SERS-fluorescence bimodal nanoprobes for in vitro imaging of the fatty acid responsive receptor GPR120†
Lifu
Xiao
 ac,
Abdul K.
Parchur
ac,
Abdul K.
Parchur
 a,
Timothy A.
Gilbertson
b and
Anhong
Zhou
*a
a,
Timothy A.
Gilbertson
b and
Anhong
Zhou
*a
aDepartment of Biological Engineering, Utah State University, 4105 Old Main Hill, Logan, Utah 84322-4105, USA. E-mail: Anhong.Zhou@usu.edu; Fax: +1-435-797-1248; Tel: +1-435-797-2863
bDepartment of Biology, Utah State University, Logan, Utah 84322-5305, USA
cDepartment of Chemistry and Biochemistry, University of Notre Dame, Notre Dame, Indiana 46556, USA
First published on 18th October 2017
Abstract
G-protein-coupled receptor 120 (GPR120), as a member of the rhodopsin family of G-protein-coupled receptors, has been shown to function as a sensor for dietary fat in the gustatory and digestive systems. Its specific role in the chemoreception of fatty acids, which is thought to be crucial in understanding the mechanism surrounding the control of fat intake and, accordingly, in the treatment of obesity, remains unclear. Here we report a novel surface-enhanced Raman spectroscopy (SERS)-fluorescence bimodal microscopic technique for detection and imaging of GPR120 in single living cells. CaMoO4:Eu3+@AuNR hybrid nanoparticles are synthesized and characterized as imaging probes. The biocompatibility and imaging capability of the probes are investigated using a model HEK293 cell line with an inducible GPR120 gene transfection. The cellular distribution of GPR120 is visualized by single-cell SERS and fluorescence imaging. A dose-dependent GPR120 response to linoleic acid treatment is revealed by SERS.
Introduction
Dietary lipids (e.g. fatty acids, triglycerides) contribute up to 40% of daily caloric intake in the Western diet, which is thought to contribute greatly to the prevalence of obesity and associated diseases.1–3 Understanding the mechanisms underlying the perception of dietary lipids thus is important to help control fat preference and develop treatments for dietary-induced obesity. G protein-coupled receptors (GPCRs) have been shown to play important roles in cellular signaling pathways that affect human sense of taste (e.g. sweet, bitter, umami).4,5 GPR120 and GPR40, members of the GPCR family, have recently been reported to mediate response to long chain fatty acids (LCFAs).6 Both GPR120 and GPR40 knock-out mice showed a diminished preference for linoleic acid (LA) and oleic acid, and diminished taste nerve responses. In addition, studies have shown that GPR120 and CD36, LCFA receptors identified in rodent taste bud cells, mediate differential Ca2+ response to fatty acids, and are differentially regulated by dietary lipids.7,8 Though several lipid receptor candidates (e.g. GPR120, GPR40, and CD36) have been found in the tongue papillae, the mechanism how these receptors act in the transduction of LCFAs remains unresolved and merits further extensive studies.3 It will be beneficial to develop an imaging technique that can visualize how these receptors respond when binding LCFAs in single cells, in order to understand their specific roles in fat chemoreception.Raman spectroscopy, by measuring the energy of molecular vibrational modes, provides an intrinsic contrast mechanism for identifying the molecular composition of cells.9–12 However, since Raman scattering is an inefficient process by nature, detecting specific membrane receptors in cells using spontaneous Raman spectroscopy is challenging. Plasmonic nanoparticles have been applied to enhance Raman signals from molecules in close proximity—known as surface-enhanced Raman spectroscopy (SERS).13 SERS is able to achieve 10–14 orders of magnitude enhancement of spontaneous Raman signals,14 which allows detection of biomolecules with ultrahigh sensitivity.15,16 To generate intense and stable SERS signals for biomedical imaging, SERS nanoprobes composed of a noble metal nanoparticle, a Raman reporter molecule, and a surface stabilizer are usually applied. Due to the advantages of high stability and biocompatibility, especially the great capability of multiplex detection, SERS nanoprobes have been applied in a variety of biomedical applications such as molecular detection,17,18 single living cell imaging,19–21 and in vivo biosensing.22,23 In addition, SERS microscopy, by generating a pseudo-color map based on relative intensities of selected Raman bands, is able to map the distribution of biomolecules such as lipids and proteins,24,25 cell surface receptors26–28 and even bacterial species29 in single living cells. However, SERS is a low-throughput (raster scan) imaging technique and it requires a relatively long acquisition time (up to seconds) due to the relatively weak signal by Raman scattering (even after surface enhancement). This limits the application of SERS in biological analyses, especially for high-throughput cell sorting and imaging.30
To overcome the limitation and expand the functionality, multimodal imaging probes have been developed to integrate SERS with other imaging modalities, such as magnetic resonance imaging (MRI),31 X-ray computed tomography (CT)32,33 and fluorescence microscopy.34–36 Among these modalities, fluorescence, due to its high-throughput imaging ability, is considered as a promising complementary technique to SERS. Fluorescence functions for quick recognition of the analytes in complex biosystems, while SERS is used to accurately detect multiplex targets in single cells. Recently, Choo's lab reported a SERS-fluorescence bimodal imaging technique to investigate multiple cancer markers co-localized in single breast cancer cells.37
Here we developed a SERS-fluorescence bimodal imaging technique for detection and imaging of GPR120 in single living cells. Europium-doped calcium molybdate nanoparticles (CaMoO4:Eu3+) showed intense red fluorescence emission under UV light excitation.38 These fluorescence-active CaMoO4:Eu3+ nanoparticles were conjugated with SERS-active gold nanorods (AuNRs) encapsulated with the Raman reporter molecule 4-mercaptobenzoic acid (MBA) to realize SERS-fluorescence dual functions. The composite nanoparticle was conjugated with an antibody for specific targeting of GPR120 expressed in living cells. By using this bimodal nanoprobe, we successfully realize specific SERS-fluorescence imaging of GPR120 at the single cell level. In addition, for the first time, we observed a linear dose-dependence of GPR120 response following linoleic acid (LA) treatments.
Materials and methods
Materials
4-Mercaptobenzoic acid (MBA), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), doxycycline hydrochloride (DOX), LA, and europium(III) nitrate hydrate (Eu(NO3)3·xH2O) were purchased from Sigma-Aldrich (St Louis, MO). Calcium nitrate tetrahydrate, (Ca(NO3)2·4H2O), ammonium molybdate (H8MoN2O4), oleic acid (OA), and 1-octadecene were purchased from Alfa Aesar (Ward Hill, MA). Blasticidin S HCl, hygromycin B, phosphate-buffered saline (PBS), 0.5% trypsin–EDTA, and LIVE/DEAD Viability/Cytotoxicity Assay Kit were purchased from Life Technologies (Carlsbad, CA). Polyethylene glycol (PEG) products, thiol PEG acid (HS–PEG–COOH, MW 5000) and methoxyl PEG thiol (mPEG–SH, MW 5000), were purchased from Nanocs Inc. (USA). The polyclonal GPR120 antibody was purchased from Santa Cruz Biotechnology Inc. (sc-99105). Ultrapure water (18 MΩ cm−1) was used in this work.Characterization techniques
Transmission electron microscopy (TEM) images and energy-dispersive X-ray (EDX) spectra were acquired using an FEI Titan 80–300 kV (S) TEM equipped with a spherical aberration (Cs) image corrector (300 kV). For the TEM measurements, the powder samples were ground and dispersed in methanol. Few drops of dispersed particles were placed on a carbon coated-copper grid and allowed to dry at room temperature. Absorption spectra were acquired using a Multiskan Spectrum spectrophotometer (Thermo Scientific). FT-IR spectra were acquired using a Varian 660-IR FT-IR spectrometer (Agilent Technologies). Luminescence spectra were acquired using a FluoroMax-3 fluorometer (Horiba Scientific). Raman spectra were measured using a Renishaw inVia Raman spectrometer, equipped with a 785 nm near-IR laser, which was focused through a 63× water immersion lens (NA = 0.90, Leica Microsystems).Synthesis of CaMoO4:Eu3+@AuNR hybrid nanoparticles
Hybrid nanoparticles were synthesized according to a previously published protocol.39 For a typical synthesis process, (i) europium-doped calcium molybdate (CaMoO4:Eu3+) nanoparticles were prepared via a simple thermolysis process at ∼309 °C. The preparation procedure can be briefly described as follows: 21 mg of Eu(NO3)3·5H2O, 50 mg of NaOH, and 1.0 g of Ca(NO3)2·4H2O were dissolved in 2 mL of distilled water. The mixture was treated with 2 mL of OA and 18 mL ODE and heated in a round-bottom flask at 80 °C for 1 h under continuous stirring. In another beaker, 0.423 g of H8MoN2O4 was dissolved in 3 mL of DI water, and 0.1 g of NaOH, 2 mL of OA, and 18 mL of ODE were added and the solution was stirred at 80 °C for 1 h. The two solutions were mixed under continuous stirring and heated at 80 °C for 30 min, and then the reaction mixture was refluxed at 309 °C for 1 h. The resulting precipitate was collected by centrifugation at ∼2500 g after washing with ethanol. The obtained precipitate was cooled at room temperature for 2 days. (ii) About 20 mg of the CaMoO4:Eu3+ NPs was dispersed in 5 mL of HCl (0.1 M) and sonicated for ∼1 h. To this ∼2 mL of diethyl ether was added and sonication was continued for further ∼0.5 h. The resulting solution was centrifuged at ∼6000 rpm for ∼15 min. The obtained precipitate was washed three times with ethanol and redispersed in ∼5 mL of PBS solution. To this, 20 mg of HS–PEG–COOH was added and sonicated for ∼1 h. The PEGylated capped NPs were collected by centrifugation and washed with PBS solution three times to remove the excess of HS–PEG–COOH present in the sample. The final precipitate obtained was redispersed in a PBS solution. (iii) For the synthesis of CaMoO4:Eu3+@AuNR hybrid nanoparticles, GNRs with 10 nm in diameter and 35 nm in length were purchased from Nanopartz, Loveland, CO, USA. First, 4 mL of the GNRs was centrifuged at ∼13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min and redispersed in PBS. Centrifugation was repeated three times to reduce the excess of cetyl trimethylammonium bromide (CTAB) present on the surface of the GNRs. GNRs (4 mL) dispersed in PBS were added to ∼1 mL of the PEGylated CaMoO4:Eu3+ NPs under continuous stirring. The resulting solution was centrifuged, and the HNPs precipitated were collected. These particles were washed with a PBS solution and redispersed in PBS.
000 rpm for 30 min and redispersed in PBS. Centrifugation was repeated three times to reduce the excess of cetyl trimethylammonium bromide (CTAB) present on the surface of the GNRs. GNRs (4 mL) dispersed in PBS were added to ∼1 mL of the PEGylated CaMoO4:Eu3+ NPs under continuous stirring. The resulting solution was centrifuged, and the HNPs precipitated were collected. These particles were washed with a PBS solution and redispersed in PBS.
Functionalization of the SERS-fluorescence bimodal probe
1 mL of as-prepared CaMoO4:Eu3+@AuNR composite nanoparticle solution was mixed with MBA solution (1 mM, 10 μL) and reacted for 30 min. Solutions of HS–PEG–COOH (1 mg mL−1, 10 μL) and mPEG–SH (1 mg mL−1, 40 μL) were sequentially added to the nanoparticle solution and incubated for 2 h. The resultant solution was then centrifuged (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 15 min) to remove excess PEG and MBA. Particles were resuspended in water. Freshly prepared EDC (10 mM, 10 μL) and NHS (25 mM, 10 μL) solutions were mixed with the nanoparticle solution and allowed to react for 30 min. The resulting solution was centrifuged and particles were resuspended in PBS. Finally, the anti-GPR120 antibody (0.2 mg mL−1, 10 μL) was added to the nanoparticle solution and incubated for 1 h. Excess antibody was removed by centrifugation. Nanoparticles were resuspended in PBS. The functionalized nanoprobe (CaMoO4:Eu3+@AuNR–MBA–Ab) was stable in solution for several days at 4 °C.
000 g, 15 min) to remove excess PEG and MBA. Particles were resuspended in water. Freshly prepared EDC (10 mM, 10 μL) and NHS (25 mM, 10 μL) solutions were mixed with the nanoparticle solution and allowed to react for 30 min. The resulting solution was centrifuged and particles were resuspended in PBS. Finally, the anti-GPR120 antibody (0.2 mg mL−1, 10 μL) was added to the nanoparticle solution and incubated for 1 h. Excess antibody was removed by centrifugation. Nanoparticles were resuspended in PBS. The functionalized nanoprobe (CaMoO4:Eu3+@AuNR–MBA–Ab) was stable in solution for several days at 4 °C.
Cell culture
Human embryonic kidney cells (HEK293 and HEK293–GPR120) were kindly provided by International Flavors & Fragrances Inc. (IFF). HEK293 cell lines transfected with an inducible GPR120 gene (HEK293–GPR120) and a constitutive CD36 gene (HEK293-CD36) were used in this context. Cells were grown in DMEM-GlutaMAX media (Life Technologies, 10569-010) supplemented with 10% Tet-free fetal bovine serum (Fisher, NC0290780). Cells were cultured in a humidified atmosphere at 37 °C with 5% CO2, and were passaged at 80–90% confluence. Blasticidin S HCl (10 μg mL−1) and hygromycin B (100 μg mL−1) were added to cell culture medium specifically for maintenance of inducible GPR120 gene. To express GPR120, HEK293–GPR120 cells were induced with DOX at 0.5 μg mL−1 for 48 h.Cell staining and viability test
The cell viability was analyzed using a LIVE/DEAD Viability/Cytotoxicity Assay Kit (Invitrogen) according to the manufacturer's instruction. Briefly, (1) cells were cultured in poly-D-lysine coated glass-bottom dishes (MatTek Corp., USA) for 24 hours; (2) cells were then washed with PBS twice; (3) 2 mL of a mixed solution of 2 μM calcein AM and 4 μM ethidium homodimer-1 (EthD-1) (both from Invitrogen) was added directly to cells, and the cells were incubated for 30 min at room temperature; (5) cells were imaged using a fluorescence microscope to analyze the relative proportion of live/dead cells. A 10× objective (0.25 NA, Olympus) was used to observe the fluorescence.Calcein AM was well retained within live cells producing green fluorescence; however, EthD-1 entered cells with damaged membranes and bonded to nucleic acids, producing a red fluorescence in dead or membrane-damaged cells. Therefore, the live/dead cells were differentiated visually.
Cellular fluorescence and SERS imaging
For fluorescence and SERS imaging experiments, cells were incubated with the functionalized nanoprobe (CaMoO4:Eu3+@AuNR–MBA–Ab) for 24 h, and rinsed with PBS 3 times before imaging experiments. For LA treatments, cells were first treated with LA for 5 min at different concentrations, and then incubated with nanoprobes for 24 h.Cellular fluorescence imaging was performed on an Olympus IX71 inverted microscope with an external 285 nm UV lamp. A 10× objective (0.75 NA, Olympus) was used to observe the fluorescence. Images were acquired and processed using DPController software (Olympus Corporation) to maintain identical light exposure for three different cell conditions.
For SERS measurements, cells were seeded on a cleaned magnesium fluoride (MgF2) optical window (United Crystals Co.) to minimize the background signal from the substrate. A 63× water immersion lens (NA = 0.90, Leica Microsystems) was used. Raman spectra between 600 and 1800 cm−1 were recorded under 3 mW laser exposure for 10 s acquisition time per spectrum. Hyperspectral Raman mapping generation was performed using Renishaw WiRE 3.3 software. Single peak intensity at 1078 cm−1 was used to generate pseudo-color SERS maps. For quantitative SERS measurements, 25 spectra from 25 cells (1 spectrum per cell) were collected on cell samples treated with LA at concentrations 0, 5, 20, 30, and 60 μM, respectively. Spectral smoothing and autofluorescence background subtraction were performed using an automated algorithm program kindly provided by BC Cancer Research Center.40 The processed spectra were exported to OriginPro 9 software for plotting. Data were reported as mean ± SE (standard error of mean). Statistical difference was analyzed by one-way analysis of variance (ANOVA).
Results
Nanoparticle structure characterization
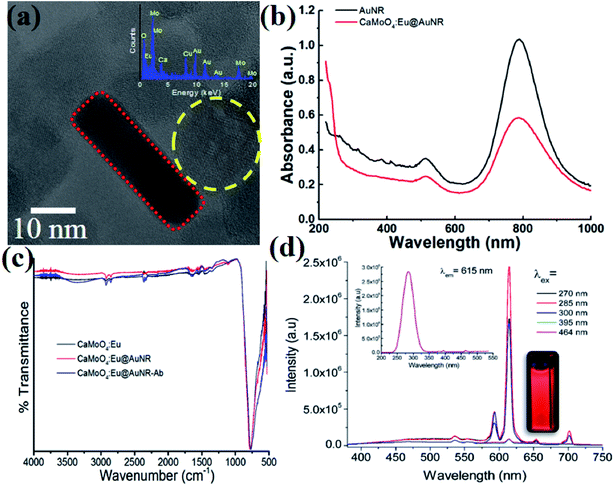
The typical TEM image of the composite CaMoO4:Eu3+@AuNR NPs is shown in Fig. 1a. It confirms the formation of hybrid nanoparticles where AuNRs were attached to the surface of CaMoO4:Eu3+ NPs. The presence of each element in the particle was confirmed by the EDX spectrum (Fig. 1a, inset). The average size of CaMoO4:Eu3+ nanoparticles was found to be ∼20 nm, whereas the average length and width of AuNRs were found to be ∼40 and 10 nm, respectively. Moreover, the hybrid nanoparticles show high dispersion for long times without precipitation. The crystalline nature of the CaMoO4:Eu3+ nanoparticles was confirmed by their X-ray diffraction pattern (data not shown). All peaks are well matched with the tetragonal structure of CaMoO4:Eu3+ (JCPDS#29-0351). The UV-vis spectra of hybrid nanoparticles in the range 200–1000 nm are shown in Fig. 1b. Strong absorption bands at 512.5 and 789 nm were observed, which are assigned to transverse and longitudinal surface plasmon resonance (SPR) bands of AuNRs, respectively. It was observed that the peak position of the SPR band at 789 nm was slightly red-shifted (∼0.8 nm) and full width at half maximum (FWHM) increased by ∼27 nm, whereas there was no shift for the SPR band at 512.5 nm and the FWHM increased by ∼14.4 nm with antibody coating on the surface of nanoparticles. Moreover, a small peak at ∼265 nm was observed for CaMoO4:Eu@AuNR NPs. This peak was ascribed to a charge transfer from the oxygen ligands to the central molybdenum atom within the MoO42− cluster (also called the Mo–O charge transfer band (CTB)), but no such peak was observed for the bare AuNRs. It was further confirmed by excitation spectra (λem = 615 nm). Also some weak peaks in the longer wavelength region 300–500 nm were ascribed to the direct 4f6–4f6 intraconfiguration transitions of Eu3+ ions (Fig. 1d, inset).41 The CaMoO4:Eu3+@AuNR NPs emit strong red fluorescence under 270, 285, 300, 395, and 464 nm excitation with peaks centered at 590 (5D0 → 7F1; magnetic dipole transition) and 615 nm (5D0 → 7F2; electric dipole transition) (Fig. 1d). The high-energy state excited (Mo–O CTB and Eu3+) electrons of Eu3+ are unstable and relaxed back to the ground states of Eu3+ ions through photon emission in the visible region. The intensity of the 5D0 → 7F2 transition is significantly higher than those of other transitions of Eu3+ ions.42 The strong luminescence of CaMoO4:Eu3+@AuNR NPs may be particularly useful for biological fluorescence labeling.Functionalization and performance of the SERS-fluorescence bimodal nanoprobe
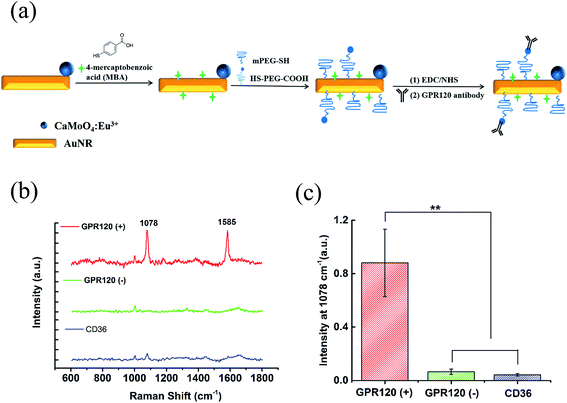
The as-prepared CaMoO4:Eu3+@AuNR nanocomposite was further functionalized to generate intense Raman signals and achieve specific targeting. Fig. 2a demonstrates the functionalization process containing three steps: (1) MBA, as a Raman reporter molecule providing strong chemical enhancement of the SERS signal and a simple SERS spectrum, was conjugated onto the nanocomposite through covalent Au–S bonding; (2) PEG linkers were coated to improve the stability and biocompatibility of the nanoprobe, as well as to minimize the non-specific binding of the nanoprobes to the cells; (3) finally, anti-GPR120 antibodies were conjugated onto the nanocomposite to realize specific GPR120 targeting in cells. The optimal dilution ratio of the GPR120 antibody was determined by immunofluorescence imaging. At a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 dilution the antibody exhibited good specificity and little background labeling of GPR120 in cells (data not shown). Thus, this ratio was used in nanoprobe functionalization and cell incubation.
250 dilution the antibody exhibited good specificity and little background labeling of GPR120 in cells (data not shown). Thus, this ratio was used in nanoprobe functionalization and cell incubation.
Fig. 1c shows the FT-IR spectra of bare CaMoO4:Eu3+, CaMoO4:Eu3+@AuNR, and antibody-conjugated CaMoO4:Eu3+@AuNR hybrid nanoparticles in the range from 500 to 4000 cm−1. Peaks at ∼1650 and 3450 cm−1 correspond to the bending and stretching vibrations of H–O–H molecules present on the surface of the hybrid nanoparticles. The characteristic peak that appeared at ∼802 cm−1 was assigned to the asymmetric stretching vibration of O–Mo–O vibration in the MoO42− tetrahedron.43 The peaks at 2923 and 2852 cm−1 indicating the C–H stretching vibrations arose from OA.44 On antibody conjugation to CaMoO4:Eu3+ nanoparticles with AuNRs and AuNR–Ab the peaks broadened. The FTIR spectra of pure HS–PEG–COOH, mPEG–HS, and the antibody are shown in Fig. S1 (see the ESI†). It was found that pure HS–PEG–COOH has several characteristic peaks from 1000 to 1700 cm−1, whereas some feeble peaks were observed at ∼2050, 1244, and 1076 cm−1 representing the antibody. Moreover, the conjugation of the antibody to the CaMoO4:Eu@AuNR nanoparticles results in some small peaks from 1000 to 1700 cm−1, which are characteristic of peaks arises from PEG and/or the antibody present on the surface of the nanoparticles.
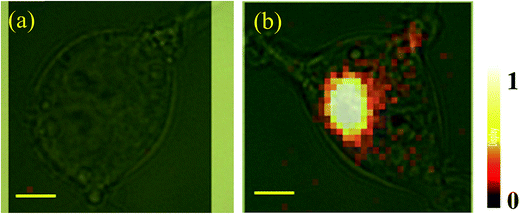
The performance of the functionalized nanoprobe (CaMoO4:Eu3+@AuNR–MBA–Ab) was assessed by SERS and fluorescence imaging. The constructed probes were incubated with cells for 24 h at a final concentration of 100 μg mL−1. Three different cell samples were used: (1) HEK293–GPR120 cells induced with DOX for 48 h (GPR120 (+)); (2) HEK293–GPR120 cells without DOX induction (GPR120 (−)); (3) constitutive HEK293-CD36 (CD36) cells. Representative SERS spectra of the nanoprobe-treated cells are shown in Fig. 2b. The typical spectrum of GPR120 (+) cells shows two enhanced peaks at 1078 and 1585 cm−1, which are assigned to the ring breathing and axial deformation modes of MBA, respectively.45,46 In contrast, spectra of GPR120 (−) and CD36 cells show nearly no Raman peaks from the reporter molecule MBA, but only regular Raman signals from cells (e.g. 1003 cm−1 from phenylalanine). The average peak intensity at 1078 cm−1 for GPR120 (+) cells is significantly higher (P < 0.001, N = 25) than those of GPR120 (−) and CD36 cells (Fig. 2c). This difference resulted from the specific targeting ability of the constructed nanoprobe CaMoO4:Eu3+@AuNR–MBA–Ab, which selectively binds to GPR120 (+) cells through specific antibody–antigen interactions, giving rise to the significantly enhanced SERS signal. This finding can also be confirmed by fluorescence images, in which the CaMoO4:Eu3+@AuNR–MBA–Ab nanoprobe-incubated GPR120 (+) cells exhibited considerable red fluorescence while the fluorescence of GPR120 (−) cells was virtually invisible (Fig. 3).
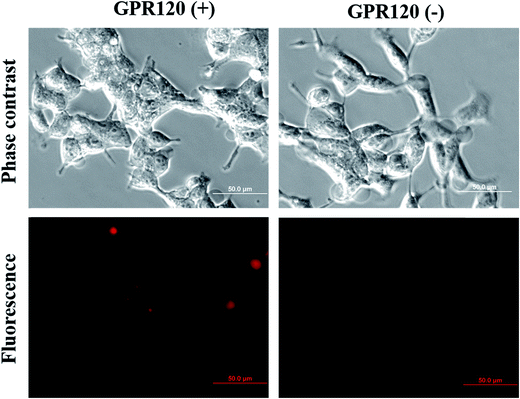 | ||
| Fig. 3 Fluorescence imaging of GPR120 (+) and GPR120 (−) cells incubated with the CaMoO4:Eu3+@AuNR–MBA–Ab nanoprobe for 24 h. | ||
The biocompatibility of the SERS-fluorescence bimodal nanoprobe CaMoO4:Eu3+@AuNR–MBA–Ab was estimated by incubating GPR120 (+) cells with the nanoprobes at different concentrations (20, 50, 80, 100, 200 μg mL−1) for 24 h, and then testing the cell viability using live/dead fluorescence imaging. As shown in Fig. S2,† there were no significant decreases in cell viability when incubating cells with nanoprobes at concentrations as high as 100 μg mL−1. Thus, this concentration (100 μg mL−1) was chosen in subsequent imaging experiments.
Cellular fluorescence and SERS microscopy
The cellular imaging capability of the bimodal nanoprobe was estimated by fluorescence and SERS measurements. Due to its high-throughput nature, fluorescence microscopy was used to quickly identify cells with high GPR120 expression levels. GPR120 (+) and GPR120 (−) HEK293 cells exhibited significant difference in fluorescence emission after incubation with the nanoprobes (Fig. 3). At the single cell level, a CD36 cell showed barely detectable red fluorescence at the central area of the cell, while a GPR120 (+) cell had strong labeling over the entire cell (Fig. S3†). Because the nanoprobes were conjugated with anti-GPR120 antibodies, they tended to bind with GPR120 but not CD36 at the cell surface. Furthermore, cellular SERS mapping was performed on the nanoprobe-treated cells. SERS images were created using the Raman intensities of peak 1078 cm−1, which is the most stable and reproducible characteristic peak from the reporter molecule (MBA). By collecting the peak intensity values all over the cell and transforming them into color values, GPR120 distribution was reflected by the brightness of the SERS image at the single cell or subcellular level. Fig. 4 shows the single-cell SERS mapping of GPR120. Apparently, strong SERS signals were recorded on GPR120 (+) cells (Fig. 4b), while SERS signals on the CD36 cells were nearly undetected (Fig. 4a). Additional SERS maps of GPR120 (+) cells are shown in Fig. S4.† SERS maps reflected the reorganized and clustered GPR120 receptors after nanoparticle binding. It has been reported that cargo binding promotes membrane receptor clustering in several different receptors.47–49 Cellular imaging results demonstrated that SERS-fluorescence bimodal microscopy is able to visualize the heterogeneous distribution of GPR120 receptors in single living cells.Detection of GPR120 under LA treatment by SERS
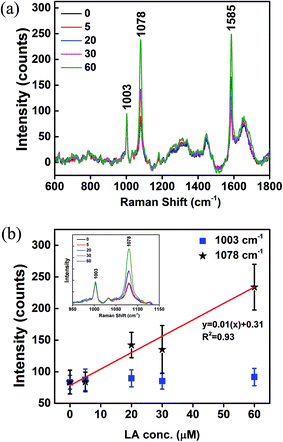
GPR120 responses to LA treatments were measured by SERS. GPR120 (+) cells were treated with LA at concentrations 0, 5, 20, 30, and 60 μM for 5 min, and then incubated with nanoprobes for 24 h. After washing off the unabsorbed nanoparticles, cells were taken for Raman measurements. Fig. 5a shows the average SERS spectra (N = 25) of the LA-treated cells. Two major intense peaks at 1078 and 1585 cm−1 were from the nanoprobes, which reflect the presence of GPR120 receptors. The peak at 1003 cm−1 was attributed to phenylalanine from cells. Elevated SERS signals from nanoprobes (1078 cm−1) were observed in high LA concentration treatments, while signals from cells themselves (1003 cm−1) were nearly unchanged (Fig. 5b, inset). SERS intensities (1003, 1078 cm−1) vs. LA concentration are plotted in Fig. 5b. A linear (R2 = 0.93) increase in 1078 cm−1 intensity was observed with increased LA concentrations. This result indicates that GPR120 activity is enhanced by LA treatment, which is consistent with previous reports using other methods.6 Furthermore, for the first time, we found that there is a linear dose-dependence of GPR120 response on LA in the 0–60 μM concentration range. These results demonstrated the potential of SERS to study the dynamic process of biomolecules in single living cells.Discussion
Recent studies have implicated that GPR120 and CD36 are candidate receptors that are involved in the gustatory perception of LCFA in taste bud cells.3 Yet their specific roles in the detection of LCFA remain to be explored. Several hypotheses are proposed to explain the situation, but the experimental results appear to be contradictory (reviewed in Besnard et al.50). An imaging technique that can visualize the behaviors of these receptors in living cells is favorable to provide direct evidence to answer these questions. Our goal in this work is to demonstrate the feasibility of the SERS-fluorescence bimodal imaging approach to achieve noninvasive live cell imaging of these fat responsive receptors. The fluorescence signal can be used as a fast detection tool for receptor recognition, while the SERS signal can be used as an accurate sensing tool for imaging localized receptor distributions.37 In addition, the outstanding capabilities of SERS for multiplex imaging51,52 would allow for the future detection of interactions between GPR120 and CD36 in the chemoreception of fatty acids.In summary, we developed a dual functional composite nanoprobe for SERS-fluorescence bimodal imaging of the fat-responsive receptor GPR120 in single living cells. The dual functional nanoprobe was composed of europium-doped calcium molybdate and gold nanorods, and further functionalized with a Raman reporter and the anti-GPR120 antibody. The functionalized nanoprobe was successfully applied for both SERS and fluorescence detection of GPR120 in vitro. The cellular distribution of GPR120 was successfully detected by single-cell SERS mapping. Taking advantages of the quantification ability of SERS, we observed an up-regulation of GPR120 by LA treatment. Moreover, a linear relationship between GPR120 activity and LA concentration in the 0–60 μM range was observed for the first time. These results demonstrate the potential of our SERS-fluorescence dual functional nanoprobes for multiplex detection of fat-responsive receptors (e.g. CD36 and GPR120) in the future.
Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was supported by National Science Foundation award #1264498 and National Institute of Health award R01DC013318.References
- T. T. Fung, E. B. Rimm, D. Spiegelman, N. Rifai, G. H. Tofler, W. C. Willett and F. B. Hu, Am. J. Clin. Nutr., 2001, 73, 61–67 CAS.
- L. Cordain, S. B. Eaton, A. Sebastian, N. Mann, S. Lindeberg, B. A. Watkins, J. H. O'Keefe and J. Brand-Miller, Am. J. Clin. Nutr., 2005, 81, 341–354 CAS.
- S. Abdoul-Azize, S. Selvakumar, H. Sadou, P. Besnard and N. A. Khan, Biochimie, 2014, 96, 8–13 CrossRef CAS PubMed.
- E. Adler, M. A. Hoon, K. L. Mueller, J. Chandrashekar, N. J. Ryba and C. S. Zuker, Cell, 2000, 100, 693–702 CrossRef CAS PubMed.
- R. F. Margolskee, J. Biol. Chem., 2002, 277, 1–4 CrossRef CAS PubMed.
- C. Cartoni, K. Yasumatsu, T. Ohkuri, N. Shigemura, R. Yoshida, N. Godinot, J. le Coutre, Y. Ninomiya and S. Damak, J. Neurosci., 2010, 30, 8376–8382 CrossRef CAS PubMed.
- C. Martin, P. Passilly-Degrace, D. Gaillard, J. F. Merlin, M. Chevrot and P. Besnard, PLoS One, 2011, 6, e24014 CAS.
- M. H. Ozdener, S. Subramaniam, S. Sundaresan, O. Sery, T. Hashimoto, Y. Asakawa, P. Besnard, N. A. Abumrad and N. A. Khan, Gastroenterology, 2014, 146, 995–1005 CrossRef CAS PubMed.
- L. Xiao, M. Tang, Q. Li and A. Zhou, Anal. Methods, 2013, 5, 874–879 RSC.
- Q. Li, E. Suasnavas, L. Xiao, S. Heywood, X. Qi, A. Zhou and S. C. Isom, J. Biophotonics, 2015, 8, 638–645 CrossRef CAS PubMed.
- Q. Li, L. Xiao, S. Harihar, D. R. Welch, E. Vargis and A. Zhou, Anal. Methods, 2015, 7, 10162–10169 RSC.
- M. Tang, Q. Li, L. Xiao, Y. Li, J. L. Jensen, T. G. Liou and A. Zhou, Toxicol. Lett., 2012, 215, 181–192 CrossRef CAS PubMed.
- P. L. Stiles, J. A. Dieringer, N. C. Shah and R. P. Van Duyne, Annu. Rev. Anal. Chem., 2008, 1, 601–626 CrossRef CAS PubMed.
- J. P. Camden, J. A. Dieringer, J. Zhao and R. P. Van Duyne, Acc. Chem. Res., 2008, 41, 1653–1661 CrossRef CAS PubMed.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- K. Kneipp, Y. Wang, H. Kneipp, L. Perelman, I. Itzkan, R. Dasari and M. Feld, Phys. Rev. Lett., 1997, 78, 1667–1670 CrossRef CAS.
- K. Kneipp, H. Kneipp, V. Kartha, R. Manoharan, G. Deinum, I. Itzkan, R. Dasari and M. Feld, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1998, 57, R6281–R6284 CrossRef CAS.
- H. Xu, E. Bjerneld, M. Kall and L. Borjesson, Phys. Rev. Lett., 1999, 83, 4357–4360 CrossRef CAS.
- K. Kneipp, A. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K. Shafer-Peltier, J. Motz, R. Dasari and M. Feld, Appl. Spectrosc., 2002, 56, 150–154 CrossRef CAS.
- J. P. Scaffidi, M. K. Gregas, V. Seewaldt and T. Vo-Dinh, Anal. Bioanal. Chem., 2009, 393, 1135–1141 CrossRef CAS PubMed.
- W. Xie, L. Wang, Y. Zhang, L. Su, A. Shen, J. Tan and J. Hu, Bioconjugate Chem., 2009, 20, 768–773 CrossRef CAS PubMed.
- A. Samanta, K. K. Maiti, K. Soh, X. Liao, M. Vendrell, U. S. Dinish, S. Yun, R. Bhuvaneswari, H. Kim, S. Rautela, J. Chung, M. Olivo and Y. Chang, Angew. Chem., Int. Ed., 2011, 50, 6089–6092 CrossRef CAS PubMed.
- H. Kang, S. Jeong, Y. Park, J. Yim, B. Jun, S. Kyeong, J. Yang, G. Kim, S. Hong, L. P. Lee, J. Kim, H. Lee, D. H. Jeong and Y. Lee, Adv. Funct. Mater., 2013, 23, 3719–3727 CrossRef CAS.
- H. Tang, X. B. Yang, J. Kirkham and D. A. Smith, Anal. Chem., 2007, 79, 3646–3653 CrossRef CAS PubMed.
- M. Richter, M. Hedegaard, T. Deckert-Gaudig, P. Lampen and V. Deckert, Small, 2011, 7, 209–214 CrossRef CAS PubMed.
- L. Xiao, S. Harihar, D. R. Welch and A. Zhou, Anal. Chim. Acta, 2014, 843, 73–82 CrossRef CAS PubMed.
- L. Xiao, H. Wang and Z. D. Schultz, Anal. Chem., 2016, 88, 6547–6553 CrossRef CAS PubMed.
- L. Xiao, K. A. Bailey, H. Wang and Z. D. Schultz, Anal. Chem., 2017, 89, 9091–9099 CrossRef CAS PubMed.
- R. M. Jarvis and R. Goodacre, Chem. Soc. Rev., 2008, 37, 931–936 RSC.
- M. Li, J. Xu, M. Romero-Gonzalez, S. A. Banwart and W. E. Huang, Curr. Opin. Biotechnol., 2012, 23, 56–63 CrossRef CAS PubMed.
- M. V. Yigit, L. Zhu, M. A. Ifediba, Y. Zhang, K. Carr, A. Moore and Z. Medarova, ACS Nano, 2011, 5, 1056–1066 CrossRef CAS PubMed.
- M. Xiao, J. Nyagilo, V. Arora, P. Kulkarni, D. Xu, X. Sun and D. P. Dave, Nanotechnology, 2010, 21, 035101 CrossRef PubMed.
- V. Amendola, S. Scaramuzza, L. Litti, M. Meneghetti, G. Zuccolotto, A. Rosato, E. Nicolato, P. Marzola, G. Fracasso, C. Anselmi, M. Pinto and M. Colombatti, Small, 2014, 10, 2476–2486 CrossRef CAS PubMed.
- Y. Wang, L. Chen and P. Liu, Chem.–Eur. J., 2012, 18, 5935–5943 CrossRef CAS PubMed.
- Z. Wang, S. Zong, W. Li, C. Wang, S. Xu, H. Chen and Y. Cui, J. Am. Chem. Soc., 2012, 134, 2993–3000 CrossRef CAS PubMed.
- X. Niu, H. Chen, Y. Wang, W. Wang, X. Sun and L. Chen, ACS Appl. Mater. Interfaces, 2014, 6, 5152–5160 CAS.
- S. Lee, H. Chon, S. Yoon, E. K. Lee, S. Chang, D. W. Lim and J. Choo, Nanoscale, 2012, 4, 124–129 RSC.
- A. K. Parchur, R. S. Ningthoujam, S. B. Rai, G. S. Okram, R. A. Singh, M. Tyagi, S. C. Gadkari, R. Tewari and R. K. Vatsa, Dalton Trans., 2011, 40, 7595–7601 RSC.
- A. K. Parchur, Q. Li and A. Zhou, Biomater. Sci., 2016, 4, 1781–1791 RSC.
- J. Zhao, H. Lui, D. I. McLean and H. Zeng, Appl. Spectrosc., 2007, 61, 1225–1232 CrossRef CAS PubMed.
- B. P. Singh, A. K. Parchur, R. S. Ningthoujam, A. A. Ansari, P. Singh and S. B. Rai, Dalton Trans., 2014, 43, 4770–4778 RSC.
- B. P. Singh, A. K. Parchur, R. S. Ningthoujam, A. A. Ansari, P. Singh and S. B. Rai, Dalton Trans., 2014, 43, 4779–4789 RSC.
- A. K. Parchur, A. I. Prasad, A. A. Ansari, S. B. Rai and R. S. Ningthoujam, Dalton Trans., 2012, 41, 11032–11045 RSC.
- J. Huang, L. Wang, X. Zhong, Y. Li, L. Yang and H. Mao, J. Mater. Chem. B, 2014, 2, 5344–5351 RSC.
- A. Michota and J. Bukowska, J. Raman Spectrosc., 2003, 34, 21–25 CrossRef CAS.
- W. Fang, Z. Wang, S. Zong, H. Chen, D. Zhu, Y. Zhong and Y. Cui, Biosens. Bioelectron., 2014, 57, 10–15 CrossRef CAS PubMed.
- A. Abulrob, Z. Lu, E. Baumann, D. Vobornik, R. Taylor, D. Stanimirovic and L. J. Johnston, J. Biol. Chem., 2010, 285, 3145–3156 CrossRef CAS PubMed.
- A. Grochmal, E. Ferrero, L. Milanesi and S. Tomas, J. Am. Chem. Soc., 2013, 135, 10172–10177 CrossRef CAS PubMed.
- B. Becker, M. R. Shaebani, D. Rammo, T. Bubel, L. Santen and M. J. Schmitt, Sci. Rep., 2016, 6, 28940 CrossRef CAS PubMed.
- P. Besnard, P. Passilly-Degrace and N. A. Khan, Physiol. Rev., 2016, 96, 151–176 CrossRef CAS PubMed.
- C. L. Zavaleta, B. R. Smith, I. Walton, W. Doering, G. Davis, B. Shojaei, M. J. Natan and S. S. Gambhir, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 13511–13516 CrossRef CAS PubMed.
- D. C. Kennedy, K. A. Hoop, L. L. Tay and J. P. Pezacki, Nanoscale, 2010, 2, 1413–1416 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ay02039b |
| This journal is © The Royal Society of Chemistry 2018 |