Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Lipopeptidomimetics derived from teixobactin have potent antibacterial activity against Staphylococcus aureus†
Georgina C.
Girt
a,
Amit
Mahindra
b,
Zaaima J. H.
Al Jabri
c,
Megan
De Ste Croix
c,
Marco R.
Oggioni
c and
Andrew G.
Jamieson
 *b
*b
aDepartment of Chemistry, University of Leicester, University Road, LE1 7RH, UK
bSchool of Chemistry, University Avenue, University of Glasgow, Glasgow, G12 8QQ, UK. E-mail: andrew.jamieson.2@glasgow.ac.uk
cDepartment of Genetics, University of Leicester, University Road, LE1 7RH, UK
First published on 27th February 2018
Abstract
A series of lipopeptidomimetics derived from teixobactin have been prepared that probe the role of residues (1–6) as a membrane anchor and the function of enduracididine. The most active compounds, with a farnesyl tail and End10 to Lys10 or Orn10 substitution have potent activity (MIC 8 μg mL−1) against S. aureus. These results pave the way for the synthesis of simple, cost-effective yet potent lipopeptidomimetic antimicrobials.
The emergence of antimicrobial resistance is becoming a global crisis, with even last-resort antibiotics giving rise to resistant strains.1 Teixobactin, discovered from previously uncultivable bacteria, gave way to a new class of antibiotics.2,3 Teixobactin appears to have multiple modes of action, preventing the rapid onset of resistance.4,5 For example inhibition of both peptidoglycan and teichoic acid synthesis is achieved through binding multiple bactoprenol-coupled cell wall precursors including lipid II.4 This key building block is involved in the rate-limiting step of peptidoglycan biosynthesis and so is essential for the production of the bacterial cell wall.5
Teixobactin is an undecapeptide antibiotic comprised of a depsipeptide macrocycle, linked to a largely hydrophobic linear peptide tail. The native compound contains four D-amino acids, and the nonproteinogenic residue L-allo-enduracididine (L-allo-End).2 The cyclic depsipeptide component of teixobactin has recently been shown to form hydrogen-bonding interactions with chloride anions and has been proposed as a pyrophosphate recognition motif.6 Lantibiotics such as nisin have also been found to form such a pyrophosphate cage;7 thus increasing membrane permeation through pore formation.8
Since the original report describing the isolation and bio-activity of teixobactin, total syntheses have been reported by the Li and Payne groups.9,10 A solution phase synthesis of the cyclic depsipeptide ring has also been reported.11 However, the synthesis of teixobactin is both laborious due to the seven step synthesis of L-allo-End and relatively expensive because four D-amino acids are required. Development of simpler, more cost-effective peptidomimetics is therefore an attractive alternative. Recently, a number of teixobactin analogues that are simpler to produce have been described. Several groups have reported the synthesis and antimicrobial activity of analogues in which the non-proteinogenic L-allo-enduracididine residue is substituted with isosteric amino acids.12–15 A Fmoc-solid phase peptide synthesis (SPPS) strategy was employed, with orthogonal protection to give the branched side chain, before cyclisation to form the 13-membered depsipeptide ring in solution. The teixobactin analogue with single End10 to Arg amino acid substitution was found to have substantial antimicrobial activity, with a minimum inhibitory concentration (MIC) value of 2 μg mL−1, yet still 8 times weaker than Teixobactin.
The Singh group investigated the role of amino acid stereochemistry through production of Arg10 analogues of teixobactin incorporating L-amino acids and acetylated at the N-terminus, all of which gave a reduction in antibiotic activity against Gram-positive bacteria.13,14 The role of each independent mutation was then clarified by the Albericio and co-workers, who found that mutation to all L-amino acids with retention of the native methylated N-terminus eradicated all antibiotic activity. Acetylation of the N-terminus as the sole alteration resulted in negligible activity with an MIC of 256 μg mL−1 for Bacillus subtilis.16
Subsequently, Nowick and co-workers reported the synthesis and antimicrobial activity of several new teixobactin analogues, mutating the enduracididine residue, the stereochemistry of various residues, cleaving the depsipeptide bond of the macrocyclic ring, and replacing part of the linear peptide tail of the Arg10 analogue with a dodecanoyl group.17 Interestingly, these results showed that mutating enduracididine to a lysine residue rather than arginine resulted in better antimicrobial activity (MIC of 0.25 μg mL−1 compared to 1 μg mL−1 against Staphylococcus epidermidis). In addition, replacement of residues 1–5 with a hydrocarbon chain gave an effective antibiotic with an MIC of 4 μg mL−1.
Structure–activity-relationship studies have also revealed the importance of polar residues using a selective lysine scan.18,19 The hydroxyl group of the Ser7 residue was also identified as significant through substitution to Ala and the alkylation state of the N-terminal Phe1 residue is important to the antibacterial activities of these analogues.
However, the functional role of the peptide tail (residues 1–6) has not been extensively studied and no lipidated peptidomimetic has been created that retains comparable activity of native teixobactin. The aim of this work was to investigate the role of residues 1–7 as a potential membrane anchor by developing lipopeptidomimetics with lipids of different lengths and evaluate the effect of substitution of End10 to amino acids with different side-chain functionality.
Isoprenoid chains are known to be effective cellular membrane anchors,20 and prenylation of cysteine residues is a commonly occurring post-translational modification (PTM) in eukaryotic proteins. The dependence on isoprenoid length for cell penetration has previously been explored.21 Prenylated peptidomimetics have the potential to serve as more cost-effective alternatives to teixobactin as antibiotic drugs.
Teixobactin contains a Ser at the 7-position, it was therefore rationalised that substitution to an cysteine residue would facilitate prenylation without significant adverse effect on activity. Our rational was that replacing the peptide tail (1–6), including three D-amino acids, with a farnesyl lipid chain would provide a lipopeptidomimetic analogue requiring fewer synthetic steps and only commercially available L-amino acids.
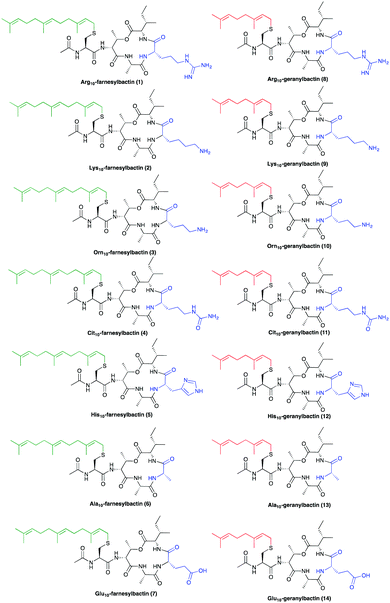
The basic guanidinium functionality of allo-enduracididine within teixobactin is important for biological activity. We therefore investigated a number of different bioisostere amino acid residues including arginine and lysine. As controls, neutral (Ala10) and acidic (Glu10) macrocyclic analogues were also prepared (see ESI†).
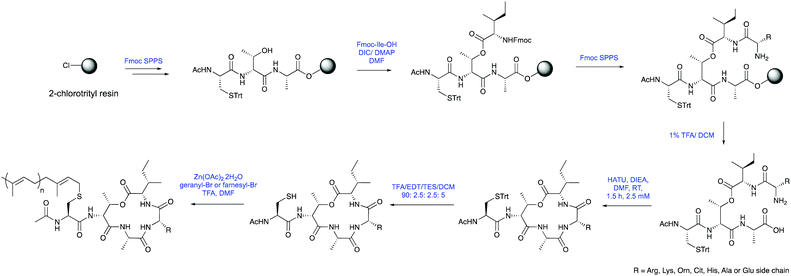
The synthesis of Arg10-farnesylbactin 1 and related analogues 2–7 was carried out via Fmoc-SPPS followed by solution-phase macrocyclisation (Scheme 1). Initially, Fmoc-Ala-OH was coupled to 2-chlorotrityl resin with DIEA in DCM, before threonine and cysteine were coupled, and the N-terminal acetylated using acetic anhydride. No capping of the hydroxyl threonine side chain was observed by LCMS on a small sample cleaved from the solid support. The depsipeptide ester linkage was then formed using Fmoc-Ile-OH, DIC and catalytic DMAP (Fig. 1).
The isoleucine Fmoc protecting group was removed and arginine (or other End10 variant Fmoc-amino acid) coupled under standard conditions, with no diketopiperizine formation detected. Following deprotection the peptide was cleaved from the solid support under mildly acidic conditions (1% TFA/DCM).
Macrocyclisation of the Ala9 C-terminal to Arg10 N-terminus was easily achieved through amide coupling using HATU and DIEA. This reaction was found to proceed extremely efficiently with >99% conversion from linear to cyclic forms, as determined by RP-HPLC. Finally, the trityl protecting group was removed from the cysteine side chain to yield a free thiol and the farnesyl chain was attached using catalytic Zn(OAc)2·2H2O and TFA. The final product was purified by RP-HPLC and characterised by HRMS, 1H NMR and TOCSY NMR.
Lipopeptidomimetic Arg10-farnesylbactin 1 was then evaluated against the Gram-positive bacterial strain Staphylococcus aureus ATCC 25923 and the Gram-negative bacterial strain Escherichia coli ATCC 25922 (Table 1). We were encouraged to observe that this compound was active (8 μg mL−1) in the minimum inhibitory concentration assays against Gram-positive but not Gram-negative bacteria and thus showed a similar trend of activity as teixobactin. In order to further investigate the effect of substituting the allo-enduracididine residue we produced a series of analogues with varying functionality at position 10 (Table 1). Lys10-farnesylbactin 2 was designed as a result of a previous report suggesting that lysine is a better mimic of the enduracididine residue than arginine.15 This proved to be the case and provided compound 2 as a more potent analogue (8 μg mL−1). Surprisingly compound 2 was also active against the Gram negative E. coli bacterial strain.
| Compound | S. aureus ATCC 25923 | E. coli ATCC 25922 | |
|---|---|---|---|
| MIC | MBC | MIC | |
| Arg10-farnesylbactin (1) | 16 | >32 | >32 |
| Lys10-farnesylbactin (2) | 8 | 16 | 16 |
| Orn10-farnesylbactin (3) | 8 | 16 | 32 |
| Cit10-farnesylbactin (4) | >32 | >32 | >32 |
| His10-farnesylbactin (5) | >32 | >32 | >32 |
| Ala10-farnesylbactin (6) | >32 | >32 | >32 |
| Glu10-farnesylbactin (7) | >32 | >32 | >32 |
| Arg10-geranylbactin (8) | >32 | >32 | >32 |
| Lys10-geranylbactin (9) | >32 | >32 | >32 |
| Orn10-geranylbactin (10) | >32 | >32 | >32 |
| Cit10-geranylbactin (11) | >32 | >32 | >32 |
| His10-geranylbactin (12) | >32 | >32 | >32 |
| Ala10-geranylbactin (13) | >32 | >32 | >32 |
| Glu10-geranybactin (14) | >32 | >32 | >32 |
Orn10-farnesylbactin 3 provides an analogue with shorter amine side-chain. This analogue has comparable activity in both the S. aureus MIC (8 μg mL−1) and MBC (16 μg mL−1) assays relative to the corresponding Lysine analogue 2.
To investigate the effect of the charge and potential for the side-chain of residue10 to form electrostatic interactions, Cit10-farnesylbactin 4 was produced which is neutral in terms of charge, however can still H-bond; His10-farnesylbactin 5 with partial positive charge; Ala10-farnesylbactin 6 in which the side-chain is removed altogether and Glu10-farnesylbactin 7 with negative charge were produced. Interestingly, all of these analogues showed a total lack of activity (Table 1).
Next, we examined whether a shorter geranyl (C8) isoprenoid chain would also provide active lipopeptidomimetic analogues. A further set of compounds without hydrocarbon tails were synthesised as controls; comprising of a cyclic tetradepsipeptide with an N-terminal acetyl cap (see Fig. S1, ESI†).
None of these compounds (Table 1, compounds 8–14), had antibiotic activity against Gram-positive bacteria S. aureus. Interestingly, the arginine, lysine and ornithine equivalents (8, 9 and 10) did not show any activity, unlike their farnesylated counterparts suggesting that slight changes in structure can result in diminished biological activity.
Of the lipopeptidomimetics investigated, only basic residues were found to have any antibiotic activity, suggesting that the positive charge and thus potentially an electrostatic interaction with the binding partner(s) is crucial for function. These results are also consistent with previous research that suggests that the mutation of L-allo-enduracididine to lysine, rather than the more structurally similar arginine, is in fact advantageous for antibiotic activity.17 The importance of the hydrophobic tail is also apparent, as the control cyclic tetradepsipeptides 15–17 were not active against either strain of bacteria (see ESI†).
In conclusion, the rational design of teixobactin lipopeptidomimetics has been achieved. Replacing the hydrophobic linear peptide tail (1–7) with isoprenoid surrogates of differing length provided biologically active analogues. A structure–activity-relationship study to investigate the function of enduracididine on antibacterial activity revealed the importance of a positively charged side-chain at residue 10. Finally, lipopeptidomimetics 1, 2 and 3 are the first examples of farnesyl lipidated teixobactin analogues with antibacterial activity, representing promising lead compounds for drug discovery.
The authors would like to thank the Universities of Leicester and Glasgow, the ERDF (IRSA) and Pepceuticals Ltd for funding. We gratefully acknowledge Mick Lee (University of Leicester) for LCMS and Gerry Griffith (University of Leicester) for NMR.
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. L. Ventola, Pharm. Ther., 2015, 40, 277–283 Search PubMed.
- L. L. Ling, T. Schneider, A. J. Peoples, A. L. Spoering, I. En-Gels, B. P. Conlon, A. Mueller, T. F. Schaberle, D. E. Hughes, S. Epstein, M. Jones, L. Lazarides, V. A. Steadman, D. R. Cohen, C. R. Felix, K. A. Fetterman, W. P. Millett, A. G. Nitti, A. M. Zullo, C. Chen and K. Lewis, Nature, 2015, 517, 455–459 CrossRef CAS PubMed.
- C. Guo, D. Mandalapu, X. Ji, J. Gao and Q. Zhang, Chem. – Eur. J., 2017 DOI:10.1002/chem.201704167.
- T. Homma, A. Nuxoll, A. B. Gandt, P. Ebner, I. Engels, T. Schneider, F. Götz, K. Lewis and B. P. Conlon, Antimicrob. Agents Chemother., 2016, 60, 6510–6517 CrossRef CAS PubMed.
- V. Ng and W. C. Chan, Chem. – Eur. J., 2016, 22, 12606–12616 CrossRef CAS PubMed.
- H. Yang, D. R. Du Bois, J. W. Zillera and J. S. Nowick, Chem. Commun., 2017, 53, 2772–2775 RSC.
- S. T. Hsu, E. Breukink, E. Tischenko, M. A. Lutters, B. de Kruijff, R. Kaptein, A. M. Bonvin and N. A. van Nuland, Nat. Struct. Mol. Biol., 2004, 11, 963–967 CAS.
- E. Breukink, H. E. van Heusden, P. J. Vollmerhaus, E. Swiezewska, L. Brunner, S. Walker, A. J. Heck and B. de Kruijff, J. Biol. Chem., 2003, 278, 19898–19903 CrossRef CAS PubMed.
- K. Jin, I. H. Sam, K. H. Po, D. Lin, E. H. G. Zadeh, S. Chen, Y. Yuan and X. Li, Nat. Commun., 2016, 7, 12394–12400 CrossRef CAS PubMed.
- A. M. Giltrap, L. J. Dowman, G. Nagalingam, J. L. Ochoa, R. G. Linington, W. J. Britton and R. J. Payne, Org. Lett., 2016, 18, 2788–2791 CrossRef CAS PubMed.
- S. Dhara, V. B. Gunjal, K. L. Handore and D. S. Reddy, Eur. J. Org. Chem., 2016, 4289–4293 CrossRef.
- Y. E. Jad, G. A. Acosta, T. Naicker, M. Ramtahal, A. El-Faham, T. Govender, H. G. Kruger, B. G. de la Torre and F. Albericio, Org. Lett., 2015, 17, 6182–6185 CrossRef CAS PubMed.
- A. Parmar, A. Iyer, C. S. Vincent, D. Van Lysebetten, S. H. Prior, A. Madder, E. J. Taylor and I. Singh, Chem. Commun., 2016, 52, 6060–6063 RSC.
- A. Parmar, S. H. Prior, A. Iyer, C. S. Vincent, D. Van Lysebetten, E. Breukink, A. Madder, E. J. Taylor and I. Singh, Chem. Commun., 2017, 53, 2016–2019 RSC.
- C. E. Schumacher, P. W. R. Harris, X.-B. Ding, B. Krause, T. H. Wright, G. M. Cook, D. P. Furkert and M. A. Brimble, Org. Biomol. Chem., 2017, 15, 8755–8760 CAS.
- S. A. H. Abdel Monaim, Y. E. Jad, G. A. Acosta, T. Naicker, E. J. Ramchuran, A. El-Faham, T. Govender, H. G. Kruger, B. G. de la Torre and F. Albericio, RSC Adv., 2016, 6, 73827–73829 RSC.
- H. Yang, K. H. Chen and J. S. Nowick, ACS Chem. Biol., 2016, 11, 1823–1826 CrossRef CAS PubMed.
- C. Wu, Z. Pan, G. Yao, W. Wang, L. Fang and W. Su, RSC Adv., 2017, 7, 1923–1926 RSC.
- K. H. Chen, S. P. Le, X. Han, J. M. Frias and J. S. Nowick, Chem. Commun., 2017, 53, 11357–11359 RSC.
- T. Al-Quadan, C. T. Price, N. London, O. Schueler-Furman and Y. A. Kwaik, Trends Microbiol., 2011, 19, 573–579 CrossRef CAS PubMed.
- J. W. Wollack, N. A. Zeliadt, J. D. Ochocki, D. G. Mullen, G. Barany, E. V. Wattenberg and M. D. Distefano, Bioorg. Med. Chem. Lett., 2010, 20, 161–163 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic methods, LCMS, HPLC, HRMS, 1D NMR, TOCSY NMR. See DOI: 10.1039/c7cc06093a |
| This journal is © The Royal Society of Chemistry 2018 |