Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Conformations and membrane-driven self-organization of rodlike fd virus particles on freestanding lipid membranes†
Anastasiia B.
Petrova‡
,
Christoph
Herold§
and
Eugene P.
Petrov
 *
*
Max Planck Institute of Biochemistry, Department of Cellular and Molecular Biophysics, 82152 Martinsried, Germany. E-mail: petrov@biochem.mpg.de
First published on 8th September 2017
Abstract
Membrane-mediated interactions and aggregation of colloidal particles adsorbed to responsive elastic membranes are challenging problems relevant for understanding the microscopic organization and dynamics of biological membranes. We experimentally study the behavior of rodlike semiflexible fd virus particles electrostatically adsorbed to freestanding cationic lipid membranes and find that their behavior can be controlled by tuning the membrane charge and ionic strength of the surrounding medium. Three distinct interaction regimes of rodlike virus particles with responsive elastic membranes can be observed. (i) A weakly charged freestanding cationic lipid bilayer in a low ionic strength medium represents a gentle quasi-2D substrate preserving the integrity, structure, and mechanical properties of the membrane-bound semiflexible fd virus, which under these conditions is characterized by a monomer length of 884 ± 4 nm and a persistence length of 2.5 ± 0.2 μm, in perfect agreement with its properties in bulk media. (ii) An increase in the membrane charge leads to the membrane-driven collapse of fd virus particles on freestanding lipid bilayers and lipid nanotubes into compact globules. (iii) When the membrane charge is low, and the mutual electrostatic repulsion of membrane-bound virus particles is screened to a considerable degree, membrane-driven self-organization of membrane-bound fd virus particles into long linear tip-to-tip aggregates showing dynamic self-assembly/disassembly and quasi-semiflexible behavior takes place. These observations are in perfect agreement with the results of recent theoretical and simulation studies predicting that membrane-mediated interactions can control the behavior of colloidal particles adsorbed on responsive elastic membranes.
1. Introduction
1.1. Membrane-mediated interactions of macromolecules and colloidal particles
Recently, membrane-mediated interactions of macromolecules and colloids bound to elastic responsive lipid membranes have attracted a lot of attention not only because they represent a challenging problem of soft-matter physics,1–4 but also because of their biological significance.5–7 Theoretical studies have demonstrated that local deformations of elastic membranes induced by the binding of colloids or macromolecules can result in attractive interactions capable of driving their clustering and self-organization.8–15 These ideas have been beautifully confirmed in simulation work, which demonstrated membrane-driven attraction and self-organization of spherical colloids on elastic membranes1,3,16–19 and elastic membrane nanotubes.20–22 A few published experimental studies23–27 addressing the behavior of spherical colloids on lipid or surfactant membranes show qualitative agreement with the theoretical and simulation work. The extended shape and bending flexibility of membrane-adsorbed particles can add new aspects to the membrane-driven self-organization. For example, local membrane deformations are expected to lead to a non-trivial behavior of a flexible polymer chain on an elastic lipid nanotube.28 Further, semiflexible chains bound to deformable elastic membranes may undergo membrane-driven collapse into compact globules, which has been demonstrated experimentally29,30 and explained theoretically.13 On the other hand, rigid rodlike particles are predicted31–39 to show membrane-driven self-organization controlled by the tension and curvature of the underlying membrane, which is believed to be relevant for understanding the biologically important issues of membrane budding and tubulation.33,35,36 Interestingly, qualitatively similar modes of behavior and self-organization have recently been observed for colloids at liquid–liquid or liquid–air interfaces—see, e.g., ref. 40–48. The important difference, however, is that interactions at interfaces are controlled by the surface tension, while membrane-mediated interactions are driven by the bending of the membrane surface.To the best of our knowledge, the aspects of membrane-mediated interactions of rodlike particles have not been investigated experimentally.
In the present paper, to address the effects of the interaction of semiflexible colloidal rods with a responsive elastic membrane, we experimentally study the behavior of filamentous virus fd electrostatically bound to freestanding cationic lipid bilayers and lipid nanotubes.
1.2. Filamentous virus fd
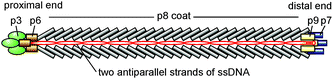
The filamentous phage fd belongs to the Ff species of filamentous bacterial viruses that infects the male strains of Escherichia coli carrying threadlike appendages called F-pili.49,50 The Ff species includes several virus strains, among which are f1, fd, M13, ZJ2, and AE2, that were isolated from sewage in the early and mid-1960s in New York;51,52 Heidelberg;53,54 Munich;55 Pangbourne, England;56 and Adelaide, Australia57—for a complete list of Ff strains, see ref. 58.It is one of the simplest organisms possible, as it consists of just a protein shell enclosing a circular single-stranded DNA of 6.4 kilobases50,59 that encodes five different types of shell proteins (Fig. 1). The capsid is mainly built from ∼2700 copies of the major coat protein p8 (encoded by gene 8), which is tightly packed to form a cylindrical shell around the circular single-stranded DNA molecule. The four minor coat proteins are present only as a few copies at the proximal and distal ends of the virus particle. As the length of the viral DNA molecule determines the size of the individual virion, filamentous virus particles feature a very narrow size distribution. The wild type virus was reported59 to have a length of about 880 nm and a diameter of 6.6 nm and show certain bending flexibility characterized by a persistence length of about 2 μm. Due to the presence of several negatively charged residues in the part of the major coat protein exposed to the environment (N-terminus), the outer surface of the protein shell of the filamentous virus is negatively charged.60,61
During the last few decades, filamentous phages have been successfully employed in cloning, phage display, nanotechnology, biosensing, and biomedicine.62–66 Recent research has convincingly demonstrated the potential of filamentous phages as a model system for soft matter physics.67
According to the canonical picture,49,50 the filamentous virus infection is initiated when the proximal end of a virion carrying the p3 protein adsorbs to the tip of an F-pilus, which, upon retraction, brings the phage to the cell surface. There, the viral protein coat disaggregates, and the viral DNA is released into the cell cytoplasm, while the coat protein associates with the cytoplasmic membrane. In spite of considerable research efforts, molecular-level details of viral coat disassembly at the cell membrane still remain elusive.
In infected cells, the filamentous virus is assembled at the cell membrane, and during the assembly, a virion is progressively extruded through it into the cell environment, so that its end distal to p3 exits the cell first.49,50 Under certain conditions, after the cylindrical shell of the p8 protein is fully constructed around the supercoiled DNA, the cap of the viral capsid may not be formed, and virion extrusion thus may proceed by further assembling the continuous cylindrical shell around the next copy of the viral DNA. In this case longer virus particles with lengths multiple of the normal virus length are extruded by cells. These multiple-length virus particles with a continuous capsid not showing any sharp bends or kinks have been reported54,56,57,68–71 for virtually all strains of Ff and were termed “polyphages”.70 The production of polyphage can be controlled by modifying the properties of both the host and virus. For example, polyphages with a length of up to ∼10 μm are much more readily formed if fd virus is grown in recA+ (as compared to recA−) E. coli strains.72 Alternatively, the probability of polyphage production is dramatically increased for mutant phages that lack a gene for the p3 protein required for the release of phages from infected cells; in this case, very long polyphages with lengths up to ∼20 μm can be formed.73
From the viewpoint of soft matter physics, polyphage particles are nothing but high-aspect ratio semiflexible rods with a diameter of 6.6 nm and contour lengths spanning the 0.88 μm to ∼10 μm range, and a persistence length of about 2 μm. The negative electric charge of virus particles should allow for their electrostatic binding to an oppositely charged surface. Furthermore, the virus can be fluorescently labeled,74 which facilitates fluorescence video-microscopy observations of single virions.
Thus, the filamentous virus polyphage interacting with freestanding cationic lipid bilayers represents a perfect experimental platform to study the behavior of rodlike semiflexible particles on elastically responsive lipid membranes, which would allow one to extend and complement the recent experimental and theoretical efforts aimed at understanding the behavior of semiflexible and rigid rodlike particles on locally deformable elastic membranes.13,29–39
1.3. Membrane-mediated interaction effects for semiflexible filaments
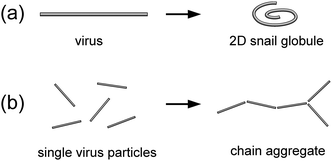
Depending on the bending flexibility of membrane-bound filamentous particles (or macromolecules), membrane-mediated interactions are expected to manifest themselves in a qualitatively different manner.On the one hand, as we have previously found in our experiments with DNA macromolecules adsorbed on freestanding cationic lipid bilayers,29,30 semiflexible chains bound to a responsive elastic membrane undergo membrane-driven collapse (coil–globule transition). Based on the experimental evidence, it was suggested29 that local membrane deformations due to electrostatic DNA binding are essential for this effect. The idea was further developed into a theoretical model13 that showed that local deformations of the membrane wrapping the DNA double helix induce membrane-mediated attraction between different segments of the membrane-adsorbed DNA polymer. This is expected to result in a membrane-driven coil–globule transition of the membrane-adsorbed macromolecule and its collapse into a dense 2D spiral (2D “snail globule”).13 This model agrees very well with the experimental observations and predicts that the membrane-driven intersegment attraction of a membrane-bound polymer can be strong enough, so that quite rigid polymers with a persistence length of up to a few micrometers can collapse into submicrometer-sized 2D globules. Thus, fd virus particles can serve as a model semiflexible polymer with high bending rigidity to test these predictions.
On the other hand, if membrane-mediated interactions are not strong enough to induce the collapse of adsorbed semiflexible filaments, they are expected to behave essentially as rigid rods. Simulation studies of rigid rodlike particles adhering to responsive elastic membranes3,31,34–36,38,39 have predicted that local membrane deformations induced by adhesion of these particles can lead to membrane-driven self-organization of the rodlike particles into linear chain aggregates. One can expect that this alternative scenario of membrane-induced effects resulting from local membrane deformations can also be explored using fd virus particles adhering to freestanding cationic lipid bilayers.
Therefore, the question naturally arises as to which of the two scenarios—membrane-driven collapse (Fig. 2a) or formation of linear chain aggregates (Fig. 2b)—will be realized for fd virus particles adsorbed on freestanding cationic lipid membranes.
1.4. Scope and structure of the paper
In the present paper, we experimentally study the interaction of monomer and polyphage fd virus particles with freestanding cationic lipid membranes mimicked by supergiant unilamellar vesicles75 and cationic lipid nanotubes.We find that fd virus readily binds to freestanding cationic lipid membranes and that the behavior of the membrane-bound filamentous phage is controlled by membrane-driven interactions and electrostatic effects.
We tune the strength of these effects by varying the membrane charge and ionic strength of the surrounding medium and find three strikingly different regimes of membrane-bound virus behavior:
(i) At low membrane charge (1 mol% cationic lipid) and low ionic strength (degassed deionized water, Debye screening length ∼1 μm), membrane-bound fd virus particles retain their integrity, dimensions, and mechanical properties, and behave as individual semiflexible filaments characterized by a virus monomer length of 884 ± 4 nm and a persistence length of 2.5 ± 0.2 μm (Section 3).
(ii) As the membrane-mediated attraction is enhanced by increasing the membrane charge, we observe that, upon binding to either a freestanding quasi-flat cationic lipid membrane or freely suspended cationic lipid nanotubes, virus particles undergo membrane-driven collapse into submicrometer-sized globules (Section 4).
(iii) When the electrostatic repulsion between fd virus particles bound to weakly charged freestanding cationic lipid membranes is screened by a surrounding medium with an ionic strength of ∼1 mM (Debye screening length ∼10 nm), membrane-mediated self-organization of virus particles takes place: membrane-bound rodlike particles form long tip-to-tip chain aggregates, which behave as semiflexible chains showing dynamic assembly–disassembly, conformational fluctuations, branching, and transient cyclization (Section 5).
2. Experimental
Here we briefly outline the experimental procedures employed in the present study. For a detailed description, see the ESI,† Section S1.Cationic supergiant unilamellar vesicles (SGUVs) with diameters >100 μm constituting a perfect model system to mimic freestanding lipid membranes were prepared by electroformation75 in degassed deionized water (ddH2O) from zwitterionic 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and cationic 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP). The cationic DOTAP lipid content was varied from 1 to 3 mol%, depending on the experiment. Under the conditions of our experiments, lipid bilayers with the above compositions are in the fluid state and are thus characterized by high lipid mobility and bending flexibility. Supported cationic lipid bilayers were prepared on freshly cleaved mica as described elsewhere.30 To facilitate fluorescence microscopy observations, red-emitting fluorescent lipid analog DiD was added to all lipid mixtures at a concentration of 0.1 mol%.
A stock sample of fd virus in 20 mM phosphate buffer, pH 7.5, predominantly contained monomeric virus particles. The sample also contained a small fraction of polyphage particles, which we denote here by fdm to reflect the relation of the polyphage length to that of the normal (monomeric) virus: Lm = mL1, where m = 2, 3, … is the multimerization degree. The amount of polyphage particles fdm with m ranging from 2 to 11 that could be found in the sample decreased sharply with the multimerization degree m. Virus particles were fluorescently labeled74 using Alexa488 green-emitting dye.
For experiments aimed at determination of the contour length and flexibility of phage particles adsorbed on weakly and moderately charged cationic lipid membranes (Sections 3 and 4), virus samples with an enriched content of polyphage particles were produced. These samples had a concentration of fd virus particles in the ∼10−12–10−13 M range and an ionic strength of the medium (I) of ∼10−7 M, which corresponds to a Debye screening length of ∼1 μm.
For experiments addressing the effects of the membrane-driven self-organization of virus particles adsorbed on weakly charged cationic bilayers (Section 5), two solutions of the monomer fd virus at a concentration of ∼10−12 M and higher ionic strengths I ∼ 10−4 M and I ∼ 10−3 M with the corresponding Debye screening lengths of ∼30 nm and ∼10 nm were produced.
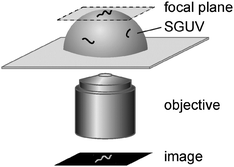
To study the interaction of fd virus with freestanding cationic lipid bilayers, virus solutions were carefully injected into chambers containing cationic SGUVs, upon which virus particles reached the membrane surface by Brownian diffusion and bound to the positively charged lipid bilayer via electrostatic attraction. By means of epifluorescence video-microscopy, we could image and track membrane-bound virus particles in the upper pole regions of SGUVs, as schematically depicted in Fig. 3. If necessary, contour lengths L, mean-square end-to-end distances 〈RE2〉, and mean-square radii of gyration 〈RG2〉 of individual virus particles were determined.
The same experimental approach was used to study the interaction of fd virus particles with moderately charged cationic lipid nanotubes. Lipid nanotubes featured a straight-line geometry and diameter not exceeding the optical resolution of our microscopy setup, and were suspended in solution at a distance of a few tens of micrometers above the coverslip.
In all experiments with freestanding cationic lipid membranes (SGUVs) and cationic lipid nanotubes, attachment of fd virus particles to the lipid bilayer was irreversible, irrespective of the conformations assumed by membrane-bound virus particles, suggesting the binding energy well in excess of the thermal energy kBT. This agrees with the estimate that can be obtained using the approach described in ref. 13, which gives ∼10kBT nm−1, or ∼104kBT for an fd virus monomer.
3. Dimensions and conformations of fd virus particles on weakly charged cationic lipid bilayers in a low ionic strength medium
In a few previous experimental studies, it has been found that fd virus particles do not interact with zwitterionic lipid bilayers either above or below their phase transition temperatures76–78—which, in our opinion, is not so surprising, taking into account the strong negative charge of the viral capsid of fd (a linear charge density of 10 e− nm−1 close to the neutral pH60). As a result, the behavior of fd virus on lipid membranes has not been previously explored: in particular, it is unclear whether, upon binding to a lipid membrane, the virus particle would preserve its integrity and its rodlike conformation, or membrane binding would immediately trigger the disassembly of the viral capsid via dissolution of the major coat protein in the lipid bilayer.The strong negative charge of fd virus uniformly spread over its capsid suggests that fd virions should bind to a positively charged lipid bilayer via electrostatic attraction. Inspired by our previous studies,29,30 we can expect that a weakly charged freestanding cationic lipid membrane would represent a gentle quasi-2D substrate preserving the structure and mechanical properties of fd virus particles.
To study the interaction of fd virus with weakly charged freestanding cationic lipid bilayers, we introduced a solution of fd virus in ddH2O with an enriched content of polyphage particles into a chamber with SGUVs containing 1 mol% cationic lipid. Under these experimental conditions, fd virus particles (both monomers and polyphages) readily bind to the surface of the cationic lipid membrane (Fig. 4).
 | ||
Fig. 4 A representative image of the top pole of an SGUV with electrostatically bound fd virus particles. Visible in the image are three fd monomers and two polyphages (dimer and trimer). Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 1. 1. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 1. | ||
Membrane-bound fd virus particles can be observed in a fluorescence microscope as semiflexible rods of various lengths performing translational and rotational Brownian motion and exhibiting conformational fluctuations, while being confined to the two-dimensional (2D) membrane surface (see Fig. 4, 5 and the ESI,† Movie 1). This dynamic behavior of the membrane-bound virus particles is expected to take place, because the lipid bilayer is in the fluid state.
The fluorescence microscopy images and movies show that the majority of the membrane-bound rodlike particles have a length of about 0.9 μm, which allowed us to identify them as fd virus monomers. We could also observe longer membrane-bound filaments with contour lengths ∼1.8 μm and above (up to ∼10 μm), with the longer particles being progressively less frequent. These longer virus particles behave as continuous elastic rods showing progressively more pronounced conformational fluctuations with increasing contour length (Fig. 5). Under these experimental conditions, membrane-bound virus particles do not show any tendency to either aggregate or fall apart into smaller subunits, which allows us to conclude that these longer semiflexible rods are individual polyphage particles.
3.1. Contour length of the membrane-bound fd virus
The contour length of fd virus has been previously determined by various experimental techniques to be about 880 nm either in bulk solution or upon deposition from solution onto various solid surfaces59,79–86 (for details on these experiments, see the ESI,† Section S2). Combining the results of these studies gives the following estimate of the contour length of fd virus monomer: L1 = 884 ± 6 nm.It is unknown, however, whether electrostatic binding to a lipid membrane affects the contour length of fd virus monomer.
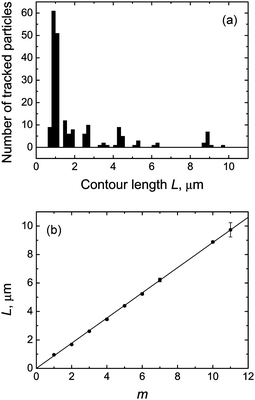
The histogram of contour lengths obtained from observations of individual particles shows pronounced peaks at ∼0.9 μm and approximate multiples of this value (Fig. 6a), thus confirming that the longer rodlike particles are indeed fd virus polyphages. One can clearly distinguish the peaks corresponding to polyphage particles with m = 2, 3, 4, 5, 6, 7, 10, and 11. (We believe that the absence of the particles with m = 8 and 9 in our observations is solely for statistical reasons.)
The contour length of fd virus monomer is obviously too short for its accurate direct determination by conventional fluorescence microscopy imaging (Fig. 5a), especially taking into account the Brownian motion of membrane-bound virus particles.
We therefore took advantage of the presence of polyphage particles in our samples, the contour lengths of which clearly show a linear dependence on the multimerization degree (Fig. 6b). The weighted least-squares fit of these data using the linear dependence Lm = mL1 gives the following value of the contour length of fd virus monomer: L1 = 884 ± 4 nm.
Thus, we find that the contour length of the fd virus monomer for virus particles electrostatically bound to a weakly charged freestanding cationic lipid membrane is in a remarkably good agreement with the values previously obtained for fd virus in bulk solvents and on solid surfaces using different experimental approaches. Furthermore, using a new experimental approach, we quantitatively determine the monomer length of the membrane-bound fd virus with an accuracy which is a factor of 3 to 18 better than achieved previously in electron microscopy and atomic force microscopy-based studies.59,79–84
3.2. Flexibility of the membrane-bound fd virus
Estimates of the persistence length lp of fd virus and the closely related M13 phage previously obtained using several experimental techniques87–92 lie in the 1.0 to 2.8 μm range. A critical analysis of these experiments (see the ESI,† Section S3) suggests that the most credible values of the virus persistence length are 2.0 ± 0.2 μm (fd),87 2.8 ± 0.7 μm (fd),89 and 2.2 ± 0.2 μm (M13).91The worm-like conformations of membrane-bound fd polyphage particles observed in our experiments (Fig. 5) qualitatively agree with the above lp estimates.
To quantitatively determine the persistence length of fd virus on lipid membranes, we again took advantage of the presence of polyphage particles in the sample, which allowed us to measure the mean-square end-to-end distance 〈RE2〉 and mean-square radius of gyration 〈RG2〉 of membrane-bound virus particles as a function of their contour length L (Fig. 7).
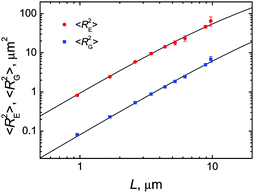 | ||
Fig. 7 Mean-square end-to-end distances 〈RE2〉 (circles) and mean-square radii of gyration 〈RG2〉 (squares) of fd virus particles electrostatically bound to weakly charged freestanding cationic lipid membranes as a function of their measured contour length. Weighted nonlinear least-squares fits to the corresponding expressions (1) and (2) for the 2D worm-like chain model, giving persistence length estimates of lp = 2.3 ± 0.2 μm (for 〈RE2〉) and 2.9 ± 0.3 μm (for 〈RG2〉), are shown with solid lines. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Surrounding medium: ddH2O. 1. Surrounding medium: ddH2O. | ||
To analyze our experimental data on 〈RE2〉 and 〈RG2〉 of membrane-bound fd virus particles, we used the 2D Kratky–Porod worm-like chain (WLC) model:93,94
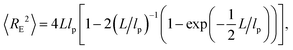 | (1) |
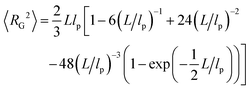 | (2) |
In the above expressions it is assumed that the persistence length lp of a semiflexible filament in 2D is related to its bending rigidity κ as follows: lp = κ/kBT (see, e.g., ref. 95), and can therefore be directly compared to the results of bulk measurements.
Weighted nonlinear least-squares fits of the 〈RE2〉 and 〈RG2〉 data using eqn (1) and (2) produced lp estimates of 2.3 ± 0.2 μm and 2.9 ± 0.3 μm, respectively. Taken together, these results give the following estimate of the persistence length of fd virus electrostatically bound to weakly charged freestanding cationic lipid membranes: lp = 2.5 ± 0.2 μm.
Thus, we find that the persistence length of fd virus electrostatically adsorbed on a weakly charged freestanding cationic lipid membrane is in a very good agreement with the most credible of the previously published values for fd and M13 phages in bulk aqueous solutions.
A question may arise on the effect of electrostatic interactions of fd virus with the oppositely charged membrane on the virus persistence length. The issue of the persistence length of a semiflexible polyelectrolyte electrostatically bound to an oppositely charged elastic responsive membrane is far from trivial—for discussion, see ref. 13. The fact that the persistence length of fd virus electrostatically bound to a weakly charged cationic lipid membrane is close to the corresponding values obtained using different techniques in bulk electrolyte solutions suggests that in our experiments the charge of the virus capsid is largely compensated by cationic lipid molecules capable of moving freely within the lipid membrane. Therefore, as long as the structure of the viral capsid remains intact, one should not anticipate any considerable effect of the membrane charge on the persistence length of the membrane-bound fd virus particles.
4. Membrane-driven collapse of fd virus particles
4.1. fd virus collapse on quasi-flat freestanding cationic lipid bilayers
To achieve a stronger adhesion of fd virus particles to the membrane, which should lead to more pronounced local membrane deformation responsible for membrane-driven interaction, we increase the membrane charge density while keeping the ionic strength of the surrounding medium low. We find that when the cationic lipid fraction exceeded 2 mol%, the observed picture changed dramatically: the membrane-bound virus particles, instead of behaving as individual semiflexible filaments, in this case assume completely or partially collapsed conformations (Fig. 8). Globules with typical sizes not exceeding a micrometer perform Brownian motion on the membrane and do not show a tendency to coalesce. The fraction of fully collapsed conformations was found to grow with the cationic lipid content in the composition of the freestanding cationic lipid membrane. | ||
Fig. 8 Representative image of fd virus particles on moderately charged freestanding cationic lipid membranes. Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. Surrounding medium: ddH2O. Scale bar: 10 μm. See also the ESI,† Movie 2. 3. Surrounding medium: ddH2O. Scale bar: 10 μm. See also the ESI,† Movie 2. | ||
The observed phenomenology is thus qualitatively very similar to the phenomenon of membrane-driven DNA collapse on freestanding cationic lipid membranes, which was reported and explained theoretically in our previous publications.13,29,30 Therefore, it seems plausible that the collapse of virus particles is a membrane-driven process taking place after the semiflexible rodlike virus particles electrostatically bind to the oppositely charged freestanding lipid membrane.
To verify whether local membrane deformations indeed play an important role in the collapse of membrane-bound virus particles, we made observations on the interaction of fd virus particles with mica-supported cationic lipid bilayers in the fluid state. The crucial difference between freestanding and supported lipid bilayers is their different ability to deform in response to the local perturbation: whereas a freestanding membrane easily deforms in response to a local perturbation, no such deformations are expected for a bilayer tightly adhering to a solid support. As a result, interactions mediated via membrane deformations should vanish for particles adsorbed on a supported bilayer. Indeed, we have found that fd virus particles bound to mica-supported cationic lipid bilayers (10 mol% cationic lipid) behaved as semiflexible filaments performing Brownian motion and conformational fluctuations, and never showed globule-like conformations (data not shown).
Single particle observations allowed us to reveal some details of the process of virus particle collapse on freestanding cationic lipid membranes.
A virus particle first attaches to the freestanding cationic lipid membrane and performs translational and rotational Brownian motion for a certain period of time, after which it may suddenly collapse into a compact globule (Fig. 9 and 10). In certain cases the collapse takes place spontaneously (Fig. 9) and starts at one of the ends of the virus particle (Fig. 9c), similar to what we have previously found for the membrane-driven collapse of DNA macromolecules.29,30 Sometimes the collapse is triggered by another membrane-bound virus particle (usually either a virus monomer or globule) that approaches one of the ends of the virus filament (Fig. 10).
 | ||
Fig. 9 Sequence of images demonstrating a spontaneous collapse of an fd2 polyphage bound to a freestanding cationic lipid membrane into a compact globule. Images correspond to the following time instants: −1.64 s (a), −0.44 (b), −0.11 (c), 0.00 (d), 0.11 (e), 0.44 (f), and 1.647 s (g). Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 3. 2. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 3. | ||
 | ||
Fig. 10 Sequence of images demonstrating a collapse of an fd4 polyphage bound to a freestanding cationic lipid membrane into a compact globule, which is triggered by an interaction of the polyphage end with a membrane-bound monomer virus particle. Images correspond to the following time instants: −4.70 s (a); −2.18 (b); −1.31 (c); −0.11 (d); 0.00 (e); 0.11 (f); and 1.31 s (g). Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 97![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 4. 3. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 4. | ||
We find that the contour lengths of the virus particles attached to the moderately charged bilayer prior to membrane-driven collapse agree within the experimental accuracy with the fd contour lengths on weakly charged cationic lipid bilayers (Section 3.1).
The rate of membrane-driven collapse of fd virus particles observed in our experiments ranges from ∼5 to ∼32 μm s−1, which is slower than, but comparable with the rate of compaction of DNA macromolecules on freestanding cationic lipid membranes30 (the rate of membrane-driven DNA compaction is 124 ± 46 kbp s−1, which, taking into account the effect of fluorescent labeling on the DNA contour length,96 amounts to ∼55 ± 20 μm s−1).
4.2. fd virus collapse on cationic lipid nanotubes
Freely suspended lipid nanotubes can be considered as an alternative variant of a freestanding membrane characterized by two principal curvatures, one of which is vanishing, while the other one is high. (This is in contrast to SGUVs which have a very low mean curvature and thus mimic a flat freestanding membrane.) To the best of our knowledge, interactions of colloidal particles and polymers with responsive lipid nanotubes have so far been addressed only via computer simulations.20–22,28We found that fd virus particles readily attach to moderately charged cationic lipid nanotubes (a cationic lipid fraction of 2 mol% and higher) via electrostatic attraction and assume a collapsed conformation (Fig. 11). A large fraction of these globules perform slow Brownian motion along the nanotube without mutual passage and thus show single-file diffusion; at the same time, some of the nanotube-bound globules are virtually immobile.
This observation of the nanotube-bound virus globules, however, does not answer the question whether the collapse of the virus particles takes place on the lipid nanotube, or the globules are first formed on the SGUVs and reach the vesicle-connected nanotubes via Brownian motion. An answer to this question can be provided if one could observe the fate of a solution-suspended virus particle after it approaches a cationic lipid nanotube close enough to be electrostatically attracted to it.
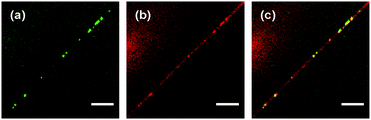
The sequence of images shown in Fig. 12 exemplifies such an event: an fd virus polyphage (most likely, m = 5) freely diffuses in the bulk solution a few micrometers above a cationic lipid nanotube (Fig. 12a–c). At some moment, the polyphage is suddenly attracted to the nanotube and immediately collapses into a globule (Fig. 12d), although the fine details of the collapse event are not available because it took place away from the focal plane of the microscope. The newly formed fd globule remains attached to the nanotube and performs Brownian motion on it (Fig. 12e–g).
 | ||
Fig. 12 Attachment of an fd virus polyphage particle to a cationic lipid nanotube followed by immediate polyphage collapse. Time interval between images: 1.20 s. Images (a)–(c): the freely diffusing polyphage is in focus, whereas the lipid nanotube is several microns away from the focal plane. Image (d): refocusing to the nanotube with the collapsed polyphage is in progress. Images (e)–(g): lipid nanotube with the attached fd polyphage globule is in the focal plane. Images represent the merge of the green (fd virus) and red (lipid nanotube) spectral channels. Fluorescent labels: Alexa488 (green, fd virus); DiD (red, lipid nanotube). Lipid composition: DOPC/DOTAP 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 5. 2. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movie 5. | ||
The formation of a tight virus globule may also occur not immediately, but rather only after some time upon attachment of a polyphage to a nanotube. We managed to follow a polyphage virus particle (m = 4) as it performed Brownian motion in the bulk medium and gradually approached a lipid nanotube, while staying approximately in the same XY-plane (Fig. 13a). When the distance between one of the ends of the virus particle and the nanotube becomes smaller than 1 μm (Fig. 13b), the polyphage quickly moves toward the lipid nanotube and attaches to it. Immediately after attachment the virus loses its stretched filament conformation, probably due to transient partial wrapping around the lipid nanotube (Fig. 13c), but after that gradually stretches itself along the nanotube (Fig. 13d and e). The length of this stretched conformation, however, is approximately twice smaller than the original length of the polyphage in the bulk solution. This conformation persists for ∼70 s, during which the virus performs translational Brownian motion along the lipid nanotube (Fig. 13f), until it encounters a small nanotube-bound virus globule (Fig. 13g). This triggers the final collapse process, which within ∼1 s transforms the polyphage particle into a compact globule (Fig. 13h and i). The newly formed virus globule then diffuses along the lipid nanotube (Fig. 13j).
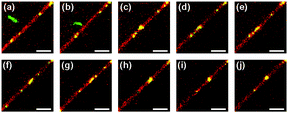 | ||
Fig. 13 Attachment of an fd virus polyphage particle to a cationic lipid nanotube and delayed polyphage collapse. Time zero is assigned to the event of attachment of the virus to the nanotube. Images correspond to the following time instants: −0.76 s (a), −0.11 (b), 0.00 (c), 0.11 (d), 0.76 (e), 69.22 (f), 71.95 (g), 72.50 (h), 73.04 (i), and 74.13 s (j). Images represent the merge of the green (fd virus) and red (lipid nanotube) spectral channels. Fluorescent labels: Alexa488 (green, fd virus); DiD (red, lipid nanotube). Lipid composition: DOPC/DOTAP 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movies 6 and 7. 2. Surrounding medium: ddH2O. Scale bar: 5 μm. See also the ESI,† Movies 6 and 7. | ||
The observed phenomenology of fd virus particle collapse on freestanding cationic lipid membranes, namely the initiation of collapse at an end of the virus particle, as well as the fact that no virus collapse is observed on solid-supported cationic lipid bilayers, is very similar to the behavior of much more flexible double-stranded DNA macromolecules on cationic lipid bilayers.13,29,30 This speaks in favor of the membrane-driven 2D snail mechanism13 of fd virus collapse that involves local membrane deformations produced upon adsorption of the charged virus on the oppositely charged lipid bilayer. The submicrometer size of the virus globules, small compared to the virus persistence length, agrees with the theoretical predictions of the membrane-driven13 and interface-driven48 compaction of adsorbed semiflexible filaments. Note that tight compaction of stiff filaments into toroids with diameters ∼5 times smaller than their persistence length has been previously observed under conditions of quasi-2D confinement.97
The picture of membrane-driven collapse may explain why some of the virus globules on lipid nanotubes show zero mobility: apparently, in these cases the virus particle collapse involves very strong deformations of the nanotube which results in its clogging, and thus suppresses the globule motion that would in this case require pushing all the fluid inside the nanotube. This explanation is supported by simulations of a flexible polymer interacting with a responsive deformable nanotube:28 in case of strong polymer adhesion, the nanotube suffers very strong deformations which in an experimental setting would have surely led to its clogging. Simulations of a polymer with a non-vanishing stiffness interacting with a deformable lipid nanotube could shed more light on the microscopic details of our observations.
4.3. Alternative scenario of membrane-driven fd virus collapse
Here we discuss a possible alternative scenario that may be responsible for the phenomenology observed in the present section.One should keep in mind that fd virus—as well as all other strains of the Ff virus species—is not a polymer in the strict sense, but rather a supramolecular assembly of coat protein subunits tightly arranged around a DNA chain.50,59 The strongly elongated molecules of the major coat protein p8 are self-assembled in a densely-packed sheath around the single-stranded DNA molecule at a small angle (13–20°) with respect to the virion axis.98
Interaction of the filamentous virus with the lipid bilayer membrane is expected to play an important role in the early stage of the infection process: in particular, it is believed that this process involves dissolution of the major coat protein in the cell membrane.49,50 In this context, the behavior of the major coat protein molecules in lipid bilayers has been addressed both experimentally and via simulations.99,100 At the same time it still remains an open question as to how the major coat protein is incorporated in the lipid membrane in vivo during the virus entry, especially that the Ff virus capsid is known to be extremely stable: it does not show any interaction with zwitterionic lipid bilayers,76–78 and can only be disrupted by strong detergents.78,101,102
On the other hand, it has been found that filamentous virus particles can undergo a strong morphological change and contract dramatically upon either treatment by ether (Pf1 phage)103,104 or interaction with the chloroform–water interface (fd, f1, and M13 phages)105–108 to form hollow spheroids (termed S-forms) with a diameter of ∼40 nm. The conversion of the virus filament to the S-form was found106,107 to take place via an intermediate structure (termed an I-form) having the shape of a cylinder with a length of ∼250 nm and a diameter of ∼15 nm. It was suggested103,104 that the transition from the filament to spheroid involves a change in the orientation of the p8 subunits from roughly parallel to the virus axis to roughly radial and may thus mimic events taking place during fd penetration into the cell.101,106–109 Based on the fact that, upon contraction into the S-form, the virus particle retains all the capsid protein, but at the same time approximately two-thirds of the viral DNA molecule is ejected outside,105 an idea was put forward that the in vitro S-form may reflect an intermediate stage of phage penetration in vivo.106,108 It should be noted, however, that, to the best of our knowledge, no evidence of the formation of S-form-like structures in vivo has been found in relevant electron microscopy-based studies.68,110–112
The question therefore arises regarding an alternative explanation for our experimental findings: namely, whether the fd virus collapse on freestanding cationic lipid membranes is related to the molecular reorganization of the viral capsid into the I- and S-forms—especially in view of certain similarities between the chloroform–water interface and the freestanding cationic lipid bilayer. Indeed, as has been found in molecular dynamics simulations,113 the orientation of chloroform and water molecules creates a uniform electric field across the interface with the positive end of the dipole pointing toward the bulk aqueous phase. As a result, electrostatic attraction of the negatively charged virus particle in the aqueous phase toward the interface should take place, similar to the attraction of the virus to the cationic lipid bilayer. Further, both the interface and freestanding cationic lipid bilayer can deform around the adhering virus particle. It is difficult, though, to come to any definite conclusions based on these properties, since the molecular mechanisms of virus contraction into the I- and S-forms by the chloroform–water interface are not known.
Previously it has been found76–78 that, while an intact filamentous virus shows no interaction with non-charged (zwitterionic) lipid bilayers, the contracted I- and S-forms of the virus are disrupted upon interaction with the same lipid membranes, most likely via incorporation of the major coat protein into the bilayer. On the other hand, in our experiments we observe that the virus in its native filamentous form readily binds to cationic lipid bilayers, and, if the membrane charge is high enough, undergoes, upon binding to the membrane, a membrane-driven collapse into a submicrometer-sized globule. We find, however, that membrane-bound virus particles in either native filamentous or collapsed globular conformations do not show any signs of capsid dissolution in the lipid bilayer.
Thus, at the moment one can only speculate whether the phenomenon of the membrane-driven compaction of the filamentous virus by cationic lipid bilayers observed in the present study may be relevant for understanding the initial stages of the phage penetration in vivo.
5. Membrane-mediated self-organization of fd virus particles
We have found that fd virus particles electrostatically adsorbed on weakly charged freestanding cationic lipid membranes in media of low ionic strength behave as individual 2D semiflexible filaments (Section 3); that is, membrane-mediated intra- and interparticle attraction in this case is negligible. On the other hand, in the case of virus particles adsorbed on moderately and strongly charged freestanding cationic lipid bilayers in media of very low ionic strength, membrane-mediated interactions are already strong enough to induce the collapse of virus particles into submicrometer-sized globules (Section 4). Therefore it would be interesting to carry out experiments in an intermediate interaction regime where, on the one hand, membrane-adsorbed virus particles would retain their filamentous conformations, but, on the other hand, interparticle membrane-mediated interactions are already significant enough, in which case membrane-adsorbed rodlike particles are expected to form chain aggregates.3,31,34–36,38,39One reason why no membrane-mediated interparticle attraction was observed for the virus particles on weakly charged freestanding cationic lipid membranes (Section 3) could be the unscreened electrostatic repulsion between the membrane-adsorbed virus particles in a medium with a very low ionic strength (dd-H2O) with a Debye screening length of ∼1 μm. Therefore, to avoid the membrane-driven collapse of membrane-adsorbed virus particles and to preserve their rodlike conformations, and at the same time to reduce their electrostatic repulsion, we carried out experiments on the interaction of fd virus particles with weakly charged freestanding cationic lipid membranes (1 mol% cationic lipid) and tuned the interparticle interactions by increasing the ionic strength of the surrounding medium. The electrostatic screening is not expected to affect the bending rigidity of the cationic membrane used in our experiment. According to our unpublished observations,114 the bending rigidity of the cationic DOPC/DOTAP bilayer with 1 mol% cationic lipid is the same as the one of the zwitterionic DOPC bilayer, which agrees with the results of previous experimental studies of the charge effect on the bending rigidity of lipid membranes (see ref. 115 for review). Furthermore, according to recent experimental evidence, salt solutions at (sub)millimolar concentrations are not expected to influence the mechanical properties of the bilayer.115,116
An increase in the ionic strength of the medium to ∼10−4 M, which corresponds to a Debye screening length of ∼30 nm, did not lead to observable changes in the behavior of fd virus particles adsorbed to a weakly charged freestanding cationic lipid bilayer compared to what has been reported in Section 3: under these conditions the membrane-bound fd virus essentially behaves as individual rodlike particles performing independent Brownian motion on the membrane surface.
When, however, the ionic strength is further increased to ∼10−3 M, which corresponds to a smaller Debye screening length of ∼10 nm and thus should further screen the electrostatic repulsion of membrane-bound virus particles, the situation changes dramatically: under these conditions we observe that the surface of the cationic SGUV membranes in the observation chamber is covered with abnormally long virus filaments frequently featuring lengths in the 10–20 μm range (Fig. 14). Sometimes these filaments form Y-shaped branched structures, like the one shown in Fig. 14d (no higher-order junctions were observed in our experiments). These long membrane-bound filaments are very dynamic: not only do they show pronounced Brownian motion and shape fluctuations, but they are continuously involved in the process of dynamic assembly and disassembly, whereby shorter filaments may join together to form a longer one, or, on the opposite, a longer filament may fall apart in several shorter ones (see the ESI,† Movies 8–11).
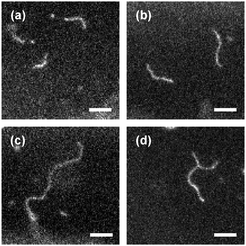 | ||
Fig. 14 Representative images of long filamentous aggregates formed by fd virus particles electrostatically bound to a weakly charged freestanding cationic lipid bilayer in a medium with an ionic strength of ∼10−3 M (Debye screening length ∼10 nm). Panels (a)–(d) correspond to four different SGUVs and are taken at different time moments. Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Scale bar: 5 μm. See also the ESI,† Movies 8–11. 1. Scale bar: 5 μm. See also the ESI,† Movies 8–11. | ||
To interpret the observed phenomenology, we first note that the experiment is carried out using a sample where polyphage particles are barely present. Second, these long membrane-bound filaments show dynamic self-assembly and disassembly – the behavior which has never been observed for polyphage particles either in our experiments (Section 3) or in previously published reports. Based on these observations we conclude that in this case we deal with the membrane-driven self-organization of monomeric fd virus particles into linear tip-to-tip chain aggregates.
The very long linear aggregates are metastable: they are self-assembled from shorter filaments and after a certain time interval may fall apart into several fragments. For example, the very long filamentous aggregate shown in Fig. 14c was originally formed by two shorter filaments joining together, and, after existing for ∼3 min, disassembled into three shorter fragments. This dynamic assembly–disassembly suggests that the energy of membrane-mediated attraction between the virus particles in the aggregate is of the order of the thermal energy kBT.
The fd linear aggregates generally behave very similar to semiflexible chains (Fig. 14) and show pronounced shape fluctuations. There is still, however, one important difference: although being semiflexible to a certain extent, the filaments are brittle. A filament may easily break up into fragments at a position where, as a result of Brownian shape fluctuations, it assumes a high local curvature (Fig. 15a–f). If after that the ends of the fragments stay close to each other, the filament may reassemble. For this to occur, however, it is not enough for the ends of the fragments to just come close enough (Fig. 15h and i): they should also be oriented approximately along the same line (Fig. 15j).
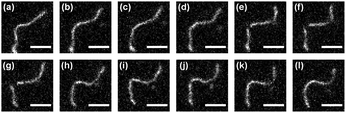 | ||
Fig. 15 Disassembly and reassembly of a long linear fd virus aggregate on a weakly charged freestanding cationic lipid membrane in a medium with an ionic strength of ∼10−3 M (Debye screening length ∼10 nm). The image sequence exemplifies the break-up of a long filament at a position with a high local curvature (a–f) and its subsequent reassembly which requires alignment of the ends of the filaments (g–l). Time interval between images: 0.33 s. Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Scale bar: 5 μm. See also the ESI,† Movie 12. 1. Scale bar: 5 μm. See also the ESI,† Movie 12. | ||
Although the linear chain aggregates of membrane-bound fd virus particles are not semiflexible in the strict sense, it nevertheless would be interesting to estimate their stiffness. Using images of several long aggregates, we estimated their persistence length lpA based on the internal mean-square end-to-end distance along the chain using the 2D WLC model. We find that lpA = 2.5 ± 0.5 μm, essentially the same as the persistence of fd virus itself (Section 3), which means that the membrane deformation-induced elastic bond between neighboring fd virus rods in the chain aggregate has a bending stiffness close to that of the virus particle.
Thus, our experimental observations of linear chain aggregates of membrane-bound virus particles showing dynamic self-assembly and disassembly and semiflexible behavior agree with the simulation work.3,31,34–36,38,39 The simulation studies predict membrane-driven self-organization of nanorods and rodlike proteins bound to responsive elastic membranes into linear tip-to-tip aggregates with interparticle attraction energies of a few kBT (see ref. 36 and 39) and semiflexible chain behavior.31,34–36,39 Similar to our experimental observations, the persistence length of these linear tip-to-tip aggregates was estimated to be in the micrometer range.31 It is worth noting that simulations have predicted that linear chain aggregates can also be formed by spherical particles bound to deformable membranes.3,18,19,36 The energetics of membrane-mediated bead interactions19 suggests that the persistence length of these linear aggregates should also lie in the range of a few micrometers.
The Y-shaped branched structures, like the one presented in Fig. 14d, are considerably less frequent than unbranched aggregates. Similar to very long filaments, Y-junctions are also metastable, with the lifetime ranging from a fraction of a second to about a minute. For example, the Y-junction structure shown in Fig. 14d, which was created upon attachment of a shorter filament to a longer one, existed for ∼50 s, after which it eventually fell apart into two filaments. Interestingly, the membrane-mediated formation of Y-junctions of linear aggregates of colloids adsorbed on elastic membranes has been reported in simulation studies of rodlike particles31,34–36,38 and spherical beads.1,3,18,19,34,36
Based on our observations we found some regularities governing the assembly and disassembly of Y-shaped filament junctions, which are illustrated by a set of images presented in Fig. 16 (see also the ESI,† Movie 13). In particular, we observe that, for a Y-junction to be formed, two filaments should not only approach one another close enough (Fig. 16a), but one of the participating filaments should be bent, and the tip of the other partner filament has to approach the bent filament from the outer side of the bend (Fig. 16b and c). Once the Y-junction is formed (Fig. 16d), the structure usually remains stable as long as all its three angles are reasonably close to 120° (Fig. 16e–g). However, when, as a result of Brownian shape fluctuations, an angle between two sides of the Y-junction opens up to approach ∼180° (Fig. 16h), the structure becomes unstable. As a result, the filament making the third side of the structure is very likely to detach (Fig. 16i and j), in which case the Y-junction ceases to exist. Thus, depending on a particular sequence of conformations the structure assumes, a “switching” event may take place, whereby a filament attaching with its tip to a bent partner filament to form a Y-junction, may, upon a break-up of the Y-junction, become a part of a newly formed longer filament, thus excluding a part of the original bent filament (Fig. 16).
 | ||
Fig. 16 Example of the assembly (a)–(e) and disassembly (f)–(j) of a Y-shaped branched aggregate of fd virus particles on a weakly charged freestanding cationic lipid membrane in a medium with an ionic strength of ∼10−3 M (Debye screening length ∼10 nm). Time interval between images in sequences (a)–(e) and (f)–(j): 0.22 s. Time interval between images (e) and (f): 0.87 s. Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Scale bar: 5 μm. See also the ESI,† Movie 13. 1. Scale bar: 5 μm. See also the ESI,† Movie 13. | ||
When a linear chain aggregate is long enough, Brownian shape fluctuations may lead to the formation of a closed loop (cyclization). Because of the dynamic character of the virus aggregates, looping is expected to have a transient character. An example of such an event is shown in Fig. 17. Here an fd virus linear chain aggregate, strongly bent as a result of conformational fluctuations (Fig. 17a), forms a closed loop (Fig. 17b). This closed-loop conformation persists for ∼1 s (Fig. 17b and e), after which the loop opens (Fig. 17f), albeit at a different position, and transforms into a bent linear filament (Fig. 17g). This observation gives another clear piece of evidence that the linear aggregates consist of individual membrane-bound fd virus particles with attraction energies of the order of kBT.
 | ||
Fig. 17 Spontaneous looping and loop-opening of a linear fd virus aggregate on a weakly charged freestanding cationic lipid membrane in a medium with an ionic strength of ∼10−3 M (Debye screening length ∼10 nm). Time interval between images: 0.33 s. Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Scale bar: 5 μm. See also the ESI,† Movie 14. 1. Scale bar: 5 μm. See also the ESI,† Movie 14. | ||
It is noteworthy that at the moments when the loop is formed and opened, its shape, rather than being circular, is tear-drop-like (Fig. 17b and f). This is in perfect agreement with the theoretical predictions for the shape of a closed loop formed by a semiflexible filament which would have the lowest bending energy.117,118 Using the contour length of the linear chain aggregate in Fig. 17, L ≈ 8.8 μm, and the above estimate of the aggregate persistence length, lpA ≈ 2.5 μm, we get the estimate of the formation energy of such a tear-drop-shaped loop117 as Eteardrop/kBT = 14.05(lpA/L) ≈ 4.0. (For comparison, in order to form a circular loop, the energy Ecircle/kBT = 2π2(lpA/L) ≈ 5.6 would be required, which makes the formation of a strictly circular loop much less likely.) Interestingly, the ratio of the chain contour length to its persistence length L/lpA ≈ 3.5 appears to be close to the optimum value for forming a tear-drop-shaped loop.119,120 Looping for shorter and longer chains is expected to be much less probable: for shorter contour lengths, the looping probability drops sharply because of the 1/L divergence of the loop energy, while for longer filaments the reduced probability of the ends finding each other is additionally strongly suppressed121 by self-avoidance in 2D.
As has been mentioned above, there is a dissymmetry between the proximal and distal ends of fd virus:50,59 the corresponding end caps are formed by p3, p6 and p7, p9 proteins, respectively. Therefore one may ask a question whether the formation of tip-to-tip aggregates of fd virus particles on the membrane is triggered by an attractive interaction of the proximal and distal ends of the neighboring virus particles, rather than being membrane-driven. If this is the case, then a fragment detached from a tip-to-tip chain aggregate should not reattach with its opposite end to its original attachment point. In our experiments, we observed several events where a portion of a long membrane-bound tip-to-tip chain aggregate first detached, then, in the course of its Brownian motion on the membrane, made a half-turn rotation, after which it reattached with its opposite end to its original attachment point and again formed a continuous long chain aggregate (Fig. 18). These observations allow us to conclude that the “polarity” of fd virus does not play an important role in the formation of linear chain aggregates of fd virus particles on the membrane and speaks strongly in favor of the membrane-driven mechanism of fd virus particle aggregation.
 | ||
Fig. 18 Detachment and opposite-end-reattachment of a fragment of a linear fd virus aggregate on a weakly charged freestanding cationic lipid membrane in a medium with an ionic strength of ∼10−3 M (Debye screening length ∼10 nm). Time interval between images: 0.22 s. Fluorescent label: Alexa488. Lipid composition: DOPC/DOTAP 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Scale bar: 5 μm. See also the ESI,† Movie 15. 1. Scale bar: 5 μm. See also the ESI,† Movie 15. | ||
The behavior we observe here for membrane-bound rodlike fd virus particles qualitatively reminds the phenomenology observed for rodlike colloidal particles at interfaces which tend to form long linear chain aggregates—see, e.g., ref. 40–43 for experimental observations and ref. 44–47 for theoretical treatment and simulations. The important difference, however, is that in these systems based on liquid–liquid or liquid–air interfaces the interactions of particles are controlled by the surface tension, while the self-organization of particles on lipid membranes is driven by the bending of the membrane surface. Furthermore, linear chains of micron-sized rodlike particles assembled by capillary forces on interfaces are typically very stiff and are characterized by a persistence length of the order of a meter.47
6. Conclusions
In this paper we studied, using single-particle fluorescence video-microscopy, the interaction of rodlike semiflexible fd virus particles with freestanding cationic lipid membranes. We found that fd virus particles can readily bind to cationic lipid bilayers via electrostatic interactions and do not show any signs of capsid dissolution in the lipid bilayer.By varying the membrane charge and ionic strength of the surrounding medium, we tuned the character of the interactions of fd virus with cationic lipid membranes, which allowed us to distinguish three different regimes of the behavior of membrane-adsorbed fd virus particles.
First, we found that a weakly charged freestanding cationic lipid bilayer in a low ionic strength medium represents a gentle quasi-2D substrate preserving the integrity, structure and mechanical properties of fd virus particles. Under these conditions, membrane-bound fd virus particles behave as individual semiflexible filaments. Analysis of single-particle conformations of membrane-bound fd polyphages allowed us to determine the length of the monomer fd virus, L1 = 884 ± 4 nm, and its persistence length, lp = 2.5 ± 0.2 μm, which are in a very good agreement with the results previously reported for fd virus in bulk media.
Second, when the membrane charge is increased, filamentous virus particles, upon binding to moderately charged freestanding cationic lipid bilayers and nanotubes, undergo membrane-driven collapse and form submicrometer-sized globules that remain attached to the bilayer or lipid nanotubes. Whether this membrane-driven collapse involves folding of the virus filament into a 2D globule conformation or proceeds via rearrangement of the major coat protein molecules in the viral capsid, or both mechanisms take place simultaneously, remains the subject of future studies.
Third, when the membrane charge is low, and mutual repulsion of membrane-bound virus particles is screened by a medium with a non-negligible ionic strength, we observed membrane-driven self-organization of membrane-bound fd virus particles into linear tip-to-tip aggregates. These aggregates, consisting of monomer fd virus particles electrostatically adsorbed to a freestanding cationic lipid membrane, exhibit a quasi-semiflexible behavior and are highly dynamic, which suggests that the interparticle attraction is of the order of the thermal energy.
To summarize, we have experimentally demonstrated that the behavior of colloidal particles adsorbed on responsive elastic membranes can be controlled by membrane-mediated interactions, which is in perfect agreement with the conclusions of recent theoretical and simulation-based studies. We believe that our results can serve as a springboard for future experimental and simulation efforts aimed at deeper understanding the physics of the interactions of colloids and polymers with elastic lipid membranes.
Conflicts of interest
There are no conflicts of interests to declare.Acknowledgements
The work was supported by the Deutsche Forschungsgemeinschaft via the Research Unit FOR 877 ‘From Local Constraints to Macroscopic Transport’. E. P. P. greatly appreciates fruitful and stimulating discussions with Dr Andrey Cherstvy (University of Potsdam). Open Access funding provided by the Max Planck Society.References
- A. Šarić and A. Cacciuto, Self-assembly of nanoparticles adsorbed on fluid and elastic membranes, Soft Matter, 2013, 9, 6677 RSC.
- C. Yolcu, R. C. Haussman and M. Deserno, The effective field theory approach towards membrane-mediated interactions between particles, Adv. Colloid Interface Sci., 2014, 208, 89 CrossRef CAS PubMed.
- M. Laradji, P. B. S. Kumar and E. J. Spangler, Exploring large-scale phenomena in composite membranes through an efficient implicit-solvent model, J. Phys. D: Appl. Phys., 2016, 49, 293001 CrossRef.
- F. Tian, T. Yue, Y. Li and X. Zhang, Simulations of cell uptake of nanoparticles: Membrane-mediated interaction, internalization pathways, and cooperative effect, in Self-Assembling Systems: Theory and Simulation, ed. L.-T. Yan, Wiley, Chichester, 2016, pp. 208–229 Search PubMed.
- M. Simunovic, G. A. Voth, A. Callan-Jones and P. Bassereau, When physics takes over: BAR proteins and membrane curvature, Trends Cell Biol., 2015, 25, 780 CrossRef CAS PubMed.
- H.-m. Ding and Y.-q. Ma, Theoretical and computational investigations of nanoparticle–biomembrane interactions in cellular delivery, Small, 2015, 11, 1055 CrossRef CAS PubMed.
- E. Rascol, J.-M. Devoisselle and J. Chopineau, The relevance of membrane models to understand nanoparticles–cell membrane interactions, Nanoscale, 2016, 8, 4780 RSC.
- K. S. Kim, J. Neu and G. Oster, Curvature-mediated interactions between membrane proteins, Biophys. J., 1998, 75, 2274 CrossRef CAS PubMed.
- A. R. Evans, M. S. Turner and P. Sens, Interactions between proteins bound to biomembranes, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2003, 67, 041907 CrossRef CAS PubMed.
- T. Auth and G. Gompper, Budding and vesiculation induced by conical membrane inclusions, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 80, 031901 CrossRef PubMed.
- A. H. Bahrami, R. Lipowsky and T. R. Weikl, Tubulation and aggregation of spherical nanoparticles adsorbed on vesicles, Phys. Rev. Lett., 2012, 109, 188102 CrossRef PubMed.
- S. Dasgupta, T. Auth and G. Gompper, Wrapping of ellipsoidal nanoparticles by fluid membranes, Soft Matter, 2013, 9, 5473 RSC.
- A. G. Cherstvy and E. P. Petrov, Modeling DNA condensation on freestanding cationic lipid membranes, Phys. Chem. Chem. Phys., 2014, 16, 2020 RSC.
- H. Tang, H. Ye, H. Zhang and Y. Zheng, Wrapping of nanoparticles by the cell membrane: the role of interactions between the nanoparticles, Soft Matter, 2015, 11, 8674 RSC.
- J. Agudo-Canalejo and R. Lipowsky, Uniform and Janus-like nanoparticles in contact with vesicles: energy landscapes and curvature-induced forces, Soft Matter, 2017, 13, 2155 RSC.
- B. J. Reynwar, G. Illya, V. A. Harmandaris, M. M. Müller, K. Kremer and M. Deserno, Aggregation and vesiculation of membrane proteins by curvature-mediated interactions, Nature, 2007, 447, 461 CrossRef CAS PubMed.
- A. Šarić and A. Cacciuto, Particle self-assembly on soft elastic shells, Soft Matter, 2011, 7, 1874 RSC.
- A. Šarić and A. Cacciuto, Soft elastic surfaces as a platform for particle self-assembly, Soft Matter, 2011, 7, 8324 RSC.
- A. Šarić and A. Cacciuto, Fluid membranes can drive linear aggregation of adsorbed spherical nanoparticles, Phys. Rev. Lett., 2012, 108, 118101 CrossRef PubMed.
- J. C. Pàmies and A. Cacciuto, Reshaping elastic nanotubes via self-assembly of surface-adhesive nanoparticles, Phys. Rev. Lett., 2011, 106, 045702 CrossRef PubMed.
- J. A. Napoli, A. Šarić and A. Cacciuto, Collapsing nanoparticle-laden nanotubes, Soft Matter, 2013, 9, 8881 RSC.
- A. Vahid and T. Idema, Pointlike inclusion interactions in tubular membranes, Phys. Rev. Lett., 2016, 117, 138102 CrossRef PubMed.
- I. Koltover, J. O. Rädler and C. R. Safinya, Membrane mediated attraction and ordered aggregation of colloidal particles bound to giant phospholipid vesicles, Phys. Rev. Lett., 1999, 82, 1991 CrossRef CAS.
- L. Ramos, T. C. Lubensky, N. Dan, P. Nelson and D. A. Weitz, Surfactant-mediated two-dimensional crystallization of colloidal crystals, Science, 1999, 286, 2325 CrossRef CAS PubMed.
- R. Sarfati and E. R. Dufresne, Long-range attraction of particles adhered to lipid vesicles, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2016, 94, 012604 CrossRef PubMed.
- C. van der Wel, A. Vahid, A. Šarić, T. Idema, D. Heinrich and D. J. Kraft, Lipid membrane-mediated attraction between curvature inducing objects, Sci. Rep., 2016, 6, 32825 CrossRef CAS PubMed.
- N. Li, N. Sahrifi-Mood, F. Tu, D. Lee, R. Radhakrishnan, T. Baumgart and K. J. Stebe, Curvature-driven migration of colloids on tense lipid bilayers, Langmuir, 2017, 33, 600 CrossRef CAS PubMed.
- A. Šarić and A. Cacciuto, Effective elasticity of a flexible filament bound to a deformable cylindrical surface, Phys. Rev. Lett., 2010, 104, 226101 CrossRef PubMed.
- C. Herold, P. Schwille and E. P. Petrov, DNA condensation at freestanding cationic lipid bilayers, Phys. Rev. Lett., 2010, 104, 148102 CrossRef CAS PubMed.
- C. Herold, P. Schwille and E. P. Petrov, Single DNA molecules on freestanding and supported cationic lipid bilayers: diverse conformational dynamics controlled by the local bilayer properties, J. Phys. D: Appl. Phys., 2016, 49, 074001 CrossRef.
- P. G. Dommersnes and J.-B. Fournier, N-body study of anisotropic membrane inclusions: Membrane mediated interactions and ordered aggregation, Eur. Phys. J. B, 1999, 12, 9 CrossRef CAS.
- X. Wen, D. Zhang, A. Chai, L. He, S. Ran and L. Zhang, Self-assembly of nanorods on soft elastic shells, Soft Matter, 2012, 8, 6706 RSC.
- N. Ramakrishnan, P. B. S. Kumar and J. H. Ipsen, Membrane-mediated aggregation of curvature-inducing nematogens and membrane tubulation, Biophys. J., 2013, 104, 1018 CrossRef CAS PubMed.
- T. Yue, X. Wang, F. Huang and X. Zhang, An unusual pathway for the membrane wrapping of rodlike nanoparticles and the orientation- and membrane wrapping-dependent nanoparticle interaction, Nanoscale, 2013, 5, 9888 RSC.
- M. Simunovic, A. Srivastava and G. A. Voth, Linear aggregation of proteins on the membrane as a prelude to membrane remodeling, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 20396 CrossRef CAS PubMed.
- M. Simunovic and G. A. Voth, Membrane tension controls the assembly of curvature-generating proteins, Nat. Commun., 2015, 6, 7219 CrossRef CAS PubMed.
- Y. Y. Zhang, Y. F. Hua and Z. Y. Deng, Driven self-assembly of hard nanoplates on soft elastic shells, Chin. Phys. B, 2015, 24, 118202 CrossRef.
- A. D. Olinger, E. J. Spangler, P. B. S. Kumar and M. Laradji, Membrane-mediated aggregation of anisotropically curved nanoparticles, Faraday Discuss., 2016, 186, 265 RSC.
- S. K. Ghosh, A. G. Cherstvy, E. P. Petrov and R. Metzler, Interactions of rod-like particles on responsive elastic sheets, Soft Matter, 2016, 12, 7908 RSC.
- J. C. Loudet, A. M. Alsayed, J. Zhang and A. G. Yodh, Capillary interactions between anisotropic colloidal particles, Phys. Rev. Lett., 2005, 94, 018301 CrossRef CAS PubMed.
- E. P. Lewandowski, M. Cavallaro, Jr., L. Botto, J. C. Bernate, V. Garbin and K. J. Stebe, Orientation and self-assembly of cylindrical particles by anisotropic capillary interactions, Langmuir, 2010, 26, 15142 CrossRef CAS PubMed.
- I. B. Liu, M. A. Gharbi, V. L. Ngo, R. D. Kamien, S. Yang and K. J. Stebe, Elastocapillary interactions on nematic films, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 6336 CrossRef CAS PubMed.
- L. Botto, E. P. Lewandowski, M. Cavallaro, Jr. and K. J. Stebe, Capillary interactions between anisotropic particles, Soft Matter, 2012, 8, 9957 RSC.
- J.-B. Fournier and P. Galatola, Anisotropic capillary interactions and jamming of colloidal particles trapped at a liquid–fluid interface, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2002, 65, 031601 CrossRef CAS PubMed.
- S. Dasgupta, M. Katava, M. Faraj, T. Auth and G. Gompper, Capillary assembly of microscale ellipsoidal, cuboidal, and spherical particles at interfaces, Langmuir, 2014, 30, 11873 CrossRef CAS PubMed.
- K. D. Danov and P. A. Kralchevsky, Capillary forces between particles at a liquid interface: general theoretical approach and interactions between capillary multipoles, Adv. Colloid Interface Sci., 2010, 154, 91 CrossRef CAS PubMed.
- L. Botto, L. Yao, R. L. Leheny and K. J. Stebe, Capillary bond between rod-like particles and the micromechanics of particle-laden interfaces, Soft Matter, 2012, 8, 4971 RSC.
- A. A. Evans, S. E. Spagnolie, D. Bartolo and E. Lauga, Elastocapillary self-folding: buckling, wrinkling, and collapse of floating filaments, Soft Matter, 2013, 9, 1711 RSC.
- D. T. Denhardt, The single-stranded DNA phages, CRC Crit. Rev. Microbiol., 1975, 4, 161 CrossRef CAS PubMed.
- D. A. Marvin, M. F. Symmons and S. K. Straus, Structure and assembly of filamentous bacteriophages, Prog. Biophys. Mol. Biol., 2014, 114, 80 CrossRef CAS PubMed.
- T. Loeb, Isolation of a bacteriophage specific for the F+ and Hfr mating types of Escherichia coli K-12, Science, 1960, 131, 932 CAS.
- N. D. Zinder, R. C. Valentine, M. Roger and W. Stoeckenius, f1, a rod-shaped male-specific bacteriophage that contains DNA, Virology, 1963, 20, 638 CrossRef CAS PubMed.
- D. A. Marvin and H. Hoffmann-Berling, Physical and chemical properties of two new small bacteriophages, Nature, 1963, 197, 517 CrossRef CAS.
- D. A. Marvin and H. Hoffmann-Berling, A fibrous DNA phage (fd) and a spherical RNA phage (fr) specific for male strains of E. coli. Part II. Physical characteristics, Z. Naturforsch., B: Anorg. Chem., Org. Chem., Biochem., Biophys., Biol., 1963, 18, 884 CAS.
- P. H. Hofschneider, Untersuchungen über “kleine” E. coli K12 Bakteriophagen: 1. und 2. Mitteilung, Z. Naturforsch., B: Anorg. Chem., Org. Chem., Biochem., Biophys., Biol., 1963, 18, 203 Search PubMed.
- D. E. Bradley, Some preliminary observations on filamentous and RNA bacteriophages, J. Ultrastruct. Res., 1964, 10, 385 CrossRef CAS PubMed.
- R. A. Panter and R. H. Symons, Isolation and properties of a DNA-containing rod-shaped bacteriophage, Aust. J. Biol. Sci., 1966, 19, 565 CrossRef CAS PubMed.
- D. A. Marvin and B. Hohn, Filamentous bacterial viruses, Bacteriol. Rev., 1969, 33, 172 CAS.
- L. A. Day, C. J. Marzec, S. A. Reisberg and A. Casadevall, DNA packaging in filamentous bacteriophages, Annu. Rev. Biophys. Biophys. Chem., 1988, 17, 509 CrossRef CAS PubMed.
- K. Zimmermann, H. Hagedorn, C. C. Heuck, M. Hinrichsen and H. Ludwig, The ionic properties of the filamentous bacteriophages Pf1 and fd, J. Biol. Chem., 1986, 261, 1653 CAS.
- K. R. Purdy and S. Fraden, Isotropic-cholesteric phase transition of filamentous virus suspensions as a function of rod length and charge, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2004, 70, 061703 CrossRef PubMed.
- G. P. Smith and V. A. Petrenko, Phage display, Chem. Rev., 1997, 97, 391 CrossRef CAS PubMed.
- J. Rakonjac, N. J. Bennett, J. Spagnuolo, D. Gagic and M. Russel, Filamentous bacteriophage: biology, phage display, and nanotechnology applications, Curr. Issues Mol. Biol., 2011, 13, 51 CAS.
- B. Cao, M. Yang and C. Mao, Phage as a genetically modifiable supramacromolecule in chemistry, materials and medicine, Acc. Chem. Res., 2016, 49, 1111 CrossRef CAS PubMed.
- I. Sorokulova, E. Olsen and V. Vodyanoy, Bacteriophage biosensors for antibiotic-resistant bacteria, Expert Rev. Med. Devices, 2014, 11, 175 CrossRef CAS PubMed.
- V. A. Petrenko and J. W. Gillespie, Paradigm shift in bacteriophage-mediated delivery of anticancer drugs: from targeted ‘magic bullets’ to self-navigated ‘magic missiles’, Expert Opin. Drug Delivery, 2017, 14, 373 CrossRef CAS PubMed.
- Z. Dogic, Filamentous phages as a model system in soft matter physics, Front. Microbiol., 2016, 7, 1013 Search PubMed.
- L. G. Caro and M. Schnös, The attachment of the male-specific bacteriophage f1 to sensitive strains of Escherichia coli, Proc. Natl. Acad. Sci. U. S. A., 1966, 56, 126 CrossRef CAS.
- W. O. Salivar, T. J. Henry and D. Pratt, Purification and properties of diploid particles of coliphage M13, Virology, 1967, 32, 41 CrossRef CAS PubMed.
- D. Pratt, H. Tzagoloff and J. Beaudoin, Conditional lethal mutants of the small filamentous coliphage M13. II. Two genes for coat proteins, Virology, 1969, 39, 42 CrossRef CAS PubMed.
- J. Lopez and R. E. Webster, Morphogenesis of filamentous bacteriophage f1: Orientation of extrusion and production of polyphage, Virology, 1983, 127, 177 CrossRef CAS PubMed.
- Z. Dogic and S. Fraden, Phase behavior of rod-like viruses and virus–sphere mixtures, in Soft Matter, vol. 2: Complex Colloidal Suspensions, ed. G. Gompper and M. Schick, Wiley-VCH, Weinheim, 2006, pp. 1–86 Search PubMed.
- J. Rakonjac and P. Model, Roles of pIII in filamentous phage assembly, J. Mol. Biol., 1998, 282, 25 CrossRef CAS PubMed.
- M. P. Lettinga, E. Barry and Z. Dogic, Self-diffusion of rod-like viruses in the nematic phase, Europhys. Lett., 2005, 71, 692 CrossRef CAS.
- C. Herold, G. Chwastek, P. Schwille and E. P. Petrov, Efficient electroformation of supergiant unilamellar vesicles containing cationic lipids on ITO-coated electrodes, Langmuir, 2012, 28, 5518 CrossRef CAS PubMed.
- M. Manning, S. Chrysogelos and J. Griffith, Insertion of bacteriophage M13 coat protein into membranes, Biophys. J., 1982, 37, 28 CrossRef CAS PubMed.
- M. Manning and J. Griffith, Association of M13 I-forms and spheroids with lipid vesicles, Arch. Biochem. Biophys., 1985, 236, 297 CrossRef CAS PubMed.
- D. Stopar, R. B. Spruijt, C. J. A. M. Wolfs and M. A. Hemminga, Mimicking initial interactions of bacteriophage M13 coat protein disassembly in model membrane systems, Biochemistry, 1998, 37, 10181 CrossRef CAS PubMed.
- H. Frank and L. A. Day, Electron microscopic observations on fd bacteriophage, its alkali denaturation products and its DNA, Virology, 1970, 42, 144 CrossRef CAS PubMed.
- D. H. Rowitch, G. J. Hunter and R. N. Perham, Variable electrostatic interaction between DNA and coat protein in filamentous bacteriophage assembly, J. Mol. Biol., 1988, 204, 663 CrossRef CAS PubMed.
- J. Greenwood, G. J. Hunter and R. N. Perham, Regulation of filamentous bacteriophage length by modification of electrostatic interactions between coat protein and DNA, J. Mol. Biol., 1991, 217, 223 CrossRef CAS PubMed.
- E. Pouget, E. Grelet and M. P. Lettinga, Dynamics in the smectic phase of stiff viral rods, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2011, 84, 041704 CrossRef PubMed.
- J. S. Wall, A high resolution scanning electron microscope for the study of single biological molecules, PhD thesis, University of Chicago, 1971 Search PubMed.
- Y. L. Lyubchenko, P. I. Oden, D. Lampner, S. M. Lindsay and K. A. Dunker, Atomic force microscopy of DNA and bacteriophage in air, water and propanol: the role of adhesion forces, Nucleic Acids Res., 1993, 21, 1117 CrossRef CAS PubMed.
- S. A. Berkowitz and L. A. Day, Mass, length, composition and structure of the filamentous bacterial virus fd, J. Mol. Biol., 1976, 102, 531 CrossRef CAS PubMed.
- J. Newman, H. L. Swinney and L. A. Day, Hydrodynamic properties and structure of fd virus, J. Mol. Biol., 1977, 116, 593 CrossRef CAS PubMed.
- T. Maeda and S. Fujime, Dynamic light-scattering study of fd virus. Application of a theory of the light-scattering spectrum of weakly bending filaments, Macromolecules, 1985, 18, 2430 CrossRef CAS.
- Y. A. Wang, X. Yu, S. Overman, M. Tsuboi, G. J. Thomas Jr. and E. H. Egelman, The structure of a filamentous bacteriophage, J. Mol. Biol., 2006, 361, 209 CrossRef CAS PubMed.
- E. Barry, D. Beller and Z. Dogic, A model liquid crystalline system based on rodlike viruses with variable chirality and persistence length, Soft Matter, 2009, 5, 2563 CAS.
- K. Beck and R. M. Duenki, Flexibility of bacteriophage M13: comparison of hydrodynamic measurements with electron microscopy, J. Struct. Biol., 1990, 105, 22 CrossRef.
- L. Song, U.-S. Kim, J. Wilcoxon and J. M. Schurr, Dynamic light scattering from weakly bending rods: Estimation of the dynamic bending rigidity of the M13 virus, Biopolymers, 1991, 31, 547 CrossRef CAS PubMed.
- A. S. Khalil, J. M. Ferrer, R. R. Brau, S. T. Kottmann, C. J. Noren, M. J. Lang and A. M. Belcher, Single M13 bacteriophage tethering and stretching, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 4892 CrossRef CAS PubMed.
- O. Kratky and G. Porod, Röntgenuntersuchung gelöster Fadenmoleküle, Recl. Trav. Chim. Pays-Bas, 1949, 68, 1106 CrossRef CAS.
- R. G. Winkler, P. Reineker and L. Harnau, Models and equilibrium properties of stiff molecular chains, J. Chem. Phys., 1994, 101, 8119 CrossRef CAS.
- J. R. C. van der Maarel, Introduction to Biopolymer Physics, World Scientific, Singapore, 2008 Search PubMed.
- K. Günther, M. Mertig and R. Seidel, Mechanical and structural properties of YOYO-1 complexed DNA, Nucleic Acids Res., 2010, 38, 6526 CrossRef PubMed.
- A. W. C. Lau, A. Prasad and Z. Dogic, Condensation of isolated semi-flexible filaments driven by depletion interactions, Europhys. Lett., 2009, 87, 48006 CrossRef.
- S. A. Overman, M. Tsuboi and G. J. Thomas Jr., Subunit orientation in the filamentous virus Ff (fd, f1, M13), J. Mol. Biol., 1996, 259, 331 CrossRef CAS PubMed.
- D. Stopar, R. Spruijt, C. J. A. M. Wolfs and M. A. Hemminga, Protein–lipid interactions of bacteriophage M13 major coat protein, Biochim. Biophys. Acta, 2003, 1611, 5 CrossRef CAS.
- M. A. Hemminga, W. L. Vos, P. V. Nazarov, R. B. M. Koehorst, C. J. A. M. Wolfs, R. B. Spruijt and D. Stopar, Viruses: incredible nanomachines. New advances with filamentous phages, Eur. Biophys. J., 2010, 39, 541 CrossRef PubMed.
- A. K. Dunker, L. D. Ensign, G. E. Arnold and L. M. Roberts, A model for fd phage penetration and assembly, FEBS Lett., 1991, 292, 271 CrossRef CAS PubMed.
- D. Stopar, R. B. Spruijt, C. J. A. M. Wolfs and M. A. Hemminga, Structural characterization of bacteriophage M13 solubilization by amphiphiles, Biochim. Biophys. Acta, 2002, 1594, 54 CrossRef CAS.
- K. Amako and K. Yasunaka, Ether induced morphological alteration of Pf-1 filamentous phage, Nature, 1977, 267, 862 CrossRef CAS PubMed.
- D. A. Marvin, Structure of the filamentous phage virion, in The Single-Stranded DNA Phages, ed. D. T. Denhardt, D. H. Dressler and D. S. Ray, Cold Spring Harbor Monograph Series, Cold Spring Harbour Laboratory Press, Plainview, NY, 1978, vol. 8, pp. 583–603 Search PubMed.
- J. Griffith, M. Manning and K. Dunn, Filamentous bacteriophage contract into hollow spherical particles upon exposure to a chloroform–water interface, Cell, 1981, 23, 747 CrossRef CAS PubMed.
- M. Manning, S. Chrysogelos and J. Griffith, Mechanism of coliphage M13 contraction: Intermediate structures trapped at low temperatures, J. Virol., 1981, 40, 912 CrossRef CAS PubMed.
- J. Lopez and R. E. Webster, Minor coat protein composition and location of the A protein in bacteriophage f1 spheroids and I-forms, J. Virol., 1982, 42, 1099 CAS.
- L. M. Roberts and A. K. Dunker, Structural changes accompanying chloroform-induced contraction of the filamentous phage fd, Biochemistry, 1993, 32, 10479 CrossRef CAS PubMed.
- J. S. Oh, D. R. Davies, J. D. Lawson, G. E. Arnold and A. K. Dunker, Isolation of chloroform-resistant mutants of filamentous phage: localization in models of phage structure, J. Mol. Biol., 1999, 287, 449 CrossRef CAS PubMed.
- A. Jacobson, Role of F pili in the penetration of bacteriophage f1, J. Virol., 1972, 10, 835 CAS.
- M. W. Bayer and M. H. Bayer, Effects of bacteriophage fd infection on Escherichia coli HB11 envelope: a morphological and biochemical study, J. Virol., 1986, 57, 258 CAS.
- M. Nakamura, K. Tsumoto, I. Kumagai and K. Ishimura, A morphologic study of filamentous phage infection of Escherichia coli using biotinylated phages, FEBS Lett., 2003, 536, 167 CrossRef CAS PubMed.
- D. K. Hore, D. S. Walker, K. MacKinnon and G. L. Richmond, Molecular structure of the chloroform–water and dichloromethane–water interfaces, J. Phys. Chem. C, 2007, 111, 8832 CAS.
- V. Czernohorsky, Diploma thesis, Technische Universität Dresden, Dresden, 2011.
- R. Dimova, Recent developments in the field of bending rigidity measurements on membranes, Adv. Colloid Interface Sci., 2014, 208, 225 CrossRef CAS PubMed.
- H. Bouvrais, L. Duelund and J. H. Ipsen, Buffers affect the bending rigidity of model lipid membranes, Langmuir, 2014, 30, 13 CrossRef CAS PubMed.
- H. Yamakawa and W. H. Stockmayer, Statistical mechanics of wormlike chains. II. Excluded volume effects, J. Chem. Phys., 1972, 57, 2843 CrossRef CAS.
- J. F. Marko, Introduction to single-DNA micromechanics, in Multiple Aspects of DNA and RNA: From Biophysics to Bioinformatics, ed. D. Chatenay, S. Cocco, R. Monasson, D. Thieffry and J. Dalibard, Elsevier, Amsterdam, 2005, pp. 211–270 Search PubMed.
- M. Fujii, J. Shimada and H. Yamakawa, Statistical mechanics of two-dimensional wormlike chains, J. Polym. Sci., Polym. Phys. Ed., 1974, 12, 1327 CrossRef CAS.
- J. Shimada and H. Yamakawa, Ring-closure probabilities for twisted wormlike chains. Application to DNA, Macromolecules, 1984, 17, 689 CrossRef CAS.
- J. T. Kindt, Pivot-coupled grand canonical Monte Carlo method for ring simulations, J. Chem. Phys., 2002, 116, 6817 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary text and 15 supplementary movies showing the behavior of fd virus particles on lipid membranes and lipid nanotubes. See DOI: 10.1039/c7sm00829e |
| ‡ Present address: Ludwig-Maximilians-Universität München, Department of Cellular Physiology, Biomedical Center Munich – BMC, 82152 Martinsried, Germany. |
| § Present address: ZELLMECHANIK DRESDEN GmbH, 01307 Dresden, Germany. |
| This journal is © The Royal Society of Chemistry 2017 |