Open Access Article
Open Access ArticleRaman-encoded, multivalent glycan-nanoconjugates for traceable specific binding and killing of bacteria†
Surendra H.
Mahadevegowda‡
a,
Shuai
Hou‡
a,
Jielin
Ma
a,
Damien
Keogh
a,
Jianhua
Zhang
a,
Asadulla
Mallick
b,
Xue-Wei
Liu
 b,
Hongwei
Duan
b,
Hongwei
Duan
 *a and
Mary B.
Chan-Park
*a and
Mary B.
Chan-Park
 *a
*a
aSchool of Chemical and Biomedical Engineering, Nanyang Technological University, 62 Nanyang Drive, Singapore 637459, Singapore. E-mail: hduan@ntu.edu.sg; mbechan@ntu.edu.sg
bDivision of Chemistry and Biological Chemistry, School of Physical and Mathematical Sciences, Nanyang Technological University, 21 Nanyang Link, Singapore 637371, Singapore
First published on 26th March 2018
Abstract
Glycan recognition plays key roles in cell–cell and host–pathogen interactions, stimulating widespread interest in developing multivalent glycoconjugates with superior binding affinity for biological and medical uses. Here, we explore the use of Raman-encoded silver coated gold nanorods (GNRs) as scaffolds to form multivalent glycoconjugates. The plasmonic scaffolds afford high-loading of glycan density and their optical properties offer the possibilities of monitoring and quantitative analysis of glycan recognition. Using E. coli strains with tailored on/off of the FimH receptors, we have demonstrated that Raman-encoded GNRs not only allow for real-time imaging and spectroscopic detection of specific binding of the glycan–GNR conjugates with bacteria of interest, but also cause rapid eradication of the bacteria due to the efficient photothermal conversion of GNRs in the near-infrared spectral window. We envision that optically active plasmonic glycoconjugates hold great potential for screening multivalent glycan ligands for therapeutic and diagnostic applications.
Introduction
Glycan recognition is commonly involved in a diverse range of biological and pathological processes such as proliferation, differentiation, immune response, and microbial infections via cell–cell and host–pathogen interactions.1 The interaction between individual carbohydrate ligands and protein receptors typically shows high specificity but low affinity, with dissociation constants in the millimolar range.2 Nature has circumvented this problem by forming multivalent carbohydrate constructs to increase their binding affinity with proteins.3 Multivalent glycoconjugates have been synthesized by the combination of carbohydrate head groups with different linkers and molecular or nanoscale scaffolds.4–7 Nanoparticles, in particular metal nanoparticles,8,9 magnetic nanoparticles,10–12 and quantum dots,13 have been employed to build multivalent glycoconjugates as they exhibit additional functionalities, which provide opportunities for magnetic separation of bacteria,10 targeting tumoral cells,14 inhibiting bacterial15 or virus infections,16,17 and detecting viruses.18 Real-time monitoring and quantitative detection of the interactions of multivalent glycan constructs with protein receptors in living systems is crucial for the design of high-affinity multivalent glycan-nanoconjugates for therapeutic and diagnostic applications. However, common analytical methods such as haemagglutination inhibition assay,19 isothermal microcalorimetry (ITC)20 and surface plasmon resonance (SPR)21 are limited to studies in solutions or at liquid–solid interfaces.Here, we use silver-coated gold nanorods (GNRs) as scaffolds to anchor glycans and establish a model system for the quantitative study of the multivalent binding between glycan-functionalized GNRs and bacteria. Due to the unique shape effect, the GNRs exhibit a strong longitudinal localized surface plasmon resonance (LSPR), which is damped via radiative and nonradiative channels.22 The radiative damping (light scattering) is easily detected using dark-field microscopy, which has been used for many bioimaging applications.23 Additionally, LSPR is accompanied by the generation of an intense electromagnetic field near the metal surface, facilitating surface enhanced Raman scattering (SERS).24–27 Finally, the nonradiative damping of the GNRs enables them to absorb light effectively and release heat into the environment. The absorption cross-section of the GNRs is four to six orders of magnitude larger than that of traditional dyes,28 making them an efficient photothermal transducer. Taken together, GNRs enable microscopic and spectroscopic detection and quantification of the binding event, and they can also be used for photothermal killing of bacteria.29
In this study, uropathogenic Escherichia coli (UPEC), which expresses the mannose-specific FimH protein on its type 1 fimbriae,30 has been chosen as a model bacterial system. UPEC causes urinary tract infection (UTI), which is one of the most prominent infectious diseases worldwide and affects 150 million people annually.31–34 The FimH subunits bind to the mannose moiety of the host cell; therefore, mannose derivatives are frequently used for targeting UPEC. We studied the specificity and affinity of such glyco-GNRs using two well-controlled UPEC strains: FimH-ON bacteria that express the FimH protein, and FimH-OFF bacteria which are FimH-deficient. We first confirmed the multivalency effect of the glycan-modified GNRs in concanavalin A (ConA) recognition. We further explored the multivalent glycan–GNR nanoconjugates for bacteria recognition. Dark-field microscopy and Raman spectroscopy clearly reveal that the GNRs specifically bind to FimH-ON bacteria. Raman spectroscopy also allows us to measure the binding affinity. Furthermore, the photothermal release of heat from the GNRs to the targeted bacteria upon near-infrared (NIR) irradiation leads to nearly 100% killing of the bacteria. Our well-defined model system provides new strategies for quantitative analysis of glycan–protein interactions in bacterial systems. Moreover, the combination of glycans and plasmonic nanoparticles is a promising platform for efficient targeted detection and killing of bacteria.
Experimental section
General methods for glycocluster synthesis and characterization
All the reactions were performed using oven-dried glassware apparatus. All thiol–ene click reactions were performed using UV light of wavelength 365 nm under an argon atmosphere. Silica gel of mesh size 35–70 μm purchased from Merck Pte Ltd was used to perform column chromatography. 1H, 13C and DEPT-135 spectra were recorded using a Bruker AV 300 MHz NMR instrument. In 1H NMR spectra, solvent residual peaks were set to 4.79 for D2O, 7.26 for CDCl3, and 3.31 for CD3OD and in proton decoupled 13C NMR spectra the middle peak of the solvent residual peak of CDCl3 was set to 77.16, and for CD3OD the middle peak was set to 49.00 ppm.Synthesis of Au nanorods
Au seeds were prepared by the reduction of HAuCl4 (0.25 mM, 5 mL) with NaBH4 (10 mM, 0.3 mL) in an aqueous cetyltrimethylammonium bromide (CTAB) solution (100 mM) and aged for 2 h before use. An aliquot of the seed solution (0.6 mL) was added to a growth solution (250 mL) containing CTAB (100 mM), HAuCl4 (0.5 mM), ascorbic acid (0.8 mM), aqueous AgNO3 (0.12 mM), and aqueous HCl (19 mM). The mixture was left undisturbed at 30 °C for 4 h. The solution was centrifuged twice (8000 rcf, 10 min) and redispersed in water to obtain a final concentration of gold equal to 0.5 mM.Synthesis of Raman dye modified Au@Ag nanorods (GNRs)
The Au nanorods were dispersed in a cetyltrimethylammonium chloride (CTAC) solution (10 mL, 10 mM). A DMF solution of the Raman dye, 4-nitrothiophenol, was added with a final concentration of 50 μM and the solution was incubated at 60 °C for 30 min. A solution of aqueous AgNO3 (0.5 mL, 10 mM) and ascorbic acid (0.5 mL, 100 mM) was added into the Au nanorod solution. The mixture was allowed to react at 60 °C for 1 h. The average Ag shell thickness was 2.2 nm. The GNRs were collected by centrifugation at 6000 rcf for 10 min and redispersed in a 0.05% sodium dodecyl sulphate (SDS) solution.Glycan-functionalization of GNRs
For a typical glycan-functionalization procedure, a solution of Man3-SH (1 mM, 50 μL) was added into 950 μL of GNR solution with vigorous stirring. The mixture was kept for 3 h before centrifugation. The glycan-functionalized GNRs were further washed twice and dispersed in water.Gel electrophoresis
The modification of the GNRs was evaluated by gel electrophoresis (Thermo Scientific EC 300XL) in 0.25% agarose gels using 1× 3-(N-morpholino)propanesulfonic acid (MOPS, pH 7.4) as the running buffer at 100 V for 10 min.Photothermal efficiency evaluation of the nanorods
The GNR dispersions of different absorbance values (0.5 and 1) were irradiated using a NIR laser (808 nm, 1 W cm−2). Water was used as a negative control. The temperature was monitored during heating. After the temperature reached equilibrium, the laser was switched off and the cooling process was monitored. Real-time thermal imaging of the samples was performed using a FLIR thermal camera. The photothermal conversion efficiency of the GNRs was calculated according to a method reported previously.35ConA recognition
ConA was prepared in a 10 mM phosphate buffer (pH 7.4) containing 2 mM Mn2+ and 2 mM Ca2+. ConA solutions of different concentrations were added into 1 mL of the Man3-GNR or Gal3-GNR solution. After incubation for 30 min, UV-vis extinction spectra of the solution were recorded. For a competitive assay, a mannose solution was first added into the Man3-GNR solution, and then the ConA solution was added to reach a final concentration of 20 nM. The extinction spectra were measured after incubation for 30 min.Isothermal calorimetric titration
Isothermal calorimetric titrations were carried out in a MICROCAL PEAQ-ITC instrument with a stirring speed of 750 rpm. Isothermal calorimetric titration was performed with 0.2 mM ConA concentration (molecular weight of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 g mol−1); methyl α-D-mannopyranoside (α-mmp) and Man3-SH were used at 2 mM concentrations in 10 mM acetate buffer of pH = 4.8 containing metal salts of 10 mM NaCl, 1 mM CaCl2 and 1 mM MnSO4. The multivalency effect of Man3-GNR in protein recognition was evaluated by titrating ligand mixtures of α-mmp and Man3-GNR with ConA.
600 g mol−1); methyl α-D-mannopyranoside (α-mmp) and Man3-SH were used at 2 mM concentrations in 10 mM acetate buffer of pH = 4.8 containing metal salts of 10 mM NaCl, 1 mM CaCl2 and 1 mM MnSO4. The multivalency effect of Man3-GNR in protein recognition was evaluated by titrating ligand mixtures of α-mmp and Man3-GNR with ConA.
Bacterial strains and culture conditions
Escherichia coli UTI89 is an E. coli strain isolated from the urine of a cystitis patient (PMID 16585510). Phase locked ON (SLC-490) and OFF (SLC-492) mutants of UTI89 were provided by the lab of Swaine Chen (unpublished). Strains SLC-490 and SLC-492 were prepared by reversing the sequence of the left inverted repeat of the fimS invertible element in the chromosome using recombineering with a dual positive–negative selection cassette (PMID 25800749 and 10829079). Escherichia coli (E. coli, UTI89) was cultured in Luria–Bertani (LB) medium for 24 h at 37 °C without shaking. After 24 h, 10 μL of the solution was subcultured statically in 1 mL of LB medium for another 4 h. The static growth is essential for the expression of FimH. The bacteria were collected by centrifugation at 1500 rcf for 10 min and washed with PBS.Dark-field imaging
A solution of Man3-GNR (0.5 nM, 50 μL) was mixed with either FimH ON or FimH OFF E. coli (108 CFU mL−1, 50 μL). After incubation for 1 h at 37 °C, the mixture was filtered using a centrifuge tube filter (pore size: 0.45 μm, 100 rcf, 10 min) to remove unbound GNRs. The bacteria were further washed twice using the same procedure and finally dispersed in 100 μL of water. 2 μL of the solution was dropped on a glass slide and covered by a coverslip. The sample was imaged using a dark-field microscope (Olympus IX71) with a 100× oil-immersed objective.Raman spectroscopy
E. coli dispersions (0.5 mL, 108 CFU mL−1) were mixed with 0.5 mL of Man3-GNR solutions with increasing concentrations (final concentrations ranging from 0 to 0.5 nM). After incubation of 1 h, unbound GNRs were removed using the same procedures as described above. The collected filtrates were redispersed in water and the Raman spectra were obtained under the excitation of a 785 nm laser (10 mW).Selective killing of E. coli UTI89 by photothermal effects of Au nanocomposites
E. coli dispersions (0.5 mL, 108 CFU mL−1) were mixed with 0.5 mL of Man3-GNR solutions with increasing concentrations (final concentrations ranging from 0 to 0.2 nM). After gently shaking for 1 h, unbound nanorods were removed using filtration as described above. The bacteria were then redispersed in 1 mL of PBS and exposed to a NIR laser (808 nm, 1 W cm−2) for 15 min. The bacteria were transferred to LB agar plates, and incubated at 37 °C for 24 h and the colony number for each plate was counted.Live/dead staining
Bacteria (108 CFU mL−1) were prepared in a similar way as stated above. The samples were then stained with the LIVE/DEAD BacLight Bacterial Viability kit for 15 min at room temperature and the bacterial viability was assessed by using an Olympus IX71 inverted microscope with a 20× objective.Transmission electron microscopy (TEM)
Man3-GNR (0.2 nM) was mixed with E. coli and incubated for 1 h. After the removal of free GNRs, the solution was exposed to a NIR laser (808 nm, 1 W cm−2). 5 μL of the diluted (100 times) bacteria culture (before or after laser treatment) was dropped and dried on a copper grid for TEM observation.Results and discussion
In this study, we used tri-thiomannoside as a sugar motif for bacterial protein targeting. These sugar clusters were synthesized by a thiol–ene click strategy;36 an ethylene glycol side chain with a terminal mercapto group was introduced via a conventional acid–amine coupling reaction using EDC/HOBT reagents and deacetylation was finally carried out to afford Man3-SH (Scheme S1, S6 and S7†). The structure of trimannoside cluster immobilized GNR (Man3-GNR) used in this study is presented in Fig. 1. The introduction of a polyethylene glycol spacer is helpful for increasing water solubility because of its ability to stabilize the assembled nanostructures in aqueous media.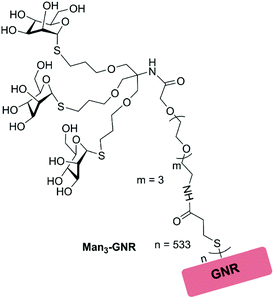 | ||
| Fig. 1 Structure of tri-thiomannoside modified GNRs (Man3-GNRs) used in this study, where n = 533 represents the number of glycoclusters covalently attached on each GNR. | ||
Synthesis and characterization of glycan-functionalized GNRs
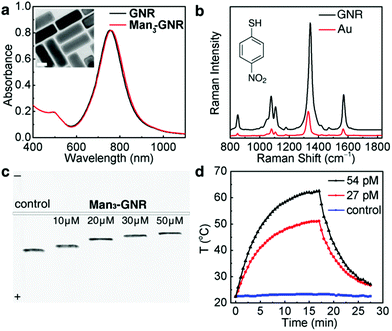
The GNRs used in this study are Raman-encoded, Ag coated Au nanorods. The Raman dye, 4-nitrothiophenol, was added during the silver deposition process (see the Experimental section); therefore, the GNRs were rendered Raman-active after the synthesis. The absorption spectrum of the obtained GNRs (Fig. 2a) was characterized using a longitudinal LSPR peak at 750 nm. After being glycan-functionalized with Man3-SH, the absorption spectrum experienced a slight red-shift to 753 nm because of the change in refractive index due to the ligands.37 There is no peak broadening after surface-modification, indicating that the obtained nanorods (denoted by Man3-GNR) are stable in water without aggregation. The average number of Man3-SH on each nanorod was determined to be 533 based on Ellman's assay (Fig. S4†). Fig. 2b shows a Raman spectrum of the Man3-GNR with characteristic Raman peaks of 4-nitrothiophenol (black line). In comparison, pure Au nanorods without silver coating show a much weaker Raman intensity, highlighting the advantage of using the core–shell GNRs.The original SDS stabilized GNRs are negatively charged with a zeta potential of −25 mV. After surface-modification, the zeta potential dropped to −19 mV. Electrophoresis results (Fig. 2c) show decreasing electrophoresis mobility with increasing concentrations of Man3-SH added into the GNR solution. As the introduction of Man3-SH does not cause aggregation of the GNRs, the reduced electrophoresis mobility should result from the decrease of surface charges. Meanwhile, the electrophoresis bands are very narrow, confirming the monodispersity and excellent stability of the glycan-functionalized GNRs. The GNRs were further tested for their photothermal properties using a NIR laser (808 nm, 1 W cm−2). Two concentrations of Man3-GNR, 27 pM and 54 pM, with absorbances of 0.5 and 1 respectively at 808 nm, were tested by a heating and cooling process (Fig. 2d). The solution was irradiated to reach equilibrium temperatures of 51 °C and 65 °C respectively for absorbances of 0.5 and 1. In comparison, pure water did not show an obvious temperature increase after laser irradiation. Based on the heating–cooling curves, the photothermal efficiency of Man3-GNR at 808 nm was estimated to be 49.3%.
Specificity and efficiency in recognition of Man3-GNR with ConA
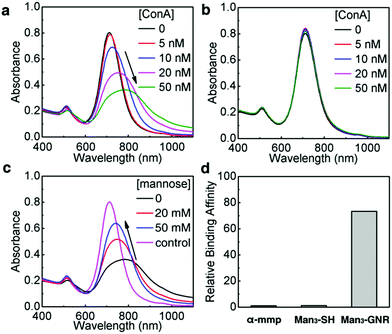
We first investigated the recognition between Man3-GNR and a model mannose-recognizing lectin, i.e. ConA.38 Absorption spectra of Man3-GNR solutions were measured after mixing with different concentrations of ConA (Fig. 3a). Binding of GNRs with ConA resulted in broadening and red-shift of the LSPR peak, a result of plasmon coupling between adjacent GNRs in the aggregates formed.39 With the increase of the ConA concentration, the nanorods experienced severer aggregation, indicated by the red-shift and broadening of the LSPR peak.40,41 To prove that the spectral change was indeed due to recognition between ConA and mannose, galactose-cluster functionalized GNRs (Gal3-GNR) were also synthesized as a control. Within the same ConA concentration range, there were no obvious changes in the absorption spectra after introducing ConA (Fig. 3b). Furthermore, mannose solutions with high concentrations (20 mM and 50 mM) were mixed with Man3-GNR for competitive binding to ConA. With the increase of the mannose concentration, lesser extents of red-shifts of the LSPR peak were observed, suggesting less aggregation of the GNRs (Fig. 3c). Dynamic light scattering (DLS) measurements were also performed to determine the aggregation states of the nanorods (Fig. S9†), which agreed well with the results obtained from the absorption spectra. These results substantiate the high specificity of Man3-GNR binding to ConA.ITC titration experiments were performed between ConA and Man3-SH to examine the binding affinity of the trimeric mannosides with ConA using methyl α-D-mannopyranoside (α-mmp) as the control; α-mmp is well-known to bind to ConA.42 The ITC titrations give an association constant of 8.79 × 103 M−1 (per mannose base) for ConA and Man3-SH, and a K1 of 7.50 × 103 M−1 with α-mmp. Notably, the addition of a low concentration of Man3-GNR (30 nM) into α-mmp (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41.6 Man3-GNR
41.6 Man3-GNR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α-mmp based on the mannose molar concentration) with ConA led to considerably increased affinity (Fig. S7†). The obtained binding constant for the two ligand mixture gave rise to the binding constant of Man3-GNR recognition with ConA of K2 = 551 × 103 M−1. This experiment confirmed the excellent multivalency effect of Man3-GNR in binding to ConA and gave the relative binding affinity of α-mmp and Man3-GNR = 1
α-mmp based on the mannose molar concentration) with ConA led to considerably increased affinity (Fig. S7†). The obtained binding constant for the two ligand mixture gave rise to the binding constant of Man3-GNR recognition with ConA of K2 = 551 × 103 M−1. This experiment confirmed the excellent multivalency effect of Man3-GNR in binding to ConA and gave the relative binding affinity of α-mmp and Man3-GNR = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 72 (Fig. 3d, mannose molar concentration bases). Furthermore, the ITC of the ligand–ligand titration between α-mmp and Man3-GNR, and Gal3-GNR with ConA confirms no binding.
72 (Fig. 3d, mannose molar concentration bases). Furthermore, the ITC of the ligand–ligand titration between α-mmp and Man3-GNR, and Gal3-GNR with ConA confirms no binding.
Confirmation of FimH-specific binding of Man3-NH-rhodamine
We next examine the selective recognition of this glycan modified GNR to bacteria (E. coli UTI89). Among the different species of pathogenic bacteria, E. coli are among the major species43,44 that initiate the damage and pathological changes in the gastrointestinal and urinary systems.45,46 UPEC is the most common pathogenic agent to cause urinary tract infections.47 The FimH expression of most of the E. coli bacteria mediates the mannose sensitive binding to target host cells.48 Fimbriae of E. coli have been shown to facilitate bacterial colonization and invasion of human bladder cells.49 A wide range of studies have been conducted on uropathogenic strains of E. coli produce type 1 fimbriae. These suggest that FimH can be a potential vaccine candidate for the prevention of infections in the urinary tract.50E. coli UTI89 bacteria were genetically engineered to obtain the FimH ON and FimH OFF mutants.51 The UTI89 FimH mutants were challenged with the glycocluster. We carried out competitive binding assay experiments by quantifying the fluorescence intensities of UTI89 bacteria treated with rhodamine dye labelled tri-thiomannoside cluster Man3-NH-rhodamine (Scheme S8†) after they were pretreated with different concentrations of α-mmp (Fig. S3†). As the mmp concentration increases, the mannose receptors would be blocked by these α-mmp molecules so that the glycocluster does not bind to the bacteria and the measured fluorescence intensities decrease as the α-mmp concentration increases. In contrast to fimbriated E. coli UTI89 strains, the non-fimbriated E. coli UTI89 upon treatment with Man3-NH-rhodamine does not show significant fluorescence intensity changes at different α-mmp concentrations. These experiments support the specific binding of the trimeric form of the mannoside cluster with FimH receptors of UTI89 bacterial strains.
Binding of Man3-GNR to fimbriated E. coli UTI89
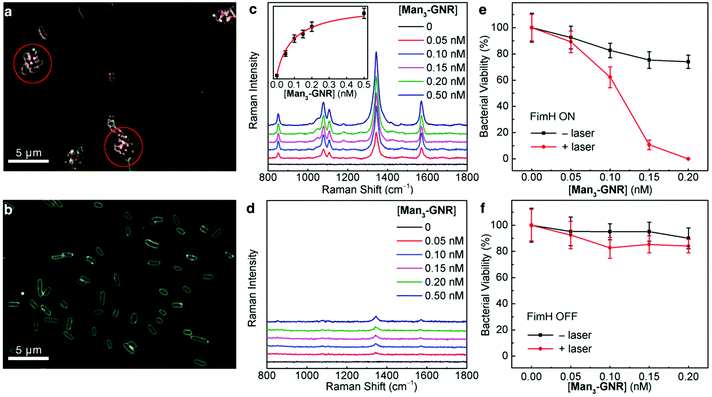
We investigated the binding of Man3-GNR to UTI89 bacteria using dark field microscopy. The nanorods appear as red dots because of their light scattering in the visible spectral range, and the bacteria were characterized by a white contour. As evidenced by the dark-field images shown in Fig. 4a, the FimH-ON bacteria were clearly clustered by Man3-GNR, while no clustering was caused for the FimH OFF bacteria (Fig. 4b). Furthermore, Gal3-GNR nanoconjugates (Schemes S9 and S10†) did not bind to the FimH-ON bacteria (Fig. S8b†), confirming the specific binding of the mannoside cluster. A competitive assay using mannose and Man3-GNR further proves that the binding site of the mannoside-cluster is the FimH protein (Fig. S8d†).SERS is a sensitive and high throughput method for quantification analysis.52,53 Different concentrations of the SERS-encoded Man3-GNR were incubated with either FimH-ON or FimH-OFF E. coli. Unbound nanorods were removed by centrifugation with a filter pore size of 0.45 μm. FimH-ON UTI89 showed a concentration dependent Raman signal whereas the FimH-OFF bacterial strain showed very weak Raman signals (Fig. 4c and d). The dependence of the SERS intensity at 1340 cm−1 on the concentration of Man3-GNR is plotted as shown in the inset of Fig. 4c. By fitting the curve according to the Hill equation,54 the binding affinity of Man3-GNR with FimH-ON UTI89 bacteria was found to be 1.08 × 1010 M−1 based on the GNR concentration (or 6.72 × 106 M−1 based on the mannose concentration). It is interesting to note that this value is two orders of magnitude larger than the binding affinity of FimH with natural high-mannose N-glycans (estimated to be 5 × 104 M−1).55 We also incubated different concentrations of FimH-ON bacteria with 0.25 nM Man3-GNR and measured the Raman spectra (Fig. S10†). E. coli at a concentration of 103 CFU mL−1 could be detected.
Selective killing of fimbriated E. coli UTI89
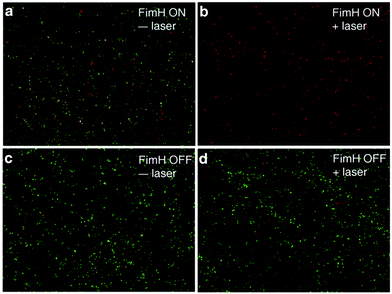
After confirming that Man3-GNR can bind to fimbriated E. coli with high affinity, we tested if selective bacterial killing is possible. Both FimH-ON and FimH-OFF bacteria were incubated with Man3-GNR for 1 h. After removing the unbound nanorods, the solution was exposed to an 808 nm laser. After irradiation for 15 min, the number of live cells was determined by plating the bacteria on LB plates, which were then incubated at 37 °C for 24 h. As shown in Fig. 4e, nearly all FimH-ON bacteria (99.98%) were dead when 0.2 nM Man3-GNR was used. Without laser irradiation, the FimH-ON E. coli were also inhibited by 26%, most likely due to the intrinsic antibacterial activity of Ag in the GNRs.56 In contrast, the viability of FimH-OFF E. coli was much higher after the same treatment (Fig. 4f). The difference in the killing efficiency of the two bacterial strains highlights the significance of targeted binding in specific antibacterial activity.Live/dead analysis using SYTO 9 and propidium iodide (PI) was performed to visualize the extent of killing of bacteria with the treatment of Man3-GNR (0.2 nM) with or without laser irradiation for both FimH-ON and FimH-OFF E. coli strains (Fig. 5). Dead cells and live cells show red and green fluorescence, respectively. The FimH-ON bacteria were almost completely stained red after laser irradiation for 15 min (Fig. 5b). Live bacteria along with some amount of killed bacteria were observed in non-treatment control and FimH-OFF E. coli strain. These results are consistent with the bacterial viability data obtained by CFU counting (Fig. 4e and f). Meanwhile, the staining assay confirms that there is membrane damage after the treatment because the cell-impermeant PI dye stained the cells.
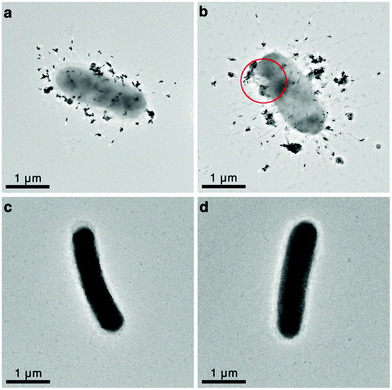
Finally, we examined the morphology changes of the UTI89 bacteria after Man3-GNR binding and laser treatment using transmission electron microscopy (TEM). Single bacteria instead of clustered bacteria were imaged for clarity. The TEM images clearly show that the FimH-ON bacteria are fimbriated (Fig. 6a and b) while the FimH OFF bacteria are not (Fig. 6c and d), and Man3-GNR specifically bind to fimbriated bacteria. Fig. 6a and b further confirm that the nanorods bind to the fimbriae rather than the cell walls of the bacteria. The number of nanorods per bacterium is over 100, which is expected to release intense heat locally to the bacteria when irradiated by light. It is evidenced in Fig. 6b that rupture of bacterial membrane occurs after laser treatment. No clear changes were observed for the FimH-OFF bacteria after laser irradiation, which is expected because bacteria without GNR attachment do not absorb NIR light.
Conclusions
We have demonstrated the specific multivalent binding of tri-thiomannoside cluster modified GNRs to fimbriated E. coli as a well-defined model system. The glycan-functionalized GNRs can be utilized for targeted detection and killing of E. coli. The glycan cluster modified GNRs showed efficient recognition with both ConA and FimH of E. coli UTI89. Dark-field imaging and SERS detection clearly differentiated FimH-ON and FimH-OFF bacterial strains. Furthermore, by employing the photothermal properties of the plasmonic nanoconjugates, specific killing of fimbriated E. coli bacteria could be achieved. Our design of multivalent glyco-nanoparticles is of potential for in vivo applications for UTI and recurrent UTI.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded and supported by Ministry of Education-Singapore (MOE2013-T3-1-002 and RG49/16), and a Singapore MOH Industry Alignment Fund (NMRC/MOHIAFCAT2/003/2014). The E. coli strains UTI89, SLC-490, and SLC-492 were kindly provided by Lu Ting Liow and Swaine Chen.Notes and references
- A. Varki, Essentials of glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 2009 Search PubMed.
- A. K. Adak, H.-J. Lin and C.-C. Lin, Org. Biomol. Chem., 2014, 12, 5563–5573 CAS.
- J. J. Lundquist and E. J. Toone, Chem. Rev., 2002, 102, 555–578 CrossRef CAS PubMed.
- M. Marradi, F. Chiodo, I. García and S. Penadés, Chem. Soc. Rev., 2013, 42, 4728–4745 RSC.
- Z. Qi, P. Bharate, C. H. Lai, B. Ziem, C. Böttcher, A. Schulz, F. Beckert, B. Hatting, R. Mülhaupt, P. H. Seeverger and R. Haag, Nano Lett., 2015, 15, 6051–6057 CrossRef CAS PubMed.
- X. Li and G. Chen, Polym. Chem., 2015, 6, 1417–1430 RSC.
- M. Gómez-García, J. M. Benito, D. Rodríguez-Lucena, J.-X. Yu, K. Chmurski, C. O. Mellet, R. G. Gallego, A. Maestre, J. Defaye and J. M. G. Fernández, J. Am. Chem. Soc., 2005, 127, 7970–7971 CrossRef PubMed.
- C.-C. Lin, Y.-C. Yeh, C.-Y. Yang, C.-L. Chen, G.-F. Chen, C.-C. Chen and Y.-C. Wu, J. Am. Chem. Soc., 2002, 124, 3508–3509 CrossRef CAS PubMed.
- F. Manea, C. Bindoli, S. Fallarini, G. Lombardi, L. Polito, L. Lay, R. Bonomi, F. Mancin and P. Scrimin, Adv. Mater., 2008, 20, 4348–4352 CrossRef CAS.
- K. El-Boubbou, C. Gruden and X. Huang, J. Am. Chem. Soc., 2007, 129, 13392–13393 CrossRef CAS PubMed.
- L. Lartigue, C. Innocenti, T. Kalaivani, A. Awwad, M. Sanchez Duque, Y. Guari, J. Larionova, C. Guérin, J.-L. Georges Montero, V. Barragan-Montero, P. Arosio, A. Lascialfari, D. Gatteschi and C. Sangregorio, J. Am. Chem. Soc., 2011, 133, 10459–10472 CrossRef CAS PubMed.
- C.-H. Lai, C.-Y. Lin, H.-T. Wu, H.-S. Chan, Y.-J. Chuang, C.-T. Chen and C.-C. Lin, Adv. Funct. Mater., 2010, 20, 3948–3958 CrossRef CAS.
- A. Cambi, D. S. Lidke, D. J. Arndt-Jovin, C. G. Figdor and T. M. Jovin, Nano Lett., 2007, 7, 970–977 CrossRef CAS PubMed.
- I. García, A. Sánchez-Iglesias, M. Henriksen-Lacey, M. Grzelczak, S. Penadés and L. M. Liz-Marzán, J. Am. Chem. Soc., 2015, 137, 3686–3692 CrossRef PubMed.
- M. Khanal, F. Larsonneur, V. Raks, A. Barras, J.-S. Baumann, F. A. Martin, R. Boukherroub, J.-M. Ghigo, C. O. Mellet, V. Zaitsev, J. M. G. Fernandez, C. Beloin, A. Siriwardena and S. Szunerits, Nanoscale, 2015, 7, 2325–2335 RSC.
- B. M. Illescas, J. Rojo, R. Delgado and N. Martín, J. Am. Chem. Soc., 2017, 139, 6018–6025 CrossRef CAS PubMed.
- A. Barras, F. A. Martin, O. Bande, J. S. Baumann, J.-M. Ghigo, R. Boukherroub, C. Beloin, A. Siriwardena and S. Szunerits, Nanoscale, 2013, 5, 2307–2316 RSC.
- J. Wei, L. Zheng, X. Lv, Y. Bi, W. Chen, W. Zhang, Y. Shi, L. Zhao, X. Sun, F. Wang, S. Cheng, J. Yan, W. Liu, X. Jiang, G. F. Gao and X. Li, ACS Nano, 2014, 8, 4600–4607 CrossRef CAS PubMed.
- M. Reynolds, M. Marradi, A. Imberty, S. Penadés and S. Perez, Chem. – Eur. J., 2012, 18, 4264–4273 CrossRef CAS PubMed.
- X. Wang, E. Matei, A. M. Gronenborn, O. Ramström and M. Yan, Anal. Chem., 2012, 84, 4248–4252 CrossRef CAS PubMed.
- C.-C. Lin, Y.-C. Yeh, C.-Y. Yang, G.-F. Chen, Y.-C. Chen, Y.-C. Wu and C.-C. Chen, Chem. Commun., 2003, 2920–2921 RSC.
- H. Chen, L. Shao, Q. Li and J. Wang, Chem. Soc. Rev., 2013, 42, 2679–2724 RSC.
- X. Xu, Y. Chen, H. Wei, B. Xia, F. Liu and N. Li, Anal. Chem., 2012, 84, 9721–9728 CrossRef CAS PubMed.
- L. A. Lane, X. Qian and S. Nie, Chem. Rev., 2015, 115, 10489–10529 CrossRef CAS PubMed.
- H. Zhang, X. Ma, Y. Liu, N. Duan, S. Wu, Z. Wang and B. Xu, Biosens. Bioelectron., 2015, 74, 872–877 CrossRef CAS PubMed.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CrossRef CAS PubMed.
- J. Langer, I. García and L. M. Liz-Marzán, Faraday Discuss., 2017, 205, 363–375 RSC.
- T. Ming, H. Chen, R. Jiang, Q. Li and J. Wang, J. Phys. Chem. Lett., 2012, 3, 191–202 CrossRef CAS.
- Y. Zhu, M. Ramasamy and D. K. Yi, ACS Appl. Mater. Interfaces, 2014, 6, 15078–15085 CAS.
- J. D. Schilling, M. A. Mulvey and S. J. Hultgren, J. Infect. Dis., 2001, 183, S36–S40 CrossRef PubMed.
- M. G. Blango and M. A. Mulvey, Antimicrob. Agents Chemother., 2010, 54, 1855–1863 CrossRef CAS PubMed.
- B. Foxman, Infect. Dis. Clin. North Am., 2014, 28, 1–13 CrossRef PubMed.
- A. L. Flores-Mireles, J. N. Walker, M. Caparon and S. J. Hultgren, Nat. Rev. Microbiol., 2015, 13, 269–284 CrossRef CAS PubMed.
- C. N. Spaulding, R. D. Klein, S. Ruer, A. L. Kau, H. L. Schreiber, Z. T. Cusumano, K. W. Dodson, J. S. Pinkner, D. H. Fremont, J. W. Janetka, H. Remaut, J. I. Gordon and S. J. Hultgren, Nature, 2017, 546, 528–532 CAS.
- H. Chen, L. Shao, T. Ming, Z. Sun, C. Zhao, B. Yang and J. Wang, Small, 2010, 6, 2272–2280 CrossRef CAS PubMed.
- A. B. Lowe, Polym. Chem., 2014, 5, 4820–4870 RSC.
- K. M. Mayer and J. H. Hafner, Chem. Rev., 2011, 111, 3828–3857 CrossRef CAS PubMed.
- J. H. Naismith and R. A. Field, J. Biol. Chem., 1996, 271, 972–976 CrossRef CAS PubMed.
- P. K. Jain, S. Eustis and M. A. El-Sayed, J. Phys. Chem. B, 2006, 110, 18243–18253 CrossRef CAS PubMed.
- S.-J. Richards, L. Otten and M. I. Gibson, J. Mater. Chem. B, 2016, 4, 3046–3053 RSC.
- X.-L. Hu, H.-Y. Jin, X.-P. He, T. D. James, G.-R. Chen and Y.-T. Long, ACS Appl. Mater. Interfaces, 2015, 7, 1874–1878 CAS.
- S. L. Mangold and M. J. Cloninger, Org. Biomol. Chem., 2006, 4, 2458–2465 CAS.
- A. Ronald, Dis.-Mon., 2003, 49(2), 71–82 CrossRef PubMed.
- J. B. Kaper, J. P. Nataro and H. L. T. Mobley, Nat. Rev. Microbiol., 2004, 2, 123–140 CrossRef CAS PubMed.
- H. M. Zowawi, P. N. A. Harris, M. J. Roberts, P. A. Tambyah, M. A. Schembri, M. D. Pezzani, D. A. Williamson and D. L. Paterson, Nat. Rev. Urol., 2015, 12, 570–584 CrossRef CAS PubMed.
- L. J. Grauke, I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams and C. J. Hovde, Appl. Environ. Microbiol., 2002, 68, 2269–2277 CrossRef CAS PubMed.
- K. J. Wright, P. C. Seed and S. J. Hultgren, Cell. Microbiol., 2007, 9, 2230–2241 CrossRef CAS PubMed.
- S. G. Stahlhut, V. Tchesnokova, C. Struve, S. J. Weissman, S. Chattopadhyay, O. Yakovenko, P. Aprikian, E. V. Sokurenko and K. A. Krogfelt, J. Bacteriol., 2009, 191, 6592–6601 CrossRef CAS PubMed.
- A. L. Flores-Mireles, J. N. Walker, M. Caparon and S. J. Hultgren, Nat. Rev. Microbiol., 2015, 13, 269–284 CrossRef CAS PubMed.
- M. A. Schembri, H. Hasman and P. Klemm, FEMS Microbiol. Lett., 2000, 188, 147–151 CrossRef CAS PubMed.
- S. L. Chen, C. S. Hung, J. S. Pinkner, J. N. Walker, C. K. Cusumano, Z. Li, J. Bouckaert, J. I. Gordon and S. J. Hultgren, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 22439–22444 CrossRef CAS PubMed.
- H.-Y. Lin, C.-H. Huang, W.-H. Hsieh, L.-H. Liu, Y.-C. Lin, C.-C. Chu, S.-T. Wang, I.-T. Kuo, L.-K. Chau and C.-Y. Yang, Small, 2014, 10, 4700–4710 CrossRef CAS PubMed.
- D. Craig, S. McAughtrie, J. Simpson, C. McCraw, K. Faulds and D. Graham, Anal. Chem., 2014, 86, 4775–4782 CrossRef CAS PubMed.
- C. Röcker, M. Pötzl, F. Zhang, W. J. Parak and G. U. Nienhaus, Nat. Nanotechnol., 2009, 4, 577–580 CrossRef PubMed.
- D. Kisiela, A. Laskowska, A. Sapeta, M. Kuczkowski, A. Wieliczko and M. Ugorski, Microbiology, 2006, 152, 1337–1346 CrossRef CAS PubMed.
- X. Ding, P. Yuan, N. Gao, H. Zhu, Y. Y. Yang and Q.-H. Xu, Nanomedicine, 2017, 13, 297–305 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8bm00139a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2018 |