expression by photocaged guanine and riboswitches†
V.
Dhamodharan
 ,
Yoko
Nomura
,
Yoko
Nomura
 ,
Mohammed
Dwidar
,
Mohammed
Dwidar
 and
Yohei
Yokobayashi
and
Yohei
Yokobayashi
 *
*
Nucleic Acid Chemistry and Engineering Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa, 904 0495, Japan. E-mail: yohei.yokobayashi@oist.jp
First published on 30th May 2018
Abstract
Optical control of biomolecules via engineered proteins or photoactive small molecules has had a profound impact on biology. However, optochemical tools to control RNA functions in living cells are relatively limited. We synthesized a photoactivatable (photocaged) guanine to modulate gene expression under riboswitch control in both mammalian cells and Escherichia coli by light.
Optogenetic technologies that enable precise temporal and spatial control of protein function by light have had a profound impact on biological research.1 Various light-responsive proteins such as ion channels, pumps, and enzymes have been engineered and used in living cells and animals.1–3 However, optical regulation of RNA functions in living cells has remained relatively underexplored. Since natural light-responsive RNAs do not exist, optical regulation of RNAs requires synthetic effectors responsive to light.
Riboswitches are gene regulatory modules that contain an RNA aptamer and an associated regulatory sequence in the untranslated regions (UTRs) of messenger RNAs (mRNAs). With appropriate mechanisms to transduce aptamer–ligand interaction into regulation of gene expression, synthetic riboswitches responsive to synthetic and natural small molecules have been reported in both prokaryotes and eukaryotes.4–7 These riboswitches can be indirectly controlled by light using photocaged riboswitch ligands. For example, Wulffen et al. reported a photocaged derivative of glucosamine-6-phosphate which functions as the natural ligand for the glmS riboswitch in vitro.8 Walsh et al. used a photocaged theophylline9 to optically activate gene expression via a synthetic riboswitch in Escherichia coli.10 An analogous attempt to regulate gene expression in mammalian cells was reported by Young et al., who synthesized and used photocaged toyocamycin,11 but the mechanism of gene expression activation by toyocamycin was attributed to nonspecific incorporation of the nucleic acid analog into cellular RNAs,12 not based on aptamer–ligand interaction. Therefore, a strictly optochemical riboswitch in mammalian cells has not been demonstrated, and photocaged theophylline remains as the only photocaged riboswitch ligand demonstrated in vivo.
To expand the limited toolbox for optochemical regulation of RNAs in living cells, we synthesized a novel photocaged guanine (pc-G or 4 in Scheme 1) and used it to control gene expression in both mammalian cells and in E. coli using light. Guanine is recognized by an RNA aptamer found in natural prokaryotic riboswitches,13 and it has been used to construct synthetic riboswitches that function in mammalian cells.14–16 The rationale for the design of the photocaged guanine (pc-G) 4 is as follows. The atomic structure of the guanine aptamer domain of the riboswitch shows that guanine forms a critically important Watson–Crick type base pairing interaction with a cytosine base in the aptamer.17 Therefore, we envisioned that masking the O6 of guanine using a photolabile group would abolish Watson–Crick as well as any potential Hoogsteen interactions with the aptamer. The methylenedioxy substituted 1-(2-nitrophenyl)ethyl group was chosen as the photolabile group because it does not produce toxic by-products upon photolysis.18 Moreover, the electron donating nature of the methylenedioxy group results in the shifting of the absorbance to a long UV range around 365 nm (Fig. S1, ESI†), thereby alleviating phototoxicity associated with short UV irradiation.
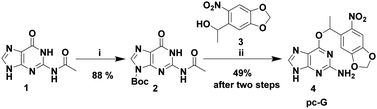 | ||
| Scheme 1 Synthesis of photocaged guanine (pc-G). Reagents and conditions: (i) Boc2O, Et3N, dry DMF, r.t., 24 h; (ii) (a) PPh3, DIAD, dry dioxane, r.t., 5 h, (b) aq. NH3, MeOH, 80 °C, 2 d. | ||
Introducing the photolabile group specifically at the O6 position requires protection of reactive N6 and N9 positions of guanine. Our initial attempts to synthesize caged guanine from the known N2,N9-diacetyl guanine were unsuccessful because of its poor solubility. Therefore, we redesigned the synthetic strategy starting from N2-acetyl guanine (1 in Scheme 1). In order to improve solubility as well as to protect the N9 position, compound 119 was reacted with Boc anhydride to furnish the N2,N9 protected form of guanine (2 in Scheme 1). As expected, introduction of a hydrophobic t-Boc group at N9 improved its solubility in solvents such as DCM and THF as compared to N2,N9-diacetyl guanine and 1 which are not soluble in these solvents. Subsequent Mitsunobu reaction of 2 and 320 provided the protected precursor to the photocaged guanine with N2-acetyl and N9-Boc groups. As the N9-Boc protection is known to be base labile,21 both the N2-acetyl and N9-boc groups were deprotected in a single step using aqueous ammonia in a sealed tube to yield pc-G (4) in 49% yield (for 2 steps). Interestingly, the photolabile group present in 4 also improved its solubility in DMSO (>100 mM) while guanine suffers from poor solubility.
Deprotection of pc-G by intense UV light (365 nm) in solution was confirmed by HPLC analysis. Exposure of pc-G (1 mM in 30% acetonitrile in water) to the UV light source (365 nm, 8 W) for 16 min almost completely deprotected pc-G (Fig. S2, ESI†). Consequently, we proceeded to use pc-G to control gene expression in mammalian cells transfected with a guanine-responsive riboswitch. We previously reported a riboswitch (GuaM8HDV) that downregulates enhanced green fluorescent protein (EGFP) expression in HEK 293 cells in the presence of guanine.15 The functional core of GuaM8HDV is the engineered aptamer–ribozyme fusion (aptazyme) inserted in the 3′ UTR of the EGFP mRNA. The self-cleavage activity of the aptazyme is greatly enhanced in the presence of guanine, thereby detaching the poly(A) tail of the mRNA and suppressing EGFP expression (Fig. 1).
The plasmid encoding the EGFP-GuaM8HDV cassette was cotransfected with an mCherry expression plasmid (as transfection efficiency control) into HEK 293 cells. Guanine (300 μM) or pc-G (300 μM) was added to the cells from stock solutions in DMSO. The same volume of DMSO was added to control cells. After 1 h, some cells were exposed to 16 min UV (365 nm) irradiation, and the cells were cultured for 2 days before fluorescence measurement. Normalized EGFP fluorescence of the control cells and the cells treated with guanine showed no significant change after UV irradiation. However, EGFP expression in the pc-G treated cells was reduced by approximately 4-fold upon UV exposure (Fig. 2A and Fig. S3, ESI†). The EGFP expression level of the pc-G and UV treated cells remained somewhat higher than that of the cells cultured in guanine, possibly due to incomplete deprotection. Use of a different photo-labile protecting group may further improve the dynamic range of gene expression. Fluorescence micrographs of the pc-G treated cells are also consistent with this result (Fig. 2B and Fig. S4, ESI†).
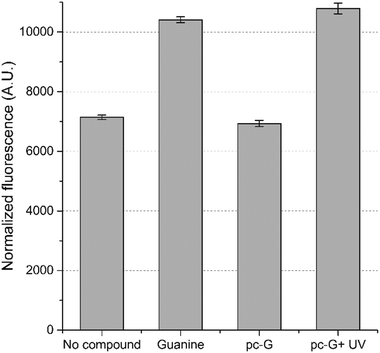
We also examined the optochemical regulation of gene expression by pc-G in E. coli. A guanine-responsive riboswitch was constructed from the adenine-activated riboswitch reported by Dixon et al.22 The uridine which makes the critical Watson–Crick base pairing interaction with the adenine ligand in the original riboswitch was mutated to cytidine to switch the ligand specificity to guanine.23 Using GFPuv as the reporter gene, we confirmed that the riboswitch activates gene expression by ∼1.5-fold in the presence of added guanine (500 μM). Comparable activation of GFPuv expression was observed when the cells were incubated with pc-G (500 μM) and exposed to UV light (16 min) (Fig. 3).
While simple chemically regulated riboswitches afford dynamic gene regulation in mammalian cells, optical gene regulation further renders precise spatial control which can be a very powerful tool in animal studies. Furthermore, optochemical control of gene expression via synthetic riboswitches requires no proteins. Genetically encoded riboswitches are also small in size (typically a few hundred bases) and function in cis making them much simpler than protein-based optogenetic switches. These characteristics make optochemical riboswitches useful for potential applications that involve viral vectors where the genetic capacity of the vectors is limited or precise tuning of trans protein factors is nontrivial.
Photocaged nucleobases have been used in the context of oligonucleotides to optically regulate nucleic acid functions such as antisense, RNAi, catalysis, and molecular sensing.24,25 However, this strategy relies on the complex synthesis of chemically modified oligonucleotides that require special cellular delivery methods. Use of synthetically accessible and cell permeable photocaged aptamer ligands to control genetically encoded riboswitches in living cells represents an alternative and flexible strategy to optically control gene expression in various cell types.
This research work was supported by the Okinawa Institute of Science and Technology Graduate University. The authors thank Dr M. C. Roy for the technical support of HPLC and HRMS analysis and Dr P. H. Patil for the NMR experiment.
Conflicts of interest
There are no conflicts to declare.Notes and references
- L. Fenno, O. Yizhar and K. Deisseroth, Annu. Rev. Neurosci., 2011, 34, 389–412 CrossRef PubMed.
- W.-C. Lin and R. H. Kramer, Bioconjugate Chem., 2018, 29, 861–869 CrossRef PubMed.
- A. S. Baker and A. Deiters, ACS Chem. Biol., 2014, 9, 1398–1407 CrossRef PubMed.
- M. Etzel and M. Mörl, Biochemistry, 2017, 56, 1181–1198 CrossRef PubMed.
- S. Ausländer and M. Fussenegger, Curr. Opin. Biotechnol., 2017, 48, 54–60 CrossRef PubMed.
- C. Berens, F. Groher and B. Suess, Biotechnol. J., 2015, 10, 246–257 CrossRef PubMed.
- M. Felletti, J. Stifel, L. A. Wurmthaler, S. Geiger and J. S. Hartig, Nat. Commun., 2016, 7, 12834 CrossRef PubMed.
- B. Wulffen, M. C. R. Buff, M. Pofahl, G. Mayer and A. Heckel, Photochem. Photobiol. Sci., 2012, 11, 489–492 Search PubMed.
- D. D. Young and A. Deiters, Bioorg. Med. Chem. Lett., 2006, 2658–2661 CrossRef PubMed.
- S. Walsh, L. Gardner, A. Deiters and G. J. Williams, ChemBioChem, 2014, 15, 1346–1351 CrossRef PubMed.
- D. D. Young, R. Garner, J. Yoder and A. Deiters, Chem. Commun., 2009, 568–570 RSC.
- L. Yen, M. Magnier, R. Weissleder, B. R. Stockwell and R. C. Mulligan, RNA, 2006, 12, 797–806 CrossRef PubMed.
- M. Mandal, B. Boese, J. E. Barrick, W. C. Winkler and R. R. Breaker, Cell, 2003, 113, 577–586 CrossRef PubMed.
- Y. Nomura, D. Kumar and Y. Yokobayashi, Chem. Commun., 2012, 48, 7215–7217 RSC.
- Y. Nomura, L. Zhou, A. Miu and Y. Yokobayashi, ACS Synth. Biol., 2013, 2, 684–689 CrossRef PubMed.
- S. Kobori, Y. Nomura, A. Miu and Y. Yokobayashi, Nucleic Acids Res., 2015, 43, e85 CrossRef PubMed.
- A. Serganov, Y.-R. Yuan, O. Pikovskaya, A. Polonskaia, L. Malinina, A. T. Phan, C. Hobartner, R. Micura, R. R. Breaker and D. J. Patel, Chem. Biol., 2004, 11, 1729–1741 CrossRef PubMed.
- J. Luo, E. Arbely, J. Zhang, C. Chou, R. Uprety, J. W. Chin and A. Deiters, Chem. Commun., 2016, 52, 8529–8532 RSC.
- D. D. Vo, C. Staedel, L. Zehnacker, R. Benhida, F. Darfeuille and M. Duca, ACS Chem. Biol., 2014, 9, 711–721 CrossRef PubMed.
- M. Wirkner, J. M. Alonso, V. Maus, M. Salierno, T. T. Lee, A. J. García and A. del Campo, Adv. Mater., 2011, 23, 3907–3910 CrossRef PubMed.
- A. Porcheddu, G. Giacomelli, I. Piredda, M. Carta and G. Nieddu, Eur. J. Org. Chem., 2008, 5786–5797 CrossRef.
- N. Dixon, J. N. Duncan, T. Geerlings, M. S. Dunstan, J. E. G. McCarthy, D. Leys and J. Micklefield, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2830–2835 CrossRef PubMed.
- M. Mandal and R. R. Breaker, Nat. Struct. Mol. Biol., 2004, 11, 29–35 Search PubMed.
- Q. Liu and A. Deiters, Acc. Chem. Res., 2014, 47, 45–55 CrossRef PubMed.
- N. Ankenbruck, T. Courtney, Y. Naro and A. Deiters, Angew. Chem., Int. Ed., 2018, 57, 2768–2798 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, UV/HPLC analysis, plasmid sequences, additional fluorescence micrographs, and NMR spectra. See DOI: 10.1039/c8cc02290a |
| This journal is © The Royal Society of Chemistry 2018 |