Open Access Article
Open Access ArticleA supramolecular self-assembly strategy for upconversion nanoparticle bioconjugation†
Yulong
Sun
 abc,
Wenjing
Zhang
acd,
Baoming
Wang
abc,
Wenjing
Zhang
acd,
Baoming
Wang
 ac,
Xiaoxue
Xu
ac,
Xiaoxue
Xu
 ac,
Joshua
Chou
ac,
Joshua
Chou
 e,
Olga
Shimoni
e,
Olga
Shimoni
 *abc,
Alison T.
Ung
*abc,
Alison T.
Ung
 c and
Dayong
Jin
c and
Dayong
Jin
 *abc
*abc
aInstitute of Biomedical Materials & Devices (IBMD), Faculty of Science, University of Technology Sydney, Ultimo, Sydney, NSW 2007, Australia
bARC Research Hub of Integrated Device for End-user Analysis at Low-level (IDEAL), Faculty of Science, University of Technology Sydney, Ultimo, NSW 2007, Australia
cSchool of Mathematical and Physical Sciences, Faculty of Science, University of Technology Sydney, Ultimo, NSW 2007, Australia
dInstitute of Advanced Materials for Nano-Bio Application, School of Ophthalmology and Optometry, Wenzhou Medical University, Wenzhou, Zhejiang, 310012, P. R. China
eSchool of Life Science, Advanced Tissue Regeneration and Drug Delivery Group, Faculty of Science, University of Technology Sydney, Ultimo, NSW 2007, Australia. E-mail: dayong.jin@uts.edu.au; olga.shimoni@uts.edu.au
First published on 19th March 2018
Abstract
An efficient surface modification for upconversion nanoparticles (UCNPs) is reported via supramolecular host–guest self-assembly. Cucurbit[7]uril (CB) can provide a hydrophilic surface and cavities for most biomolecules. High biological efficiency, activity and versatility of the approach enable UCNPs to be significantly applied in bio-imaging, early disease detection, and bio-sensing.
Surface functionalization and bioconjugation hold the key to driving many purpose-synthesized inorganic nanoparticles into real-world applications. Lanthanide ion doped upconversion nanoparticle (UCNP) is a good example1 as it possesses a range of diverse applications including: molecular probes, biosensors, drug carriers and light transducers for drug delivery,2 light activated therapy,3 single molecular bio-sensing,4 super-resolution nanoscopy5 and disease diagnostics.6
However, the hydrophobic surface of UCNPs is one of the main challenges for biomedical applications. It remains a bottleneck challenge for the community to explore practical strategies to functionalize the UCNP surfaces for conversion of hydrophilicity and further bioconjugation to meet many cellular and molecular specific needs. The most direct approach to convert hydrophobic UCNPs into a hydrophilic surface is by removing the hydrophobic oleic acid (OA) molecules on the surface using an acid solution, then coating the hydrophilic molecules with a reactive group for further modification.7 However, the bare UCNPs tend to be extremely unstable causing hydrolysis, ripping, corrosion and aggregation during the exchange process under acidic conditions.8 Direct ligand-exchange can be applied to displace the native ligands according to the order of binding strength.9 For example, phosphate was used for replacing the surface OA, because its binding strength is higher than the carboxylate on the surface of UCNPs.10 Several (co)polymers with carboxylate and/or phosphate have been utilized where each polymer molecule would react with multiple binding sites. This resulted in a much higher interaction than that of the ligands with a single binding site.11–14 However, it is difficult to control all the binding groups from each polymer molecule to attach orderly and selectively to the UCNP without inducing aggregation of the nanoparticles. Other strategies, such as oxidation of OA,15–17 amphiphilic interaction,18 and silica/metal shell coating,19–22 have been explored to render UCNPs with a hydrophilic surface.
Although the above methods have achieved varying degrees of success, their subsequent conjugation of biomolecules onto the functionalised surface (e.g., EDC reaction)23 remains a challenge, often resulting in low-yield modified biomolecules due to repulsion, steric hindrance effects, and depleted bioactivities of the modified biomolecules, which hinders their stability and functionality caused by the change in the conformation or blockage of the targeted functional groups.
In this study, we report a one-step integrated approach to convert the hydrophobicity of UCNPs (OA–UCNPs) into hydrophilicity via interfacial ligand exchange (Fig. 1). The new method is based on the host–guest self-assembly. Cucurbit[7]uril (CB[7]) has been chosen in this work as the host molecule to efficiently replace the surfactant molecules of OA24 and form CB[7] coated UCNPs (CB–UCNPs). The glycoluril molecular structure (Fig. 1) in CB[7] forms the circular wall frame of the ring, and the seven ketones with high electronegativity sit on the top and bottom of the ring structure to form the molecular host structure. This unique structure allows CB[7] to easily entirely replace OA due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the urea functional group that can form resonance delocalization of N lone pair electrons to form polar interactions on both edges of the CB[7] molecular ring. Either one of these edges can undergo selective self-assembly on the surface of UCNP as indicated in Fig. 1. The molecular architecture of CB[7] comprises a hydrophobic host-environment which allows the host–guest recognition of non-polar guest molecules such as adamantane. This approach can achieve successful ligand exchange without acid washing. One of the main benefits of using CB[7] over polymer molecules is that it can avoid particle aggregation. Moreover, this will allow for selective and controlled self-assembly of biomolecules onto the surface of UCNP via the CB[7] host–guest inclusion of a specific non-polar guest appended-molecule such as adamantane (Fig. 1) without the need of an organic reaction. This modular approach to the construction of bioconjugated UCNPs can overcome the difficulty often encountered with the attachment of biomolecules and allows for a broad range of structural biomolecules that can be applied.25 As the cavity of CB[7] is directly perpendicular to the particle surface,7 during self-assembly, the biomolecules will align and radiate outward on the surface of the UCNP due to the positive surface charge.
O of the urea functional group that can form resonance delocalization of N lone pair electrons to form polar interactions on both edges of the CB[7] molecular ring. Either one of these edges can undergo selective self-assembly on the surface of UCNP as indicated in Fig. 1. The molecular architecture of CB[7] comprises a hydrophobic host-environment which allows the host–guest recognition of non-polar guest molecules such as adamantane. This approach can achieve successful ligand exchange without acid washing. One of the main benefits of using CB[7] over polymer molecules is that it can avoid particle aggregation. Moreover, this will allow for selective and controlled self-assembly of biomolecules onto the surface of UCNP via the CB[7] host–guest inclusion of a specific non-polar guest appended-molecule such as adamantane (Fig. 1) without the need of an organic reaction. This modular approach to the construction of bioconjugated UCNPs can overcome the difficulty often encountered with the attachment of biomolecules and allows for a broad range of structural biomolecules that can be applied.25 As the cavity of CB[7] is directly perpendicular to the particle surface,7 during self-assembly, the biomolecules will align and radiate outward on the surface of the UCNP due to the positive surface charge.
 | ||
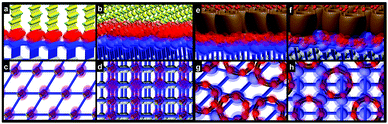
| Fig. 1 Cucurbit[7]uril-based one-step hydrophilic ligand exchange approach without reaction conjugation via host–guest self-assembly. | ||
To evaluate the efficiency of the replacement and the binding behavior of CB-UCNPs, we calculated the binding site and mode of OA–UCNPs and CB–UCNPs via crystallographic analysis (more details in Section 4 of the ESI†). In OA–UCNPs, two types of OA (OA–COOH and OA–COO−) can bind onto the surface of bare UCNPs (see Table S1 in the ESI†). OA–COO−, when compared with OA–COOH, can form two equal oxygen–yttrium bonds because of the resonance effect, which is stronger than an yttrium–oxygen bond and a hydrogen bond from OA–COOH. Due to the distance of the two yttrium atoms, both oxygens in OA–COO− bind to the same yttrium atom. The optimised binding modes on both crystal facets are shown in Fig. 2a–d. In comparison with the binding mechanics of OA–UCNPs, there is strong charge interaction between the negatively charged edge of CB[7] molecules and the positively charged surface of UCNP. The array of negatively charged oxygen atoms that are aligned to the edges of the CB[7] molecules contributes to the binding (Fig. 2e–h and Fig. S23 in ESI†). Therefore, the binding of CB[7] was calculated to be much stronger than that found between OA and the UCNP (see Section 4.3 in the ESI†).
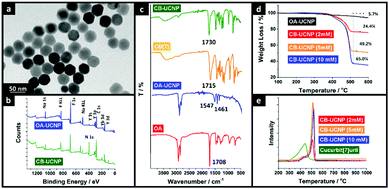
The TEM image of CB–UCNPs (Fig. 3a) shows a hexagonal morphology, consistent monodispersity and homogeneous size, which is the same as OA–UCNPs (Fig. S14 in ESI†). The image illustrates that one-step ligand exchange retains the morphology of UCNPs. The average size of the nanoparticles is around 37 nm before and after ligand exchange, according to the distributions of OA–UCNPs and CB–UCNPs, which were calculated and are displayed in Fig. S15 and S16 (ESI†). Fig. 3b shows the X-ray photoelectron spectra (XPS) to determine the surface properties. Compared with the XPS of OA–UCNPs (blue in Fig. 3b), the nitrogen (N 1s) from the glycoluril units in CB[7] was identified in CB–UCNPs (green in Fig. 3b). More XPS data on other elements can be found in Section 3.4 of the ESI.† Fourier transform infrared (FT-IR) spectroscopy was also used to investigate the surface of UCNPs. Fig. 3c shows the stretching vibration of the carbonyl group in free OA molecules at 1708 cm−1. After being stabilized on the surface of UCNPs, the absorption peak shifted and appeared as two bands: at 1547 cm−1 (the asymmetric stretching vibration of the carbonyl group) and 1461 cm−1 (the symmetric stretching vibration of the carbonyl group). This indicates that all the acidic terminal groups of OA are carboxylate (–CO2–) rather than carboxylic acid (–COOH). It further reinforces our calculation of the optimized binding modes on the surface of UCNPs. After OA was exchanged with CB[7], the stretching vibration of C–H (∼3005 cm−1) in the sp2 carbon of OA was completely absent (see Fig. S4 in the ESI†), proving that the OA on the surface of UCNP has been completely removed. This is further reinforced by the presence of a strong and sharp carbonyl stretching vibration at 1730 cm−1. This band was identified to be the asymmetric stretching vibration of CB[7] (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) which was shifted to higher values (∼15 cm−1) when bound to the surface of UCNPs. These changes corroborated the interaction of CB[7] with the UCNP surface through the carbonyl groups. The higher values suggest a further deviation from the planarity of the carbonyl bond compared with the N–C–N plane when the CB[7] is anchored to the nanoparticle surface, which can attribute to a less effective interaction of CB[7] with the surface of the nanoparticle. Thermogravimetric analysis (TGA) curves and derivative thermogravimetric analysis (DTGA) were used for verifying the attachment of CB[7] molecules to the surface of UCNPs (Fig. 3d) and for determining the best ligand-exchange concentration of CB[7] (Fig. 3d and e). The detection range for weight loss was between 100 °C to 600 °C, and all the samples showed weight loss below 180 °C resulting from the physical loss of free water. In Fig. 3d, the loss of 5.7% of the weight from 100 °C to 450 °C in OA–UCNPs was due to the removal of the OA on the surface of the UCNPs. However, the weight loss of CB–UCNPs shifted from 400 °C to 550 °C. To determine the optimal concentration of CB[7] for preparing CB–UCNPs, TGA and DTGA results from samples with different concentrations of CB[7] are displayed in Fig. 3d and e. The weight losses from 350 °C to 550 °C of CB–UCNP with ligand-exchange with 2 mM, 5 mM and 10 mM concentrations are 24.4%, 49.2% and 65.0%, respectively. As already shown in Fig. 3e, after CB[7] attached to the surface of UCNPs, the derivative weight loss peak moves to the higher temperature as free molecules. Fig. 3e shows that all the main peaks of CB–UCNPs (2 mM, 5 mM, and 10 mM) shift to 488 °C, 491 °C, and 498 °C, respectively. This means that CB[7] molecules were stabilized on the surface of UCNPs. However, another peak in both 5 mM and 10 mM samples was identified at the same position of free CB[7], which proved that many free CB[7] molecules still exist in the system. This will affect the self-assembly behavior in subsequent experiments, so the 2 mM concentration was selected for all samples in Fig. 3a–c.
O) which was shifted to higher values (∼15 cm−1) when bound to the surface of UCNPs. These changes corroborated the interaction of CB[7] with the UCNP surface through the carbonyl groups. The higher values suggest a further deviation from the planarity of the carbonyl bond compared with the N–C–N plane when the CB[7] is anchored to the nanoparticle surface, which can attribute to a less effective interaction of CB[7] with the surface of the nanoparticle. Thermogravimetric analysis (TGA) curves and derivative thermogravimetric analysis (DTGA) were used for verifying the attachment of CB[7] molecules to the surface of UCNPs (Fig. 3d) and for determining the best ligand-exchange concentration of CB[7] (Fig. 3d and e). The detection range for weight loss was between 100 °C to 600 °C, and all the samples showed weight loss below 180 °C resulting from the physical loss of free water. In Fig. 3d, the loss of 5.7% of the weight from 100 °C to 450 °C in OA–UCNPs was due to the removal of the OA on the surface of the UCNPs. However, the weight loss of CB–UCNPs shifted from 400 °C to 550 °C. To determine the optimal concentration of CB[7] for preparing CB–UCNPs, TGA and DTGA results from samples with different concentrations of CB[7] are displayed in Fig. 3d and e. The weight losses from 350 °C to 550 °C of CB–UCNP with ligand-exchange with 2 mM, 5 mM and 10 mM concentrations are 24.4%, 49.2% and 65.0%, respectively. As already shown in Fig. 3e, after CB[7] attached to the surface of UCNPs, the derivative weight loss peak moves to the higher temperature as free molecules. Fig. 3e shows that all the main peaks of CB–UCNPs (2 mM, 5 mM, and 10 mM) shift to 488 °C, 491 °C, and 498 °C, respectively. This means that CB[7] molecules were stabilized on the surface of UCNPs. However, another peak in both 5 mM and 10 mM samples was identified at the same position of free CB[7], which proved that many free CB[7] molecules still exist in the system. This will affect the self-assembly behavior in subsequent experiments, so the 2 mM concentration was selected for all samples in Fig. 3a–c.
As mentioned above, the water-soluble CB–UCNPs provide a number of cavities which can bind with biomolecules via supramolecular host–guest self-assembly. Most of the biomolecules can assemble on the CB-UCNPs as long as the molecule has (1) a positive charge portion (charge transfer with upper ketones) and/or (2) a hydrophobic portion (hydrophobic interaction with hydrophobic cavities).26 If the biomolecule lacked both of the required properties, or if its active sites are neither positive nor hydrophobic, then the biomolecules can be modified via simple organic reactions (see Section 2.5.2 in the ESI†) which will allow it to bind into the cavities.
Compared with EDC amide coupling, self-assembly has shown several advantages in bio-conjugation.27 Self-assembly is based on intermolecular forces without requiring any organic reaction for conjugation.24 When CB–UCNPs were mixed with guest molecules, the guest molecules were shown to undergo host–guest recognition and then attached into the CB–UCNP cavities. Although there is no formation of covalent bonds in the process the binding strength is sufficiently strong to form a stable assembly on the surface of the nanoparticles.28 The binding mechanism is shown in Fig. S27 of the ESI.†
Most biological conjugations utilize EDC amide coupling because of the mild reaction conditions.29 However, the strong positive surface charge of the nanoparticle impedes the reaction process. It is therefore difficult for the biomolecules to get close to the nanoparticle surface due the repulsive force created by the positive charge of the nanoparticle surface. The positive charge could also attract the conjugated molecules to bind directly to the surface resulting in the misalignment of the assembly or even changing the conformation of the biomolecules.30 To investigate the binding behavior of the CB–UCNPs, an IgG conjugate was chosen for this study. The IgG was coupled to a molecule with high binding strength, 1-adamantane acid, at the N-terminus of the protein via an amide coupling reaction. This creates a biomolecule that contains a small guest-molecule (1-adamantanecarboxyl, ADC) which strategically appended at the N-terminus of the protein to form IgG–ADC. The ADC moiety is designed to anchor to the surface of CB–UCNP via the host–guest inclusion complex. This structural design would reduce the undesired interactions of the biomolecules with each other or to the surface of the UCNP. This approach would encourage the self-assembly of the biomolecules in an organized fashion.
The IgG–ADC would be expected to self-assemble on the surface of CB–UCNPs to form CB–UCNP@IgG–ADC after mixing with the CB–UCNP dispersion. In comparison to the EDC binding that used 10 times higher IgG concentration than IgG–ADC, the binding efficiency of the self-assembly of IgG–ADC is more than twice that of EDC amide coupling (Fig. 4a). The accessibility of CB–UCNPs for bioconjugation was also assessed. The binding efficiency was found to be four times higher than that of the EDC coupling method (Fig. 4b). The binding rate of self-assembly was also evaluated by allowing the IgG–ADC to interact with CB-UCNPs at different mixing times. At each time point, the resulting CB–UCNP@IgG–ADC was collected for analysis. The half-life time of IgG–ADC binding to CB–UCNP is about thirty minutes (Fig. 4c). The shelf-life of the conjugation was tested and is shown in Section 4.11 of the ESI.† The half-life of the shelf-life is almost a week (ca. 167 h). Only 6.6% free IgG was shown in the solution, indicating the disassembly at a very slow rate after 8 d. This behavior shows the enormous potential for CB–UCNPs as a reagent in early disease detection. To display the targeting behavior of CB–UCNP@IgG–ADC, cytotoxicity by MTT assay with a HeLa cell line has shown the nanoparticles to be biocompatible (see Fig. S30 in the ESI†). To demonstrate the biological specificity of the CB–UCNPs, E-cadherin IgG bound to CB–UCNP was tested by immunofluorescence imaging under excitation of 980 nm laser. From Fig. 4d, it is clear that the E-cadherin IgG bound CB–UCNP was able to target the E-cadherin transmembrane protein of the HeLa cells.
 | ||
| Fig. 4 Characterizations of bioconjugation on the surface of CB–UCNPs via supramolecular self-assembly, compared with conventional binding. (a) Conjugation amount and (b) availability of functional groups on the surface of CB–UCNPs and UCNPs–COOH (see section 2.3 in ESI†); (c) binding rate of IgG–ADC on the surface of CB–UCNPs and (d) cell imaging of CB–UCNP@IgG–ADC (blue) targeting on the surface of HeLa cells. The actin cytoskeleton of the cell was stained with Phalloidin–FITC (green) and the nucleus stained with DAPI (yellow). | ||
In conclusion, CB[7] has shown to have dual roles in both an efficient ligand exchange process and supramolecular conjugation of biomolecules. It converts UCNPs from a hydrophobic surface into a hydrophilic surface and creates an array of specific anchor points for the attachment of biomolecules via host–guest inclusion. This offers an advance in simplicity, stability, and high yield, compared with EDC bioconjugation. The binding behavior of CB–UCNPs to a selected biomolecule, e.g., IgG antibodies, shows the advantages of this approach in binding capability, availability, conjugation rate and cell targeting. The scope of the applications of CB–UCNPs in the field of bioconjugation to create molecular tools that can be used in early disease detection would be wide and significant.
We acknowledge the financial support provided by the Australian Research Council (ARC) Future Fellowship Scheme (FT 130100517), Chancellor Postdoctoral Research Fellowship (PRO16-1920-2017), and National Health and Medical Research Council (NHMRC) Dementia Research Fellowship (Grant APP1101258).
Conflicts of interest
There are no conflicts to declare.Notes and references
- B. Zhou, B. Shi, D. Jin and X. Liu, Nat. Nanotechnol., 2015, 10, 924–936 CrossRef CAS PubMed.
- J. Shen, L. Zhao and G. Han, Adv. Drug Delivery Rev., 2013, 65, 744–755 CrossRef CAS PubMed.
- N. M. Idris, M. K. Jayakumar, A. Bansal and Y. Zhang, Chem. Soc. Rev., 2015, 44, 1449–1478 RSC.
- C. L. Wang, X. M. Li and F. Zhang, Analyst, 2016, 141, 3601–3620 RSC.
- Y. Liu, Y. Lu, X. Yang, X. Zheng, S. Wen, F. Wang, X. Vidal, J. Zhao, D. Liu, Z. Zhou, C. Ma, J. Zhou, J. A. Piper, P. Xi and D. Jin, Nature, 2017, 543, 229–233 CrossRef CAS PubMed.
- P. Huang, W. Zheng, S. Y. Zhou, D. T. Tu, Z. Chen, H. M. Zhu, R. F. Li, E. Ma, M. D. Huang and X. Y. Chen, Angew. Chem., Int. Ed., 2014, 53, 1252–1257 CrossRef CAS PubMed.
- L. Frances-Soriano, M. Gonzalez-Bejar and J. Perez-Prieto, Nanoscale, 2015, 7, 5140–5146 RSC.
- S. Lahtinen, A. Lyytikäinen, H. Päkkilä, E. Hömppi, N. Perälä, M. Lastusaari and T. Soukka, J. Phys. Chem. C, 2017, 121, 656–665 CAS.
- L. N. Sun, T. Liu, Y. N. Qiu, J. L. Liu, L. Y. Shi and O. S. Wolfbeis, Microchim. Acta, 2014, 181, 775–781 CrossRef CAS.
- J. Lu, Y. Chen, D. Liu, W. Ren, Y. Lu, Y. Shi, J. Piper, I. Paulsen and D. Jin, Anal. Chem., 2015, 87, 10406–10413 CrossRef CAS PubMed.
- R. Naccache, F. Vetrone, V. Mahalingam, L. A. Cuccia and J. A. Capobianco, Chem. Mater., 2009, 21, 717–723 CrossRef CAS.
- J. C. Boyer, M. P. Manseau, J. I. Murray and F. van Veggel, Langmuir, 2010, 26, 1157–1164 CrossRef CAS PubMed.
- V. Voliani, M. Gonzalez-Bejar, V. Herranz-Perez, M. Duran-Moreno, G. Signore, J. M. Garcia-Verdugo and J. Perez-Prieto, Chem. – Eur. J., 2013, 19, 13538–13546 CrossRef CAS PubMed.
- L. Xia, X. G. Kong, X. M. Liu, L. P. Tu, Y. L. Zhang, Y. L. Chang, K. Liu, D. Z. Shen, H. Y. Zhao and H. Zhang, Biomaterials, 2014, 35, 4146–4156 CrossRef CAS PubMed.
- Z. G. Chen, H. L. Chen, H. Hu, M. X. Yu, F. Y. Li, Q. Zhang, Z. G. Zhou, T. Yi and C. H. Huang, J. Am. Chem. Soc., 2008, 130, 3023–3029 CrossRef CAS PubMed.
- H. Hu, M. X. Yu, F. Y. Li, Z. G. Chen, X. Gao, L. Q. Xiong and C. H. Huang, Chem. Mater., 2008, 20, 7003–7009 CrossRef CAS.
- H. P. Zhou, C. H. Xu, W. Sun and C. H. Yan, Adv. Funct. Mater., 2009, 19, 3892–3900 CrossRef CAS.
- L. L. Li, R. B. Zhang, L. L. Yin, K. Z. Zheng, W. P. Qin, P. R. Selvin and Y. Lu, Angew. Chem., Int. Ed., 2012, 51, 6121–6125 CrossRef CAS PubMed.
- H. Hu, L. Q. Xiong, J. Zhou, F. Y. Li, T. Y. Cao and C. H. Huang, Chem. – Eur. J., 2009, 15, 3577–3584 CrossRef CAS PubMed.
- L. Sudheendra, V. Ortalan, S. Dey, N. D. Browning and I. M. Kennedy, Chem. Mater., 2011, 23, 2987–2993 CrossRef CAS PubMed.
- B. A. Dong, S. Xu, J. A. Sun, S. Bi, D. Li, X. Bai, Y. Wang, L. P. Wang and H. W. Song, J. Mater. Chem., 2011, 21, 6193–6200 RSC.
- C. Zhang and J. Y. Lee, J. Phys. Chem. C, 2013, 117, 15253–15259 CAS.
- Y. Shi, B. Y. Shi, A. V. E. Dass, Y. Q. Lu, N. Sayyadi, L. Kautto, R. D. Willows, R. Chung, J. Piper, H. Nevalainen, B. Walsh, D. Y. Jin and N. H. Packer, Sci. Rep., 2016, 6, 11 CrossRef PubMed.
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Isaacs, Angew. Chem., Int. Ed., 2005, 44, 4844–4870 CrossRef CAS PubMed.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. Del Barrio and O. A. Scherman, Chem. Rev., 2015, 115, 12320–12406 CrossRef CAS PubMed.
- A. R. Urbach and V. Ramalingam, Isr. J. Chem., 2011, 51, 664–678 CrossRef CAS.
- A. Fragoso, J. Caballero, E. Almirall, R. Villalonga and R. Cao, Langmuir, 2002, 18, 5051–5054 CrossRef CAS.
- A. Corma, H. Garcia, P. Montes-Navajas, A. Primo, J. J. Calvino and S. Trasobares, Chemistry, 2007, 13, 6359–6364 CrossRef CAS PubMed.
- K. A. Totaro, X. L. Liao, K. Bhattacharya, J. I. Finneman, J. B. Sperry, M. A. Massa, J. Thorn, S. V. Ho and B. L. Pentelute, Bioconjugate Chem., 2016, 27, 994–1004 CrossRef CAS PubMed.
- R. A. Sperling and W. J. Parak, Philos. Trans. R. Soc., A, 2010, 368, 1333–1383 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, preparation and synthesis, materials characterizations, binding behaviours, and cell study. See DOI: 10.1039/c8cc00708j |
| This journal is © The Royal Society of Chemistry 2018 |