 Open Access Article
Open Access ArticleStructural basis for controlling the enzymatic properties of polymannuronate preferred alginate lyase FlAlyA from the PL-7 family†
Hui-Min
Qin‡
ab,
Takuya
Miyakawa‡
a,
Akira
Inoue
c,
Ryuji
Nishiyama
c,
Akira
Nakamura
a,
Atsuko
Asano
a,
Takao
Ojima
c and
Masaru
Tanokura
 *ab
*ab
aLaboratory of Basic Science on Healthy Longevity, Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo, Tokyo 113-8657, Japan. E-mail: amtanok@mail.ecc.u-tokyo.ac.jp
bCollege of Biotechnology, Tianjin University of Science and Technology, No. 29, 13th Avenue, Tianjin, 300457, China
cLaboratory of Marine Biotechnology and Microbiology, Graduate School of Fisheries Sciences, Hokkaido University, 3-1-1 Minato-cho, Hakodate, 041-8611, Japan
First published on 2nd January 2018
Abstract
FlAlyA is an endolytic enzyme with a preference for polymannuronate. The crystal structure and mutagenesis studies elucidated that the structural variations at outer uronate-binding subsites +2, +3 and −2 control the enzymatic properties of PL-7 family enzymes. Lys158 mutations changed the pH dependency and enhanced the production of mono- and disaccharides.
Brown seaweeds, a large group of mostly marine multicellular algae, exhibit several key features of an ideal feedstock for the production of biofuels and renewable commodity chemical compounds.1,2 Among these, alginate is a cell wall polysaccharide composed of a linear block copolymer of two uronic acids, β-D-mannuronate (M) and its C5 epimer α-L-guluronate (G). The uronic acids are arranged in various sequences, with uniform regions of M-block, G-block, and/or a mixture of MG-block.3 Alginate oligosaccharides are released from alginate through the action of alginate lyases and function as a bifidus factor and an elicitor of plant growth.4,5 Enzymatic degradation and modification of alginate are required to aid the development of alginate oligosaccharides with biological functions and physicochemical properties.
Alginate depolymerization is catalysed by diverse alginate lyases via a β-elimination reaction of the 4-O-glycosidic bond. Alginate lyases have been classified into the polysaccharide lyase (PL)-5, 6, 7, 14, 15, 17, and 18 families based on the Carbohydrate-Active enZymes (CAZy) database (http://www.cazy.org/).6 Enzymes belonging to the PL-7, 14 and 18 families are endolytic alginate lyases with β-jelly roll folds.7–9 The substrate specificity, mode of action and tertiary structure are different among these alginate lyase families.10 In particular, the PL-7 family enzymes have been well-investigated in bacteria, and several structures have been reported.11,12 Structural studies provide useful information about the substrate recognition and depolymerization mode of action. The A1-II′ structure proposes the catalytic mechanism for the PL-7 family enzymes.11 A tyrosine residue located near subsite +1 abstracts the C5 proton and donates it to the O4 atom as a general base and a general acid, respectively. Zobellia galactanivorans AlyA5 is a unique exolytic lyase with a large loop inserted into the catalytic groove,12 and a tryptophan residue serves as a hydrophobic wall, which regulates substrate recognition. Major PL-7 alginate lyases are specific for poly(G),12 whereas PA1167 and A1-II′ show the highest activity toward poly(MG).13 In addition, some enzymes showing poly(M) specificity have been found in Photobacterium sp. and Flavobacterium sp.14,15 Therefore, the PL-7 family contains alginate lyases with diverse substrate specificities, but the mechanistic bases controlling substrate specificity still remain unclear.
A novel alginate lyase from the Flavobacterium sp. strain UMI-01, FlAlyA (EC 4.2.2.3), efficiently degrades poly(M) and poly(MG) but only moderately degrades poly(G),15 and it shows an over 20-fold enhancement in the endolytic activity for the degradation of alginate compared with commercial alginate lyases. Herein, we present the crystal structure of FlAlyA at 1.54 Å resolution (Fig. 1a and Table S1, ESI†). It possesses a conserved β-jelly roll fold, which was composed of two antiparallel β-sheets. Some β-strands comprising sheet A twisted somewhat to form an electropositive-rich concavity where the substrates were indicated to be located (Fig. S1, ESI†). Structural superposition of the substrate-binding sites of FlAlyA with A1-II′ indicated that it was possible for alginate oligosaccharides to bind in the deep concavity of sheet A by salt bridges or hydrogen bonds (Fig. 1b).
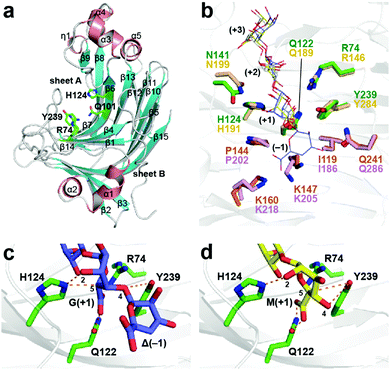 | ||
| Fig. 1 FlAlyA structure and residue arrangements at subsites +1 and −1. (a) Ribbon representation of the overall FlAlyA structure. (b) Superposed structures of FlAlyA (green and orange) and A1-II′ (wheat and pink) from Sphingomonas sp. (PDB code: 2ZAA). Alginate oligosaccharides coloured blue, white and yellow are ΔGMG (PDB code, 2ZAA), GGG (2ZAB) and MMG (2ZAC), respectively. Proposed binding modes of G (c) and M (d) groups at subsite +1 of the FlAlyA structure. | ||
FlAlyA subsite +1 was composed of five residues (Arg74, Gln122, His124, Asn141 and Tyr239), which are the conserved residues in A1-II′ (Fig. 1b and Fig. S2, ESI†).11 Asn141 is the sole residue that is substituted with different types of hydrophilic resides (Gln, Asp and Ser) in other PL-7 family enzymes (Fig. S2, ESI†). To evaluate the catalytic residues and reaction modes, we compared the FlAlyA model structure bound with ΔGMG (Δ means a 4-deoxy-α-L-erythro-hex-4-enuronosyl group) and MMG (Fig. 1c and d). In both binding models, Gln122 interacts with the C6 carboxylate at subsite +1, and, therefore, the residue could remove or neutralize the negative charge of the C6 carboxylate. In the binding model of ΔGMG (Fig. 1c), His124 and Tyr239 are located near the C5 proton and O4 atom of the first G group positioned at subsite +1, respectively. Therefore, His124 acts as a general base to abstract the C5 proton, and Tyr239 donates a proton to the O4 atom. On the other hand, the C5 proton faced towards Tyr239 in the binding model of MMG, where the first M group was positioned at subsite +1 (Fig. 1d), suggesting that Tyr239 acts as both the general base and general acid to transfer the C5 proton of the M group to the O4 atom. These structural observations show that FlAlyA catalyses the β-elimination reaction using different combinations of the two catalytic residues His124 and Tyr239, which may depend on the type of uronate group at subsite +1. The catalytic activity of FlAlyA toward alginate was extremely decreased upon mutations of His124 and Tyr239 (Table S2, ESI†). In addition, H124N/Y239F showed much lower activity. These results support that these residues participate in the catalytic reaction. Table S3 (ESI†) shows the specific activity of the mutants toward poly(M), poly(G) and poly(MG). The specific activity toward poly(M) was remarkably decreased upon H124N mutation, as well as toward poly(G). Therefore, these data do not directly support the proposed catalytic role of His124. The side chain of His124 is also located near the hydroxyl group at the C2 position of the first M group (+1) in the MMG binding model (Fig. 1d). As another possible role of His124 in the catalytic reaction, the residue may contribute to adjust the orientation of the uronate group at subsite +1 through interaction with the C2-hydroxyl group.
In the ΔGMG-bound model of FlAlyA, the non-reducing end group (Δ) at subsite −1 was surrounded by Ile119, Pro144, Lys147, Lys160 and Gln241 (Fig. 1b). Some residues were different between poly(G)-specific PL-7 alginate lyases (AlyPG, AlyA1 and KpAlyA) and other alginate lyases with a preferred substrate specificity toward poly(M) and poly(MG) (FlAlyA, A1-II′ and PA1167). FlAlyA Pro144 and Lys160 were conserved in A1-II′ (Pro202 and Lys218) and PA1167 (Pro115 and Arg135, which is another positively charged residue), whereas poly(G)-specific enzymes tended to adopt acidic residues (Asp and Glu) at the same positions (Fig. S2, ESI†). The ΔGMG-bound model of FlAlyA showed that Pro144 and Lys160 were located in close proximity to the carboxyl group of the non-reducing end group (Δ) (Fig. 1b). The substitutions at the acidic residues may cause conformational changes to the uronate group at subsite −1 due to the electrostatic repulsion between their carboxyl groups. Tables S2 and S3 (ESI†) show a considerably decreased activity of FlAlyA upon P144E or K160E mutation to a similar extent in the mutation of catalytic residues, which suggests that the binding mode of the uronate group at subsite −1 is controlled by the local surface charge to enhance an efficient catalytic reaction toward all types of alginate polysaccharides. On the other hand, all of the residues comprising subsite −1 were conserved between FlAlyA and A1-II′ (Fig. 1b and Fig. S2, ESI†). Although FlAlyA efficiently cleaves poly(M) and poly(MG) rather than poly(G) (Table S3, ESI†), unlike the broad specificity of A1-II′,13,15 the superposed structures show that the residue arrangements at subsites +1 and −1 do not seem to affect the different substrate specificities of FlAlyA and A1-II′.
FlAlyA adopted a closed concavity for substrate binding by extending two loops of Phe61–Ser73 (L3 loop) and Gly136–Pro144 (L6 loop) over subsite +2 (Fig. 2 and Fig. S1, ESI†). These two loops were connected by a hydrogen bond between Asn69 and Asp139 on the L3 and L6 loops, respectively (Fig. 2). The conformation of the L6 loop was quite different from that of the corresponding loop in A1-II′, and the interaction between Asn69 and Asp139 was not conserved in the A1-II′ structure. For the activity of A1-II′, the flexibility of the loop is required for the regulation of substrate binding.11 The L3 loop is located closer to the uronate group at subsite +2 than the corresponding loop of A1-II′ (Fig. 2a and b). In addition, a non-conserved residue Thr70 (Ser142 in A1-II′) lies on the L3 loop of FlAlyA. These structures seem to form a tight contact between the L3 loop and the carboxyl group of the uronate group at subsite +2.
Among the residues at the bottom of subsite +2, Arg78, Tyr233 and Lys235 were conserved in A1-II′ (Arg150, Tyr278 and Lys280) (Fig. 2a and b). The side-chain of Tyr233 lies in close proximity to the C2 hydroxyl group of the G group at subsite +2 in the GGG-binding model of FlAlyA (Fig. 2b), whereas the M group directed the C2 hydroxyl group toward a distal space from Tyr233 (Fig. 2a). The space of subsite +2 is narrow, which may restrict the direction of substrate entry together with the L3 and L6 loops. Other residues surrounding the subsites +2 and +3 (Lys95, Gln97 and Glu105 for A1-II′) were substituted with those with short side chains in the FlAlyA structure (Thr12, Thr14 and Ser28) (Fig. 2c and Fig. S2, ESI†), which may provide an expanded space for the substrate entry into the active site of FlAlyA through subsite +2.
To evaluate the structural findings with respect to the substrate specificity of FlAlyA, we designed several mutants at subsites +2 and +3 and measured their activities toward alginate, poly(M), poly(G) and poly(MG). Table S2 (ESI†) shows the activities of the mutants toward alginate at subsites +2 and +3. The activity was not influenced by the mutations of Thr70 (T70S) and Ser28 (S28E/D). All other tested mutants at subsites +2 (D139A and Y233F) and +3 (T12K/R and T14Q/N) showed decreased activities compared with the wild-type enzyme. The D139A mutation breaks a hydrogen bond connecting the L3 and L6 loops. A decreased activity in the D139A mutant indicates that a tunnel-like structure is required to enhance the activity of FlAlyA. In addition, the T70S/D139A double mutant showed an ∼1.5-fold higher activity than the D139A mutant (Table S2, ESI†). Since the single mutant T70S also showed higher activity than the wild-type, the T70S mutation contributes to the recovery of the activity. The side-chain methyl group of Thr70 may cause an unfavorable contact with the carboxyl group of the substrate (Fig. 2a and b). Table S4 (ESI†) shows the activities of T70S, D139A and T70S/D139A mutants toward poly(M), poly(G) and poly(MG), respectively. Although the substrate specificity is not affected by the T70S and D139A mutations, the T70S/D139A mutation decreased the relative activity of poly(G) toward poly(M). Therefore, the structure composed of the L3 and L6 loops is also required to moderate the substrate specificity of FlAlyA. Among the tested mutants, the Y233F mutant showed an increase in the relative activity of poly(G) or poly(MG) toward poly(M) (Table S4, ESI†), which does not contradict the structural findings (Fig. 2a and b). Contrary to a structural expectation, the poly(G) preference was not improved by the quintet mutation of FlAlyA (T12K/T14Q/S28E/T70S/D139A) that mimics the residues at subsites +2 and +3 of A1-II′ (Table S4, ESI† and Fig. 2c), whereas this mutant showed an ∼1.9-fold higher activity toward poly(MG) than the wild-type enzyme. Based on the specific activities of the single mutants (Table S4, ESI†), we concluded that the specificity toward poly(MG) was increased upon T12K and T14Q mutations as compared with toward poly(M) and poly(G), respectively. These results suggest that subsites +2 and +3 contribute towards controlling the substrate specificity of FlAlyA, although the mechanism is still complicated.
There is no structural information at subsite −2 reported in the PL-7 family of enzymes. We predicted subsite −2 based on the ΔGMG-binding model of FlAlyA. The model showed that Arg117, Tyr149, Lys158 and Asp180 were located near subsite −2 (Fig. 3a), which suggests that they contribute by interacting with an uronate group at subsite −2. The superposed structures of FlAlyA and ZgAlyA5 showed that FlAlyA subsite −2 overlapped with the side chains of ZgAlyA5 Trp313. ZgAlyA5 shows a large insertion in the sequence, and Trp313 upon insertion blocked subsite −2 to exert exolytic activity.12 Unlike the depolymerization mode of ZgAlyA5, FlAlyA shows endolytic activity and degrades alginate into disaccharides as a final product (Fig. 3b).
To mimic the role of ZgAlyA5 Trp313 in the FlAlyA structure, we substituted Lys158 with a tryptophan residue and measured the degraded products by TLC. As shown in Fig. 3b, the spots for disaccharides and 4-deoxy-L-erythro-5-hexoseulose uronic acid (DEH) were increased upon reaction of alginate with the K158W mutant, although the K158W mutant does not show complete exolytic activity. The mutant could block the catalytic groove at one end but the effect may not be enough as compared with ZgAlyA5, in which Trp313 and its harbouring structural element (shown as cyan in Fig. 3a) are more suitable to block at subsite −2. In addition, the DEH production activity was enhanced under acidic pH conditions (Fig. 3c). The pH profile showed that the overall activity of FlAlyA was the highest in the pH range 6.8 to 8.0, and the maximum activity was shifted to the pH range 6.0 to 6.8 upon K158W mutation (Fig. 3d and Table S5, ESI†). The pKa values of His124 and Tyr239 are not directly changed upon K158W mutation because the catalytic residues are located far from Lys158. However, Lys158 is located in close proximity to Asp180 and may function to neutralize the negative charge of Asp180. The acidic residue also forms a salt bridge with Lys160 that weakens the negative charge of the C6 carboxylate at subsite −1 (Fig. 3a). K158W mutation may impair the ability of Lys160 and thus require the protonation of Asp180 under low pH conditions. Our results show that neutralization of the C6 carboxylate at subsite −1 may be required for the progression of the β-elimination reaction of FlAlyA. HPLC analysis also confirmed the increased activity of the K158W mutant for disaccharide production in each optimal pH range. The disaccharide yields of ΔG and ΔM were improved upon mutation (Table S6 and Fig. S3, ESI†). Compared with the wild-type FlAlyA, the peaks of tri- and tetrasaccharides were decreased by the reaction of the K158W mutant. This result suggests that the mutant may be able to cleave tri- and tetrasaccharides to produce DEH and disaccharides more effectively than the wild-type.
Other Lys158 mutants, K158Y, K158F, K158L, K158H and K158N, were tested to determine which mutant exerted efficient activity for disaccharide production. All tested mutants showed higher activity than the wild-type enzyme under acidic pH conditions (Table S5, ESI†). In particular, the activity was enhanced by the K158N mutation (∼2.7-fold) at a pH value of 6.0 (Fig. 3d). After the alginate was degraded for 12 hours by FlAlyA mutants under the optimal pH conditions, the polysaccharide products were analysed by HPLC. All tested mutants showed improved disaccharide yields of ΔG and ΔM (Fig. S3, ESI†), and substitutions to Trp, Tyr and Phe showed over 2-fold increase in the ΔG product (Table S6 and Fig. S3, ESI†). In addition, K158W and K158Y mutants showed a more than 4-fold increase in activity in the production of ΔM compared to the wild-type enzyme. These results suggest that the introduction of a bulky side chain (Trp, Tyr or Phe) into position 158 effectively blocks the extension of the substrate to subsite −3. Additionally, the aromatic side chains may strengthen the π–π stacking interaction with a double bond-containing ring of the non-reducing end group (Δ) at subsite −2. Asp180 was another residue showing improved activity at subsite −2. D180E mutation elevated the activity (∼1.4-fold) but did not change the pH dependency (Fig. 3d) and the production of disaccharides (Table S6 and Fig. S4, ESI†).
In summary, we reported the crystal structure of FlAlyA, which is the first structure of a poly(M)-preferred alginate lyase belonging to the PL-7 family. The structural studies showed that two loops (L3 and L6 loops) were key structural bases for controlling the activity and substrate specificity as a lid for subsite +2. In particular, Lys158 at subsite −2 is a target residue that changes the enzymatic properties. The structural and functional characterization of FlAlyA augments the current understanding of substrate recognition and structure based rational design.
This work was supported by the Tohoku Marine Science Project from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan and the National Natural Science Foundation of China (31771911) and Tianjin (16JCQNJC09200). The synchrotron-radiation experiments were performed on beamline NE12A at the Photon Factory, Tsukuba, Japan (Proposal No. 2013G652).
Conflicts of interest
There are no conflicts to declare.Notes and references
- A. J. Wargacki, E. Leonard, M. N. Win, D. D. Regitsky, C. N. S. Santos, P. B. Kim, S. R. Cooper, R. M. Raisner, A. Herman and A. B. Sivitz, Science, 2012, 335, 308–313 CrossRef CAS PubMed.
- H. Takeda, F. Yoneyama, S. Kawai, W. Hashimoto and K. Murata, Energy Environ. Sci., 2011, 4, 2575–2581 CAS.
- T. Y. Wong, L. A. Preston and N. L. Schiller, Annu. Rev. Microbiol., 2000, 54, 289–340 CrossRef CAS PubMed.
- A. Darvill, C. Bergmann, F. Cervone, G. De Lorenzo, K. Ham, M. Spiro, W. York and P. Albersheim, Biochem. Soc. Symp., 1994, 60, 89–94 CAS.
- Y. Yonemoto, H. Tanaka, T. Yamashita, N. Kitabatake, Y. Ishida, A. Kimura and K. J. Murata, J. Ferment. Bioeng., 1993, 75, 68–70 CrossRef CAS.
- V. Lombard, H. Golaconda Ramulu, E. Drula, P. M. Coutinho and B. Henrissat, Nucleic Acids Res., 2014, 42, D490–D495 CrossRef CAS PubMed.
- H.-M. Qin, T. Miyakawa, A. Inoue, R. Nishiyama, A. Nakamura, A. Asano, Y. Sawano, T. Ojima and M. Tanokura, J. Biol. Chem., 2017, 292, 2182–2190 CrossRef CAS PubMed.
- K. Ogura, M. Yamasaki, T. Yamada, B. Mikami, W. Hashimoto and K. Murata, J. Biol. Chem., 2009, 284, 35572–35579 CrossRef CAS PubMed.
- S. Dong, T.-D. Wei, X.-L. Chen, C.-Y. Li, P. Wang, B.-B. Xie, Q.-L. Qin, X.-Y. Zhang, X.-H. Pang and B.-C. Zhou, J. Biol. Chem., 2014, 289, 29558–29569 CrossRef CAS PubMed.
- B. Zhu and H. Yin, Bioengineered, 2015, 6, 125–131 CrossRef CAS PubMed.
- K. Ogura, M. Yamasaki, B. Mikami, W. Hashimoto and K. Murata, J. Mol. Biol., 2008, 380, 373–385 CrossRef CAS PubMed.
- F. Thomas, L. C. Lundqvist, M. Jam, A. Jeudy, T. Barbeyron, C. Sandström, G. Michel and M. Czjzek, J. Biol. Chem., 2013, 288, 23021–23037 CrossRef CAS PubMed.
- O. Miyake, A. Ochiai, W. Hashimoto and K. Murata, J. Bacteriol., 2004, 186, 2891–2896 CrossRef CAS PubMed.
- M. Malissard, C. Duez, M. Guinand, M.-J. Vacheron, G. Michel, N. Marty, B. Joris, I. Thamm and J.-M. Ghuysen, FEMS Microbiol. Lett., 1993, 110, 101–106 CrossRef CAS PubMed.
- A. Inoue, K. Takadono, R. Nishiyama, K. Tajima, T. Kobayashi and T. Ojima, Mar. Drugs, 2014, 12, 4693–4712 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Complete experimental details and additional figures. See DOI: 10.1039/c7cc06523j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |


