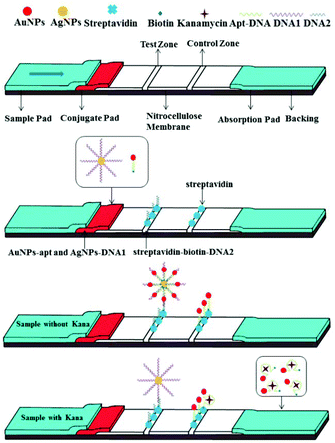
An aptamer and functionalized nanoparticle-based strip biosensor for on-site detection of kanamycin in food samples†
Jing
Liu
,
Jingyi
Zeng
,
Yaping
Tian
and
Nandi
Zhou
 *
*
The Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, China. E-mail: zhounandi@jiangnan.edu.cn
First published on 8th November 2017
Abstract
A lateral flow strip biosensor for fast, sensitive, low-cost and on-site detection of kanamycin was developed by using kanamycin-specific aptamer-modified gold nanoparticles (AuNPs-apt) as a probe and oligonucleotide DNA1-modified silver nanoparticles (AgNPs-DNA1) as a signal amplification element. Through the complementary sequences of DNA1 and the aptamer, the AgNP-DNA1-apt-AuNPs complex can be formed and further captured on the test zone of the strip, where a capture probe DNA2 complementary to the 3′-terminal of DNA1 was immobilized. In the presence of kanamycin, it can competitively bind to the aptamer, and then inhibit the formation of the complex and the accumulation of AuNPs on the test zone. AuNPs-apt can finally be captured on the control zone via the specific binding between biotin and streptavidin. The assay avoids multiple incubation and washing steps and can be completed within 10 min. By observing the color change of the test zone, a qualitative detection for kanamycin can be achieved by the naked eye, with the visual limit of 35 nM. Meanwhile, a linear detection range of 1–30 nM with a low detection limit of 0.0778 nM for quantitative analysis can be achieved by using a scanning reader. The lateral flow strip biosensor exhibited high specificity and stability. Moreover, it was applied to detect kanamycin in various food samples, indicating its great potential in field testing.
Introduction
Antibiotics, a class of secondary metabolites, have anti-pathogen and other activities. Because antibiotics can effectively inhibit detrimental bacteria and prevent a series of infections,1 they have been widely used in the development of modern animal husbandry. As a broad spectrum of aminoglycoside antibiotic, kanamycin has been used as human and veterinary medicine due to its inhibition of the growth of Gram-negative and Gram-positive bacteria.2 However, the excessive use of kanamycin leads to its accumulation in animal-derived food and its ultimate uptake by humans,3 which will cause serious side effects, such as ototoxicity and nephrotoxicity.4 For consumer protection, the European Union has established maximum residue limits (MRLs) of 150 μg kg−1 for kanamycin in milk.5 Nowadays, a variety of techniques have been developed for detection of antibiotic residues including kanamycin, e.g. ELISA,6 capillary zone electrophoresis (CZE),7 HPLC,8 surface plasmon resonance (SPR)9 and electrochemical analysis.10 Although these conventional methods can detect kanamycin accurately, their application is still limited due to the need for specialized equipment, complicated operation, being susceptible to interference, time-consuming, and low sensitivity.11,12 Therefore, it is necessary to establish a rapid, specific and sensitive method for on-site detection of kanamycin and other antibiotic residues.The use of lateral flow test strip biosensors is a promising approach for on-site rapid detection of a variety of targets owing to their long-term stability, short assay time, low cost and no need for complicated equipment and skilled technicians.13,14 The first report of strip analysis was proposed in 1956 by Plotz and Singer.15 Since then, test strips have been widely developed and applied in the field of clinical diagnosis, and environment and food monitoring.16–18 So far, most of these strip biosensors have used antibodies or specific enzymes as target-recognition elements. The use of antibodies or enzymes may encounter some issues, such as poor pH and thermo-stability, high cost, or lack of antibodies with good performance or suitable enzymes, etc. Aptamers (apt) generally indicate short single-stranded DNA or RNA with high affinity for the targets, which are obtained by in vitro screening known as SELEX.19 Due to their high specificity, high stability, and being easy to modify and regenerate over antibodies,20,21 aptamers have been widely used to fabricate biosensors for various small molecules, proteins and cells.22–25 Since the report of a kanamycin-specific aptamer,26 it has been used as a recognition element in a variety of aptamer-based kanamycin biosensors.27–30 Research on the application of the aptamers in the fabrication of strip biosensors will provide alternative strategies for fast detection of diverse targets, and may have great prospects in drug analysis, disease prevention and targeted therapy.31–34 Actually, aptamer-based strips have already exhibited excellent characteristics, such as a wide detection spectrum, high stability and low cost.35,36 Compared with these antibody-based strips, the test strips based on aptamers have an equivalent or even higher sensitivity.37 Such test strips have been applied in the detection of nucleic acids,38,39 toxins,37,40 ions,41,42 small molecular compounds,43,44 proteins,45 and microbials.46 However, to the best of our knowledge, aptamer-based strip biosensors for the detection of antibiotic residues have not yet been reported.
In this study, we developed an aptamer and functionalized nanoparticle-based dry-reagent strip biosensor for potential on-site detection of kanamycin. In this strip biosensor, gold nanoparticles (AuNPs) were used as a color probe, and silver nanoparticles (AgNPs) were used as a signal amplification element. The strip can be used to detect kanamycin in various food samples either qualitatively by the naked eye or quantitatively by using a scanning reader.
Experimental
Materials and reagents
Kanamycin, streptavidin, tris-(2-carboxyethyl)-phosphine hydrochloride (TCEP) and dATP were purchased from Sangon Biotech Co., Ltd (Shanghai, China). Silver nitrate was obtained from Sigma-Aldrich (St Louis, MO, USA). Hydrogen tetrachloroaurate hydrate (HAuCl4·4H2O), sodium citrate, sodium borohydride (NaBH4) and sodium desoxycholate were procured from Sinopharm Chemical Reagent Co., Ltd (Shanghai, China). Sample pads, conjugate pads, nitrocellulose membrane, absorption pads and adhesive backing were purchased from Jieyi Biotechnology Co., Ltd (Shanghai, China). Other chemicals were of analytical grade. All solutions were prepared using ultrapure water (18.2 MΩ cm) obtained from a Millipore water purification system.All the oligonucleotides including kanamycin-specific aptamers, DNA1 and DNA2 were synthesized and HPLC-purified by Sangon Biotech Co., Ltd. The specific sequences of the aptamer, DNA1 and DNA2 are as follows:
Aptamer: 5′-HS-(CH2)6-TGGGGGTTGA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) -biotin-3′26
-biotin-3′26
DNA1: 5′-HS-(CH2)6-TCAG![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GTCCAACGTCAGATCC-3′
GTCCAACGTCAGATCC-3′
DNA2: 5′-biotin-CCGATGGATCTGACGT-3′
The underlined and italic letters represent the complementary sequences.
Synthesis and modification of gold nanoparticles (AuNPs) and silver nanoparticles (AgNPs)
The preparation of AuNPs is according to the procedure reported previously.47 100 mL of 0.01% (w/v) HAuCl4 was put into a 250 mL round bottom flask, stirred and heated. After boiling for 2 min, 3.5 mL of 1% (w/v) sodium citrate was added rapidly with stirring. The mixture was continuously stirred and heated for 15 min, and then stirred at room temperature for another 30 min. The as-prepared AuNPs were concentrated 5 times by heating and evaporating the excessive water until a final volume of 20 mL was reached. After cooling to room temperature, the prepared AuNPs solution was stored at 4 °C in the dark.The preparation of AgNPs is according to the previously reported method,48 using silver nitrate as a raw material, sodium borohydride as a reducing agent and sodium deoxycholate as a stabilizer and a chiral inducer. Firstly, 100 mL of 1 mM AgNO3 and 100 mL of 2 mM sodium deoxycholate were mixed and aged at pH 7.0 for 24 h. Then 0.858 mL of freshly prepared 20 mM NaBH4 was added dropwise into an ice bath with vigorous stirring. After 10 min reaction, AgNPs were prepared and then stored at 4 °C in the dark.
After the preparation of the previous nanoparticles, oligonucleotides were covalently immobilized on the surface of the nanoparticles via metal–S bonds. The preparation of the aptamer-modified AuNPs (AuNPs-apt) and DNA1-modified AgNPs (AgNPs-DNA1) is according to previously reported procedures.49 Briefly, 15 μL of 1 mM TCEP was mixed with 30 μL of 100 μM aptamer or DNA1 to activate the thiol group of aptamer/DNA1. Then the mixture was added into 1 mL of the prepared AuNPs/AgNPs and stirred at room temperature for 1 h. After that, 30 μL of 15 mM dATP was added to the solution and stirred for 35 min, followed by the addition of 20 μL of 1 M NaCl and incubation first at room temperature for 30 min and then at 4 °C for 6 h to increase the stability of the modified nanoparticles. Finally, the unbound oligonucleotides were removed by centrifugation at 8000 rpm for 20 min at 4 °C. And the pellet was re-suspended in 1 mL of 20 mM Na3PO4 containing 0.5% Tween-20, 5% BSA, 10% sucrose and 0.5% Triton X-100. The modified nanoparticles were stored at 4 °C in the dark.
Preparation of streptavidin–biotin–DNA2 conjugate
DNA2 was designed as a capture probe to capture the AgNP-DNA1-apt-AuNPs complex on the test zone. To immobilize DNA2 on the test zone, a streptavidin–biotin–DNA2 conjugate was prepared using biotinylated DNA2 and streptavidin via the high affinity between streptavidin and biotin. Streptavidin was dissolved in 10 mM PBS (pH 7.4) to a concentration of 1 mg mL−1. Then 5 μL streptavidin and 20 μL of 10 μM DNA2 were mixed and incubated at 4 °C for 2 h to allow complete binding between streptavidin and biotinylated DNA2. After that, the prepared streptavidin–biotin–DNA2 conjugate was further incubated with 10 μL of 5 mM biotin to completely block the remaining binding sites of streptavidin. The excessive DNA2 and biotin were removed by centrifugation with a ultrafiltration tube (30 kDa). The prepared streptavidin–biotin–DNA2 was stored at 4 °C.Assembly of the lateral flow strip biosensor
The strip biosensor consists of a sample pad, a conjugate pad, a nitrocellulose membrane (NC membrane), an absorption pad and PVC adhesive backing.After cutting into the appropriate size, the conjugate pad and the sample pad were immersed in the treatment solution (0.1 M Tris-HCl buffer containing 1% NaCl, 0.5% Tween-20, 1% BSA, 2% sucrose and 1% Triton X-100, pH 8.2) for 30 min, and then dried at 45 °C. Then 10 μL of 5 μM AuNPs-apt and 10 μL of 2 μM AgNPs-DNA1 (the concentrations refer to DNA on the functionalized nanoparticles) were spread evenly on the conjugate pad, which was then dried in a thermostatic chamber at 37 °C for 2 h.
To prepare the strip biosensor, the NC membrane and the pads were overlapped and pasted on the adhesive backing (Scheme 1). Then 1 μL of a streptavidin–biotin–DNA2 conjugate was used to draw a line on the NC membrane near the conjugate pad to construct the test zone (T zone). And 1 μL of 0.5 mg mL−1 streptavidin was used to draw a line on the NC film near the absorption pad to construct the control zone (C zone). The distance between the T zone and the C zone was fixed at 5 mm. After drawing the T and C zones on the NC membrane, the strips were dried at 37 °C for 1 h. Finally, the assembled strip biosensors were stored in tin foil bags at 4 °C.
Detection of kanamycin using the strip biosensor
Kanamycin was diluted with saline sodium citrate (SSC) containing 1% BSA (pH 7.0) to different concentrations as the test solution. The test strips were placed in the cartridges. 100 μL of the test solution was added into the sample wells on the sample pad. Due to the siphon action of the capillary, the test solution will migrate through the whole strip from the sample pad to the absorption pad. This procedure can be completed within 10 min. Red bands can be observed on the T zone and C zone, due to the accumulation of AuNPs. Thus the evaluation can be achieved visually. For quantitative analysis of kanamycin, the optical density (expressed as peak area) of the T zone was determined using a scanning reader (Jieyi Biotechnology Co., Ltd, Shanghai, China).Detection of kanamycin in real food samples
To verify the performance of the strip biosensor for real food samples, kanamycin was added into honey, milk and milk powder to prepare artificially kanamycin-contaminated samples. For honey samples, the artificially contaminated honey was directly diluted 10-fold with SSC (for milk and milk powder samples see the ESI†). The detection was then carried out with the test strips as described above.Results and discussion
Principle of the strip biosensor for kanamycin
The configuration and principle of detection of the aptamer and functionalized nanoparticle-based lateral flow strip biosensor are shown in Scheme 1. AuNPs-apt and AgNPs-DNA1 have already been modified on the conjugate pad before the assembly of the strip. The streptavidin–biotin–DNA2 conjugate and free streptavidin were immobilized on the T zone and C zone of the NC membrane, respectively. Since DNA1 is complementary to the aptamer at its 5′-terminal and complementary to DNA2 at its 3′-terminal, the AgNP-DNA1-apt-AuNPs complex can be formed and further captured on the test zone of the strip through hybridization between these complementary oligonucleotides in the absence of kanamycin. Thus a clear deep red band can be observed on the T zone due to the accumulation of AuNPs. The excessive AuNPs-apt will further migrate to the C zone on the strip and be captured through the binding of biotin at the 3′-end of the aptamer and streptavidin immobilized on the C zone. Therefore a red band can also be observed on the C zone. In the presence of kanamycin, it binds specifically to the aptamer modified on AuNPs, dehybridizes the duplex between the aptamer and DNA1, and then releases AuNPs-apt from the AgNP-DNA1-apt-AuNPs complex. The released AuNPs-apt can be captured on the C zone through biotin–streptavidin interaction. Due to the reduced accumulation of AuNPs, light red color will appear on the T zone. The shade of the color on the T zone is negatively correlated to the concentration of kanamycin. Therefore, a strip exhibiting two red bands represents a negative result, whereas a strip exhibiting a lighter or completely faded T zone represents a positive result.Synthesis and modification of AuNPs and AgNPs
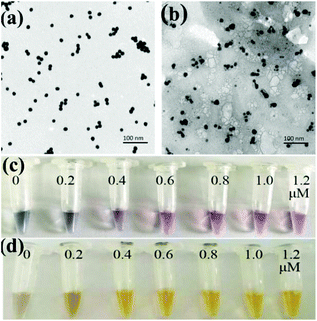
The prepared AuNPs and AgNPs were characterized by using transmission electron microscopy (TEM) and a UV-vis spectrophotometer. As shown in Fig. 1a and b, the prepared AuNPs and AgNPs were well dispersed and had uniform particle sizes of 13 nm and 20 nm, respectively. The typical adsorption peaks of AuNPs and AgNPs at 520 nm and 420 nm, respectively, can be also observed in UV-vis spectra (Fig. S1†).The nanoparticles were then functionalized with oligonucleotides to obtain AuNPs-apt and AgNPs-DNA1. The concentration of the aptamer or DNA1 used to modify the nanoparticles can influence the coverage of these oligonucleotides on the surface of the nanoparticles and the performance of the strip sensor. The modified oligonucleotides are not only responsible for the recognition of the target kanamycin or the complementary sequence, but also for maintaining the stability of the nanoparticles. It is well known that bare AuNPs or AgNPs aggregate and change their color at high concentration of NaCl, and the modification of nucleic acids on the surface of these nanoparticles can greatly improve their stability and maintain their dispersity.50 Therefore, the stability of the modified nanoparticles in NaCl solution was chosen as an index to optimize the concentration of the oligonucleotides during the functionalization of the nanoparticles. The prepared AuNPs were incubated with different concentrations of the aptamer, and then NaCl was added into the solution to a final concentration of 20 mM. As shown in Fig. 1c, when the concentration of the aptamer was lower than 0.6 μM, the color of AuNPs-apt turned grey after the addition of NaCl, indicating the aggregation of AuNPs. Therefore, 0.6 μM aptamer was chosen to modify AuNPs. Similarly, the prepared AgNPs were incubated with different concentrations of DNA1, and then NaCl was introduced. As shown in Fig. 1d, after incubation with DNA1 with concentrations higher than 0.4 μM, the obtained AgNPs-DNA1 remained stable in NaCl solution. Thus 0.4 μM DNA1 was chosen to modify AgNPs.
Optimization of the parameters
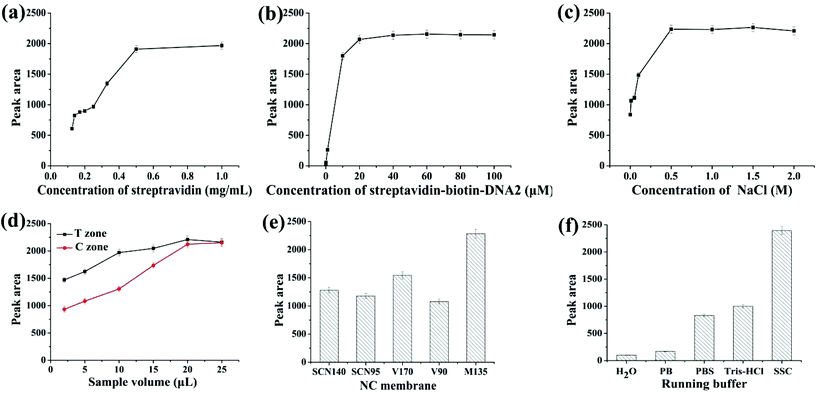
In order to achieve high sensitivity and signal-to-noise ratio, low detection limits and high stability, the parameters for strip preparation, including the concentrations of streptavidin, the streptavidin–biotin–DNA2 conjugate and NaCl, the amount of AuNPs-apt and AgNPs-DNA1, the type of NC membrane and running buffer on the conjugate pad, were optimized. The conditions used in the preparation of the strip can ensure the appearance of clear deep red bands on the T and C zones.Streptavidin immobilized on the C zone is responsible for capturing excessive AuNPs-apt that cannot form the AgNP-DNA1-apt-AuNPs complex and be captured on the T zone, or AuNPs-apt released from the complex due to the binding of kanamycin to the aptamer. Streptavidin solutions with different concentrations were immobilized on the C zone, and then SSC buffer was applied on the strip, and the color density of the C zone was determined by using the colloidal gold scanning reader. As shown in Fig. 2a, the optical density of the C zone, expressed in peak area, increases with the concentration of streptavidin. When the concentration of streptavidin is higher than 0.5 mg mL−1, the peak area almost remains constant. Thus, 0.5 mg mL−1 streptavidin solution was chosen to construct the C zone.
A streptavidin–biotin–DNA2 conjugate was immobilized on the T zone to capture the AgNP-DNA1-apt-AuNPs complex through the hybridization between the complementary sequences of DNA1 and DNA2. Thus the coverage of streptavidin–biotin–DNA2 on the T zone is a critical factor to ensure the efficient capture of the complex and the optical density of the T zone. Different concentrations of the streptavidin–biotin–DNA2 conjugate ranging from 0.001 to 100 μM (refer to the concentration of DNA2) were used to construct the T zone, whereas the other conditions remained constant. The results are shown in Fig. 2b. With the increase of the concentration of the conjugate, the peak area of the T zone increases, and the color density almost remains stable when the concentration reaches 20 μM. Therefore, 20 μM streptavidin–biotin–DNA2 conjugate was chosen to construct the T zone.
The concentration of NaCl used during the preparation of the T and C zones has a great influence on the color shown on the zones after detection. Generally, on the one hand, if the concentration of salt is too high, the color seen on the strip is too dark. On the other hand, if the concentration of salt is too low, the strip cannot show a clear band due to the hydrophilicity of the NC membrane.51 The streptavidin–biotin–DNA2 conjugate and streptavidin were, respectively, prepared with different concentrations of NaCl in the range of 0–2 M, whereas the other experimental conditions remained unchanged. The relationship between the peak area of the T zone and the concentration of NaCl is shown in Fig. 2c. When the concentration of NaCl is lower than 0.5 M, the color band on the strip is divergent, and the peak area is reduced. When the concentration is higher than 0.5 M, obvious bands appear on the strip, and the increase of the peak area levels off. Thus, 0.5 M was chosen as the optimal concentration of NaCl to prepare the streptavidin–biotin–DNA2 conjugate and streptavidin.
The amount of AuNPs-apt and AgNPs-DNA1 applied on the conjugate pad is the key factor in the optical density of the bands on the T and C zones, and thus was investigated. Equal volumes of AuNPs-apt and AgNPs-DNA1 were first mixed and then different volumes of the mixture were applied on the conjugate pad. From Fig. 2d, it can be seen that the optical intensity of the T and C zones is proportional to the applied volume of the mixture of AuNPs-apt and AgNPs-DNA1, and substantially remains constant once the volume reaches 20 μL. Thus the optimal volume of AuNPs-apt and AgNPs-DNA1 applied on the conjugate pad is 20 μL.
Different types of NC membranes were used to prepare the strips and their influence on detection was investigated. Several frequently used NC membranes, including Sartorius CN140 (SCN140), Sartorius CN95 (SCN95), Vivid 170 (V170), Vivid 90 (V90) and Millipore 135 (M135), were employed to assemble the strips, and the color intensity of the T zone under the same experimental conditions was determined (Fig. 2e). Among them, the Millipore 135 membrane showed the best performance and was chosen to fabricate the strip biosensor.
The running buffer used in detection has a huge impact on the sensitivity and specificity, and therefore was also optimized. Five different running buffers (H2O; 10 mM phosphate buffer, pH 7.0; 10 mM phosphate buffer saline, pH 7.0; 10 mM Tris-HCl, pH 7.0; and 15×SSC (0.225 M sodium citrate containing 2.25 M NaCl, pH 7.0) containing 1% BSA) were selected as candidates. As shown in Fig. 2f, the most suitable running buffer is SSC containing 1% BSA.
The performance of the strip biosensor
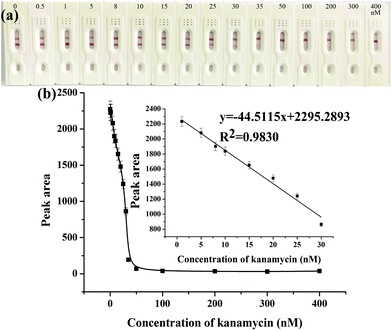
After the optimization of the above experimental parameters, the strip biosensor was fabricated under the optimal conditions and was used to detect kanamycin qualitatively by the naked eye or quantitatively by using a scanning reader.The sensitivity and detection limit of the strip biosensor were evaluated by using a series of kanamycin standard solutions within the concentration range of 0–400 nM. As shown in Fig. 3a, in the absence of kanamycin, a clear red color can be observed on both the T zone and the C zone. With the increase of the concentration of kanamycin, the color density of the C zone is basically unchanged, whereas the red band on the T zone is gradually shallower. As the concentration of kanamycin reaches 35 nM, the T zone completely fades. Therefore, the visual detection limit of the strip sensor is estimated to be 35 nM. Furthermore, after the determination of the optical density of the T zone, the relationship between the peak area of the T zone and the concentration of kanamycin was plotted and is shown in Fig. 3b. The peak area is gradually decreased with the increase of the concentration of kanamycin, and is no longer changed when the concentration of kanamycin is higher than 50 nM. This is in accordance with the observation. Within the concentration range of 1–30 nM, a linear relationship can be derived between the peak area and the concentration of kanamycin, with the linear regression equation y = −44.5115x + 2295.2893, R2 = 0.9830, where x represents the concentration of kanamycin (nM), and y represents the peak area of the T zone. The detection limit was estimated to be 0.0778 nM (S/N = 3), which indicates the high sensitivity of the strip biosensor.
To verify the accuracy of the strip biosensor, a widely recognized method in antibiotic detection, HPLC was carried out simultaneously. Different concentrations of kanamycin solution (3, 10 and 15 mM) were prepared and analyzed by using HPLC and the aptamer-based strip biosensor, respectively. The results are shown in Table 1. Both methods exhibit excellent accuracy. Although the RSD value of the strip is higher than that of HPLC, it is still good enough for general applications. Moreover, the sensitivity of the test strip is much higher than that of the HPLC method.
| Test strip biosensor | HPLC | |||
|---|---|---|---|---|
| Concentration of kanamycin (mM) | Average detection value (mM) | RSD (%) | Average detection value (mM) | RSD (%) |
| 3 | 3.09 | 2.08 | 2.95 | 0.59 |
| 10 | 10.12 | 4.41 | 10.05 | 0.25 |
| 15 | 14.91 | 3.78 | 15.03 | 0.32 |
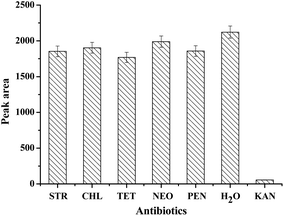
To evaluate the specificity of the prepared strip biosensor, it was challenged by a series of other antibiotics, including streptomycin (STR), chloramphenicol (CHL), tetracycline (TET), neomycin (NEO), penicillin (PEN) and water. The concentration of the antibiotics was fixed at 100 nM. All the detection conditions were the same, and the optical density of the T zone was determined and compared, as shown in Fig. 4. A significant fade on the T zone only appears in the presence of kanamycin. For other antibiotics, the peak area of the T zone shows only a slight decrease as compared to that of the control (water). Therefore, the strip biosensor is highly specific for kanamycin.
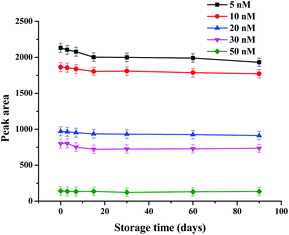
The stability of the strip biosensor was also checked. The strips were stored at 4 °C under dry conditions. They were used to detect different concentrations of kanamycin (5, 10, 20, 30 and 50 nM) after 3, 7, 15, 30, 60 and 90 days’ storage. The results are shown in Fig. 5. No significant change in the peak area occurs after storage for 3 months (the decrease of the peak area was within 0.73–8.37%), indicating satisfactory stability of the strip biosensor.
Detection of kanamycin in real samples
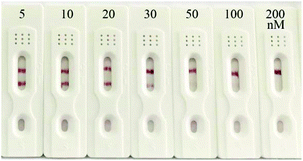
To evaluate the practicability of the prepared strip biosensor, artificially contaminated food samples, including honey, milk and milk powder, were analyzed by using the strips. Different concentrations of kanamycin (5, 10, 20, 30, 50, 100 and 200 nM) were introduced into kanamycin-free honey, and the honey samples were detected directly after dilution 10-fold with SSC. Then the samples were applied to the strips, and the results are shown in Fig. 6 (the results for milk and milk powder samples are shown in Fig. S2 and S3†). The visual detection limits for kanamycin in honey, milk and milk powder samples were estimated to be 50 nM, 100 nM and 200 nM, respectively. Thus the prepared strip biosensor can be used to detect actual food samples. And the detection limits are basically equivalent to those of the antibody-based strip for kanamycin.52 However, different matrices of the food samples do influence the sensitivity of detection to a certain extent. For example, the matrix of milk and milk powder can interfere in the detection, and the pretreatment can lead to a slight loss of the target. Thus the visual detection limits for milk and milk powder samples are higher than that of the standard solution. Nevertheless, the matrix of honey has a negligible effect on the detection. No pretreatment is required before detection, and the detection limit is even similar to that of the standard solution.Conclusions
In summary, a lateral flow strip biosensor for kanamycin was successfully developed employing aptamer-based molecular recognition and nanoparticle-based coloration and signal-amplification. After the optimization of key parameters, the strip exhibits excellent sensitivity, as well as high specificity and high stability. The operation of the detection is simple and can be completed in 10 min. Moreover, the strip biosensor can be applied to detect kanamycin in different food samples. Thus it may have great prospects in application scenarios where on-site detection is required.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the National Natural Science Foundation of China (no. 31271860) and the Fundamental Research Funds for the Central Universities (JUSRP51402A) for financial support of this work.References
- A. Kling, C. Chatelle, L. Armbrecht, E. Qelibari, J. Kieninger, C. Dincer, W. Weber and G. Urban, Anal. Chem., 2016, 88, 10036–10043 CrossRef CAS PubMed.
- Q. G. Liao, B. H. Wei and L. G. Luo, Microchim. Acta, 2016, 184, 627–632 CrossRef.
- R. Y. Robati, A. Arab, M. Ramezani, F. A. Langroodi, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2016, 82, 162–172 CrossRef CAS PubMed.
- R. Z. Li, Y. Liu, L. Cheng, C. Z. Yang and J. D. Zhang, Anal. Chem., 2014, 86, 9372–9375 CrossRef CAS PubMed.
- Q. G. Liao, B. H. Wei and L. G. Luo, Microchim. Acta, 2017, 184, 627–632 CrossRef CAS.
- C. Li, Y. Y. Zhang, S. A. Eremin, O. Yakup, G. Yao and X. Y. Zhang, Food Chem., 2017, 227, 48–54 CrossRef CAS PubMed.
- C. Z. Yu, Y. Z. He, G. N. Fu, H. Y. Xie and W. E. Gan, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 333–338 CrossRef CAS PubMed.
- C. Sheu, S. C. Chen and C. C. Lo, J. Environ. Sci. Health, Part B, 2010, 45, 478–484 CrossRef CAS PubMed.
- F. Fernandez, D. G. Pinacho, F. Sanchez-Baeza and M. P. Marco, J. Agric. Food Chem., 2011, 59, 5036–5043 CrossRef CAS PubMed.
- R. Liu, Z. H. Yang, Q. Guo, J. C. Zhao, J. Ma, Q. Kang, Y. F. Tang, Y. Xue, X. H. Lou and M. He, Electrochim. Acta, 2015, 182, 516–523 CrossRef CAS.
- Y. Zhu, P. Chandra, K. M. Song, C. Ban and Y. B. Shim, Biosens. Bioelectron., 2012, 36, 29–34 CrossRef CAS PubMed.
- H. Z. Wang, Y. Wang, S. Liu, J. H. Yu, Y. N. Guo, Y. Xu and J. D. Huang, Biosens. Bioelectron., 2016, 80, 471–476 CrossRef CAS PubMed.
- L. M. Hu, K. Luo, J. Xia, G. M. Xu, C. H. Wu, J. J. Han, G. G. Zhang, M. Liu and W. H. Lai, Biosens. Bioelectron., 2017, 91, 95–103 CrossRef CAS PubMed.
- B. H. Park, S. J. Oh, J. H. Jung, G. Choi, J. H. Seo, D. H. Kim, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2017, 91, 334–340 CrossRef CAS PubMed.
- C. M. Plotz and J. M. Singer, Am. J. Med., 1956, 21, 888–892 CrossRef CAS PubMed.
- W. J. Liu, M. F. Zhang, X. Y. Liu, A. Sharma and X. T. Ding, Biosens. Bioelectron., 2017, 96, 213–219 CrossRef CAS PubMed.
- Q. Zhang, Q. Y. Qu, S. S. Chen, X. W. Liu and P. W. Li, Food Chem., 2017, 231, 295–300 CrossRef CAS PubMed.
- Z. P. Liang, W. Z. Ha, Z. L. Xiao, H. T. Lei, Y. D. Shen, Y. M. Sun, H. Wang, J. Y. Yang and Z. L. Xu, Food Anal. Methods, 2017, 10, 2444–2453 CrossRef.
- T. Kunii, S. Ogura, M. Mie and E. Kobatake, Analyst, 2011, 136, 1310–1312 RSC.
- K. H. Leung, H. Z. He, D. S. H. Chan, W. C. Fu, C. H. Leung and D. L. Ma, Sens. Actuators, B, 2013, 177, 487–492 CrossRef CAS.
- X. Sun, F. L. Li, G. H. Shen, J. D. Huang and X. Y. Wang, Analyst, 2014, 139, 299–308 RSC.
- S. Cheng, B. Zheng, M. Z. Wang, M. H. W. Lam and X. W. Ge, Talanta, 2013, 115, 506–511 CrossRef CAS PubMed.
- W. H. Zhou, P. J. Huang, J. Ding and J. W. Liu, Analyst, 2014, 139, 2627–2640 RSC.
- Y. Y. Xu, T. Han, X. Q. Li, L. H. Sun, Y. J. Zhang and Y. S. Zhang, Anal. Chim. Acta, 2015, 891, 298–303 CrossRef CAS PubMed.
- J. J. Li, M. Xu, H. P. Huang, J. J. Zhou, E. S. Abdel-Halimb, J. R. Zhang and J. J. Zhu, Talanta, 2011, 85, 2113–2120 CrossRef CAS PubMed.
- K. M. Song, M. Cho, H. Jo, K. Min, S. H. Jeon, T. Kim, M. S. Han, J. K. Ku and C. Ban, Anal. Biochem., 2011, 415, 175–181 CrossRef CAS PubMed.
- C. S. Wang, C. Liu, J. B. Luo, Y. P. Tian and N. D. Zhou, Anal. Chim. Acta, 2016, 936, 75–82 CrossRef CAS PubMed.
- M. Chen, N. Gan, Y. Zhou, T. H. Li, Q. Xu, Y. T. Cao and Y. J. Chen, Sens. Actuators, B, 2017, 242, 1201–1209 CrossRef CAS.
- Y. Y. Xu, T. Han, X. Q. Li, L. H. Sun, Y. J. Zhang and Y. S. Zhang, Anal. Chim. Acta, 2015, 891, 298–303 CrossRef CAS PubMed.
- Y. Y. Xu, L. H. Sun, X. C. Huang, Y. Y. Sun and C. H. Lu, Anal. Methods, 2016, 8, 726–730 RSC.
- N. Cheng, Y. C. Xu, Y. B. Luo, L. Y. Zhu, Y. T. Zhang, K. L. Huang and W. T. Xu, Chem. Commun., 2017, 53, 4222–4225 RSC.
- G. D. Liu, X. Mao, J. A. Phillips, H. Xu, W. H. Tan and L. W. Zeng, Anal. Chem., 2009, 81, 10013–10018 CrossRef CAS PubMed.
- M. Jauset-Rubio, M. Svobodova, T. Mairal, C. McNeil, N. Keegan, M. S. El-Shahawi, A. S. Bashammakh, A. O. Alyoubi and C. K. O'Sullivan, Anal. Chem., 2016, 88, 10701–10709 CrossRef CAS PubMed.
- J. H. Chen, Z. Y. Fang, P. C. Lie and L. W. Zeng, Anal. Chem., 2012, 84, 6321–6325 CrossRef CAS PubMed.
- G. Y. Shen, S. B. Zhang and X. Hu, Clin. Biochem., 2013, 46, 1734–1738 CrossRef CAS PubMed.
- W. B. Shim, M. J. Kim, H. Mun and M. G. Kim, Biosens. Bioelectron., 2014, 62, 288–294 CrossRef CAS PubMed.
- L. B. Wang, W. W. Ma, W. Chen, L. Q. Liu, W. Ma, Y. Y. Zhu, L. G. Xu, H. Kuang and C. L. Xu, Biosens. Bioelectron., 2011, 26, 3059–3062 CrossRef CAS PubMed.
- Y. Gao, X. L. Deng, W. Wen, X. H. Zhang and S. F. Wang, Biosens. Bioelectron., 2017, 92, 529–535 CrossRef CAS PubMed.
- X. Mao, H. Xu, Q. X. Zeng, L. W. Zeng and G. D. Liu, Chem. Commun., 2009, 21, 3065–3067 RSC.
- L. B. Wang, W. Chen, W. W. Ma, L. Q. Liu, W. Ma, Y. Y. Zhao, Y. Zhu, L. G. Xu, H. Kuang and C. L. Xu, Chem. Commun., 2011, 47, 1574–1576 RSC.
- L. Yao, J. Teng, M. Y. Zhu, L. Zheng, Y. H. Zhong, G. D. Liu, F. Xue and W. Chen, Biosens. Bioelectron., 2016, 85, 331–336 CrossRef CAS PubMed.
- Y. L. Wang, L. M. Wang, J. J. Xue, J. B. Dong, J. Cai, X. D. Hua, M. H. Wang, C. Z. Zhang and F. Q. Liu, PLoS One, 2017, 12, 1–13 CAS.
- V. C. Ozalp, D. Cam, F. J. Hernandez, L. I. Hernandez, T. Schafer and H. A. Oktem, Analyst, 2016, 141, 2595–2599 RSC.
- C. Zhu, Y. Zhao, M. M. Yan, Y. F. Huang, J. Yan, W. H. Bai and A. L. Chen, Anal. Bioanal. Chem., 2016, 408, 4151–4158 CrossRef CAS PubMed.
- X. Mao, T. E. Du, Y. Y. Wang and L. L. Meng, Biosens. Bioelectron., 2015, 65, 390–396 CrossRef CAS PubMed.
- W. Wu, S. M. Zhao, Y. P. Mao, Z. Y. Fang, X. W. Lu and L. W. Zeng, Anal. Chim. Acta, 2015, 861, 62–68 CrossRef CAS PubMed.
- X. F. Gao, L. P. Xu, T. T. Wu, Y. Q. Wen, X. L. Ma and X. J. Zhang, Talanta, 2016, 146, 648–654 CrossRef CAS PubMed.
- W. T. Liu, H. Lai, R. Huang, C. T. Zhao, Y. M. Wang, X. C. Weng and X. Zhou, Biosens. Bioelectron., 2015, 68, 736–740 CrossRef CAS PubMed.
- W. L. Zhou, W. J. Kong, X. W. Dou, M. Zhao, Z. Ouyang and M. H. Yang, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2016, 1022, 102–108 CrossRef CAS PubMed.
- C. Y. Qin, W. Wen, X. H. Zhang, H. S. Gu and S. F. Wang, Analyst, 2015, 140, 7710–7717 RSC.
- T. M. Nargang, M. Runck, D. Helmer and B. E. Rapp, Eng. Life Sci., 2016, 16, 525–531 CrossRef.
- Y. Q. Chen, Z. Q. Wang, Z. H. Wang, S. S. Tang, Y. Zhu and X. L. Xiao, J. Agric. Food Chem., 2008, 56, 2944–2952 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental of detection of real samples and Fig. S1–S3. See DOI: 10.1039/c7an01476g |
| This journal is © The Royal Society of Chemistry 2018 |