Open Access Article
Open Access ArticleLectin inspired polymers based on the dipeptide Ser-Asp for glycopeptide enrichment†
B.
Zhang
a,
R. Z.
Yu
b,
Y. H.
Yu
a,
C.
Peng
a,
R.
Xie
a,
Y.
Zhang
a and
J. Y.
Chen
 *a
*a
aDepartment of Gastrointestinal Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China. E-mail: union.bzhang@aliyun.com
bMedical Department, Chinese People's Liberation Army 210 Hospital, Dalian, Liaoning 116015, China
First published on 1st October 2018
Abstract
Lectin inspired polymers were prepared through modification of silica microspheres with Ser-Asp (SD). This functional polymer showed distinct adsorption and retention towards different disaccharides and demonstrated high-efficiency enrichment of glycopeptides.
The covalent binding of glycans on proteins not only regulates the important biological processes but also reflects the state of cells. Typically, enzymes are equipped with certain glycans for recognition sites in gene transcription or protein translation.1,2 Aberrant protein glycosylation can change the physicochemical properties or structures of biomolecules, and then trigger diseases, e.g., N-glycosylated tau protein can only be detected in biofluids in Alzheimer's disease.3 Therefore, it is important to identify protein glycosylation, which provides information about the protein sequence, glycosylation sites and glycan structures. However, it is difficult to characterize protein glycosylation with state-of-the-art mass spectrometry (MS) because of the low abundance of glycopeptides.4
In the last decade, several enrichment methods for glycopeptides have been developed, including lectin affinity chromatography,5–7 hydrophilic interaction chromatography,8,9 boronic acid affinity chromatography,10,11etc. Among these, lectin affinity chromatography is widely used to enrich glycopeptides/glycoproteins.5 Lectin recognizes specific carbohydrate moieties through the reversible binding between the anomeric hydroxyl groups on carbohydrates and the hydrophilic groups from amino acid residues in lectin protein.12,13 Such interactions are interpreted as the synergistic effects of multiple forces, including hydrogen bonding, CH–π interactions, ion pairing, van der Waals forces, etc.14,15 Even though the single peptide–carbohydrate interaction is weak, the multiple binding sites form a network and enhance these interactions. These peptide–carbohydrate interactions inspire us to design artificial lectins for glycopeptide enrichment.
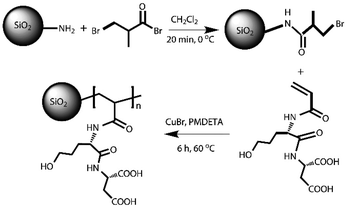
In this paper, a lectin inspired polymer was prepared through modification of anino-silica microspheres with polymerized Ser-Asp (SD) (denoted as polySD-SiO2 in Fig. 1a). The Ser-Asp was selected to mimic the amino acid residues in lectins and the polymer chain is used to amplify the stereoselective recognition between Ser-Asp and glycans. The bio-inspired material polySD-SiO2 was synthesized by surface initiated atom transfer radical polymerization (Scheme 1) because this method is robust and easy to prepare, and uses commercially available, inexpensive catalysts and initiators.16–19
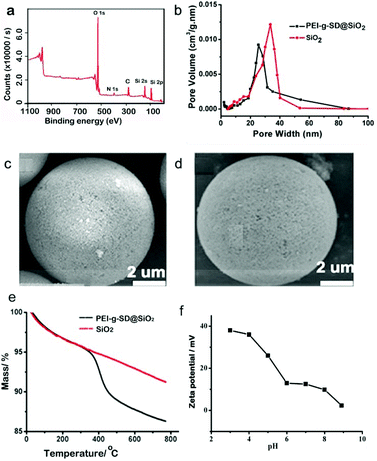
The synthesized materials were characterized by high resolution scanning electron microscopy (HRSEM), zeta potential analysis and thermogravimetry analysis (TGA), and used to enrich glycopeptides from different samples. HRSEM images reveal that polySD-SiO2 materials preserve a similar morphology to that of silica microspheres. These data proved that polySD-SiO2 materials are not degraded after coating with polymers on the silica surface. The nitrogen sorption isotherm of polySD-SiO2 showed a typical type IV curve (Fig. S1†). The Barrett–Joyner–Halenda (BJH) pore size of polySD-SiO2 was 26 nm compared with 33 nm of silica microspheres without modification (Fig. 1b), indicating a 3.5 nm polymer coated on the silica sphere. The elemental analysis and X-ray photoelectron spectroscopy data (Fig. 1a) imply the presence of carbon, nitrogen and oxygen on the polySD-SiO2. TGA analysis was used to further measure the relative composition of polySD-SiO2 (Fig. 1e). The sharp decline above 400 °C resulted from the weight loss of the polymer layer. The zeta-potentials of polySD-SiO2 under different pH conditions were measured. As shown in Fig. 1f, the surface charge of polySD-SiO2 is zero at solution pH 5.1, and negative at a pH higher than 5.1. The tunable surface charges are ascribed to the naturally smart zwitterions of Ser-Asp chemical composition. These results suggest that polySD-SiO2 was successfully modified on the silica microspheres.
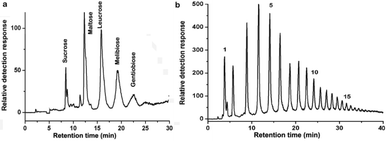
In order to exhibit the distinct discrimination of polySD-SiO2 toward saccharides, the chromatographic separation of disaccharides and fructo-oligosaccharides was carried out. Because of the subtle differences in stereochemistry, chemoselective discrimination of disaccharides is very challenging. Five disaccharides (sucrose, maltose, leucrose, melibiose, and gentiobiose) with subtle differences in structure could be well separated with the column (Fig. 2a). The application of this column to oligosaccharides with different polymerization degrees was also investigated. The components of fructo-oligosaccharides were separated into 26 constituents with different polymerization degrees, as labelled by numbers (Fig. 2b). The fructo-oligosaccharides with low to high polymerization degrees could be gradually eluted from the adsorbent using a solution with high to low acetonitrile content, which matches well with the typical characteristics of hydrophilic interaction chromatography. These data clearly demonstrate the powerful discrimination capacities of the polySD-SiO2-based column toward different saccharides. This column effectively addresses the problem of saccharide separation and can be widely applied in carbohydrate chemistry.
The enrichment conditions were optimized. The loading solution was 80% CH3CN/0.1% FA because most of the non-glycopeptides and all glycopeptides retained well under these conditions (Fig. S2†). The elution solution was 40% CH3CN/H2O/5 mM NH4HCO3 because of the weaker retention of glycopeptides at pH 8.0 and a lower CH3CN content (Fig. S3†). Tryptic digests of bovine fetuin were chosen as the model sample, attributing to its definite glycosylation sites and glycan structures. In order to mimic the complexity of the biosamples, the tryptic digests of bovine fetuin and bovine serum albumin were mixed at different molar ratios and treated with polySD-SiO2. After enrichment with polySD-SiO2, up to 43 glycopeptides were detected from the mixture of fetuin and BSA at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, in sharp contrast to no detectable glycopeptides in the mixture before enrichment (Fig. S4†). For comparison, only 7 glycopeptides were captured from the same sample by commercial zwitterionic hydrophilic interaction liquid chromatography (ZIC-HILIC) (Fig. 3c). Moreover, the selectivity of the material toward glycopeptides remained even in the case of fetuin and BSA at a molar ratio of 1
100, in sharp contrast to no detectable glycopeptides in the mixture before enrichment (Fig. S4†). For comparison, only 7 glycopeptides were captured from the same sample by commercial zwitterionic hydrophilic interaction liquid chromatography (ZIC-HILIC) (Fig. 3c). Moreover, the selectivity of the material toward glycopeptides remained even in the case of fetuin and BSA at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 (Fig. 3b). It should be noted that some glycopeptides enriched with polySD-SiO2 originated from the impurities of BSA, as reported in the literature,9 indicating the ability of polySD-SiO2 to enrich trace glycopeptides from a vast amount of interferents. These results demonstrate the high selectivity of polySD-SiO2 for glycopeptides. The high selectivity of polySD-SiO2 toward glycopeptides might be attributed to the reason that the monomer Ser-Asp binds glycans via hydrogen bonds and the multiple sites of Ser-Asp in the polymer further enhance their interaction.
3000 (Fig. 3b). It should be noted that some glycopeptides enriched with polySD-SiO2 originated from the impurities of BSA, as reported in the literature,9 indicating the ability of polySD-SiO2 to enrich trace glycopeptides from a vast amount of interferents. These results demonstrate the high selectivity of polySD-SiO2 for glycopeptides. The high selectivity of polySD-SiO2 toward glycopeptides might be attributed to the reason that the monomer Ser-Asp binds glycans via hydrogen bonds and the multiple sites of Ser-Asp in the polymer further enhance their interaction.
In addition to its high selectivity toward glycopeptides, polySD-SiO2 also exhibited high recovery and adsorption capacity for glycopeptides, which are two important parameters to assess adsorbents. A stable-isotope dimethyl labelling method20 was performed to determine the recovery of polySD-SiO2 toward a standard glycopeptide (peptide sequence: KVANKT, N: glycosylation site). The recovery rate was 86.6 ± 5.0% (n = 5), contributing to the strong binding of glycopeptides on materials during sample loading and the thorough elution of targets from materials. The adsorption capability of polySD-SiO2 toward the standard glycopeptide was 83 mg g−1. These results demonstrated the high efficiency of polySD-SiO2 materials for glycopeptide enrichment.
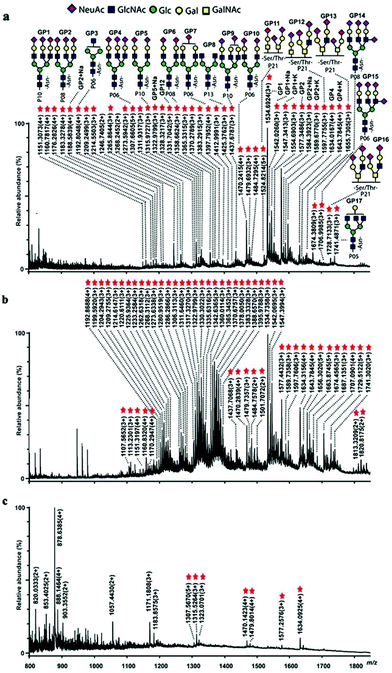
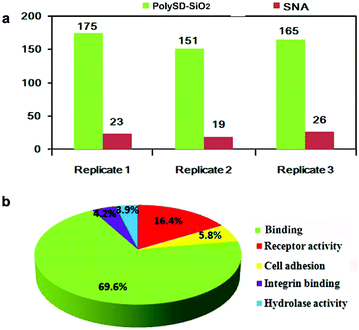
The established enrichment strategy was further applied to enrich glycopeptides from the HeLa S3 cell lysate. Three technical replicates were performed using 20 μg of total protein. With a false discovery rate of 1% at both the peptide and site levels, 175, 151, and 165 unique glycosylation sites were characterized after treatment with polySD-SiO2, respectively (Fig. 4a). Furthermore, substantial overlap of the identified glycosylation sites could be found between two replicates (Fig S5 in the ESI†). The average selectivity of polySD-SiO2 toward glycopeptides was 68% for three technique replicates. The detailed information of the identified glycosylation sites is shown in Table S1.† For comparison, Sambucus nigra agglutinin II (SNA), a lectin, was used to enrich glycopeptides from the same samples. The numbers of identified glycosylation sites were 23, 19 and 26, respectively. These numbers are substantially less than those obtained with our materials. This result could explain that functional monomer Ser-Asp in poly SD-SiO2 could effectively bind various glycans in the glycopeptides, while the SNA is specific to only one subset of glycans. Gene ontology analysis indicated that 69.6% of the matched glycoproteins were annotated as binding proteins, and other glycoprotiens were involved in the cell adhesion and receptor activity (Fig. 4b). These data exemplify the excellent potential of our material for use in glycoproteomic analysis.
Conclusions
In conclusion, lectin inspired materials showed high selectivity for glycopeptides, and could enrich low abundance glycopeptides from both the fetuin/BSA mixture and real biosamples. This material could also discriminate disaccharides with a slight difference in their structures. These features well match the demand for fine discrimination of glycans and in-depth analysis of glycoprotoeomics. As a biomimetic and bioinspired approach employed in this study, lectin inspired materials pave the way for designing artificial saccharide receptors with high specificity.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (81472487 and 81773197).Notes and references
- M. Aebi, R. Bernasconi, S. Clerc and M. Molinari, Trends Biochem. Sci., 2010, 35, 74–82 CrossRef CAS PubMed.
- R. Kadirvelraj, J.-Y. Yang, J. H. Sanders, L. Liu, A. Ramiah, P. K. Prabhakar, G.-J. Boons, Z. A. Wood and K. W. Moremen, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4637–4642 Search PubMed.
- S. A. Yuzwa, A. H. Cheung, M. Okon, L. P. McIntosh and D. J. Vocadlo, J. Mol. Biol., 2014, 426, 1736–1752 CrossRef CAS PubMed.
- D. F. Zielinska, F. Gnad, J. R. Wisniewski and M. Mann, Cell, 2010, 141, 897–907 CrossRef CAS PubMed.
- M. Chen, X. Shi, R. M. Duke, C. I. Ruse, N. Dai, C. H. Taron and J. C. Samuelson, Nat. Commun., 2017, 8, 15487 CrossRef CAS PubMed.
- E. Ruiz-May, S. Hucko, K. J. Howe, S. Zhang, R. W. Sherwood, T. W. Thannhauser and J. K. C. Rose, Mol. Cell. Proteomics, 2014, 13, 566–579 CrossRef CAS PubMed.
- E. Song, R. Zhu, Z. T. Hammond and Y. Mechref, J. Proteome Res., 2014, 13, 4808–4820 CrossRef CAS PubMed.
- X. Li, Y. Xiong, G. Qing, G. Jiang, X. Li, T. Sun and X. Liang, ACS Appl. Mater. Interfaces, 2016, 8, 13294–13302 CrossRef CAS PubMed.
- G. Qing, X. Li, P. Xiong, C. Chen, M. Zhan, X. Liang and T. Sun, ACS Appl. Mater. Interfaces, 2016, 8, 22084–22092 CrossRef CAS PubMed.
- Z. Bie, Y. Chen, H. Li, R. Wu and Z. Liu, Anal. Chim. Acta, 2014, 834, 1–8 CrossRef CAS PubMed.
- X. Li, H. Liu, G. Qing, S. Wang and X. Liang, J. Mater. Chem. B, 2014, 2, 2276–2281 RSC.
- Y. Hang, X.-P. He, L. Yang and J. Hua, Biosens. Bioelectron., 2015, 65, 420–426 CrossRef CAS PubMed.
- J. M. Propster, F. Yang, S. Rabbani, B. Ernst, F. H. T. Allain and M. Schubert, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4170–E4179 CrossRef CAS PubMed.
- W. I. Weis, K. Drickamer and W. A. Hendrickson, Nature, 1992, 360, 127–134 CrossRef CAS PubMed.
- W. I. Weis, R. Kahn, R. Fourme, K. Drickamer and W. A. Hendrickson, Science, 1991, 254, 1608–1615 CrossRef CAS PubMed.
- H. W. Ma, J. H. Hyun, P. Stiller and A. Chilkoti, Adv. Mater., 2004, 16, 338–341 CrossRef CAS.
- L. Hua, M. Y. Liu, L. C. Mao, Q. Huang, H. Y. Huang, G. J. Zeng, J. W. Tian, Y. Q. Wen, X. Y. Zhang and Y. Wei, Appl. Surf. Sci., 2018, 434, 204–221 CrossRef.
- L. Hua, J. Wu, M. Y. Liu, L. C. Mao, H. Y. Huang, Q. Wang, Y. F. Dai, Y. Q. Wen, X. Y. Zhang and Y. Wei, J. Colloid Interface Sci., 2017, 508, 396–404 CrossRef PubMed.
- L. C. Mao, X. H. Liu, M. Y. Liu, L. Hua, D. Z. Xu, R. M. Jiang, Q. Huang, Y. Q. Wen, X. Y. Zhang and Y. Wei, Appl. Surf. Sci., 2017, 419, 188–196 CrossRef CAS.
- F. Wang, R. Chen, J. Zhu, D. Sun, C. Song, Y. Wu, M. Ye, L. Wang and H. Zou, Anal. Chem., 2010, 82, 3007–3015 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8an01258j |
| This journal is © The Royal Society of Chemistry 2018 |