Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Antibiotic functionalised polymers reduce bacterial biofilm and bioburden in a simulated infection of the cornea†
Natalya
Doroshenko
a,
Stephen
Rimmer
 b,
Richard
Hoskins
b,
Prashant
Garg
c,
Thomas
Swift
b,
Richard
Hoskins
b,
Prashant
Garg
c,
Thomas
Swift
 b,
Hannah L. M.
Spencer
b,
Rianne M.
Lord
b,
Hannah L. M.
Spencer
b,
Rianne M.
Lord
 b,
Maria
Katsikogianni
b,
Maria
Katsikogianni
 b,
David
Pownall
b,
Sheila
MacNeil
b,
David
Pownall
b,
Sheila
MacNeil
 d,
C. W. Ian
Douglas
a and
Joanna
Shepherd
d,
C. W. Ian
Douglas
a and
Joanna
Shepherd
 *a
*a
aSchool of Clinical Dentistry, University of Sheffield, Sheffield, S10 2TA, UK. E-mail: j.shepherd@sheffield.ac.uk
bDepartment of Chemistry and Biosciences, University of Bradford, Bradford, BD7 1DP, UK
cKallam Anji Reddy Campus, L V Prasad Marg, Banjara Hills, Hyderabad 500 034, Telangana, India
dDepartment of Materials Science and Engineering, University of Sheffield, Sheffield, S3 7HQ, UK
First published on 4th June 2018
Abstract
Microbial keratitis can arise from penetrating injuries to the cornea. Corneal trauma promotes bacterial attachment and biofilm growth, which decrease the effectiveness of antimicrobials against microbial keratitis. Improved therapeutic efficacy can be achieved by reducing microbial burden prior to antimicrobial therapy. This paper assesses a highly-branched poly(N-isopropyl acrylamide) with vancomycin end groups (HB-PNIPAM-van), for reducing bacterial attachment and biofilm formation. The polymer lacked antimicrobial activity against Staphylococcus aureus, but significantly inhibited biofilm formation (p = 0.0008) on plastic. Furthermore, pre-incubation of S. aureus cells with HB-PNIPAM-van reduced cell attachment by 50% and application of HB-PNIPAM-van to infected ex vivo rabbit corneas caused a 1-log reduction in bacterial recovery, compared to controls (p = 0.002). In conclusion, HB-PNIPAM-van may be a useful adjunct to antimicrobial therapy in the treatment of corneal infections.
Introduction
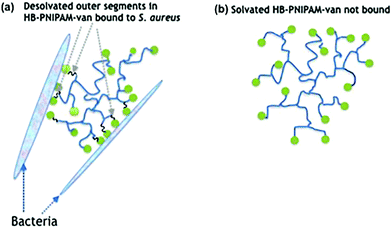
Biofilm formation is central to the colonisation of materials and tissues by infective bacteria.1 The adhesion of bacterial cells to surfaces (whether biotic or abiotic) is the initial stage of formation of biofilms,2 which are bacterial communities attached to a surface encased in a biopolymer matrix.3 It is estimated that 80% of all clinical infections are biofilm related4 and the particular clinical problem they pose is their high levels of antimicrobial non-responsiveness compared with planktonic bacteria. This non-responsiveness is not generally due to acquisition of resistance genes but instead arises from altered physiological and physical features of the community.5,6 The biomolecule-enriched matrix provides a penetration barrier to antimicrobials7 and subpopulations of bacteria in biofilms (known as persister cells) grow more slowly and are less metabolically active than their planktonic counterparts.5 Thus biofilm infections are difficult to treat and therefore strategies that prevent biofilm formation or disrupt established biofilms may offer valuable adjuncts to antimicrobial therapy.8–10 Biofilm disruption can be achieved by applying complexing agents or surfactant molecules,11–13 such as low molar mass14 or polymer amphiphilles.15,16 Antimicrobial peptides and lipopeptides are also amphiphiles and they are known to disrupt biofilms.17,18In previous work we showed that highly-branched poly(N-isopropyl acrylamide) (HB-PNIPAM) functionalised at the chain ends with vancomycin (HB-PNIPAM-van) responded after binding to bacteria by desolvation of a fraction of the polymer segments.19–22 Upon attachment of bacteria to the vancomycin chain ends, the polymer passes through a phase transition that involves desolvation of segments. This induces a change in the conformation of the polymer from a coiled to a globular state (Fig. 1), which in this context manifests macroscopically as the formation of polymer–bacteria aggregates. Similarly, results were reported with a polymyxin variant of the same polymers (HB-PNIPAM-pmx) on binding to the Gram negative species, Pseudomonas aeruginosa.23 Although often described as a hydrophobic phase, the globular desolvated state is amphiphilic and retains water after the main desolvation event. In this context, we considered that bacteria being bound within the desolvated polymer structures would disrupt biofilms in the same way that other amphiphiles have been shown to act. However, the disruptive action would be expected to be enhanced as the polymer competes with the extracellular macromolecules of the biofilm to bind to the resident bacteria. In support of this concept, previous work by us has also shown that both HB-PNIPAM-van and HB-PNIPAM-pmx can dislodge bacteria from in vitro infected dermal tissue models.20
As an exemplar of this principle we examined the effect of the HB-PNIPAM-van polymer on biofilms associated with infection of the eye, microbial keratitis. Microbial keratitis is a sight-threatening disease, and is recognized by the World Health Organization as a major cause of corneal-related blindness.24 The disease can have a bacterial, fungal, viral or protozoal cause and the most common bacteria are Staphylococcus aureus and Pseudomonas aeruginosa,25 although streptococcal and enterobacterial causes are also known. Since ophthalmic biofilm infections, also implicated in cases of endophthalmitis and crystalline keratopathy,26 are so difficult to treat, surgical removal of the infected tissue resulting in corneal blindness often remains the only permanent treatment option.27
In this work, we evaluate the effect of HB-PNIPAM-van on the ability of S. aureus to attach and form a biofilm on a plastic substrate and its ability to remove bacteria form a simulated infection model of keratitis using ex vivo rabbit corneas.
Results and discussion
HB-PNIPAM-van binds S. aureus but not P. aeruginosa
A HB-PNIPAM-van polymer was prepared. The material had a degree of branching of 0.042, Mn 1937 kDa, Mw = 2269 kDa, and the chain end functionality was 0.08 mg vanc per mg polymer, (see ESI† for full details). HB-PNIPAM-van has been previously shown to interact with S. aureus using an “aggregation assay” that showed the formation of large bacterial aggregates in the presence of HB-PNIPAM-van.19,20 In the work reported here, electron microscopy was used to investigate the polymer–bacteria interaction. Fig. 2a shows an image of S. aureus alone dispersed evenly on a surface. Incubation of HB-PNIPAM-van with S. aureus for 4 hours resulted in the formation of large bacterial aggregates with average dimensions of 13 μm. Further images of aggregates formed by HB-PNIPAM-van only and HB-PNIPAM-van with S. aureus over shorter timescales can be seen in ESI, Fig. S5 and S6.†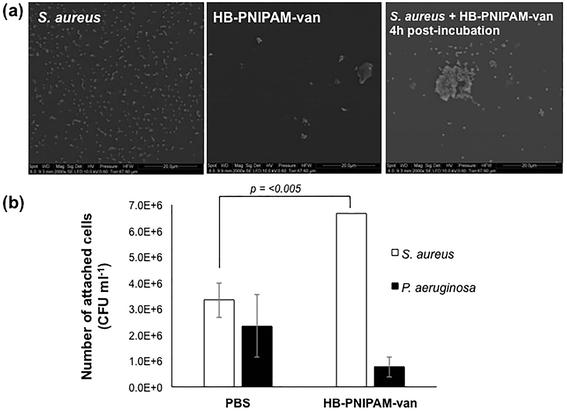 | ||
| Fig. 2 Incubation of S. aureus with HB-PNIPAM-van results in formation of aggregates. (a) Electron micrographs showing the formation of S. aureus and HB-PNIPAM-van aggregates. Scale bar, 20 μm. More images of polymer–bacteria aggregates can be seen in ESI, Fig. S5 and S6.† (b) The average number of viable S. aureus and P. aeruginosa cells attached to HB-PNIPAM-van-treated or PBS-treated (control) microplate wells. Error bars; standard error of the mean, n = 9 (3 independent experiments, 3 replicates for each experiment). | ||
To quantify the degree of polymer–bacteria interaction, viable counts of attached bacteria in wells to which HB-PNIPAM-van was anchored to the surface via an anti-vancomycin antibody were determined. For comparison attachment of a Gram-negative species, P. aeruginosa, which should not bind significantly to HB-PNIPAM-van, was assessed. The value for S. aureus attachment to the control wells (anti-vancomycin but no polymer) was higher than expected but this was probably due to protein A-mediated binding to the mouse IgM. Nonetheless, viable count data showed a significant, 60% rise in S. aureus cell attachment (p = 0.0038) to polymer-coated wells as opposed to the uncoated control (PBS) (Fig. 2b). Moreover, there was no significant difference in the attachment of P. aeruginosa to PBS or HB-PNIPAM-van treated surfaces, confirming earlier findings that HB-PNIPAM-van exhibits binding specificity for S. aureus.20
HB-PNIPAM-van inhibits cell attachment and biofilm formation
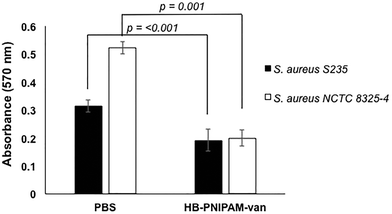
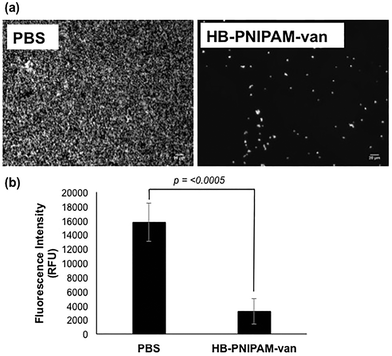
To determine the effect of HB-PNIPAM-van on S. aureus biofilm formation, total biofilm biomass of 24 h biofilms formed in the presence of HB-PNIPAM-van was measured using crystal violet staining. HB-PNIPAM-van caused a 38% and 61% reduction in biofilm biomass formed by S. aureus S-235 and S. aureus NCTC 8325-4 respectively, when compared to the PBS control (Fig. 3). It is important to highlight that the concentration of HB-PNIPAM-van used in this assay was 10 times lower (0.5 mg ml−1) than in the other experiments presented in this paper but was still sufficient to cause a statistically significant inhibition of biofilm formation for S. aureus S-235 (p = 0.0008) and S. aureus NCTC 8325-4 (p = 0.001). Interestingly, HB-PNIPAM-van caused a larger reduction in the biofilm biomass of S. aureus NCTC 8325-4, which was the better biofilm-forming strain.To determine how HB-PNIPAM-van interferes with biofilm formation, Syto9-stained S. aureus S-235 was pre-incubated with HB-PNIPAM-van and added to a microplate to initiate attachment. The results showed that a much lower proportion of S. aureus attached to the micro-plate when the bacteria were pre-treated with HB-PNIPAM-van as opposed to treatment with PBS (Fig. 4a). Furthermore, the micrographs were confirmed by quantification of fluorescence intensity of attached bacteria, which showed that polymer pre-treated cells attached at a statistically significantly lower level than untreated cells (p = <0.001) (Fig. 4b). A similar experiment in which the fluorescence intensity of different concentrations of S. aureus (cells per ml) was measured (ESI, Fig. S7†) revealed that 109 cells per ml resulted in a fluorescence intensity reading of 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 687 RFU. The difference in fluorescence intensity from the bacterial cells treated with PBS or HB-PNIPAM-van treated surfaces was 12
687 RFU. The difference in fluorescence intensity from the bacterial cells treated with PBS or HB-PNIPAM-van treated surfaces was 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 583 RFU (Fig. 4b). This suggests that HB-PNIPAM-van inhibited the attachment of approximately 3 × 108 cells per ml.
583 RFU (Fig. 4b). This suggests that HB-PNIPAM-van inhibited the attachment of approximately 3 × 108 cells per ml.
HB-PNIPAM-van changes biofilm architecture
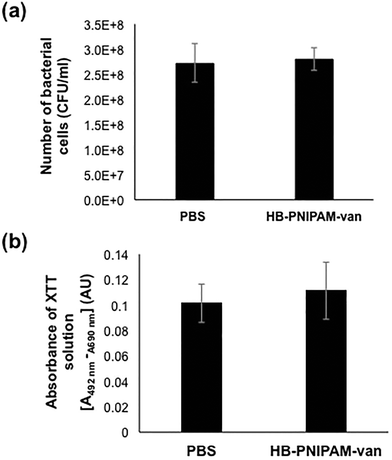
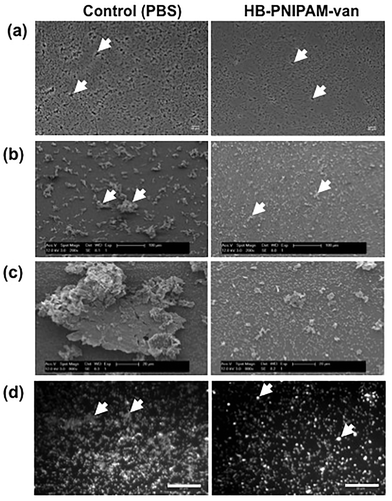
To assess the effects of HB-PNIPAM-van on pre-formed biofilms, the total number of bacteria left after treatment with HB-PNIPAM-van and their metabolic activity was quantified from biofilms treated with PBS or HB-PNIPAM-van for 24 hours. The results showed no significant differences in the number of recovered viable bacteria from biofilms treated with PBS (2.7 × 108 (±6.7 × 107) CFU ml−1) or HB-PNIPAM-van (2.8 × 108 (±3.9 × 107) CFU ml−1) (Fig. 5a), indicating that HB-PNIPAM-van did not kill bacteria in this system. Similarly, there was no difference in the metabolic activity of bacteria between the PBS and HB-PNIPAM-van treated biofilms (Fig. 5b) suggesting that HB-PNIPAM-van has no affect on proliferation of bacteria within biofilms. However when PBS and polymer-treated biofilms were analysed by microscopy, differences in the structure of the biofilm were observed (Fig. 6). Phase contrast microscopy showed that compared to biofilms exposed to HB-PNIPAM-van, PBS-treated biofilms were formed of larger aggregates (Fig. 6a). Similarly, SEM data revealed that control biofilms contained fewer but larger aggregates than the polymer-treated biofilm (Fig. 6b). High magnification images of the aggregates observed in both biofilm samples also showed that control biofilms consisted of much larger aggregates (Fig. 6c). Since the bacterial population between PBS-treated and polymer-treated biofilms is similar (Fig. 5a), it is possible that bacteria residing in control biofilms produce a greater abundance of biofilm matrix molecules which form the basis of the larger aggregates observed in control biofilms. Given that HB-PNIPAM-van binds bacteria, the findings presented in Fig. 6c, suggest that the polymer may have prevented S. aureus cells from either being integrated into the already formed aggregates or from actively forming their own aggregates, and so HB-PNIPAM-van may promote the segregation of S. aureus as individual cells (Fig. 6c).Data from fluorescence experiments, in which PBS and polymer-treated biofilms were stained with the fluorescently labelled lectin (WGA), showed that polymer-treated biofilms appeared thinner and more dispersed compared to control biofilms, which developed as a thick collection of conjoined aggregates (Fig. 6d). Since poly-N-acetylglucoasmine (PNAG) is an abundant component of S. aureus biofilms and WGA binds to N-acetylglucosamine residues within bacteria and the biofilm matrix, the data suggest that the abundance of PNAG was reduced in the biofilm exposed to HB-PNIPAM-van, given the absence of larger areas of WGA fluorescence (Fig. 6d). Collectively these findings imply that HB-PNIPAM-van alters the biofilm structure and possibly the arrangement of bacteria within the biofilm.
HB-PNIPAM-van reduces the bioburden in a corneal infection model
To test the potential of HB-PNIPAM-van as a treatment for tissue infections, HB-PNIPAM-van or PBS (control treatment) were applied to deliberately wounded and infected ex vivo rabbit corneas for a period of 4 hours. Afterwards the corneas were washed and the remaining bacteria enumerated. The data showed that corneas exposed to HB-PNIPAM-van contained significantly fewer (p < 0.002) bacteria than corneas treated with PBS (Fig. 7a). Since the application of HB-PNIPAM-van to infected corneas resulted in a 10-fold reduction in concentration (CFU ml−1) of bacteria, this demonstrates that HB-PNIPAM-van effectively reduces the bioburden in a corneal infection.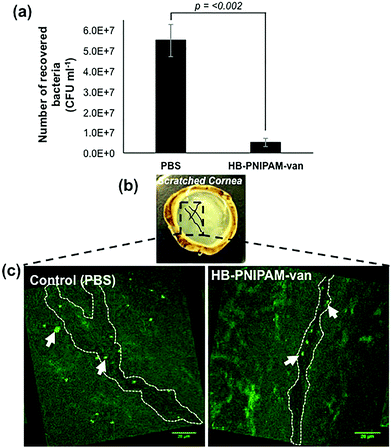 | ||
| Fig. 7 HB-PNIPAM-van reduces bacterial load in a corneal infection model. (a) The number of S. aureus cells recovered from S. aureus infected ex vivo rabbit corneas following a 4 h treatment with HB-PNIPAM-van or PBS (control). Error bars; standard error of the mean, n = 9 (3 independent experiments, 3 replicate corneas per an experiment). (b) Photograph of an ex vivo rabbit cornea which has been scratched with several criss-cross lines using a scalpel. Scratches are illustrated in the photograph with black solid lines. The box drawn in black dashed lines represents areas of interest which formed the basis of confocal microscopy analysis; i.e. the scratch sites were predicted to harbour most bacteria and so confocal images of these particular sites of the cornea were taken. (c) Representative confocal microscopy images of infected ex vivo rabbit corneas following a 4 h treatment with HB-PNIPAM-van or PBS (control). White dashed outline illustrates the shape of the scratch made during the infection stage of the experiment. Background fluorescence is auto-fluorescence from the cornea. White arrows indicate bright circular aggregates of GFP-labelled S. aureus, which are confined to the scratch area. Raw confocal microscopy images can be seen in ESI, Fig. S4.† | ||
In a similar experiment, PBS and polymer-treated corneas were examined by microscopy, specifically focusing on the area around the wound made to the cornea during the infection process, as this was the area of the cornea previously shown to harbour the most bacteria.28 As seen in Fig. 7b & c, in the PBS-treated cornea, GFP-labelled S. aureus formed large aggregates not only in the location of the wound, but throughout the entire cornea. Alternatively, the cornea exposed to HB-PNIPAM-van treatment appeared to contain much fewer S. aureus aggregates and these appeared smaller in size and were mostly localised around the surface of the scratch wound (Fig. 7b & c). Some green background autofluorescence from the corneal cells is also visible.
To address any potential cytotoxicity issues, all polymeric materials were found to be non-toxic to human epithelial (and other tested) cell types following 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) cell viability tests (see ESI, Table S3†).
Discussion
The aim of this study was to explore the potential of HB-PNIPAM-van to influence biofilm formation and already formed biofilms both on abiotic surfaces and in an infected tissue model. This study builds on the work of Shepherd et al. (2011),20 which showed that HB-PNIPAM-van could bind but not kill bacteria in an infected skin wound model and shows the generality of the approach. The findings presented here provide more information on how the HB-PNIPAM-van interacts with S. aureus and how the polymer affects the formation and development of S. aureus biofilms.The current work shows that HB-PNIPAM-van exhibited activity against S. aureus but not P. aeruginosa (Fig. 2b), which is consistent with the binding mechanism of vancomycin; the Gram-negative outer membrane provides a penetration barrier to large molecules like vancomycin29 and HB-PNIPAM-van, so preventing binding to the vancomycin target. Furthermore, effective binding between HB-PNIPAM-van and S. aureus (an interaction which caused a significant reduction in bacterial numbers in infected cornea, Fig. 7) occurred within a relatively short time frame (4 hours), suggesting that HB-PNIPAM-van could be a viable candidate for topical application. Using a tissue-engineered human skin model of a burn wound, it was previously shown that treatment with HB-PNIPAM-van for 1 hour caused a significant reduction of the bacterial bioburden in the wound,20 thus further demonstrating the versatility of this polymer. Biofilm formation of S. aureus in nutrient rich media was inhibited in the presence of HB-PNIPAM-van (Fig. 3) and pre-incubation of S. aureus cells with HB-PNIPAM-van inhibited their attachment to an abiotic surface (Fig. 4). Many bacteria live on surfaces as biofilms; contact lens-associated microbial keratitis is characterized by biofilm growth on the lens, whereas in endophthalmitis, staphylococci adhere to intraocular lenses leading to infection of the intraocular cavity.26 Treatment with HB-PNIPAM-van may be of benefit in such cases as the anti-fouling properties of HB-PNIPAM-van may prevent the bacteria on the implant from spreading, attaching and colonizing the cornea and thus HB-PNIPAM-van may retard the development of implant associated corneal infections. However, this requires further exploration.
Important findings from this study show that HB-PNIPAM-van is able to reduce the binding of bacteria to surfaces and alter the structure of the remaining biofilm (Fig. 6) in an established corneal infection (Fig. 7) Compared to a biofilm treated with PBS, HB-PNIPAM-van treated biofilms consisted of smaller bacterial aggregates and a more dispersed biofilm matrix showed that HB-PNIPAM-van is able to bind bacteria from an established corneal infection (Fig. 7), and that it altered the structure of the remaining biofilm (Fig. 6). Compared to a biofilm treated with PBS, HB-PNIPAM-van treated biofilms consisted of smaller bacterial aggregates. Such modified biofilm structures may allow greater penetration of antimicrobials into the depths of the biofilm and achieve greater killing. Collectively these data suggest that HB-PNIPAM-van could be given as an adjunctive topical treatment alongside antimicrobials. In an established corneal infection, HB-PNIPAM-van could eliminate enough bacteria from the site of injury and subsequently reduce the overload on the eye's natural defence systems. The dispersion of the biofilm matrix created by the polymer and the lower bacterial concentration in the eye following polymer treatment could improve the success rate of subsequent antimicrobial therapy, using pre-existing agents such as Moxifloxacin.
Whilst it is clear that the effects of polymers and other materials on biofilms is complex, time dependent and species dependent, the data support the hypothesis that HB-PNIPAM with selective ligands at the chain ends forms specific aggregates with the target bacteria, and that these aggregates disrupt the normal formation of biofilms. The aggregates are large amphiphilic structures and their formation appears in this case to promote the dispersion of the biofilm as the bacteria preferentially attach to the polymer rather than the extracellular macromolecules of the biofilm or the underlying surface.
Experimental
Materials
N-Isopropylacrylamide, NIPAM (97%), N-hydroxysuccinimide, NHS (98%), dicyclohexyl carbodiimide, DCC (98% purity), azobiscyanovaleric acid, ACVA (98%), and vancomycin hydrochloride hydrate were obtained from Sigma Aldrich. 4-Vinylbenzylpyrrole carbodithioate (1) was synthesised and purified according to the previously reported method.30 Dioxane (Analar grade), diethyl ether (anhydrous), dimethyl formamide (Analar grade), ethanol (Analar grade), acetone (Analar grade) and hydrochloric acid (35% wt/wt) were obtained from VWR Ltd.Highly-branched polymer (HB-PNIPAM)
NIPAM (20 g, 0.17 moles), 1 (1.84 g, 0.0071 moles) and ACVA (1.98 g, 0.0071 moles) were dissolved in 140 ml of 1,4-dioxane. The solution was degassed and the polymerization conducted under nitrogen at 60 °C for 48 hours. The polymers were purified by precipitation (twice) in diethyl ether. Sample yield 23.1 g, monomer conversion = 97%. The polymer was then dissolved in ethanol and the lower molar mass fraction was removed using ultra-filtration through a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 molar mass cut off membrane. 1H NMR (400 MHz, DMSO) (ppm): d 0.9–1.1 (6H, s, –N(CH3)2), d 1.3–1.7 (2H, br m, –CH2–CH–Ar–), d 1.8–2.2 (2H, br m, –CH2–CH–CO–NH–) and (1H, br m, CH2–CH–CONH–), d 3.3 (H2O-polymer bound), d 3.9 (1H, br s, (CH3)2CH–), d 6.4 (H2, br s, N-pyrrole–H), d 6.6–7.6 (br m, –Ar–), d 7.7 (2H, br s, N-pyrrole–H), d 12.2 (br m COOH). FTIR peak from COOH at 1713 cm−1. The polymer molar mass moments after ultra-filtration via methanol size exclusion chromatography were Mn 1937.2 kDa, Mw 2269.9 kDa, Mz 2698.2 kDa, LCST (deionised H2O) 18.6 °C.
000 g mol−1 molar mass cut off membrane. 1H NMR (400 MHz, DMSO) (ppm): d 0.9–1.1 (6H, s, –N(CH3)2), d 1.3–1.7 (2H, br m, –CH2–CH–Ar–), d 1.8–2.2 (2H, br m, –CH2–CH–CO–NH–) and (1H, br m, CH2–CH–CONH–), d 3.3 (H2O-polymer bound), d 3.9 (1H, br s, (CH3)2CH–), d 6.4 (H2, br s, N-pyrrole–H), d 6.6–7.6 (br m, –Ar–), d 7.7 (2H, br s, N-pyrrole–H), d 12.2 (br m COOH). FTIR peak from COOH at 1713 cm−1. The polymer molar mass moments after ultra-filtration via methanol size exclusion chromatography were Mn 1937.2 kDa, Mw 2269.9 kDa, Mz 2698.2 kDa, LCST (deionised H2O) 18.6 °C.
Polymer chain end modification
The chain ends of the highly branched polymers were converted to carboxylic acid by means of 3 additions of ACVA (20 fold molar excess relative to RAFT agent in the initial polymerisation feed). HB-PNIPAM (22 g) was dissolved in DMF (150 ml). ACVA (19 g, 0.068 moles) was added 3 times after 24 hours intervals. The solution was maintained at 65 °C under a nitrogen atmosphere. The polymer was precipitated into diethyl ether. The solid was dissolved in ethanol and concentrated by ultra-filtration and then rotary evaporated to remove the solvent. 17.7 g of derivatised polymer was obtained. FTIR COOH 1713 cm−1. LCST 19.3 cm−1. The carboxylic acid groups were converted to succinimide by reaction of carboxylic acid chain ends with N-hydroxysuccinimide. Highly branched polymer with carboxylic acid chain ends (17.7 g) was dissolved in DMF (150 ml). N-Hydroxy succinimide (3.03 g, 0.027 moles) and dicyclohexyl carbodiimide (5.5 g, 0.027 moles) were added and the solution stirred overnight. The mixture was filtered and the polymer precipitated into diethyl ether. The solid was dissolved in ethanol and concentrated by ultra-filtration (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 molar mass cut off membrane) and then rotary evaporated to give 11.8 g of polymer, 67% yield. FTIR (succinimide) 1741 cm−1, 1783 cm−1. LCST 19.7 °C. Succinimide derivatized polymer (3 g) was dissolved in water (150 ml) over ice. A solution of vancomycin (144 mg, 9.7 × 10−5 moles) in phosphate buffered saline pH 9.5 (5 ml) was added. The solution was stirred for 3 days at room temperature and purified by ultrafiltration and then freeze dried. FTIR (ESI, Table S1†) shows no identified succinimide or COOH peaks. Mn 4402 kDa, Mw 5439 kDa, Mz 6380.6 kDa, RH (DOSY, D2O) Rhn 8.22, Rhw 8.24, Rhz 8.26 nm. LCST (deionised H2O) 32.5 °C.
000 g mol−1 molar mass cut off membrane) and then rotary evaporated to give 11.8 g of polymer, 67% yield. FTIR (succinimide) 1741 cm−1, 1783 cm−1. LCST 19.7 °C. Succinimide derivatized polymer (3 g) was dissolved in water (150 ml) over ice. A solution of vancomycin (144 mg, 9.7 × 10−5 moles) in phosphate buffered saline pH 9.5 (5 ml) was added. The solution was stirred for 3 days at room temperature and purified by ultrafiltration and then freeze dried. FTIR (ESI, Table S1†) shows no identified succinimide or COOH peaks. Mn 4402 kDa, Mw 5439 kDa, Mz 6380.6 kDa, RH (DOSY, D2O) Rhn 8.22, Rhw 8.24, Rhz 8.26 nm. LCST (deionised H2O) 32.5 °C.
Polymer characterisation
All 1H NMR spectra (ESI, Table S2†) were measured and recorded on a Bruker AC400 which was operated at with D2O as the solvent. FTIR measurements were carried out on a Thermo Scientific Nicolet iS10 FTIR Spectrometer. Solid samples were dried in a vacuum oven overnight before use. Polymer molar mass averages were obtained by size exclusion chromatography using Agilent Polargel columns with methanol as the eluent, using previously published methodologies.31 This used 3× Agilent Polargel columns connected to an Agilent 1260 Infinity system with a refractive index, ultraviolet light and viscometric detector, and was calibrated using linear polyNIPAM standards with a methanol eluent at 1 ml min−1 flow rate. Polymer hydrodynamic radii (RH) were calculated using diffusion ordered spectroscopy (DOSY) NMR in D2O at 298 K. Differential Scanning Calorimetry (DSC) was carried out using a TA Instruments MicroDSC in deionised water (1 mg ml−1) polymer solution in the range 2 to 60 °C at heating rate of 1 °C min−1. Raw data (NMR, FTIR, GPC) are contained in the supplementary material (Fig. S1–S3, Tables S1 and S2†). Polymer samples were stored as a dry powder at −18 °C.Bacterial culture
Four strains of S. aureus were employed; a reference strain NCTC 8325-4, strain NCTC 6571, strain SH1000-GFP (provided by the Foster lab, University of Sheffield) and strain S-235, a local clinical isolate. In addition P. aeruginosa strain SOM-1, a local clinical isolate was used. All strains were cultured in brain heart infusion (BHI) broth (Oxoid, UK) at 37 °C overnight and stored at 4 °C on BHI agar (Sigma-Aldrich, UK). The number of bacterial cells in an overnight culture was enumerated by microscopic counting and diluted to the concentrations required for each experiment.Electron microscopy analysis of HB-PNIPAM-van and bacteria binding
Overnight bacterial cultures of S. aureus S-235 were diluted with H2O to concentrations of 1 × 108 CFU ml−1. Afterwards, 10 μl of each sample was pipetted on to a cylindrical glass slide and left to stand for 2 hours at room temperature to facilitate attachment. If required, 100 μl of 0.5 mg ml−1 polymer solution was added to the bacterial sample and left to stand for 4 hours at room temperature before aspiration. Bacteria were fixed using 2.5% glutaraldehyde (GA) (Sigma Aldrich, Cat no. G5882) for 20 minutes. Following aspiration of the GA, samples were washed twice with 100 μl of H2O and then dehydrated via a graded 100 μl ethanol series (10, 30, 50, 70, 90, 100% ethanol, 10 minutes each) before re-aspiration. Samples were then sputter coated in gold (4 nm) under a 10 mA current for 1 minute and imaged under low vacuum (pressure of 0.59–0.63 Torr) using a scanning electron microscope (Quanta 400 FEI).Quantification of bacteria binding to HB-PNIPAM-van
Monoclonal mouse anti-vancomycin antibody (M997129; Abcam, UK) (100 μl per well) was diluted in coating buffer (sodium bicarbonate, 0.035 M, sodium carbonate, 0.015 M and sodium azide, 0.003 M, adjusted to pH 9.6) at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in 96-well ELISA microplates and incubated at 4 °C overnight. The plates were then washed 4 times with 200 μl PBS-Tween (1 μl Tween 20 per 200 ml of PBS) and HB-PNIPAM-van (5 mg ml−1 in PBS) was added to some wells while PBS was added to others to act as a control. The plates were incubated at RT overnight, washed 4 times with 200 μl of PBS and 100 μl per well of bacterial suspension was added and incubated at room temperature (RT) overnight. S. aureus S-235 or P. aeruginosa suspensions were prepared at 7 × 108 cells per ml in PBS.
500 in 96-well ELISA microplates and incubated at 4 °C overnight. The plates were then washed 4 times with 200 μl PBS-Tween (1 μl Tween 20 per 200 ml of PBS) and HB-PNIPAM-van (5 mg ml−1 in PBS) was added to some wells while PBS was added to others to act as a control. The plates were incubated at RT overnight, washed 4 times with 200 μl of PBS and 100 μl per well of bacterial suspension was added and incubated at room temperature (RT) overnight. S. aureus S-235 or P. aeruginosa suspensions were prepared at 7 × 108 cells per ml in PBS.
After removal of any unattached bacteria by washing 4 times in PBS, attached bacteria were dislodged by thoroughly scraping and vigorously pipetting 100 μl of PBS in each well. Serial dilutions were then made and viable counting performed following spotting (10 μl) of each dilution onto BHI agar and incubation at 37 °C overnight.
An alternative method was also employed in which an overnight culture of bacteria was diluted to 7 × 108 bacteria per ml, washed twice in PBS and re-suspended in a solution of FilmTracer™ FM® 1–43 Green Biofilm Cell Stain (Invitrogen, UK) prepared according to manufacturer's instructions for 20 min at RT. After washing thoroughly in PBS 100 μl of the stained bacterial suspension was added to each coated well (as above) the plates incubated at RT overnight. Plates were washed 4 times with PBS to remove unattached bacteria and residual fluorescence in the wells was measured using a plate-reader (Infinite M200, Tecan, UK) operated by Magellan software (excitation/emission: 472/580 nm).
The effect of HB-PNIPAM-van on bacterial attachment
An overnight culture of S. aureus S-235 was diluted to 109 bacteria per ml, washed twice and resuspended in PBS. Then, in fresh Eppendorf tubes, the bacterial suspension was mixed with HB-PNIPAM-van (5 mg ml−1) or PBS at a final concentration of 5 × 108 cells per ml. The tubes were incubated for 4 hours at 37 °C to encourage HB-PNIPAM-van and bacteria binding. Afterwards, the PBS and HB-PNIPAM-van-S. aureus mixtures or the PBS-bacteria controls were added to 96-well microplates and incubated for 2 hours at 37 °C to initiate attachment. Then, the plate was washed 3 times with PBS and the samples stained with SYTO® 9 from LIVE/DEAD™ BacLight™ bacterial viability kit (Invitrogen, UK), (prepared according to manufactures instructions). After washing to remove any residual stain, fluorescence was measured using a Tecan plate reader (excitation/emission: 488/520 nm). Microscopic images were also acquired using the Zeiss Axiovert 200 M inverted fluorescence microscope and the AxioVision software.The effect of HB-PNIPAM-van on biofilm formation
Overnight cultures of S. aureus S-235 and S. aureus NCTC 8325-4 were diluted and added to fresh BHI in a 24-well microplate at a final concentration of 5 × 108 bacteria per ml. HB-PNIPAM-van solution (500 μg ml−1 in PBS) was added to wells while PBS alone was added to polymer-free wells as controls. Biofilms were formed in at 37 °C for 24 hours with gentle shaking (35 rpm), after which wells were washed with PBS and stained with 0.1% (w/v) crystal violet solution for 20 min. The plate was washed thoroughly with water and 200 μl of 95% ethanol was added for 20 min to release the stain from the biofilm bacteria. The absorbance of the resultant alcohol solution was measured at 595 nm using the plate-reader mentioned previously.The effect of HB-PNIPAM-van on biofilm cell growth and biofilm structure
To examine the effects of HB-PNIPAM-van on growth of attached cells and biofilm appearance, S. aureus S-235 biofilms were formed as described above. HB-PNIPAM-van (5 mg ml−1) in PBS was added to the microplate wells and incubation continued for a further 24 hours at 37 °C without shaking. Viable counts from wells were obtained after washing, scraping, serial dilution and plating on BHI agar as described above. The metabolic activity of biofilm cells was assessed using the cell proliferation kit II (XTT) (Roche). Briefly, to S. aureus S235 biofilms 100 μl of XTT solution was added (prepared according to manufacturer's instructions), the plates were covered in foil and incubated at 37 °C for 30 min. Following incubation, the XTT solution was transferred into a fresh plate and absorbance readings were recorded at 450 nm with a reference wavelength of 690 nm.The effects of HB-PNIPAM-van on biofilm structure were examined by fluorescence microscopy and scanning electron microscopy (SEM). For fluorescence microscopy, 24 h S. aureus S235 biofilms were grown in 96-well plates as above and stained with 5 μg ml−1 of wheat germ agglutinin (WGA) (Texas Red® – X conjugate, Thermo Fisher Scientific, UK) for 15 minutes at 37 °C. In parallel some biofilms were viewed by phase contrast microscopy and this plus the fluorescence images were collected using Zeiss Axiovert 200 M inverted fluorescence microscope and AxioVision software. For scanning electron microscopy S. aureus S235, 24 h biofilms were grown in BHI on circular 13 mm diameter cover slips and treated with PBS or HB-PNIPAM-van for 24 hours. Biofilms were then fixed in 3% glutaraldehyde, 0.1 M sodium cacodylate (pH 7.2) and stained, firstly with 0.15% Alcian blue and then with 2% osmium tetroxide in 0.1 M sodium cacodylate (pH 7.2) according to the method of Howlin et al. (2015).32 Finally, samples were critically point dried and sputter coated with gold (Edwards S150B). Images were acquired under full vacuum using a Philips XL-20 scanning electron microscope.
Corneal infection model
Ex vivo corneas were obtained from wild brown rabbits (Blackface Meat Company, Dumfries, Scotland) and their excision, decontamination and infection procedures were performed according to the protocol described by Pinnock et al., (2016).28 After the development of infection, HB-PNIPAM-van was made up in antibiotic-free Dulbecco's modified eagle's medium (DMEM) to 5 mg ml−1 and 100 μl of this or PBS (control) was added to the corneas and incubated for 4 hours at 37 °C. Afterwards, the corneas were homogenised and numbers of bacteria were measured by viable counting as previously described.28 Three corneas were used for each condition per experiment and the experiment was repeated 3 times.In experiments involving confocal microscopy, corneas were infected with GFP-labelled S. aureus SH1000 according to the procedures described above. After treatment with PBS or HB-PNIPAM-van, corneas were washed twice with PBS and attached to the bottom of a small petri dish (35 mm in diameter) using a tissue adhesive glue (Vetbond™, 3 M Science). Then, the petri dishes were filled with approximately 4 ml of PBS and the corneas were analysed using a Zeiss LSM510 NLO microscope, running AIM software and a Zeiss 63×/1.0 water immersion lens. During preliminary studies, it was discovered that the cornea exhibited some auto-fluorescence at the GFP emission wavelength (Fig. 7), however this was of low intensity and did not prevent imaging of the bacteria. The raw images are shown in ESI, Fig. S4.† The confocal images were processed using ImageJ where the brightness of both the auto-fluorescence and bacterial fluorescence was artificially enhanced for illustration purposes as this enabled visualisation of the bacteria on the corneal surface. There were 2 corneas for each condition and the experiment was repeated twice.
Conclusions
HB-PNIPAM-van altered the biofilm structure on a surface, inhibited bacterial cell attachment and reduced the bacterial load in a simulated infection of the cornea. These findings suggest that HB-PNIPAM-van offers a new approach to limiting the development of corneal infections and maybe a useful adjunct to antimicrobial treatment.Conflicts of interest
There are no conflicts to declareAcknowledgements
The authors wish to thank, Jason Heath, Colin Gray and Chris J. Hill for technical support. This work was supported by the Medical Research Council and the Department of Biotechnology, India under grant number, MR/N50188/2.References
- S. McCarty, E. M. Jones, S. Finnegan, E. Woods, C. A. Cochrane and S. L. Percival, Chapter Eighteen - Wound Infection and Biofilms. In Biofilms in Infection Prevention and Control, Academic Press, Boston, 2014, pp. 339–358 Search PubMed.
- M. E. Davey and G. A. O'Toole, Microbial biofilms: from ecology to molecular genetics, Microbiol. Mol. Biol. Rev., 2000, 64(4), 847–867 CrossRef PubMed.
- S. S. Branda, S. Vik, L. Friedman and R. Kolter, Biofilms: the matrix revisited, Trends Microbiol., 2005, 13(1), 20–26 CrossRef PubMed.
- P. D. Fey, Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections, Curr. Opin. Microbiol., 2010, 13(5), 610–615 CrossRef PubMed.
- K. I. M. Lewis, Minireview: Riddle of Biofilm Resistance, Antimicrob. Agents Chemother., 2001, 45(4), 999–1007 CrossRef PubMed.
- B. R. Levin and D. E. Rozen, Non-inherited antibiotic resistance, Nat. Rev. Microbiol., 2006, 4(7), 556–562 CrossRef PubMed.
- N. Doroshenko, B. S. Tseng, R. P. Howlin, J. Deacon, J. A. Wharton, P. J. Thurner, B. F. Gilmore, M. R. Parsek and P. Stoodley, Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin, Antimicrob. Agents Chemother., 2014, 58(12), 7273–7282 CrossRef PubMed.
- N. Barraud, D. J. Hassett, S.-H. Hwang, S. A. Rice, S. Kjelleberg and J. S. Webb, Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa, J. Bacteriol., 2006, 188(21), 7344–7353 CrossRef PubMed.
- I. Kolodkin-Gal, D. Romero, S. Cao and J. Clardy, D-amino acids trigger biofilm disassembly, Science, 2010, 328(5978), 627–629 CrossRef PubMed.
- B. Boles and A. Horswill, Staphylococcal, biofilm disassembly, Trends Microbiol., 2011, 19(9), 449–455 CrossRef PubMed.
- I. M. Banat, M. A. D. De Rienzo and G. A. Quinn, Microbial biofilms: biosurfactants as antibiofilm agents, Appl. Microbiol. Biotechnol., 2014, 98(24), 9915–9929 CrossRef PubMed.
- O. Schreiberová, P. Hedbávná, A. Čejková, V. Jirků and J. Masák, Effect of surfactants on the biofilm of Rhodococcus erythropolis, a potent degrader of aromatic pollutants, New Biotechnol., 2012, 30(1), 62–68 CrossRef PubMed.
- J. Luo, W. Lv, Y. Deng and Y. Sun, Cellulose–Ethylenediaminetetraacetic Acid Conjugates Protect Mammalian Cells from Bacterial Cells, Biomacromolecules, 2013, 14(4), 1054–1062 CrossRef PubMed.
- S. S. E Silva, J. W. P. Carvalho, C. P. Aires and M. Nitschke, Disruption of Staphylococcus aureus biofilms using rhamnolipid biosurfactants, J. Dairy Sci., 2017, 100(10), 7864–7873 CrossRef PubMed.
- P. S. Yavvari, S. Gupta, D. Arora, V. K. Nandicoori, A. Srivastava and A. Bajaj, Clathrin-Independent Killing of Intracellular Mycobacteria and Biofilm Disruptions Using Synthetic Antimicrobial Polymers, Biomacromolecules, 2017, 18(7), 2024–2033 CrossRef PubMed.
- D. S. Uppu, S. Samaddar, C. Ghosh, K. Paramanandham, B. R. Shome and J. Haldar, Amide side chain amphiphilic polymers disrupt surface established bacterial bio-films and protect mice from chronic Acinetobacter baumannii infection, Biomaterials, 2016, 74, 131–143 CrossRef PubMed.
- J. S. Khara, S. Obuobi, Y. Wang, M. S. Hamilton, B. D. Robertson, S. M. Newton, Y. Y. Yang, P. R. Langford and P. L. R. Ee, Disruption of drug-resistant biofilms using de novo designed short α-helical antimicrobial peptides with idealized facial amphiphilicity, Acta Biomater., 2017, 57(Supplement C), 103–114 CrossRef PubMed.
- J. Coronel-León, A. M. Marqués, J. Bastida and A. Manresa, Optimizing the production of the biosurfactant lichenysin and its application in biofilm control, J. Appl. Microbiol., 2016, 120(1), 99–111 CrossRef PubMed.
- J. Shepherd, P. Sarker, K. Swindells, C. W. I. Douglas, S. MacNeil, L. Swanson and S. Rimmer, Binding bacteria to highly branched poly(N-isopropyl acrylamide) modified with vancomycin induces the coil-to-globule transition, J. Am. Chem. Soc., 2010, 132(6), 1736–1737 CrossRef PubMed.
- J. Shepherd, P. Sarker, S. Rimmer, L. Swanson, S. MacNeil and I. Douglas, Hyperbranched poly(NIPAM) polymers modified with antibiotics for the reduction of bacterial burden in infected human tissue engineered skin, Biomaterials, 2011, 32(1), 258–267 CrossRef PubMed.
- P. Sarker, K. Swindells, C. W. I. Douglas, S. MacNeil, S. Rimmer and L. Swanson, Förster resonance energy transfer confirms the bacterial-induced conformational transition in highly-branched poly(N-isopropyl acrylamide with vancomycin end groups on binding to Staphylococcus aureus, Soft Matter, 2014, 10(31), 5824–5835 RSC.
- P. Teratanatorn, R. Hoskins, T. Swift, C. W. I. Douglas, J. Shepherd and S. Rimmer, Binding of bacteria to poly (N-isopropylacrylamide) modified with vancomycin: Comparison of behavior of linear and highly branched polymers, Biomacromolecules, 2017, 18(9), 2887–2899 CrossRef PubMed.
- P. Sarker, J. Shepherd, K. Swindells, I. Douglas, S. MacNeil, L. Swanson and S. Rimmer, Highly branched polymers with polymyxin end groups responsive to Pseudomonas aeruginosa, Biomacromolecules, 2011, 12(1), 1–5 CrossRef PubMed.
- S. Resnikoff, D. Pascolini, D. Etya'ale, I. Kocur, R. Pararajasegaram, G. P. Pokharel and S. P. Mariotti, Global data on visual impairment in the year 2002, Bull. W. H. O., 2004, 82(11), 844–851 Search PubMed.
- M. Green, A. Apel and F. Stapleton, Risk Factors and Causative Organisms in Microbial Keratitis, Cornea, 2008, 27(1), 22–27 CrossRef PubMed.
- M. J. Elder, F. Stapleton, E. Evans and J. K. Dart, Biofilm-related infections in Ophthalmology, Eye, 1995, 9, 102–109 CrossRef PubMed.
- K. K. Jefferson, What drives bacteria to produce a biofilm, FEMS Microbiol. Lett., 2004, 236(2), 163–173 CrossRef PubMed.
- A. Pinnock, N. Shivshetty, S. Roy, S. Rimmer, I. Douglas, S. MacNeil and P. Garg, Ex vivo rabbit and human corneas as models for bacterial and fungal keratitis, Graefes Arch. Exp. Ophthalmol., 2016, 255, 333–342 CrossRef PubMed.
- I. A. D. Lessard and C. T. Walsh, VanX, a bacterial D-alanyl-D-alanine dipeptidase: Resistance, immunity, or survival function, PNAS, 1999, 96, 11028–11032 CrossRef.
- S. Hopkins, S. R. Carter, J. W. Haycock, N. J. Fullwood, S. MacNeil and S. Rimmer, Sub-micron poly(N-isopropylacrylamide) particles as temerpaertaure responsive vehicles for the detachment and delivery of human cells, Soft Matter, 2009, 5, 4928–4937 RSC.
- T. Swift, R. Hoskins, R. Telford, R. Plenderleith, D. Pownall and S. Rimmer, Analysis using size exclusion chromatography of poly(N-isopropylacrylamide) using methanol as an eluent, J. Chromatogr. A, 2017, 1508, 16–23 CrossRef PubMed.
- R. P. Howlin, M. J. Brayford, J. S. Webb, J. J. Cooper, S. S. Aiken and P. Stoodley, Antibiotic-loaded synthetic calcium sulfate beads for prevention of bacterial colonization and biofilm formation in periprosthetic infections, Antimicrob. Agents Chemother., 2015, 59(1), 111–120 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8bm00201k |
| This journal is © The Royal Society of Chemistry 2018 |