Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microfluidic formation of proteinosomes†
Martina
Ugrinic
a,
Adrian
Zambrano
 b,
Simon
Berger
a,
Stephen
Mann
c,
T.-Y. Dora
Tang
*bc and
Andrew
deMello
b,
Simon
Berger
a,
Stephen
Mann
c,
T.-Y. Dora
Tang
*bc and
Andrew
deMello
 *a
*a
aDepartment of Chemistry & Applied Biosciences, ETH Zurich, Vladimir Prelog Weg 1, 8093 Zurich, Switzerland. E-mail: andrew.demello@chem.ethz.ch
bMax Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstraße 108, 01307 Dresden, Germany. E-mail: tang@mpi-cbg.de
cCentre for Protolife Research, School of Chemistry, University of Bristol, Bristol, BS8 1TS, UK
First published on 6th December 2017
Abstract
Herein we describe a novel microfluidic method for the generation of proteinosome micro-droplets, based on bovine serum albumin and glucose oxidase conjugated to PNIPAAm chains. The size of such water-in-oil droplets is regulated via control of the input reagent flow rate, with generated proteinosome populations exhibiting narrower size distributions than those observed when using standard bulk methodologies. Importantly, proteinosomes transferred from an oil to an aqueous-environment remain intact, become fully hydrated and exhibit an increase in average size. Moreover, functional proteinosomes prepared via microfluidics exhibit lower Km values and higher enzymatic activities than proteinosomes produced by bulk methodologies.
A central aim in synthetic biology is the creation of artificial cells or cell-like objects, a feat generally accomplished by compartmentalizing minimalistic biological reactions in micron-scale water filled environments, thus mimicking the compartmentalisation and isolation of complex processes characteristic of cells and higher organisms.1 Compartmentalisation can be achieved by spontaneous self-assembly of polyelectrolytes to form chemically enriched membrane free droplets. Alternatively, membrane bound systems, such as surfactant stabilised water-in-oil emulsions2 or water–water systems based on inorganic nanoparticles (colloidosomes),3 polymer (polymersomes),4 lipids (liposomes)5 or protein–polymer conjugates (proteinosomes)6 have also been demonstrated.
In the majority of these compartments, the membrane acts as a barrier to contain various complex biochemical reactions, such as PCR,7 cell-free gene expression8,9 or more targeted directed evolution experiments,10 as well as a whole host of chemical reactions.11 However, the membrane typically remains inert unless additional components are added to impart activity.12 Proteinosomes offer an alternative route to membrane activation, where chemically modified enzymes can be rendered amphiphilic by the addition of poly(N-isopropylacrylamide) (PNIPAAm) chains and then used to generate a stable water-in-oil emulsion. The self-assembled protein-conjugates can further be chemically cross-linked and transferred into water to generate micron-sized water-in-water compartments with an enzymatically active membrane. The biological activity of a proteinosome can further be tailored by encapsulating a variety of enzymes, yielding a route towards compartmentalised, multi-step enzyme cascades. Critically, proteinosomes are stable over many weeks, can be externally stimulated by creating temperature or chemical gradients,12 and can be engineered to generate complex nested structures,13 capable of performing multi-step enzymatic reactions. However, despite their robustness and versatility as artificial cellular compartments, proteinosomes generated using standard bulk methodologies exhibit a large variability in their overall size and volume. Accordingly, methods that provide for facile control of proteinosome size and population size distribution (and thus mass transfer kinetics and diffusion rates) are highly desirable.
Herein, we address these limitations using a microfluidic device, capable of generating monodisperse, micron-sized water-in-oil compartments, stabilised either by cross-linked cationized bovine serum albumin/PNIPAAm (BSA-NH2/PNIPAAm) or glucose oxidase/PNIPAAm (GOx-NH2/PNIPAAm) conjugates, in a high-throughput manner. We demonstrate, that water-in-water proteinosomes generated using our microfluidic device retain encapsulated enzymes and are able to initiate multi-step enzyme cascades with high efficiency.‡
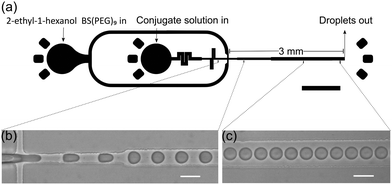
Water-in-oil emulsions stabilized by either BSA-NH2/PNIPAAm or enzymatically active GOx-NH2/PNIPAAm membranes, encapsulating horseradish peroxidase (HRP), were prepared using a microfluidic flow-focusing junction (Fig. 1a).
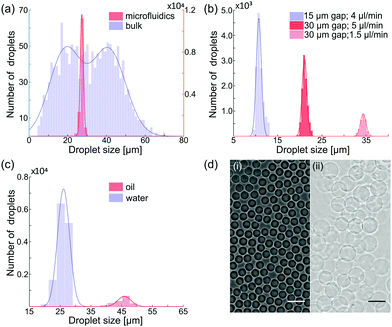
The microfluidic device was fabricated in polydimethylsiloxane, following conventional soft lithography protocols and incorporated a 3 mm long collection channel, gradually expanding from 30 to 70 μm width. The disperse and continuous phase inputs at the flow junction were 30 and 15 μm wide respectively. All channels were 36 μm in height and fluidic resistors at the aqueous phase inlet were positioned upstream of the flow focusing junction to ensure stable droplet formation. High-speed bright-field imaging confirmed that water-in-oil droplet formation at the flow focusing junction was both stable and reproducible (Fig. 1b, c and ESI†). Analysis of droplets generated using flow rates of 1 and 3.5 μl min−1 for the disperse (BSA-NH2/PNIPAAm) and continuous phase respectively, yielded an average diameter of 27.1 μm (±1.0 μm) (Fig. 2a). Comparisons with proteinosomes produced using bulk methodologies showed a significant decrease in the RSD (relative standard deviation) from 51.8% to 3.6%. Likewise, water-in-oil emulsions formed with GOx-NH2/PNIPAAm yielded droplets with an average diameter of 25.7 μm (±1.8 μm) and a RSD of 7% (Fig. 2c), confirming that the microfluidic method is applicable to different protein-conjugates (i.e. BSA and GOx). Average droplet size was constrained by the dimensions of the flow focusing junction; for example, droplets of approximately 30 μm were produced using a junction width of 30 μm, whilst a 15 μm wide flow-focusing junction led to a decrease in the average droplet size to approximately 15 μm (Fig. 2b). Flow rates between the 30 μm and 15 μm channels were not directly comparable due to a difference in channel dimensions, where the latter device comprised a 3 mm long collection channel which varied from 15 to 70 μm, a channel height of 20 μm and a 10 μm input channel for the continuous phase. As expected, droplet size could also be regulated by varying the flow rates of the input flows. Specifically, an increase in the flow rate of the continuous phase led to smaller droplets, whilst a reduction in the flow rate of the continuous phase led to the production of large droplets with RSDs consistently below 7% (Fig. S1, ESI†). Thus, by rational design of the channel geometry and regulation of the flow rate, we were able to use this methodology to controllably vary the size of the droplets and maintain a high degree of monodispersity, whilst generating more than 1 million droplets in 30 minutes (see ESI†), using only 30 μl of the polymer–protein conjugate solution. In addition, the process can be scaled up by operating multiple devices in parallel (32 devices are integrated on a single glass slide, Fig. S2, ESI†) or by increasing the volume of conjugate solution in the inlet channel and running the device for longer periods of time.
Whilst the generation of water-in-oil droplets provides a unique route to generating synthetic cells, production of water-in-water droplets offers the opportunity to utilise the interface as part of a reaction system, leading to increased biological functionality. Consequently, PEG-bis (N-succinimidyl succinate) (BS(PEG)9) was included in the continuous phase to chemically crosslink protein conjugates at the surface of the water-in-oil droplets. After collection and 2 hours of incubation, the monodisperse water-in-oil emulsion was transferred into an ethanol/water mix and subsequently into Milli-Q® water prior to imaging. Image analysis confirmed that the formed droplets were chemically cross-linked, with the diameters increased by 167% for BSA-proteinosomes and 63% for GOx-proteinosomes due to full hydration (Fig. 2c and d). Moreover, droplets were found to be stable for periods in excess of one month, with no structural changes or aggregation, and retention of the encapsulated enzyme (Fig. S3a, ESI†).
With a view to generating complex biological systems with hierarchically organized enzyme reactions, we assessed the encapsulation efficiency of horseradish peroxidase within our water-in-oil droplets. Fluorescently labelled horseradish peroxidase (HRP-FITC) added to the aqueous phase, was encapsulated in GOx-NH2/PNIPAAm stabilised water-in-oil droplets either via standard bulk methodologies or using the microfluidic device. Fluorescence images demonstrate a homogeneous distribution of FITC in the water-in-oil droplets (Fig. 3a(i)) produced in the microfluidic device, confirming the utility of the microfluidic method for the uniform encapsulation of enzymes. In addition, transfer of HRP encapsulated proteinosomes into water showed retention of the fluorescent HRP within the protocells (Fig. 3a(ii)) for over a month (Fig. S3b, ESI†). Subsequently, the activity of cross-linked GOx-NH2/PNIPAAm proteinosomes containing HRP in water was assessed by determining the Michaelis–Menten constant, Km, for a glucose oxidase and horseradish peroxidase assay (Fig. S6, ESI†). Km was determined via the fluorescence increase observed upon oxidation of Amplex Red to Resorufin (λmax = 587 nm), as a function of glucose concentration, and with an excess of glucose oxidase and HRP (ESI,† Materials and methods and Fig. S8). For microfluidically produced proteinosomes a Km of 6 mM (±1.3 mM) for GOx/HRP was determined (Fig. 3b), approximately half the value found for proteinosomes generated via standard bulk methodologies, Km = 12.6 mM (±2.2 mM). Correspondingly, microfluidically generated proteinosomes show a significantly higher enzyme activity, compared to those produced in bulk. Importantly, in both instances the Km value is greater than for non-cross-linked GOx-NH2/PNIPAAm conjugates (Km = 3.1 mM ± 1.1 mM) and native HRP (Km = 4 mM ± 1 mM) indicating that conjugate cross-linking at the membrane leads to more inefficient substrate binding attributable to either change in structure of the binding site or a reduction in the mass transfer of substrates into and within the proteinosomes.
In summary, we have presented a novel and high-throughput microfluidic platform for the production of proteinosomes, constructed from protein–polymer conjugates. Critically, the approach provides for significantly narrower size distributions of the formed proteinosomes and enhanced encapsulation properties when compared to bulk techniques. Furthermore, we demonstrate that proteinosomes produced via microfluidics are functional, through the enhanced reaction between glucose oxidase and horseradish peroxidase, compared to the bulk method. Such excellent size control and improved activity opens up new possibilities in the design of complex multi-compartmentalised structures for in vitro simulation of complex biological systems.
We thank Dr Sebastian Sauch and Dr Petra Uhlmann (Leibniz-Institut für Polymerforschung, Dresden) for RAFT agent synthesis and Dr Albena Lederer (Leibniz-Institut für Polymerforschung, Dresden) for PNIPAAm characterisation. NMR support was provided by Annett Lohmann and Dr Andre Nadler. TYDT and AZ acknowledge financial support from the MPG and MaxSynBio Consortium, jointly funded by the Federal Ministry of Education and Research (Germany) and the Max Planck Society, TYDT and SM also acknowledge the BrisSynBio (a BBSRC/EPSRC synthetic Biology research centre) for financial support (Grant number (BB/L01386X/1)). AdM acknowledges partial support from a National Research Foundation (NRF) grant funded by the Ministry of Science, ICT and Future Planning of Korea, through the Global Research Laboratory Program (Grant number 2009-00426).
Open Access funding provided by the Max Planck Society.
Conflicts of interest
There are no conflicts to declare.Notes and references
- E. Andrianantoandro, S. Basu, D. K. Karig and R. Weiss, Mol. Syst. Biol., 2006, 2, 1–14 CrossRef PubMed.
- R. K. Kumar, M. Li, S. N. Olof, A. J. Patil and S. Mann, Small, 2013, 9, 357–362 CrossRef CAS PubMed.
- M. Li, D. C. Green, J. L. R. Anderson, B. P. Binks and S. Mann, Chem. Sci., 2011, 2, 1739 RSC.
- R. J. R. W. Peters, I. Louzao and J. C. M. van Hest, Chem. Sci., 2012, 3, 335–342 RSC.
- S. Deshpande, Y. Caspi, A. E. C. Meijering and C. Dekker, Nat. Commun., 2016, 7, 10447 CrossRef CAS PubMed.
- X. Huang, M. Li, D. C. Green, D. S. Williams, A. J. Patil and S. Mann, Nat. Commun., 2013, 4, 1–9 RSC.
- S. S. Mansy, J. P. Schrum, M. Krishnamurthy, S. Tobé, D. A. Treco and J. W. Szostak, Nature, 2008, 454, 122–125 CrossRef CAS PubMed.
- T.-Y. Dora. Tang, D. van Swaay, A. deMello, J. L. Ross Anderson and S. Mann, Chem. Commun., 2015, 51, 11429–11432 RSC.
- D. van Swaay, T. Y. D. Tang, S. Mann and A. DeMello, Angew. Chem., Int. Ed., 2015, 54, 8398–8401 CrossRef CAS PubMed.
- T.-Y. Dora Tang, C. Rohaida Che Hak, A. J. Thompson, M. K. Kuimova, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2014, 6, 527–533 CrossRef CAS PubMed.
- X. Huang, M. Li and S. Mann, Chem. Commun., 2014, 50, 6278 RSC.
- P. Zhou, X. Liu, G. Wu, P. Wen, L. Wang, Y. Huang and X. Huang, ACS Macro Lett., 2016, 5, 961–966 CrossRef CAS.
- X. Liu, P. Zhou, Y. Huang, M. Li, X. Huang and S. Mann, Angew. Chem., Int. Ed., 2016, 55, 7095–7100 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc08466h |
| ‡ The protein–polymer conjugate solution (4 mg ml−1 of BSA-NH2/PNIPAAm or GOx-NH2/PNIPAAm in HEPES buffer) with or without 0.6 mg ml−1 FITC tagged HRP or native HRP, and the continuous phase (2-ethyl-1-hexanol) containing 0.2 vol% BS(PEG)9 were delivered into the microfluidic device using two syringe pumps (neMESYS Syringe Pumps, CETONI GmbH, Korbussen, Germany). Water-in-oil droplets stabilised by the protein polymer were then formed at a flow-focusing junction. The flow rate was varied, but typically the continuous phase flow rate was kept between 1 to 5 μl min−1, whilst the protein–polymer conjugate flow rate was kept constant at 1 μl min−1. Water-in-oil droplets were collected from the device and transferred, through dialysis, from the oil phase into a mixture of ethanol/water, and then into Milli-Q® water via stepwise dilution of ethanol over 16 hours. Optical microscopy images of the proteinosomes were acquired before (water–oil) and after transfer into water, and 30 images were analysed using a custom-made MATLAB script (MathWorks, Switzerland). The enzymatic activity of a two-step GOx/HRP enzyme cascade comprised of either native GOx with native HRP, free GOx-PNIPAAm (0.025 mg ml−1) and native horseradish peroxidase (0.004 mg ml−1) or a cross-linked GOx-NH2/PNIPAAm (4 mg ml−1) membrane encapsulating native HRP (0.6 mg ml−1) (generated by microfluidics or by standard bulk methodologies) in HEPES buffer (pH 7.5) with Amplex Red (0.1 mM) was characterised via production of Resorufin after the addition of D-glucose. The Michaelis–Menten constant, Km, was obtained by varying the glucose concentration and measuring the reaction kinetics at 27 °C in a Spark 20 M well plate reader spectrophotometer (Tecan AG, Männedorf, Switzerland) using excitation and emission wavelengths of 572 ± 5 nm and 587 ± 5 nm respectively. To extract Km values, initial reaction rates were plotted against their corresponding glucose concentration and the curve fitted to the Michaelis–Menten model. |
| This journal is © The Royal Society of Chemistry 2018 |