Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Palladium-unleashed proteins: gentle aldehyde decaging for site-selective protein modification†
Robin L.
Brabham
 a,
Richard J.
Spears
a,
Richard J.
Spears
 a,
Julia
Walton
a,
Julia
Walton
 a,
Swati
Tyagi
b,
Edward A.
Lemke
b and
Martin A.
Fascione
a,
Swati
Tyagi
b,
Edward A.
Lemke
b and
Martin A.
Fascione
 *a
*a
aDepartment of Chemistry, University of York, Heslington Road, YO31 5DD, York, UK. E-mail: martin.fascione@york.ac.uk
bEMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany
First published on 24th January 2018
Abstract
Protein bioconjugation frequently makes use of aldehydes as reactive handles, with methods for their installation being highly valued. Here a new, powerful strategy to unmask a reactive protein aldehyde is presented. A genetically encoded caged glyoxyl aldehyde, situated in solvent-accessible locations, can be rapidly decaged through treatment with just one equivalent of allylpalladium(II) chloride dimer at physiological pH. The protein aldehyde can undergo subsequent oxime ligation for site-selective protein modification. Quick yet mild conditions, orthogonality and powerful exposed reactivity make this strategy of great potential in protein modification.
Aldehydes are a powerful yet underutilised tool for bioorthogonal chemistry, where the high electrophilicity coupled with good stability and low abundance in nature make them an attractive handle for protein modification.1 Bioorthogonal reactions involving aldehydes have been developed to take advantage of the unique reactivity of this functional group2–8 and an impressive array of bioconjugates are synthetically accessible, including antibody–drug conjugates,9 protein–protein conjugates10 and labelled live cells.11 Access to such aldehydes, however, can be an impediment to their usage, with incorporation methods either requiring enzyme recognition sequences, location at a protein terminus, or both. Use of formylglycine-generating enzyme (FGE), for example, will only form an aldehyde on the side chain of a cysteine in a CXPXR sequence (Fig. 1a).12 Some strategies are less flexible for aldehyde positioning: periodate-mediated oxidative cleavage of serine or threonine residues (Fig. 1b)13 or transamination of glycine residues14 occurs only at such N-terminal residues, whilst tubulin tyrosine ligase (“Tub”) requires a Tub tag on a protein C-terminus to append tyrosine derivatives such as m-formyl-L-tyrosine 1 (Fig. S1, ESI†).15
The technique of unnatural amino acid (UAA) mutagenesis has become a standard tool in chemical biology. Use of the pyrrolysine (Pyl) tRNACUA/pyrrolysyl-tRNA synthetase (RS) pair from several species of archaeal methanogens for amber stop codon (TAG) suppression has allowed access to proteins containing a wide range of non-canonical functionality, including alkenes,16–18 alkynes,19–21 azides,22 and aryl halides,23 with generally excellent levels of site specificity; indeed, UAA mutagenesis has become a widely utilised tool in chemical biology.24 Notably, the aldehyde-containing UAA m-formyl-L-phenylalanine 2 (Fig. S1, ESI†) has been genetically encoded using an engineered pylRS variant from Methanosarcina mazei.25 The wild-type Methanosarcina barkeri pyrrolysine tRNA-RS pair genetically encodes 2-thiazolidine derivative ThzK, with the methyl ester 3 used as a suitable precursor for incorporation into proteins (Fig. 1c).26 The thiazolidine group has seen use in peptide synthesis as an aldehyde-based protecting group for 1,2-aminothiols such as cysteine with various deprotection strategies,27–29 and a 4-thiazolidine derivative has been used as a caged pseudo-N-terminal cysteine for protein CBT condensation ligations,30 showing the potential for a 2-thiazolidine to be used to smuggle an aldehyde group into a protein. Notably, a thiazolidine will decage to yield a glyoxyl group, a highly reactive aldehyde to the extent that more reliable, reproducible and standardised modification methodologies have been established for this protein aldehyde than any other.1 As reactive carbonyls have been documented as forming undesired adducts with 1,2-aminothiols in biological media,31,32 the use of a protection/deprotection strategy avoids such side reactions which may suppress genetic incorporation or conjugation yields. Indeed, UAA mutagenesis has been shown to exhibit excellent synergy with decaging strategies, where photodecaging33 and metal-mediated decaging34–36 reactions have expanded the paradigm of functionality which can be genetically encoded. In this work a rapid yet mild palladium glyoxyl-decaging strategy is presented which reveals protein aldehydes from surface-exposed ThzK residues in a protein without the need for enzyme recognition sequences, using just a single equivalent of palladium (Fig. 1c). The new aldehydes are subsequently shown to be amenable to site-selective modification.
Green fluorescent protein (GFP) and superfolder green fluorescent protein (sfGFP) are highly useful test systems for protein modification due to ease of visualisation and highly optimised expression systems for use with UAA mutagenesis. Two GFP mutants containing amber stop codons at surface-exposed sites were selected: sfGFP(N150TAG)37 and GFP(Y39TAG).38 An advantage of using GFP is the facile confirmation and assaying of successful stop codon suppression through green fluorescence of harvested cell pellets.39 Genetic encoding of ThzK has previously made use of the M. barkeri pyrrolysine tRNA-RS pair,26 although the promiscuity of the corresponding M. mazei pair is generally sufficiently similar to the extent that either pair would be likely to genetically encode ThzK.30 Protected ThzK 3 was synthesised in four straightforward steps via4–9 with a cumulative yield of ca. 45% (Scheme S1, ESI†) following literature precedent.26 Separate saponification is unnecessary as this amino acid can be delivered into protein expression systems in a mildly basic stock solution, exposing the C-terminus as needed for translation. Using an adapted general method for amber stop codon suppression in GFP expression,38 both mutants were individually expressed with the supplementation of 3 at 1.6 mM in growth media and the green fluorescence of the harvested cell pellets confirmed the presence of full-length protein, demonstrating that ThzK can be encoded by the M. mazei pyrrolysine tRNA-RS pair. Following nickel affinity purification of the cell lysate, both GFP(Y39ThzK) 10 and sfGFP(N150ThzK) 11 could be isolated and their purity confirmed by SDS-PAGE and ESI-FTICR-MS (Fig. S2 and S3, ESI†).
Following successful ThzK incorporation, 10 and 11 were used as test beds for novel decaging strategies to yield GFP(Y39GlyoxylK) 12 and sfGFP(N150GlyoxylK) 13 (Fig. 2a). Inspired by examples of palladium-mediated deprotection in aqueous conditions,35,36 including thiazolidine cleavage,40 we opted to screen commercially available palladium sources as potential biologically compatible reagents for unmasking of glyoxyl aldehydes. Palladium complexes 14–17 were trialled at 37 °C, pH 7.4 in order to maintain biocompatibility over a range of concentrations and time intervals. These complexes are a mixture of Pd(0) and Pd(II)41 and at 100 equivalents have been used previously to decage thiazolidine peptides.40 Addition of 100 equiv. Pd(OAc)214 and allylpalladium(II) chloride dimer 16 led to protein precipitation, although some decaging was observed with 16. PdCl2(amphos)215 was found to be unreactive, neither decaging nor inducing precipitation. Success was first achieved using tris(dibenzylideneacetone)dipalladium(0) 17, with complete decaging observed after 48 hours. Further optimisation of conditions led to complete decaging within 24 hours. Curiously, under these conditions decaging of ThzK-containing 10 largely afforded glyoxyl-containing 12 as the aldehyde form, whilst glyoxyl aldehydes in aqueous conditions generally exist in the hydrated form. On closer inspection, the observation that allylpalladium(II) chloride dimer 16 induced precipitation of GFP seemed unusual given its successful use for other types of decaging with GFP.36 Subsequently, further screening using this palladium source showed that at concentrations greater than 0.5 mM, 16 will cause GFP to precipitate, but not at lower concentrations. Hence further screening was carried out using fewer equivalents with an identical concentration of GFP(Y39ThzK) 10 and pleasingly full decaging of the ThzK group to a glyoxyl could be observed within one hour using just one equivalent of allylpalladium(II)chloride dimer 16 (Fig. 2b) at room temperature. Notably, use of tris(dibenzylideneacetone)dipalladium(0) 17 under the same conditions afforded no detectable decaging. One equivalent of 16 strikes the balance between minimal protein precipitation and maximum extent of decaging, with substoichiometric quantities of palladium resulting in poorer conversion. These optimised conditions were then applied to sfGFP(N150ThzK) 11 (Fig. 2c) and complete decaging was also achieved with no protocol alterations necessary, despite the altered position of the ThzK moiety within the protein scaffold. In this example, the decaged glyoxyl exists almost exclusively as the hydrate, indicating that differences in the microenvironment surrounding the thiazolidine may affect the electrophilicity and reactivity of the resulting glyoxyl species. The final optimised procedure using allylpalladium(II) chloride dimer 16 is simple and rapid, requiring only limited exposure to a very low loading of palladium reagent.
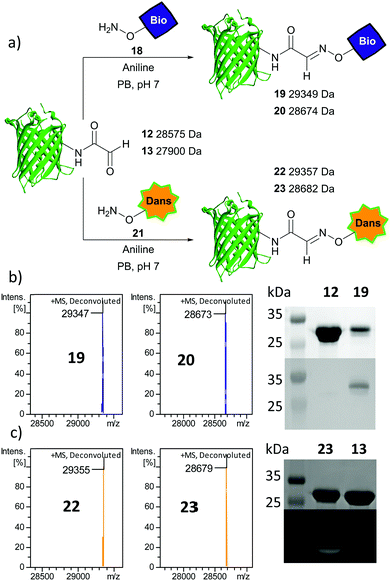
As further confirmation of the exposed aldehyde reactivity, aniline-catalysed oxime ligation was performed upon protein glyoxyl species 12 and 13 using an aminooxy biotin probe 18 to afford biotinylated proteins 19 and 20 respectively (Fig. 3a). Pleasingly, complete ligation was observed with both proteins within 24 h, with a Western blot confirming the incorporation of the biotin probe in 19 (Fig. 3b). Further oxime ligation was performed using the fluorescent aminooxy dansyl probe 21 with 12 and 13 to afford dansylated proteins 22 and 23 respectively. Again full conversion was observed and through denatured protein in-gel fluorescence the presence of a dansyl group in 23 could be unequivocally visualised (Fig. 3c). Irrespective of differences in protein and aldehyde/hydrate distribution, the aldehydes uncaged by the method reported here can be modified to completion following established protocols.
In summary, a new way to uncage a genetically encoded glyoxyl aldehyde precursor at physiological pH has been demonstrated using stoichiometric Pd(II), facilitating access to internally-modified proteins through aldehyde ligations without the need for an enzyme recognition sequence and hence minimising structural perturbations. This method requires only short reaction times under gentle conditions and the resulting aldehyde can be modified in completion. It is hoped that this latest addition to the chemical biologist's toolbox will open up opportunities for creating exciting new bioconjugates, achieving a greater understanding of complex biological systems.
We thank Dr Ed Bergstrom and the CoEMS for support with protein MS. This work was supported by the University of York (R. L. B.) and an EPSRC DTG studentship (EP/M506680/1, R. J. S.).
Conflicts of interest
R. L. B., R. J. S., & M. A. F. are authors on PCT/GB/2017/052896 application which in one claim covers Pd-mediated decaging.References
- R. J. Spears and M. A. Fascione, Org. Biomol. Chem., 2016, 14, 7622–7638 CAS.
- X. Ning, R. P. Temming, J. Dommerholt, J. Guo, D. B. Ania, M. F. Debets, M. A. Wolfert, G. J. Boons and F. L. van Delft, Angew. Chem., Int. Ed., 2010, 49, 3065–3068 CrossRef CAS PubMed.
- R. P. Temming, L. Eggermont, M. B. van Eldijk, J. C. van Hest and F. L. van Delft, Org. Biomol. Chem., 2013, 11, 2772–2779 CAS.
- L. Purushottam, S. R. Adusumalli, M. Chilamari and V. Rai, Chem. Commun., 2017, 53, 959–962 RSC.
- M. Chilamari, L. Purushottam and V. Rai, Chemistry, 2017, 23, 3819–3823 CrossRef CAS PubMed.
- D. Chen, M. M. Disotuar, X. Xiong, Y. Wang and D. H. Chou, Chem. Sci., 2017, 8, 2717–2722 RSC.
- T. Sasaki, K. Kodama, H. Suzuki, S. Fukuzawa and K. Tachibana, Bioorg. Med. Chem. Lett., 2008, 18, 4550–4553 CrossRef CAS PubMed.
- M.-J. Han, D.-C. Xiong and X.-S. Ye, Chem. Commun., 2012, 48, 11079–11081 RSC.
- R. A. Kudirka, R. M. Barfield, J. M. McFarland, P. M. Drake, A. Carlson, S. Banas, W. Zmolek, A. W. Garofalo and D. Rabuka, ACS Med. Chem. Lett., 2016, 7, 994–998 CrossRef CAS PubMed.
- J. E. Hudak, R. M. Barfield, G. W. de Hart, P. Grob, E. Nogales, C. R. Bertozzi and D. Rabuka, Angew. Chem., Int. Ed., 2012, 51, 4161–4165 CrossRef CAS PubMed.
- M. Colombo, S. Sommaruga, S. Mazzucchelli, L. Polito, P. Verderio, P. Galeffi, F. Corsi, P. Tortora and D. Prosperi, Angew. Chem., Int. Ed., 2012, 51, 496–499 CrossRef CAS PubMed.
- I. S. Carrico, B. L. Carlson and C. R. Bertozzi, Nat. Chem. Biol., 2007, 3, 321–322 CrossRef CAS PubMed.
- K. F. Geoghegan and J. G. Stroh, Bioconjugate Chem., 1992, 3, 138–146 CrossRef CAS PubMed.
- J. M. Gilmore, R. A. Scheck, A. P. Esser-Kahn, N. S. Joshi and M. B. Francis, Angew. Chem., Int. Ed., 2006, 45, 5307–5311 CrossRef CAS PubMed.
- D. Schumacher, J. Helma, F. A. Mann, G. Pichler, F. Natale, E. Krause, M. C. Cardoso, C. P. R. Hackenberger and H. Leonhardt, Angew. Chem., Int. Ed., 2015, 54, 13787–13791 CrossRef CAS PubMed.
- Y. Kurra, K. A. Odoi, Y. J. Lee, Y. Yang, T. Lu, S. E. Wheeler, J. Torres-Kolbus, A. Deiters and W. R. Liu, Bioconjugate Chem., 2014, 25, 1730–1738 CrossRef CAS PubMed.
- J. Li, S. Jia and P. R. Chen, Nat. Chem. Biol., 2014, 10, 1003–1005 CrossRef CAS PubMed.
- K. Lang, L. Davis, J. Torres-Kolbus, C. Chou, A. Deiters and J. W. Chin, Nat. Chem., 2012, 4, 298–304 CrossRef CAS PubMed.
- K. Lang, L. Davis, S. Wallace, M. Mahesh, D. J. Cox, M. L. Blackman, J. M. Fox and J. W. Chin, J. Am. Chem. Soc., 2012, 134, 10317–10320 CrossRef CAS PubMed.
- Y. S. Wang, X. Fang, A. L. Wallace, B. Wu and W. R. Liu, J. Am. Chem. Soc., 2012, 134, 2950–2953 CrossRef CAS PubMed.
- S. Wallace and J. W. Chin, Chem. Sci., 2014, 5, 1742–1744 RSC.
- Y. Ge, X. Fan and P. R. Chen, Chem. Sci., 2016, 7, 7055–7060 RSC.
- H. Xiao, F. B. Peters, P. Y. Yang, S. Reed, J. R. Chittuluru and P. G. Schultz, ACS Chem. Biol., 2014, 9, 1092–1096 CrossRef CAS PubMed.
- R. Brabham and M. A. Fascione, ChemBioChem, 2017, 18, 1973–1983 CrossRef CAS PubMed.
- A. Tuley, Y. J. Lee, B. Wu, Z. U. Wang and W. R. Liu, Chem. Commun., 2014, 50, 7424–7426 RSC.
- X. Bi, K. K. Pasunooti, J. Lescar and C. F. Liu, Bioconjugate Chem., 2017, 28, 325–329 CrossRef CAS PubMed.
- M. Jbara, S. Laps, S. K. Maity and A. Brik, Chem. – Eur. J., 2016, 22, 14851–14855 CrossRef PubMed.
- M. Haj-Yahya, K. S. Ajish Kumar, L. A. Erlich and A. Brik, Pept. Sci., 2010, 94, 504–510 CrossRef CAS PubMed.
- G. S. Creech, C. Paresi, Y. M. Li and S. J. Danishefsky, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2891–2896 CrossRef CAS PubMed.
- D. P. Nguyen, T. Elliott, M. Holt, T. W. Muir and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 11418–11421 CrossRef CAS PubMed.
- I. E. Gentle, D. P. De Souza and M. Baca, Bioconjugate Chem., 2004, 15, 658–663 CrossRef CAS PubMed.
- J. J. Ottesen, M. Bar-Dagan, B. Giovani and T. W. Muir, Pept. Sci., 2008, 90, 406–414 CrossRef CAS PubMed.
- D. P. Nguyen, M. Mahesh, S. J. Elsasser, S. M. Hancock, C. Uttamapinant and J. W. Chin, J. Am. Chem. Soc., 2014, 136, 2240–2243 CrossRef CAS PubMed.
- H. W. Ai, J. W. Lee and P. G. Schultz, Chem. Commun., 2010, 46, 5506–5508 RSC.
- J. Wang, S. Zheng, Y. Liu, Z. Zhang, Z. Lin, J. Li, G. Zhang, X. Wang, J. Li and P. R. Chen, J. Am. Chem. Soc., 2016, 138, 15118–15121 CrossRef CAS PubMed.
- J. Li, J. Yu, J. Zhao, J. Wang, S. Zheng, S. Lin, L. Chen, M. Yang, S. Jia, X. Zhang and P. R. Chen, Nat. Chem., 2014, 6, 352–361 CrossRef CAS PubMed.
- S. J. Miyake-Stoner, C. A. Refakis, J. T. Hammill, H. Lusic, J. L. Hazen, A. Deiters and R. A. Mehl, Biochemistry, 2010, 49, 1667–1677 CrossRef CAS PubMed.
- T. Plass, S. Milles, C. Koehler, C. Schultz and E. A. Lemke, Angew. Chem., Int. Ed., 2011, 50, 3878–3881 CrossRef CAS PubMed.
- T. S. Young, I. Ahmad, J. A. Yin and P. G. Schultz, J. Mol. Biol., 2010, 395, 361–374 CrossRef CAS PubMed.
- M. Jbara, S. K. Maity, M. Seenaiah and A. Brik, J. Am. Chem. Soc., 2016, 138, 5069–5075 CrossRef CAS PubMed.
- M. Jbara, S. K. Maity and A. Brik, Angew. Chem., Int. Ed., 2017, 56, 10644–10655 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7cc07740h |
| This journal is © The Royal Society of Chemistry 2018 |