Advanced cell culture platforms: a growing quest for emulating natural tissues
Marziye
Mirbagheri
abc,
Vahid
Adibnia
c,
Bethany R.
Hughes
ab,
Stephen D.
Waldman
ab,
Xavier
Banquy
c and
Dae Kun
Hwang
 *ab
*ab
aDepartment of Chemical Engineering, Faculty of Engineering & Architectural Science, Ryerson University, Toronto, Ontario M5B 2K3, Canada. E-mail: dkhwang@ryerson.ca
bKeenan Research Center, Li Ki Shing Knowledge Institute, St. Michael's Hospital, Toronto, Ontario M5B 1W8, Canada
cFaculty of Pharmacy, Université de Montréal, C.P. 6128, Succursale Centre Ville, Montreal, Quebec H3C 3J7, Canada
First published on 1st November 2018
Abstract
In the body, cells inhabit within a complex three-dimensional (3D) extracellular matrix that provides physical and chemical signals to regulate the cell fate. Cultured cells in Petri dishes and tissue culture flasks (2D) receive completely different environmental cues compared to natural tissues, causing radical alterations in cell morphology and function. Three-dimensional culture models have been able to revolutionize biomedical applications by better emulating natural tissues. However, sample handling and high-throughput screening can be challenging with 3D cell culture. Moreover, most 3D matrices are unable to quantify intracellular mechanics due to their structurally undefined surface characteristics. Therefore, highly structured surfaces (2½D) comprising various micro- and nano-patterns were introduced to address these limitations. The topographical substrates have also been shown to retain in vivo cell functionalities, such as proliferative capacity. Here, we review recent advancements in modulation of surface patterns that have been able to control cell adhesion in two or three dimensions, and their impacts on the cell behavior. Finally, we provide a comparison between 2D, 2½D and 3D systems and present several clinical applications of non-planar substrates.
1 Introduction
Cell culture experiments have been mainly performed on two-dimensional (2D) platforms, such as multi-well plates, Petri dishes, and tissue culture flasks, in which the cells are plated onto a rigid planar surface. These conventional 2D substrates—with ease of use and high cell viability—have notably improved our understanding of basic cell function. However, they are notoriously unable to appropriately imitate the cellular microenvironment in vivo, leading to physiologically irrelevant cell behaviors in most cases. Cells inherently respond to chemical (e.g., surface chemistry and material composition) and physical (e.g., mechanical and structural properties) signals from their surroundings at all length scales,1–5 analyze the information and decide to undergo any of the cellular fate processes (division, apoptosis, migration and/or differentiation).In their natural environment, nearly all cells reside within a complex 3D fibrous meshwork, known as the extracellular matrix (ECM).4,6 The 3D nanostructure of the ECM, which is specific to each cell type, supports the cells and guides their function.7 The loss of 3D cues, disorganized cell–cell interactions, high oxygen tension, and high growth factor concentrations in 2D cell culture can considerably alter the inter- and intra-cellular behavior.8,9
The critical limitations of 2D cell culture can lead to several practical complications. For example, in end-stage destructive joint diseases, such as osteoporosis, osteoarthritis and bone tumors, the bone quality and bone formation are decreased while the bone resorption is increased. Current therapies involve the replacement of damaged tissue by an implant. The life span of these implants and, thus, the patients' life quality greatly depend on the initial bone tissue response.7 Surface topography is a key parameter in controlling this initial response10 by modulating the interactions between tissue and the implant.11 Many researchers have already proved the beneficial effects of biomaterials that mimic the bone surface roughness on osteoblast proliferation and adhesion.12–18 The shortcomings of 2D planar platforms have been repeatedly demonstrated in other studies as well.19,20 According to Lee et al.,20 the cytotoxicity testing of CdTe nanoparticles on HepG2 cells is dramatically different between 2D and 3D cultures, which can be problematic in preclinical drug screening, where in vitro analyses determine the toxicity of a target drug.21 Therefore, 2D planar platforms not only fail to reproduce the 3D cellular microenvironment in the body, but also can mislead our perception of cell responses.
To overcome these limitations, 3D cell culture methods including spheroids, microcarriers, and tissue-engineered models were introduced.22–33 While microcarriers are mainly used for cellular expansion in vitro, spheroids and 3D scaffold-based models have diverse applications in tissue engineering, cancer research and fundamental studies of cellular function.34 Spheroids are highly dense tissue analogs that are self-assembled due to the tendency of adherent cells to aggregate. Three-dimensional matrices are highly porous substrates that can support cell function by homogeneously surrounding the cell with extracellular materials.35 In tissue engineering, primary cells from patients adhere and proliferate on the surface of a 3D scaffold, generating the ECM components of the living tissue.
Cellular behavior in 3D cultures is relatively closer to natural tissue, specifically, the cell morphology. For example, human fibrochondrocytes have distinct 3D round or oval shapes in vivo and on 3D scaffolds, whereas they exhibit stretched, fibroblast-like morphologies in 2D culture flasks.36 Cell metabolism and functionality, including proliferation, differentiation and gene expression, are also significantly different between 2D and 3D models.19,34,36 This can be substantially important in clinical applications, such as mesenchymal stem cell (MSC) transplantation for treating osteogenesis imperfecta,37 where the loss of proliferative capacity of MSCs in 2D culture can lead to poor engraftment and limited cell survival upon transplantation into the body.38 Therefore, 3D systems are mainly favorable to 2D structures for cell culture and tissue models. Nonetheless, several challenges remain.
With 3D cell culture, sample handling and imaging39 as well as high-throughput screening35 can be challenging, demanding technological innovations to fully benefit from the third dimension.40,41 Furthermore, due to structurally undefined surface characteristics, most 3D scaffolds are unable to quantify intracellular mechanics.9 In recent years, highly structured surfaces composed of various patterns, also known as 2½ dimensional (2½D) objects, have been developed to promote our understanding of cell behavior. On these substrates, adhered cells conform to their specific surface patterns, whether simple, such as grooves, or complex, such as plant topographies.42 This can significantly improve drug testing and disease modeling, in which tissue models should adequately represent natural cell organizations to obtain physiologically relevant information.
This review highlights recent developments in modulation of surface topography, and its impacts on cell behavior and function. While most surface topographies induce 2D planar cell morphologies, we also explain advanced 2½D substrates that have been able to control cell adhesion and cell shape in three dimensions. Recall that here any topographical pattern (e.g., groove, post, pit, and other complex designs) on a flat substrate is recognized as 2½D. Moreover, we discuss the advantages and disadvantages of 2½D substrates in comparison with 2D and 3D models. Finally, we introduce several contemporary applications of various non-planar cell culture platforms.
2 Cell culture on structured surfaces
Numerous soft and hard materials43–46 are utilized in biomedical and healthcare industries for applications such as tissue engineering,47 medical implants,48 and drug delivery.49,50 Particularly, many types of natural (e.g., collagen, alginate and fibrin) and synthetic polymers (e.g., acrylamide, poly(ethylene glycol) (PEG), poly(dimethyl siloxane) (PDMS), and poly(methyl methacrylate) (PMMA)) have been used for tissue engineering and cell transplantation.51 For example, in a recent breakthrough, European researchers were able to regenerate the entire human epidermis using transgenic stem cells,52 providing high hopes for treatment of the so-far incurable Junctional Epidermolysis Bullosa (JEB) disease. In this research, the keratinocyte cells were cultured onto fibrin gels, allowing for preparation of larger grafts from the same number of clonogenic cells as would be needed for plastic-cultured grafts.52,53Such clinical studies are abundant and what they all have in common is the importance of cell interactions with the scaffolding material, which can directly determine the cell response. In recent years, it has been extensively documented that surface topography in particular can greatly influence the cell behavior.42,54–56 In this section, first we review topographical substrates that lead to planar cell morphologies in which cells migrate in a 2D plane. Next, advanced 2½D structures that induce a crossover from 2D to 3D cell behavior are discussed. Here, in addition to physical patterning, surfaces may have been treated chemically to control cell adhesion.44,57,58 Since the focus of this review is on physical cues, the impact of chemical surface modifications is discussed only when found relevant. Moreover, throughout this article, the material types that are used to construct the topographical substrates or 3D tissue-engineered models are specified, as they can be important factors in modulating cell–material interactions.59,60
Physical topographies that induce planar cell morphology
Diverse techniques including photo-,61,62,240 soft,63,64 colloidal65 and electron beam lithography,66 laser patterning,67 and dry etching66,68 have been used to fabricate topographical surfaces using various materials, such as polymers, silicon oxide, and metals.69–71 Surface topography is especially important for synthetic polymers due to the lack of biological recognition on their surfaces.72Schulte et al.73 demonstrated how topographical patterns on PEG can solely manipulate cellular behavior without biofunctionalization. According to their study, cells on smooth polymer surfaces showed no pronounced stress fibre network (Fig. 1(a)) with completely round bodies (Fig. 1(b)) and an inclination to form small clusters (Fig. 1(c)).
 | ||
| Fig. 1 Mouse fibroblast cell line (L929) cultured on starPEG hydrogels with smooth surface after 24 h (a–c), post patterns after 4 h (d and e), and line patterns with groove widths of 5 μm after 48 h (f and g) and 10 μm after 24 h (h) of incubation. Immuno-staining of F-actin (a, d and h) and cell nucleus (a and h), and electron micrographs of dried cells (b, c and e–g). Scale bars: 5 μm. Reproduced with permission.73 | ||
However, on patterned surfaces with posts, protrusions were formed (Fig. 1(d)), and the cell body flattened around the surface structure (Fig. 1(e)) with a noticeably reduced cluster formation. Similarly, single cells on top of (Fig. 1(f)) or within (Fig. 1(g)) the grooves showed enhanced spreading and a larger cell–surface contact area compared to smooth surfaces (Fig. 1(h)). These observations indicated that fibroblast cells adhered and spread on imprinted topographies, while the smooth surfaces were nonadhesive.
The cell morphological and functional responses to surface patterns greatly depend on the cell type74 as well as the pattern type and dimensions (several examples are summarized in Table 1). The effects of micro- and nano-grooves on different cells have been widely investigated.75–78 On these linear cues, cells usually align and migrate in the groove direction. However, the cell response is strongly governed by the groove width and depth.
| Pattern | Dimension (μm) | Method | Materials | Surface treatment | Cell type | Culture time | Effect | Ref. |
|---|---|---|---|---|---|---|---|---|
| a Square arrays with dots displaced randomly by up to 50 nm on x and y axes from their position in a true square. b Abbreviations are poly(L-lactic acid) (PLLA), tissue culture polystyrene (TCPS), and a co-polymer of poly(ethylene oxide) and poly(butylene terephtalate) (PEOT/PBT). | ||||||||
| Grooves | 5 < width < 10 | Replica molding | PEG | — | Mouse fibroblast | 24 h | Enhanced adhesion, spreading, and cell alignment and reduced cluster formation. | 73 |
| Depth: 5 | ||||||||
| Grooves | 20 < width | Replica molding | PEG | — | Mouse fibroblast | 24 h | Cells reacted similar to those on a smooth surface. | 73 |
| Depth: 5 | ||||||||
| Grooves | 0.1 < width <1 | Replica molding | Polystyrene | — | Rat dermal fibroblast | 4 (24) h | Enhanced cell alignment. | 77 |
| 0.075 (0.035) < depth <0.35 | ||||||||
| Grooves | Width: 6 | Photolithography and dry etching | PMMA | — | Baby Hamster Kidney fibroblast (aka BHK) | — | Poor cell alignment. | 62 |
| Depth: 0.56 | ||||||||
| Grooves | Width: 6 | Photolithography and dry etching | PMMA | — | MDCK (epithelial cell) | — | ∼100% cell alignment. | 62 |
| Depth: 0.56 | ||||||||
| Grooves | Width: 6 | Photolithography and dry etching | PMMA | — | Baby Hamster Kidney fibroblast (aka BHK) | — | ∼100% cell alignment. | 62 |
| Depth: 2 | ||||||||
| Grooves | Width: 8 | Hot-embossing imprint lithography | Polyimide | Fibronectin | Osteoblast | — | Cells aligned according to the mechanical topography rather than the chemical patterns. | 76 |
| Depth: 4 | ||||||||
| Micropillars (undeformable) | Height: 10 | Photolithography and replica molding | PDMS | Fibronectin | 3T3 fibroblast | 6–24 h | Cells on top of the posts had more elongated and branched shapes, fewer focal adhesions and slower movements. | 81 |
| Diameter: 5 | ||||||||
| Spacing: 5 | ||||||||
| Micropillars (undeformable) | Height: 0.8 | Photolithography and replica molding | PDMS | Fibronectin | Malignant fibroblast | 1–9 h | As oppose to healthy cells, fewer cancer cells were elongated and they exhibited a disorganized motility. | 111 |
| Diameter: 1 | ||||||||
| Spacing: 1.6 | ||||||||
| Square micropillars (undeformable) | Height: 4.5–14 | Replica molding | PLLA, TCPS, PEOT-PBT, and PDMSb | — | C2C12 muscle cell | 1–4 days | Improved initial cell attachment but reduced proliferation for hydrophobic materials (PDMS). However, better proliferation on hydrophilic materials. | 112 |
| Length: 5 | ||||||||
| Spacing: 2–26 | ||||||||
| Nanopillars (undeformable) | Diameter: 0.2 | Nanoimprint lithography | UV resin | — | Human osteoblast | 3 days | Long filopodia extensions and lamellipodia formation. | 113 |
| Spacing: 0.7 | ||||||||
| Sharp-tip nanoposts | Height: 0.05–0.6 | Interface lithography and DRIE | Silicon | — | Human foreskin fibroblast | 3 days | Elongated cells on medium height features, but lower cell viability and proliferation than on smooth surfaces, which became more pronounced with height. The cells also became smaller and rounder with height. | 97 |
| Spacing: 0.23 | ||||||||
| Sharp-tip nanogrates | Height: 0.05–0.6 | Interface lithography and DRIE | Silicon | — | Human foreskin fibroblast | 3 days | Lower cell viability and proliferation than on smooth surfaces. Elongated cells in the grate direction, with enhanced cell alignment for taller grates. | 97 |
| Width: 0.23 | ||||||||
| Nanogrates | Height: 0–0.35 | Nanoimprint photoimprint and lithography | Poly(styrene), PMMA and a cross-linked dimethacrylate | — | Murine preosteoblast | 24 h | Physical topography induced higher cell alignment than surface chemistry. Cell alignment increased with grate height and trough width. | 114 |
| Pitch: 0.42 and 0.8 | ||||||||
| Hexagonal and disordered nanopit arrays | Diameter: 0.12 | Electron beam lithography | PMMA | — | Human osteoprogenitor and MSC | 21 days | Enhanced osteogenesis compared to planar surface, specifically on DSQ50 arrays.a | 98 |
| Depth: 0.1 | ||||||||
| Spacing: 0.3 | ||||||||
| Square array of nanopits | Diameter: 0.12 | Electron beam lithography and hot embossing | PCL | — | Human MSC | 28 days | Prolonged maintenance of MSCs phenotype and multipotency, due to a reduced osteogenic differentiation but improved MSC markers retention. | 99 |
| Depth: 0.1 | ||||||||
| Spacing: 0.3 | ||||||||
| Orthogonal and hexagonal array of nanopits | Diameter: 0.035, 0.075 and 0.12 | Nanoimprinting and hot embossing | PMMA and PCL | — | Human fibroblast and rat epitenon | — | Reduced cell adhesion on ordered arrays compared to planar surfaces. | 100 |
| Spacing: 0.1, 0.2 and 0.3 | ||||||||
| Micropits | Diameter: 7, 15 and 25 | Photolithography | Quartz | — | Human fibroblast | 24 h | Slightly improved cell proliferation, and higher cell motility rates on 7 μm diameter pits. Cells can enter, divide in and exit 25 μm diameter pits. | 103 |
| Spacing: 20 and 40 | ||||||||
| Depth: 4.8 | ||||||||
| Square micropits | Length: 2 | Photolithography | Quartz and polyimide | Collagen on quartz | Neutrophil | — | Cell adhesion was stronger on quartz than polyimide or collagen-coated quartz. Cell motility was higher on collagen-coated-quartz smooth surfaces. Micropits with 10 μm spacing also improved the motility. | 104 |
| Depth: 0.21 | ||||||||
| Spacing: 6–14 | ||||||||
| Membranes with micro- and nano-sized columnar pores | Diameter: 0.1–3 | — | Polycarbonate | — | Corneal epithelial cell | 21 days | Superior stratification on pores with diameters in the range of 0.1–0.8 μm. | 115 |
Experiments with fibroblast show that groove width smaller than the cell size provokes a stronger cell response,73 while there is a cut-off value below which the cells no longer recognize the patterns.77 For example, rat dermal fibroblasts obtained from the ventral skin of male Wistar rats and cultured on nanogrooves experience a threshold feature size of 35 nm.77 In contrast, cell alignment has been shown to be proportional to groove depth (Fig. 2), with an upper threshold above which no significant improvements have been observed.62,73 For example, for MDCK cell lines cultured on groove structures with groove width ∼12 μm, the upper threshold for groove depth is reported to be ∼1.1 μm.62 The groove depth has also been reported to be more influential than its width.62,77
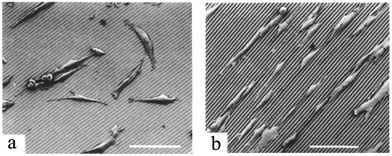 | ||
| Fig. 2 Scanning electron micrographs (SEM) of BHK cells on PMMA microgrooves with 6 μm width and 300 nm (a) or 2 μm (b) depth. Cell alignment improves with deeper grooves. Scale bars: 120 μm. Reproduced with permission.62 | ||
On a certain 2½D substrate, the cell response may also vary with time,77 emphasizing the need for sufficiently prolonged culture time to establish the long-term cell behavior in medical applications, such as implants. Guided cell alignment can be crucial for disease modeling and drug testing, where cell disruption due to a certain illness is analyzed to identify potential drugs. Here, the model tissue should appropriately represent the natural cell organization and function. In many natural tissues, cells are elongated and structurally aligned.79,80 More clinical applications of these platforms are discussed in Section 4.
Another topographical feature that has been used to investigate cellular behavior is micropillar or micropost. Similar to grooves, the pillar size and spacing influence cell organization. Ghibaudo et al.81 studied fibroblast migration onto chemically identical substrates composed of micropillars. In this study, the surfaces were uniformly coated with fibronectin to promote cell adhesion, and the pillars were too stiff to be significantly deformed by cell traction forces.82,83 Cell morphologies on pillars with a height of ∼2 μm were close to those on a flat surface (Fig. 3(a)), while on taller pillars they adopted more elongated and branched shapes (Fig. 3(b)). Whereas cells migrated on top of the posts for small pillar spacings, they encountered alternating flat and rough surfaces for wider spacings (Fig. 3(c)). Furthermore, cell migration patterns were changed from a fast and random movement on flat surfaces to a slower and persistent movement on post arrays, allowing their translocation from pillar to pillar. Compared to flat surfaces, fewer actin stress fibers and focal adhesions were observed, and they appeared more aggregated and more stable over time on the pillars. This guidance of focal adhesion formation was also observed in other studies.84,85 More information on focal adhesions and their role in transmitting extracellular physical or topographic signals can be found in ref. 85. A potential application of these platforms is for cellular phenotype discrimination, since various cell types react differently to pillar topographies.
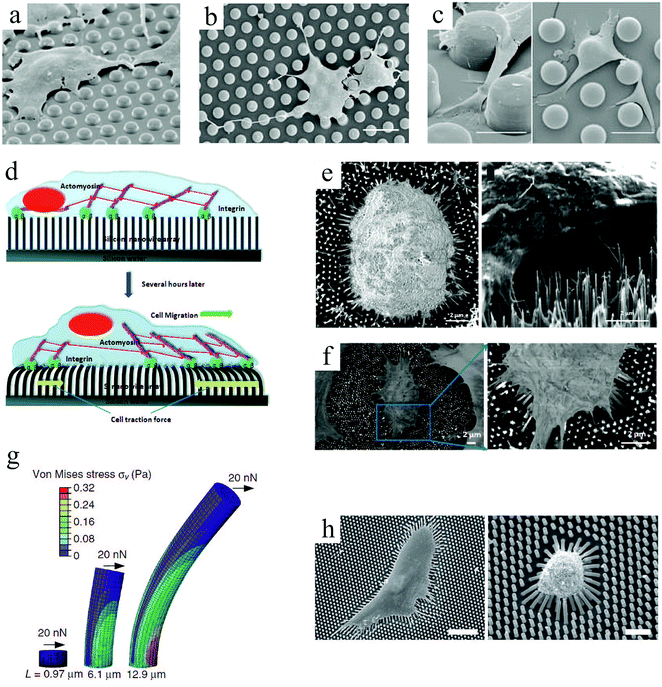 | ||
| Fig. 3 (top) SEM images of fibroblast cell response to arrays of undeformable micropillars with height H, diameter D and spacing S. To promote adhesion, the entire surfaces were coated with fibronectin: (a) H = 2 μm, D = 5 μm, and S = 5 μm; (b) H = 10 μm, D = 5 μm, and S = 5 μm; (c) H = 10 μm, D = 10 μm, and S = 10 μm.81 (d) Schematic representation of cell adhesion to silicon nanowire arrays and the traction forces generated by cell's actin cytoskeleton. SEM images of normal mechanocytes (e) and cancerous L929 (f) cells bending silicon nanowires through their contraction forces. Scale bars: 2 μm.86 (g) Graphical depiction of a finite-element method analysis of micropost with heights (L) deflected in response to a lateral traction force (F) of 20 nN. (h) SEM micrographs of human MSCs plated onto PDMS micropost arrays with L = 6.10 (left) and 12.9 μm (right). For this study, only the post tips were coated with fibronectin. Scale bars: 100 μm.87 Reproduced with permission. | ||
Studies of cellular mechanotransduction has also greatly benefited from post patterns. Analyzing the cell responses to physical cues suggests that when cells adhere to a surface, they sense their environment by mechanically probing it.88 Adherent cells exert contractile forces, also known as traction forces, on the substrate using their actin–myosin cytoskeleton to propel themselves.89–91 Traction forces regulate cell's motility and morphology,91 thus playing a crucial role in cell signaling, proliferation, differentiation, migration and other biological processes, such as angiogenesis, embryogenesis, inflammation and wound healing. Specifically, assessing the contractility of human stem cell-derived cardiomyocytes83 can help devise better strategies for heart repair,92 disease modeling,93 and drug discovery.94 One of the most important functional characteristics of a cardiomyocyte is its ability to produce contractile forces, as mechanical contraction plays a key role in blood circulation throughout the body. For example, measuring cardiomyocyte contractility can be of interest in studying cardiac diseases associated with cardiomyopathies (impaired contractility).
Microfabricated arrays of pillars have been extensively used to manipulate and measure the mechanical interactions between cells and their environment.82,83,95,96 Using silicon nanowires, Li et al.86 compared the mechanical behavior of normal, benign and malignant mammalian cells (Fig. 3(d)–(f)). According to their results, cancer cells exhibit a larger traction force than normal cells by 20 and 50% for the malignant and benign cells, respectively. These comparative techniques can be beneficial for disease diagnosis.
Intuitively, taller posts are more susceptible to bending in response to a horizontal traction force. Experimentally and numerically, Fu et al.87 demonstrated the relationship between post height and deflection due to traction forces exerted by human MSCs, and its consequent impact on cell behavior (Fig. 3(g) and (h)). In contrast to soft (tall) microposts, cells were well spread with highly organized actin stress fibers and large focal adhesions on rigid (short) posts. Therefore, they established a coupling between cell shape, focal adhesion and cytoskeletal tension in response to substrate rigidity.
Pillar arrays in these studies had relatively flat tips. Choi et al.97 investigated the behavior of human foreskin fibroblasts on sharp-tip nanoposts and nanogrates with various heights. They observed a significantly lower cell viability and proliferation on sharp-tip nanostructures compared to the smooth planar surfaces, and this effect was more pronounced on nanoposts versus nanogrates and with increasing height of both patterns. Whereas cells elongated on medium height nanoposts, they became smaller and rounder on taller posts, indicating poor cell adhesion (Fig. 4(a)–(d)). Grate structures also provoked elongated cell morphology in the grate direction; however, in contrast to posts, cell alignment and elongation were more pronounced on taller features (Fig. 4(e)–(h)). This is in agreement with previous observations of enhanced cell alignment with groove depth (ridge height).62,73
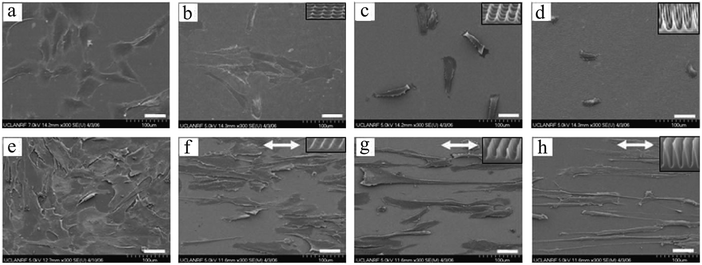 | ||
| Fig. 4 SEM micrographs of fibroblast cells cultured on smooth surfaces (a: control for posts, and e: control for grates), needle-like nanoposts (height increasing from b to d), and blade-like nanogrates (height increasing from f to h). Insets show higher magnification of the sample's nanotopography, and arrows (↔) in panels (f–h) indicate the nanograte directions. Scale bars: 50 μm. Reproduced with permission.97 | ||
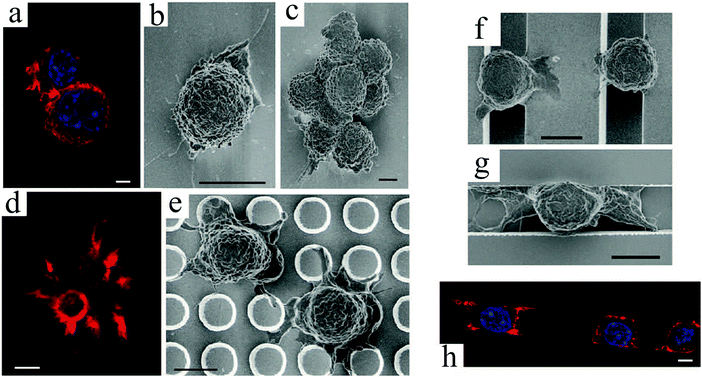
The last type of ordered surface topography is nano- or micro-pit. Dalby et al.98 studied the interaction of two cell types, osteoprogenitors and MSCs, when cultured on nanopits of different symmetry, including orthogonal and hexagonal arrays. Compared to cell culture on planar surfaces, human osteoprogenitor cell density decreased, especially on hexagonal arrays. Measuring the expression of bone-specific ECM proteins by osteoprogenitors, namely osteopontin (OPN) and osteocalcin (OCN), a slight increase in production of these proteins was observed on symmetric pit arrays, with a larger difference for the square configuration (Fig. 5(a)–(c) and (f)–(h)). On the other hand, increasing the randomness of nanopit arrangement resulted in denser cell populations with increased levels of OPN and OCN, specifically on disordered square arrays with dots displaced randomly by up to 50 nm on x and y axes from their position in a true square (DSQ50), as shown in Fig. 5(d), (e), (i) and (j).
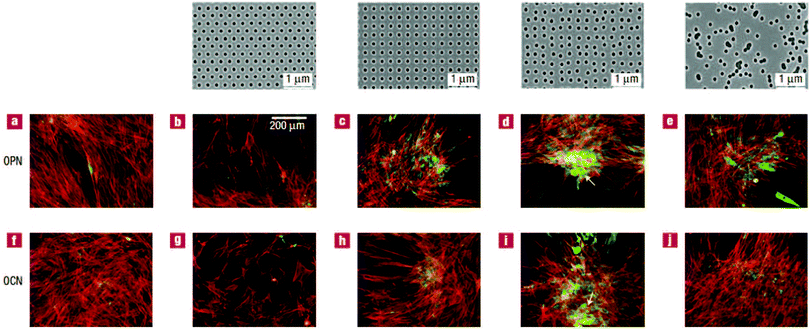 | ||
| Fig. 5 OPN and OCN staining of osteoprogenitors after 21 days of culture. (top) Images of nanopits with diameter 120 nm, depth 100 nm, and absolute or average center to center spacing 300 nm in hexagonal, square, displaced (DSQ50) and random configurations (respectively, from left to right). Osteoprogenitors cultured on the control planar surface with lack of positive OPN and OCN (a and f); hexagonal array with poor cell adhesion (b and g); square configuration with reduced cell density but slightly increased positive OPN and OCN compared to the control (c and h); DSQ50 showing bone nodule formation and enhanced OPN and OCN expression (d and i); random arrangement with good cell population and improved OPN and OCN production (e and j). Actin = red and OPN/OCN = green. Scale bar, as shown in panel b, is the same for panels a–j. Reproduced with permission.98 | ||
Similarly, MSCs cultured on DSQ50 exhibited intense cell aggregation and early bone nodule formation with both OPN- and OCN-positive regions. In contrast to osteoprogenitors, OPN and OCN expressions by MSCs were lost on completely random arrays (similar to the far-right, top panel in Fig. 5).
Later, McMurray et al.99 demonstrated how culturing cells on orthogonal arrays of nanopits can allow a long-term maintenance of MSC phenotype and multipotency, in contrast to cells cultured on displaced arrangements such as DSQ50. In this study, cells on symmetric orthogonal arrays retained expression of MSC markers, i.e., STRO-1 and ALCAM (activated leukocyte cell adhesion molecule, CD166), while no osteogenic markers (OPN and OCN) were observed. Therefore, following a prolonged culture (28 days), MSCs could be removed from the square pit arrays using trypsin, plated onto a coverslip and treated with differentiation media to promote osteogenesis or adipogenesis, indicating that cells not only expressed MSC markers but also were multipotent. This again highlights the impact of surface topography on differentiative capacity of MSCs with crucial importance in cell transplantation therapies, where tissue regeneration upon reinfusion depends on MSC differentiation into tissue-specific cell types.
In another study, Curtis et al.100 reported that ordered nanopit arrays (orthogonal or hexagonal) in polycaprolactone (PCL) or polymethylmethacrylate (PMMA) can considerably decrease cell adhesion compared to planar surfaces. As illustrated in Fig. 6, cell adhesion on PCL surfaces with pit patterns was very minimal, except for the smallest pits with diameter 35 nm. Perhaps on these small patterns, cells ability to distinguish the holes diminishes. A similar conclusion was drawn in previous studies.101,102 Moreover, comparing the cells’ orientation on square or hexagonal pit arrays, they observed that not only were the orientations nonrandom but they were also influenced by different configurations, thus suggesting that cells are able to distinguish between different symmetries.
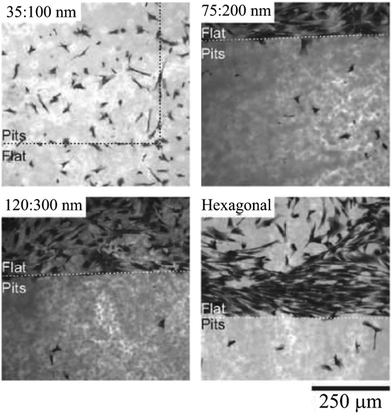 | ||
| Fig. 6 Rat epitenon cells cultured (24 h) on PCL substrates composed of flat surfaces and pit patterns. In top, left corner of each image, diameter:center-to-center spacing of nanopits is indicated. Cells are shown in black. Cell adhesion on pit arrays is considerably lower than that on flat surfaces, with relatively larger adhesion for smaller pits. Reproduced with permission.100 | ||
It is widely established that cell response can significantly vary with length scale. While the aforementioned studies analyzed cell behavior on nanopits, Berry et al.103 investigated fibroblast attachment and motility on micropits. They reported a clear dependence of cell response on pit size and spacing. Cell proliferation slightly increased only on the smallest pits with diameter 7 μm (a maximum difference of 6%), with even higher proliferation on closer pit spacing. The largest pit diameters (25 μm) allowed the cells to enter the pits while sustaining their viability. Their results indicated that not only were cells able to enter these pits, but they could also freely exit them, in contrast to earlier observations for macrophages that were trapped inside such pits.2 There was even evidence of cells actually dividing inside the 25 μm pits prior to the daughter cell emerging. On the smallest pits, however, a majority of cells moved over the pits regardless of the spacing. Furthermore, cells on the smallest pits moved faster, perhaps by using the pit edges as footholds to gain mechanical adhesion.104
A possible application of pit patterns is to establish appropriate pore sizes for 3D scaffolds in tissue engineering that will enhance cell penetration (ingrowth)105 whilst maintaining cell viability and proliferation. Microwells, if classified as pit patterns with large pit diameters such that cells can colonize inside the pits, have also been explored for high-throughput studies of cancer progression and cancer drug discovery106–109 as well as for tissue repair and regeneration applications.110
A number of irregular topographies have also been the subject of several studies. Among the most popular ones is the surface roughness, which is usually quantified by measuring the protrusions and depressions on a surface.55,116–120 A myriad of evidence show that surface roughness can play an important role in cell function, for example increasing cell adhesion and growth.121 Informative reviews are available on this subject.9,44
In an interesting work by Unadkat et al.,122 mathematical algorithms were used to design a library of nonbiased and random surface features on poly(lactic acid) with more than 2000 different topographies. As demonstrated in Fig. 7, a multitude of MSC cell morphologies, including elongated, branched, and round, was obtained. Using high-throughput screening, they illustrated MSC proliferation and osteogenic differentiation on the surface patterns.
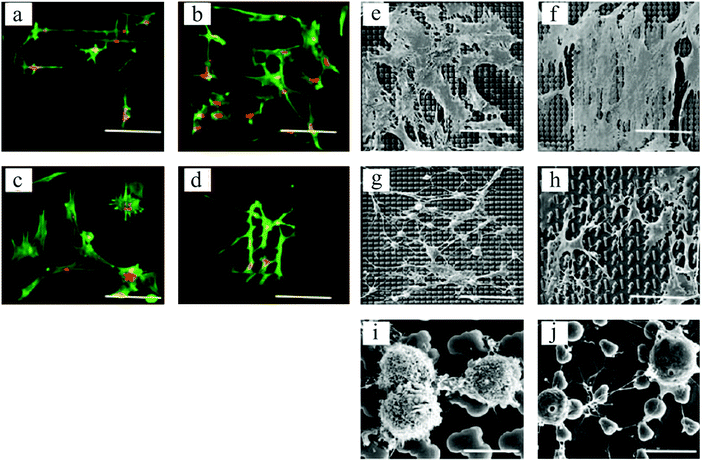 | ||
| Fig. 7 Fluorescent microscopic images of spread and elongated MSCs, with alignments according to various topographical features (a–d). Actin stained with Alexa Fluor 488 phalloidin = green, and nucleus stained with TOTO-3 = red. SEM images of cell morphologies on different topographies (e–h), and round cells on two distinct platforms exhibiting different cell membrane textures (i and j). Scale bars: 90 μm (a–h) and 10 μm (i and j). Reproduced with permission.122 | ||
The 2½D platforms reviewed in this section induce different cell morphologies, and furnish valuable information about cell metabolism and function compared to the oversimplified 2D planar surfaces (see Table 1 for more examples). Nonetheless, cells on these substrates generally lay flat and migrate in a 2D plane similarly to those on a smooth surface. Fig. 8(a) and (b) demonstrates an example of such behavior by comparing the human corneal epithelial cell shapes on a flat control and a nanogrooved surface. The only exception is for micropits that allow a certain level of 3D migration when cells enter the pits103 (Fig. 8(c)).
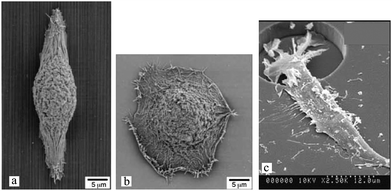 | ||
| Fig. 8 SEM images of human corneal epithelial cells cultured on (a) nanogrooved (width: 400 nm, depth: 600 nm, and ridge width: 70 nm) and (b) flat control silicon oxide substrates,123 and (c) fibroblasts on a micropit-patterned quartz surface (diameter: 25 μm and spacing: 40 μm).103 Reproduced with permission. | ||
Devices that can capture more of the 3D complexity in natural tissues are more desirable for understanding the cell behavior in vivo. In the following, we will discuss novel 2½D platforms that have been able to induce cell movement and morphology in 3 dimensions.
Advanced topographical substrates to induce 3D cell behavior
Various physical and chemical treatments have been employed to create 2½D scaffolds with more resemblance to topographical complexity in natural ECM,124 and to manipulate cell adhesion and shape in 3 dimensions.68,125,126Using direct laser writing (DLW), Klein et al.127 fabricated a substrate consisting of pillars that were connected by beams at two different heights by 10 μm offset (Fig. 9(a)).
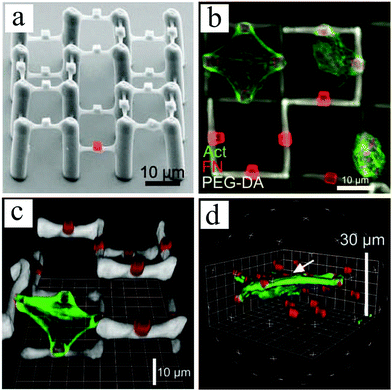 | ||
| Fig. 9 (a) SEM image of a 2½D polymer scaffold composed of a protein-repellent PEG-DA framework, which has embedded photoresist Ormocomp cubes on the PEG-DA beams (one cube is highlighted in red). Primary chicken fibroblasts cultivated on the scaffold are immunostained for fibronectin (red) and f-actin (green). Top view (b) and 3D construction (c and d) of confocal image stacks illustrating adherent cells to one or more fibronectin-positive Ormocomp cubes at the same or different heights. Reproduced with permission.127 | ||
Poly(ethylene glycol) diacrylate (PEG-DA) crosslinked with pentaerythritol tetraacrylate (PETA) was selected for the main skeleton of the scaffold, as PEG-DA is known for its protein-repellent properties that prevent cell attachment. To control cell adhesion and ECM distribution, protein-binding Ormocomp cubes were embedded on PEG-DA beams. Ormocomp is a biocompatible photoresist. Upon incubation of these scaffolds with fibronectin, this ECM protein bound preferentially to the Ormocomp cubes, as shown by immunofluorescence labelling in Fig. 9. Therefore, chicken fibroblasts cultured on these platforms adhered and spread on a single or multiple Ormocomp cubes, particularly revealing a true 3D growth pattern when spreading between cubes on beams at different heights (Fig. 9(c) and (d)).
Greiner et al.128 studied the growth of several cell types on fibronectin-coated 2½D platforms fabricated also using DLW (Fig. 10 (left)). As illustrated in Fig. 10, the cells adapted true 3D geometries on these platforms. It was also demonstrated that cell and nuclear volumes may change between 2D and 3D cell morphology, depending on the cell type. Whereas epithelial cells retained similar cell and nuclear volumes regardless of the cell morphology, fibroblasts displayed significantly increased cell and nuclear volumes for the 3D cell shapes compared to the 2D. They attributed these differences to the tissue-specific cell arrangements in vivo, where fibroblasts reside within loose connective tissues and epithelial cells within densely-packed flat layers. Nevertheless, the nucleus-to-cell (N/C) volume ratios remained constant for all cell types and culture conditions. N/C ratio alteration is a widely used metric in cancer detection.129
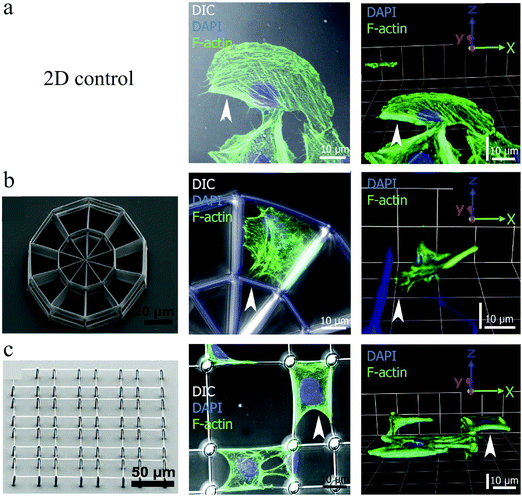 | ||
| Fig. 10 Buffalo rat liver cell growth on a 2D control surface (a), and a stiff (b) and a soft (c) 2½D scaffold, which were all coated with fibronectin to control cell adhesion. Left panels indicate the SEM images of culturing structures, the middle panels present the maximum projections of confocal image stacks, and the right panels are 3D constructions of confocal images. F-Actin = green and nucleus = blue. Reproduced with permission.128 | ||
In the aforementioned studies, an ECM protein (i.e., fibronectin) was used to control binding sites and, therefore, manipulate cell adhesion to induce 3D morphologies. Using a similar strategy, Ochesner et al.68 were able to provide a 3D shape control of single cells in microwell arrays. In their study, cell adhesion was limited within the microwells by passivation of the upper flat surface, while backfilling the wells with adhesive proteins or lipid bilayers.
Another creative use of adhesive proteins for cell adhesion was demonstrated by Richter et al.,130 who produced 2½D cell instructive platforms by incorporating two distinct ECM binding proteins in a passivating framework. They illustrated that cells with different sets of integrin receptors preferentially adhered to one of the proteins. However, since the binding sites were in the same height, cells spread in a 2D plane. Positioning the distinct binding proteins at different levels can possibly be a strategic approach to obtain 3D cell shapes.
In a recent work by our group,131 surface wrinkling was implemented as a physical technique to precisely distribute adhesion sites and induce 3D cell morphologies. Using photolithography and microfluidics, arrays of PEG-DA posts were created. By controlling the UV exposure time, a thin layer of partially cured polymer was formed on the posts, which wrinkled when exposed to plasma. The wrinkling mechanism is extensively explained by Kim et al.132 Whereas fibroblast cells adhered to the flat surface when the posts were smooth, they preferentially conformed to the wrinkled posts exhibiting 3D morphologies (Fig. 11(a) and (b)). It has also been shown in previous studies that cells prefer the flat surface in arrays of smooth pillars.81 Interestingly, some cells on adjacent wrinkled posts appeared to form bridges, as shown in Fig. 12(c).
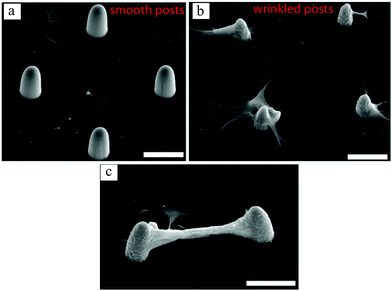 | ||
| Fig. 11 SEM images of bovine fibroblast cell growth on smooth (a) and wrinkled (b and c) posts. Comparing (a and b), cells preferred the flat surface when posts were smooth, whereas with the wrinkled posts they attached to and spread on the posts, conforming their 3D curvature. Some cells even formed bridges between the wrinkled posts, creating an overhanging connection between them (c). Scale bars: 30 μm. Reproduced with permission.131 | ||
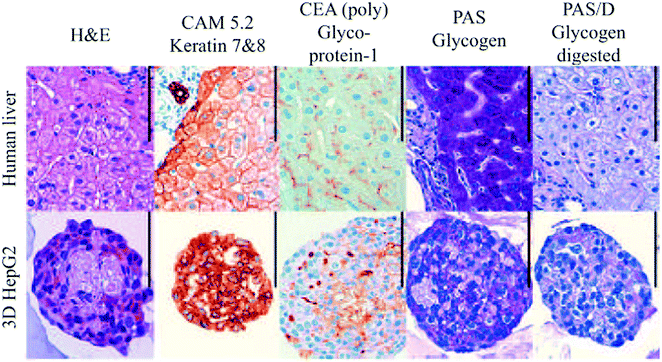 | ||
| Fig. 12 Histological examination of human liver and HepG2 spheroids. Scale bar: 100 μm. Reproduced with permission.139 | ||
In contrast to 2½D platforms reviewed in the previous section, studies on advanced surface topographies that are capable of producing 3D cell geometries have just begun in the past decade. Nonetheless, they have provided exciting opportunities to elucidate the cell metabolism and function in more detail, and help advance engineered microenvironments to better emulate in vivo conditions.
3 A comparison with three-dimensional cell culture systems
In this section, we first briefly introduce different 3D cell culture techniques and explain their differences with 2D monolayer models. Next, the advantages and disadvantages of 3D cultures are weighed against 2½D platforms for human bone-marrow-derived MSCs, as one of the most investigated cell types using these techniques in the literature.3.1 3D vs. 2D (planar surfaces)
Three-dimensional ex vivo cell culture models can be categorized as spheroids or microtissues, microcarriers, and tissue-engineered models. Microcarriers are mainly used to expand anchorage-dependent cells in vitro, providing several advantages compared to 2D models such as enhancing in vitro differentiation30 with high reproducibility and scale-up potential.31 However, due to their relatively less diverse applications, we will mainly focus on spheroids and 3D tissue-engineered models in this review.Cellular spheroids are simple 3D tissue analogs that are self-assembled due to the tendency of adherent cells to aggregate. The term spheroid is more commonly used because most tissue aggregates are spherical, whereas a more general term, namely multi-cellular microtissues or simply microtissues, describes other shapes as well.133,134 Several single culture or co-culture techniques, such as pellet culture, spinner culture, hanging drop, rotating wall vessel (RWV) and microfluidics, have been used to create microtissues.32,33 Regardless of the fabrication method, spheroids with a high cell density create more in vivo-like microenvironments by forming complex cell–cell and cell–ECM interactions in addition to establishing diffusion barriers to soluble components (e.g., nutrients, oxygen, growth factors and wastes). These biomimetic characteristics are particularly crucial for cancer drug discovery applications. The molecular gradient of nutrients and oxygen can cause necrosis and hypoxia toward the center of the spheroids, and the enhanced intercellular communications can regulate cell function and fate.32 Similarly, solid tumors often show hypoxia/necrosis, slow proliferation and drug diffusion barriers that contribute to cancer drug resistance.135 These features are not properly reflected in monolayer cell cultures in which cells proliferate rapidly, show high drug activity and have equal access to high concentrations of nutrients and oxygen.
Several other studies have demonstrated the benefits of spheroids over 2D monolayers.136–138 For example, as illustrated in Fig. 12, hepatocyte spheroids exhibit liver-like functionalities with improved differentiation and reduced proliferation in contrast to 2D monolayers of hepatocytes, which rapidly dedifferentiate and lose critical hepatocyte functions.139
Nonetheless, various challenges in spheroid culture procedures still remain. Conventional methods for spheroid production, such as spinner cultures, hanging drop, and rotating systems, can be labor-intensive, time-consuming, and are unable to maintain spheroid size uniformity on large scales,140,141 whereas more sophisticated platforms142 can be more complicated and require specially trained users for fabrication and operation. Table 2 provides a list of advantages and disadvantages of spheroids compared to 2D models.
| Cellular characteristics | 2D monolayer | 3D cell spheroids | 3D tissue-engineered models | Ref. |
|---|---|---|---|---|
| Morphology | Flat and stretched monolayer | Aggregates of natural 3D shapes at the interior of spheroid | Can assume stellate, round or elongated morphologies. Regardless of the shape they are homogeneously surrounded by the ECM | 135, 139, 143 and 144 |
| Adhesion | Cells adhere to the supporting surface | Cell-to-cell adhesion is dominant | Both cell–cell and cell–ECM adhesion can be observed | 144 and 145 |
| Viability | Cell viability is acceptable | Cell viability is improved or is similar to 2D | 3D cultures can significantly increase cell viability | 32, 136, 137, 139, 146 and 147 |
| Stage of cell cycle | Cells are more likely to be at the same cycle due to being evenly exposed to the medium | May contain proliferating, quiescent, hypoxic and necrotic cells. Larger spheroids show more necrosis in the center due to hypoxia | — | 139 |
| Proliferation | Often proliferate faster than in vivo | Lower or comparable proliferation rate compared to 2D | Lower, comparable or higher proliferation rate compared to 2D | 139, 143, 148–150 |
| Differentiation | Often reduces the differentiative capacities of stem cells | Can enhance the differentiation potential of multipotent cells | Can preserve the differentiative capacities and provide a more reliable understanding of differentiation mechanisms | 34, 136, 151–153 |
| Gene and protein expression | Gene and protein expression levels generally differ from in vivo models | Gene and protein expression profiles are closer to natural tissues | Can furnish similar gene and protein expressions as in in vivo conditions | 34, 139 and 154 |
| Cellular interactions | Cell–surface interactions prevail rather than cell–cell and cell–ECM interactions | Cellular and cell–ECM interactions are promoted | Interactions of the cell cytoskeleton and the ECM can significantly vary between 2D and 3D scaffold-based models, impacting various cell activities, such as apoptosis | 34, 138 and 139 |
| Directed growth | Cells grow randomly | Cellular aggregates are mainly spherical, whereas other shapes were obtained in limited studies | Can be customized to direct cellular growth in various patterns, such as elongated or heterogeneous cell distributions | 133, 134, 155 and 156 |
| Sample handling and imaging | Simple and well suited for high-throughput screening | Sample handling is more complicated while large number of spheroids can be provided for high-throughput drug testing. | Sample handling and high-throughput screening is more challenging | 35, 39 and 157 |
| Scale up and reproducibility | Less expensive for large-scale studies and easier to reproduce | Depending on the fabrication technique, spheroid properties can be reproducible or greatly variable | Reproducibility can be challenging and scaffold's properties may vary from batch to batch for naturally derived scaffolds | 135, 139, 141, 158 and 159 |
| Complexity | Involve the simplest procedures | Often labor-intensive, time-consuming, more expensive and more complicated | More expensive and more complicated compared to 2D | 141 |
| Drug testing | Drugs appear effective but the results are physiologically irrelevant | Cells often show more resistance to therapeutics and better represent in vivo conditions | Cells responses are more reliable, showing more resistance to drugs or providing better prediction of drug cytotoxicity and efficacy | 135 and 160–162 |
| Wound healing | Lower effectiveness compared to 3D models, as simulating natural conditions is almost impossible | Spheroids have been shown to promote angiogenesis and wound healing compared to single cells recovered from 2D cultures | These models can allow a more reliable studying of wound healing by better evaluating the cell contractility and matrix compaction as well as simulating the mechanical forces in tissue in vivo | 163 and 164 |
In 3D tissue-engineered models, cells are grown in 3D matrices or scaffolds that are prepared ex vivo using natural and/or synthetic materials. Generally, it is preferable to use the materials extracted from living tissue ECM that are specific to the cells under study. Commercially available gelatinous protein mixtures, such as Matrigel®, and decellularized tissues used for treating cardiovascular diseases165 and diabetes166 as well as for reconstruction medicine167 are practical examples of this method. However, such matrices are often difficult to customize and may vary from batch to batch.158 Alternatively, attention has been paid to synthetic polymers that can form 3D networks with microscopic pores. Polymer selection depends on cell adhesion, biocompatibility, wettability and structural properties such as porosity, pore geometry, transport and degradation kinetics. Detailed reviews on artificial 3D cell culture scaffolds and their characteristics can be found elsewhere.4,39,158,168–174 Spatial organization of cells in the media directly depends on their microstructural geometry. Therefore, perfecting the material structure to mimic the tissue is a key step in regulating cell growth and promoting the functionality of specific cell types to produce targeted outcomes that are unachievable in 2D planar models.
An example of artificial 3D scaffolds that direct cell proliferation and differentiation is a 3D self-assembled network of peptide amphiphiles introduced by Silva et al.,175 which was used to grow murine neural progenitor cells. The self-assembly was triggered by injection of cell culture medium, causing the peptides to aggregate into nanofibers with high aspect ratios, thereby forming a solid hydrogel that mechanically supported the medium (Fig. 13(a)). The molecular design of the scaffolds promoted neurite sprouting and direct neurite growth, inducing very rapid cell differentiation into neurons.
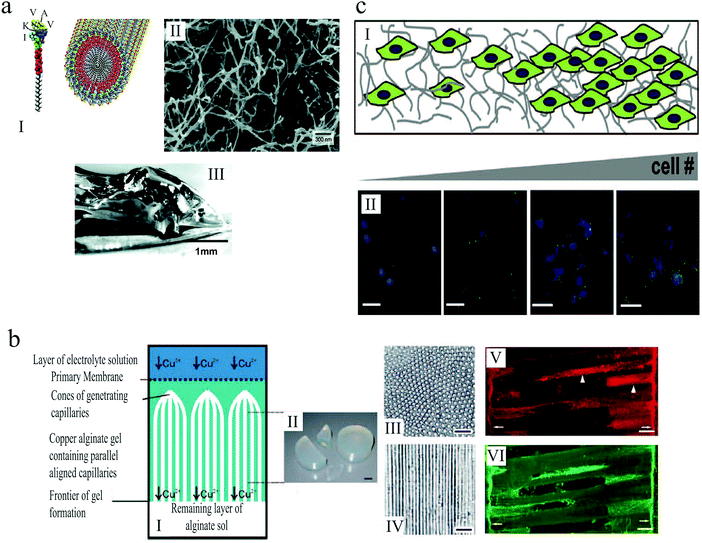 | ||
| Fig. 13 (a) (I) Schematic illustration of the self-assembly of peptide amphiphile molecules into nanofibers. (II) SEM micrograph of the nanofiber network after adding the cell medium. (III) Macroscopic image of the gel formed by adding peptide solution to the cell culture medium.175 (b) (I) Illustration of anisotropic capillary alginate gel formation. (II) Macroscopic image of the gel. (III) Cross section of the gel. (IV) Longitudinal section of the gel. (V and VI) Confocal fluorescent micrographs show regenerating axons in the alginate capillaries. Scale bars: 1 cm (b, II), 100 μm (b, III and IV), and 50 μm (b, V and VI).155 (c) (I) Schematic illustration of gelatin hydrogels designed by microfluidics that contain a gradient in cell number. (II) Increase in hypoxia inducible factor 1 gene expression (green) with increasing cell (blue) density across the gel. Scale bar: 50 μm.156 Reproduced with permission. | ||
Alginate-based scaffolds have been used to promote directed axon regeneration in vitro and in adult rat spinal cord lesions in vivo.155 The hydrogel microstructure contained self-assembled anisotropic capillaries that were formed during the gel synthesis (Fig. 13(b)). After seeding with adult neural progenitor cells, which are known to improve axon regeneration, these gels were implanted into acute cervical spinal cord lesions in rats and integrated into the spinal cord parenchyma without inflammatory responses. In accordance with in vitro findings, the anisotropic structure of these implants induced directed axon regeneration across the artificial scaffold, as shown in Fig. 13(b)(V) and (VI). Moreover, the combined cell- and artificial scaffold-based therapy further improved neuron restoration. The alginate-based hydrogels were presented as a promising strategy for nerve regrowth following spinal cord injury, when the targeted neuron reinnervation depends on longitudinally directed regrowth of transected axons.
Tumor microenvironments are temporally and spatially heterogeneous, and it is unclear how the gradations in biophysical cues can impact malignant phenotype and response to therapy. Three dimensional platforms capable of mimicking this complexity can provide valuable information about tumor etiology, growth and spreading. In an attempt to replicate the heterogeneity of tumor ECMs, Pedron et al.156 generated 3D hydrogels with controlled spatial gradient of matrix and cellular content (Fig. 13(c)). Investigating glioblastoma multiforme (GBM), as a common form of brain tumor in adults, they demonstrated how gradients in cell density can lead to heterogeneous gene expression. Moreover, they reported that a spatially graded matrix stiffness can significantly alter cell proliferation and morphology.
Several other novel hydrogel designs for cell culture have been reported in the literature176–182 that demonstrate the superiority of 3D cultures to 2D planar substrates. Table 2 also furnishes a thorough comparison between 3D scaffold-based and 2D monolayer models. Ultimately, an ideal environment for 3D cell culture requires a control of structural and physicochemical properties on length scales from nano to macro, and time scales from seconds to weeks. Therefore, the next step in effectively mimicking the ECM and guiding cell behavior is to develop materials whose properties can be tuned according to the time and length scale of the cell development. The conventional fabrication approaches, such as lithography, molding and solvent casting, are unable to sufficiently control the scaffold properties. These include nanoscale surface modifications, such as biofunctionalization; microstructural details, such as pore size and shape, porosity, and pore distribution; macroscale architecture, such as the external appearances of artificial devices. Hence, a user-controlled and replicable technique seems essential. 3D bioprinting has recently emerged to address these shortcomings.
Bioprinting is a rapidly growing computer-aided technology that can create complex tissue architectures by patterning a cell-encapsulating bioink.183 Owing to its significantly improved resolution and cheaper cost in the recent years, it is implemented in tissue engineering to create 3D cell-laden materials in which the bioink contains the cells,184,185 or to produce scaffolds onto which cells are seeded.186 Due to its versatile patterning of cells and scaffolding materials, bioprinting has been able to revolutionize biomedical applications, such as organ-on-a-chip devices,187 cancer research studies,188,189 and pharmaceutical analyses.190 Organ printing, which uses this technology to prototype living human organs (Fig. 14), offers new promise for solving organ transplantation crisis.191–194
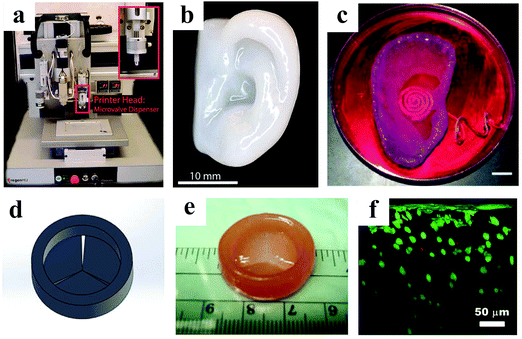 | ||
| Fig. 14 A 3D bioprinter (a) was used to prototype a human ear (b).195 (c) Image of a 3D printed bionic ear during in vitro culture. Scale bar: 1 cm.196 Panels (d, e and f) show a heart valve model designed by Solidworks, the bioprinted valve conduit, and the cross-sectional view of live/dead cells from surface to >300 μm depth, respectively.197 Reproduced with permission. | ||
Bioprinted structures offer various desirable features to the field of tissue engineering. Spatial patterning of complex 3D environments can be relatively easy using 3D printing.183,198 Bioprinting also allows for co-culturing of multiple cell-types, which can be attractive for 3D modeling of neural tissues185,199 that contain various glial cell types.183 Moreover, it is known for its ability to control the scaffold porosity and produce interconnected pores,200 which can greatly enhance cell attachment and ingrowth,201 both desirable in tissue engineering applications.202,203 For more information about different aspects of bioprinting, such as the choice of material and bioprinter, challenges, and future perspectives of this technology, the reader is encouraged to see detailed reviews available elsewhere.183,185,194,204–206
3.2 3D vs. 2½D (non-planar surfaces)
Two-dimensional monolayer cultures are usually used as control experiments when developing 3D culture systems and investigating their impacts on cellular functions. Therefore, direct comparison between 2D and 3D models, reported in Section 3.1, could be readily found in the literature. However, topographical substrates are not usually directly compared with 3D systems in a single study, requiring a broader and more careful survey to acquire this comparative information. In addition, an accurate comparison can be conducted only between similar cell types that are derived from the same tissue origin. For example, Baksh et al.207 demonstrated that human MSCs derived from umbilical cord and bone marrow may exhibit different proliferative and differentiative capacities. Therefore, we will only focus on bone-marrow-derived human MSCs in this section, since they have been more extensively investigated using all culture systems.Bone-marrow-derived MSCs are multipotential cells which are capable of self-renewing and differentiating to a number of cell types, including osteocytes, chondrocytes, adipocytes, hepatocytes, myocytes, neurons, and cardiomyocytes. For tissue engineering applications, MSCs are often expanded and/or differentiated in vitro prior to in vivo transplantation. Conventional 2D monolayer methods normally fail to maintain and/or support efficient cellular functions such as differentiation and replication. On the other hand, topographical substrates, spheroids and 3D tissue constructs have been shown to improve MSC differentiation while providing proliferation and viability similar to or higher than 2D models, according to several studies summarized in Table 3. However, the cell characteristics as well as biomaterial properties can greatly impact the outcome. For example, 3D-polycaprolactone/hydroxyapatite (PCL/HAp) scaffolds have been shown to increase the proliferation of bone-marrow-derived MSCs compared to 2D monolayers, whereas adipose-derived MSCs exhibited similar replication rate on 3D scaffolds and monolayer cultures.208 In another study, MSC viability on nanofibrous topographies with random fibre orientations, which was assumed to better mimic the structure of the native bone ECM, was higher than that on aligned fibres.209
| Structure/fabrication | Cell morphology | Viability | Proliferation | Differentiation | Ref. |
|---|---|---|---|---|---|
| a Cell functions in this example are in comparison with their spheroid counterparts. | |||||
| 2½D substrates | |||||
| Square array of PMMA nanopits | Flat and polygonal morphology | — | Higher proliferation and cell phenotype/multipotency retention on ordered arrays than 2D planar control and displaced nanopits | Higher osteoblastic differentiation on displaced arrays than 2D flat and ordered nanopits | 98 and 99 |
| Silicon nanowires | More spherical and less elongated morphologies with sturdy protrusions | Cell survivability was close to monolayers | — | Shortest nanowires significantly promoted osteogenic differentiation than 2D monolayers and longer nanowires | 210 |
| PCL nanofiber substrates | Aligned according to fibre orientation. Cells on aligned fibers were elongated and spindle-shaped, whereas they were mostly round on random-oriented fibers | Higher viability on random fibres than aligned fibres and 2D culture plates | Improved proliferation on random fibres compared with aligned orientations and monolayers | Osteochondrogenic differentiation was significantly increased on random fibres | 209 |
| 3D cell spheroids | |||||
| Spinner flasks and RWV | Continuous layer of elongated MSCs on the outer surface with irregularly shaped cells in the spheroid interior | Comparable viability to monolayer cultures | 4.3% or 3.5% proliferating cells for spinner and RWV, which was comparable to monolayers | Increased osteogenic and adipogenic differentiation in spheroids, where the latter was more significant in RWV | 211 |
| Hydrogel encapsulated spheroids formed using the hanging drop technique | — | Entrapped spheroids exhibit greater resistance to apoptosis than entrapped cells from monolayer cultures | Proliferation did not differ between entrapped spheroids and dissociated cells | Similar osteogenic potential for spheroids and their monolayer counterparts, when encapsulated in fibrin gels | 136 |
| Photolithography and micropatterning techniques | 3D cell aggregates | High viability was obtained for at least 30 days | — | Adipogenic and osteogenic differentiation was more efficient in spheroids than monolayers | 212 |
| 3D tissue-engineered models | |||||
| 3D PCL nanofibrousa scaffolds | Flat fibroblast-like cells on the surface with round, chondrocyte-like cells in middle, surrounded by abundant cartilaginous matrix | — | Cell proliferation was higher than in cell pellets | Chondrogenic differentiation was similar to cell pellets | 213 |
| β-Tricalcium phosphate (TCP) ceramics | — | Mean values of metabolic activity was larger with porosity, however, the comparison was elusive due to high standard deviations | Proliferation was proportional to porosity in 3D scaffolds, and significantly higher than monolayer culture | 3D culture provided higher osteogenic differentiation than 2D monolayer, but it was independent of porosity | 150 |
| 3D-Polycaprolactone/hydroxyapatite (PCL/HAp) scaffold | — | Cells were highly viable | Higher proliferation rate than 2D monolayer | ALP activity and mineral deposition were higher on the 3D scaffold than 2D culture plates, indicating higher osteogenic differentiation | 208 |
Comparing 2½D and 3D systems, spheroids might accommodate higher cell culture capacity in cell aggregates than 2½D substrates and 3D scaffolds with limited culture spaces.212 However, the biomaterial-based culture methods often improve cell proliferation (Table 3), while spheroids usually yield similar/lower proliferation rate compared to 2D monolayers.136,139,211 On the other hand, MSC differentiation can be improved using all three methods, thus, the optimal selection should be based on the final application. Due to their structurally controlled surface characteristics, topographical substrates can provide more detailed information about intercellular mechanics by tailoring and manipulating cell–cell interactions. Similarly, they may be more desirable for high-throughput screening and imaging, since cells are far more accessible for optical characterization when they are plated onto a surface than confined within the pores of a 3D scaffold with high thickness.
While both spheroids and scaffold-based models can be good sources for obtaining a supply of specifically differentiated MSCs or providing valuable information about MSC differentiation, one might be more desirable than the other depending on the purpose of the culture. In tissue engineering applications, such as cartilage repair, spheroids usually suffer from small size and uniformly weak mechanical properties,213 and these limitations can be addressed using 3D scaffold-based models. Spheroids, however, have been rather more attractive for other applications such as cancer research by better mimicking the human solid tumor microenvironments, where necrotic, quiescent and proliferating cells are present.34 In some cases, a combination of both methods can be more effective. For example, hydrogel encapsulated spheroids have been shown to exhibit greater resistance to apoptosis than entrapped dissociated cells.136 Nonetheless, a judicious selection of the cell culture method is a trade-off between the efficacy and performance of the model, sample handling, complexity, reproducibility and economical factors.
4 Applications
Cell-based devices, analyses and therapies are increasingly being used in clinical applications. Here, we describe some recent examples, although many others can be found in the literature.4.1 Cell therapy
Cell therapy has been widely used in a variety of clinical contexts. Stem cell transplantation is a promising therapeutic strategy for several illnesses, such as neurological214 and cardiovascular215 diseases, type 1 diabetes,216 and osteogenesis imperfecta.37,217Prior to implantation, the stem cells isolated from a donor are expanded in vitro using conventional 2D tissue culture techniques.218 However, it has been demonstrated that certain cell-specific properties of stem cells are lost over time in 2D culture to the point they no longer reflect in vivo cell behavior.38,219 These cells are usually unable to maintain long-term tissue regeneration upon reinfusion into the body, highlighting the necessity of improved methods for in vitro cell culture. For example, 2½D99 and 3D211 cultures have been shown to enhance osteogenic and adipogenic differentiation potential in MSCs when compared with 2D monolayer cultures.
Recently, 3D scaffolds were used to regenerate an entire, fully functional epidermis on a seven-year-old child suffering from a life-threatening form of JEB.52 For this treatment, the keratinocyte cells were cultured on 3D fibrin scaffolds, allowing the preparation of larger grafts from the same number of cells as would be needed for 2D plastic-cultured grafts.52,53
4.2 Vaccine production
In 1949, the key scientific discovery by Enders et al.220 that poliovirus could be grown in cell culture led to the development of poliovirus vaccine, and to significant advances in producing other virus vaccines in the following years. Prior to this, the few available vaccines were produced in animal systems, such as embryonated eggs for influenza and yellow fever viruses.221Currently, many viruses are still being produced in animal systems due to reliably high yields. However, these methods are time consuming and expensive. In addition, in some cases, such as embryonated eggs for seasonal influenza, the vaccines are developed more than 6 months in advance of the flu season to allow for adequate time to produce the required quantities. This lag can be problematic if the selected strains do not match the predominant ones during the winter. Unfortunately, people with egg allergies cannot use vaccines that are produced in eggs. Therefore, finding an alternative approach, such as in vitro cell culture, can be attractive.222 In this regard, Tree et al.223 introduced the 3D culture of MDCK cells using microcarriers224 as a potential technique for producing influenza virus A vaccine strains. Although they suggested that a comparable yield of influenza virus A production could be obtained between scaled-up 3D cell cultures and embryonated eggs, the differences may be larger for a different strain of influenza virus. Thus, the search for an efficient approach is still ongoing.
4.3 Disease modeling
Engineered tissues can furnish a powerful tool to probe the disruption in cell organization and function in response to a disease.225 However, to obtain biologically relevant models, the cell arrangement should be closely related to the tissue organization in vivo. For example, human embryonic stem cells can be directed into cardiac lineages, such as cardiomyocytes,226 for disease modeling. Whereas ventricular cardiomyocytes are naturally aligned, human embryonic stem cell-derived cardiomyocytes are heterogenous and randomly organized. It has been demonstrated that biomimetic culture platforms comprising physical topographies, such as wrinkles227 and grooves,228 can stimulate cardiomyocyte cell alignment and, therefore, lead to physiologically relevant responses.228The dynamic cell alignment can be an important factor when modeling the progression of various diseases. For example, during heart failure, the highly aligned cardiac muscle fibres become disorganized, causing progressive fibrosis and ventricular dilatation. While many of the in vitro models using surface topographies are static, a tunable culture platform that can dynamically manipulate the temporal and spatial cell alignments can be attractive. Lam et al.229 were the first to introduce reversible cell alignment by compression-induced dynamic lamellar patterns on PDMS substrates. However, their pattern versatility was limited to one direction. To address this limitation, Guvendiren and Burdick230 reported the fabrication of strain-responsive buckling patterns on PDMS substrates that could control the human MSC organization with a higher complexity and versatility. In their study, multidirectional lamellar patterns appeared and disappeared when relaxing and stretching the substrates, respectively, in various directions and angles. They observed that the preferential alignment of MSCs in the presence of lamellar topography was completely eliminated by stretching the platform, producing a disorganized cell arrangement. Such platforms can have considerable potential for targeted disease modeling and drug discovery.
4.4 Drug screening
Drug development and pharmacokinetics studies using animal models are costly, time consuming and arguably unethical when using an extensive number of animals.231 Therefore, the early stages of drug discovery rely on toxicity testing using in vitro cell-based assays, to at least exclude poor therapeutic candidates before animal testing.94,232 To increase their predictive accuracy, the cell-based systems are expected to effectively represent essential aspects of in vivo conditions. However, a substantial number of new therapeutic agents fail in late-stage human drug testing,233 calling for better in vivo-mimetic culture platforms.The cell behavior is greatly regulated by the cell's genetic codes and its communications with neighboring cells.234 Therefore, the spatial position of cells is crucial for their functionality235 and their response to a therapeutic agent. For example, multicellular tumor spheroids have been shown to provide more physiologically relevant models for anticancer drug screening compared to monolayer cultures, because the intercellular adhesion in tumor spheroids can assist tumor cells in evading the cytotoxic effects of anticancer drugs.236 Cell patterning on engineered extracellular environments and, then replicating the cultured cells in miniaturized regular arrays can improve cell-based drug testing.232,237 For example, tumor-on-a-chip systems comprising arrays of tumor spheroids have been presented by several researchers to promote cancer drug discovery238 by providing a large number of uniform tumor spheroids for enhanced drug screening.239
5 Summary and outlook
Cells in their biological ECM reside within a complex 3D network, in which every physical and chemical detail is essential for cell functionality. The oversimplified monolayer cell cultures have appreciably promoted our understanding of cell function. Nonetheless, they are notoriously inadequate for successful clinical applications. Adapting to their new environment, cultured cells lose specific properties over time and deviate from their in vivo behaviors. Thus, there has been an increasing demand for culture platforms that better emulate biological conditions.A wide range of ordered and random topographical surfaces (identified as 2½D substrates), capable of better mimicking the natural ECM, and their impact on cell behavior were detailed in this review. With a relatively simple fabrication procedure, 2½D platforms show great potential in advancing biomedical applications by retaining in vivo cell functionalities, such as their proliferative capacity. This can be significantly important for cell transplantation and tissue engineering, where the cells should display a long-term regeneration upon reinfusion into the body. In addition, topographical structures with tunable characteristics that induce 3D cell morphology furnish exciting opportunities for better understanding cellular response to environmental cues. Stimuli-responsive platforms are another type of novel artificial ECMs that are expected to contrive the dynamic investigation of cellular responses. Despite these advances, major challenges remain as innumerate factors simultaneously contribute to cell behavior. Nonetheless, recent advances in micro- and nanofabrication techniques have paved the route to studying a single cell population and to address these challenges.
Topographical substrates are also more desirable for cell imaging and high-throughput screening in comparison with 3D models, in which optical-readouts can be particularly cumbersome. Nonetheless, in certain applications, such as cancer research and producing implantable tissue constructs, spheroids and 3D porous scaffolds can be better options as they better emulate in vivo ECMs. However, a judicious selection between topographical surfaces and 3D models depends on the efficiency and performance of the models to achieve a targeted outcome as well as on sample handling, complexity, reproducibility and economical factors.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
Support from Bourses d'excellence TransMedTech (to M. M. and V. A.), FRQNT (The Fonds de recherche du Québec – Nature et technologies to V. A.), CRC (Canada Research Chairs program to X. B. and D. K. H.) and NSERC (Natural Sciences and Engineering Research Council of Canada to X. B. and D. K. H. (Discovery Grant 2017-04489)) is gratefully acknowledged.References
- M. E. Lukashev and Z. Werb, ECM signalling: Orchestrating cell behaviour and misbehaviour, Trends Cell Biol., 1998, 8, 437–441 CrossRef CAS PubMed.
- A. S. G. Curtis and C. D. W. Wilkinson, Reactions of cells to topography, J. Biomater. Sci., Polym. Ed., 1998, 9, 1313–1329 CrossRef CAS PubMed.
- M. J. Bissell, A. Rizki and I. S. Mian, Tissue architecture: The ultimate regulator of breast epithelial function, Curr. Opin. Cell Biol., 2003, 15, 753–763 CrossRef CAS PubMed.
- J. Lee, M. J. Cuddihy and N. A. Kotov, Three-dimensional cell culture matrices: State of the art, Tissue Eng., Part B, 2008, 14, 61–86 CrossRef CAS PubMed.
- B. Geiger, J. P. Spatz and A. D. Bershadsky, Environmental sensing through focal adhesions, Nat. Rev. Mol. Cell Biol., 2009, 10, 21–33 CrossRef CAS PubMed.
- L. Wang and R. L. Carrier, in Advances in Biomimetics, ed. A. George, InTech, 2011, ch. Biomimetic topography: Bioinspired cell culture substrates and scaffolds Search PubMed.
- M. M. Stevens and J. H. George, Exploring and engineering the cell surface interface, Science, 2005, 310, 1135–1138 CrossRef CAS PubMed.
- J. El-Ali, P. K. Sorger and K. F. Jensen, Cells on chips, Nature, 2006, 442, 403–411 CrossRef CAS PubMed.
- A. M. Ross, Z. Jiang, M. Bastmeyer and J. Lahann, Physical aspects of cell culture substrates: Topography, roughness, and elasticity, Small, 2012, 8, 336–355 CrossRef CAS PubMed.
- C. J. Bettinger, R. Langer and J. T. Borenstein, Engineering substrate topography at the micro- and nanoscale to control cell function, Angew. Chem., Int. Ed., 2009, 48, 5406–5415 CrossRef CAS PubMed.
- B. Kasemo and J. Gold, Implant surfaces and interface processes, Adv. Dent. Res., 1999, 13, 8–20 CrossRef CAS PubMed.
- T. J. Webster, C. Ergun, R. H. Doremus, R. W. Siegel and R. Bizios, Enhanced functions of osteoblasts on nanophase ceramics, Biomaterials, 2000, 21, 1803–1810 CrossRef CAS PubMed.
- Y. Wan, Y. Wang, Z. Liu, X. Qu, B. Han, J. Bei and S. Wang, Adhesion and proliferation of OCT-1 osteoblast-like cells on micro- and nano-scale topography structured poly(L-lactide), Biomaterials, 2005, 26, 4453–4459 CrossRef CAS PubMed.
- E. Palin, N. H. Liu and T. J. Webster, Mimicking the nanofeatures of bone increases bone-forming cell adhesion and proliferation, Nanotechnology, 2005, 16, 1828–1835 CrossRef CAS.
- M. J. Dalby, D. McCloy, M. Robertson, H. Agheli, D. Sutherland, S. Affrossman and R. O. C. Oreffo, Osteoprogenitor response to semi-ordered and random nanotopographies, Biomaterials, 2006, 27, 2980–2987 CrossRef CAS PubMed.
- M. J. Dalby, D. McCloy, M. Robertson, C. D. Wilkinson and R. O. Oreffo, Osteoprogenitor response to defined topographies with nanoscale depths, Biomaterials, 2006, 27, 1306–1315 CrossRef CAS PubMed.
- P. T. de Oliveira, S. F. Zalzal, M. M. Beloti, A. L. Rosa and A. Nanci, Enhancement of in vitro osteogenesis on titanium by chemically produced nanotopography, J. Biomed. Mater. Res., Part A, 2007, 80, 554–564 CrossRef PubMed.
- A. C. De Luca, M. Zink, A. Weidt, S. G. Mayr and A. E. Markaki, Effect of microgrooved surface topography on osteoblast maturation and protein adsorption, J. Biomed. Mater. Res., Part A, 2015, 103, 2689–2700 CrossRef CAS PubMed.
- J. E. Barralet, L. Wang, M. Lawson, J. T. Triffitt, P. R. Cooper and R. M. Shelton, Comparison of bone marrow cell growth on 2D and 3D alginate hydrogels, J. Mater. Sci.: Mater. Med., 2005, 16, 515–519 CrossRef CAS PubMed.
- J. Lee, G. D. Lilly, R. C. Doty, P. Podsiadlo and N. A. Kotov, In vitro toxicity testing of nanoparticles in 3D cell culture, Small, 2009, 5, 1213–1221 CAS.
- S. Breslin and L. O'Driscoll, Three-dimensional cell culture: the missing link in drug discovery, Drug Discovery Today, 2013, 18, 240–249 CrossRef CAS PubMed.
- S. Even-Ram and K. M. Yamada, Cell migration in 3D matrix, Curr. Opin. Cell Biol., 2005, 17, 524–532 CrossRef CAS PubMed.
- T. Elkayam, S. Amitay-Shaprut, M. Dvir-Ginzberg, T. Harel and S. Cohen, Enhancing the drug metabolism activities of C3A–A human hepatocyte cell line–by tissue engineering within alginate scaffolds, Tissue Eng., 2006, 12, 1357–1368 CrossRef CAS PubMed.
- J. Barrila, A. L. Radtke, A. Crabbé, S. F. Sarker, M. M. Herbst-Kralovetz, C. M. Ott and C. A. Nickerson, Organotypic 3D cell culture models: using the rotating wall vessel to study host-pathogen interactions, Nat. Rev. Microbiol., 2010, 8, 791–801 CrossRef CAS PubMed.
- C. R. Thoma, M. Zimmermann, I. Agarkova, J. M. Kelm and W. Krek, 3D cell culture systems modeling tumor growth determinants in cancer target discovery, Adv. Drug Delivery Rev., 2014, 69–70, 29–41 CrossRef CAS PubMed.
- D. D. McKinnon, D. W. Domaille, J. N. Cha and K. S. Anseth, Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3D cell culture systems, Adv. Mater., 2014, 26, 865–872 CrossRef CAS PubMed.
- Y. C. Lu, W. Song, D. An, B. J. Kim, R. Schwartz, M. Wu and M. Ma, Designing compartmentalized hydrogel microparticles for cell encapsulation and scalable 3D cell culture, J. Mater. Chem. B, 2015, 3, 353–360 RSC.
- S. Utech, R. Prodanovic, A. S. Mao, R. Ostafe, D. J. Mooney and D. A. Weitz, Microfluidic generation of monodisperse, structurally homogeneous alginate microgels for cell encapsulation and 3D cell culture, Adv. Healthcare Mater., 2015, 4, 1628–1633 CrossRef CAS PubMed.
- V. van Duinen, S. J. Trietsch, J. Joore, P. Vulto and T. Hankemeier, Microfluidic 3D cell culture: from tools to tissue models, Curr. Opin. Biotechnol., 2015, 35, 118–126 CrossRef CAS PubMed.
- L. Y. Sun, D. K. Hsieh, W. S. Syu, Y. S. Li, H. T. Chiu and T. W. Chiou, Cell proliferation of human bone marrow mesenchymal stem cells on biodegradable microcarriers enhances in vitro differentiation potential, Cell Proliferation, 2010, 43, 445–456 CrossRef CAS PubMed.
- S. Sart, S. N. Agathos and Y. Li, Engineering stem cell fate with biochemical and biomechanical properties of microcarriers, Biotechnol. Prog., 2013, 29, 1354–1366 CrossRef CAS PubMed.
- R. Z. Lin and H. Y. Chang, Recent advances in three-dimensional multicellular spheroid culture for biomedical research, Biotechnol. J., 2008, 3, 1172–1184 CrossRef CAS PubMed.
- T.-M. Achilli, J. Meyer and J. R. Morgan, Advances in the formation, use and understanding of multi-cellular spheroids, Expert Opin. Biol. Ther., 2012, 12, 1347–1360 CrossRef CAS PubMed.
- M. Ravi, V. Paramesh, S. R. Kaviya, E. Anuradha and F. D. Paul Solomon, 3D cell culture systems: Advantages and applications, J. Cell. Physiol., 2015, 230, 16–26 CrossRef CAS PubMed.
- B. Derby, Printing and prototyping of tissues and scaffolds, Science, 2012, 338, 921–926 CrossRef CAS PubMed.
- G. K. Tan, D. L. M. Dinnes, P. T. Myers and J. J. Cooper-White, Effects of biomimetic surfaces and oxygen tension on redifferentiation of passaged human fibrochondrocytes in 2D and 3D cultures, Biomaterials, 2011, 32, 5600–5614 CrossRef CAS PubMed.
- E. M. Horwitz, D. J. Prockop, P. L. Gordon, W. W. K. Koo, L. A. Fitzpatrick, M. D. Neel, M. E. McCarville, P. J. Orchard, R. E. Pyeritz and M. K. Brenner, Clinical responses to bone marrow transplantation in children with severe osteogenesis imperfecta, Blood, 2001, 97, 1227–1231 CrossRef CAS PubMed.
- M. A. Baxter, R. F. Wynn, S. N. Jowitt, J. E. Wraith, L. J. Fairbairn and I. Bellantuono, Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion, Stem Cells, 2004, 22, 675–682 CrossRef CAS PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. K. Stelzer, The third dimension bridges the gap between cell culture and live tissue, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS PubMed.
- M. Y. Lee, R. A. Kumar, S. M. Sukumaran, M. G. Hogg, D. S. Clark and J. S. Dordick, Three-dimensional cellular microarray for high-throughput toxicology assays, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 59–63 CrossRef CAS PubMed.
- T. G. Fernandes, S. J. Kwon, S. S. Bale, M. Y. Lee, M. M. Diogo, D. S. Clark, J. M. S. Cabral and J. S. Dordick, Three-dimensional cell culture microarray for high-throughput studies of stem cell fate, Biotechnol. Bioeng., 2010, 106, 106–118 CAS.
- G. Fontana, J. Gershlak, M. Adamski, J. S. Lee, S. Matsumoto, H. D. Le, B. Binder, J. Wirth, G. Gaudette and W. L. Murphy, Biofunctionalized plants as diverse biomaterials for human cell culture, Adv. Healthcare Mater., 2017, 6, 1601225 CrossRef PubMed.
- J. E. Puskas and Y. Chen, Biomedical application of commercial polymers and novel polyisobutylene-based thermoplastic elastomers for soft tissue replacement, Biomacromolecules, 2004, 5, 1141–1154 CrossRef CAS PubMed.
- N. M. Alves, I. Pashkuleva, R. L. Reis and J. F. Mano, Controlling cell behavior through the design of polymer surfaces, Small, 2010, 6, 2208–2220 CrossRef CAS PubMed.
- I. Milo
![[s with combining tilde]](https://www.rsc.org/images/entities/char_0073_0303.gif) ev, Metallic materials for biomedical applications: Laboratory and clinical studies, Pure Appl. Chem., 2010, 83, 309–324 Search PubMed.
ev, Metallic materials for biomedical applications: Laboratory and clinical studies, Pure Appl. Chem., 2010, 83, 309–324 Search PubMed. -
I. Milo
![[s with combining tilde]](https://www.rsc.org/images/entities/char_0073_0303.gif) ev, in Surface treatments of titanium with antibacterial agents for implant applications, ed. S. Djokić, Springer International Publishing, Cham, 2016, pp. 1–87 Search PubMed.
ev, in Surface treatments of titanium with antibacterial agents for implant applications, ed. S. Djokić, Springer International Publishing, Cham, 2016, pp. 1–87 Search PubMed. - C. M. Agrawal and R. B. Ray, Biodegradable polymeric scaffolds for musculoskeletal tissue engineering, J. Biomed. Mater. Res., Part A, 2001, 55, 141–150 CrossRef CAS.
- J. A. Shibli, S. Grassi, L. Cristina de Figueiredo, M. Feres, E. Marcantonio, G. Iezzi and A. Piattelli, Influence of implant surface topography on early osseointegration: A histological study in human jaws, J. Biomed. Mater. Res., Part B, 2007, 80, 377–385 CrossRef PubMed.
- M. Goldberg, R. Langer and X. Jia, Nanostructured materials for applications in drug delivery and tissue engineering, J. Biomater. Sci., Polym. Ed., 2007, 18, 241–268 CrossRef CAS PubMed.
- H. Hillaireau and P. Couvreur, Nanocarriers' entry into the cell: relevance to drug delivery, Cell. Mol. Life Sci., 2009, 66, 2873–2896 CrossRef CAS PubMed.
- B. Dhandayuthapani, Y. Yoshida, T. Maekawa and D. S. Kumar, Polymeric Scaffolds in Tissue Engineering Application: A Review, Int. J. Polym. Sci., 2011, 2011, 290602 Search PubMed.
- T. Hirsch, et al., Regeneration of the entire human epidermis using transgenic stem cells, Nature, 2017, 551, 327–332 CrossRef CAS PubMed.
- G. Pellegrini, R. Ranno, G. Stracuzzi, S. Bondanza, L. Guerra, G. Zambruno, G. Micali and M. De Luca, The control of epidermal stem cells (holoclones) in the treatment of massive full-thickness burns with autologous keratinocytes cultured on fibrin, Transplantation, 1999, 68, 868–879 CrossRef CAS PubMed.
- A. S. Andersson, F. Bäckhed, A. von Euler, A. Richter-Dahlfors, D. Sutherland and B. Kasemo, Nanoscale features influence epithelial cell morphology and cytokine production, Biomaterials, 2003, 24, 3427–3436 CrossRef CAS PubMed.
- R. L. Price, K. Ellison, K. M. Haberstroh and T. J. Webster, Nanometer surface roughness increases select osteoblast adhesion on carbon nanofiber compacts, J. Biomed. Mater. Res., Part A, 2004, 70, 129–138 CrossRef PubMed.
- L. Altomare, N. Gadegaard, L. Visai, M. C. Tanzi and S. Farè, Biodegradable microgrooved polymeric surfaces obtained by photolithography for skeletal muscle cell orientation and myotube development, Acta Biomater., 2010, 6, 1948–1957 CrossRef CAS PubMed.
- K. Kolind, K. W. Leong, F. Besenbacher and M. Foss, Guidance of stem cell fate on 2D patterned surfaces, Biomaterials, 2012, 33, 6626–6633 CrossRef CAS PubMed.
- S. Turunen, A. M. Haaparanta, R. Äänismaa and M. Kellomäki, Chemical and topographical patterning of hydrogels for neural cell guidance in vitro, J. Tissue Eng. Regener. Med., 2013, 7, 253–270 CrossRef CAS PubMed.
- K. S. Masters and K. S. Anseth, Cell–material interactions, Adv. Chem. Eng., 2004, 29, 7–46 CAS.
- H. S. Dhowre, S. Rajput, N. A. Russell and M. Zelzer, Responsive cell–material interfaces, Nanomedicine, 2015, 10, 849–871 CrossRef CAS PubMed.
- P. Clark, P. Connolly, A. S. G. Curtis, J. A. T. Dow and C. D. W. Wilkinson, Topographical control of cell behaviour: I. Simple step cues, Development, 1987, 99, 439–448 CAS.
- P. Clark, P. Connolly, A. S. G. Curtis, J. A. T. Dow and C. D. W. Wilkinson, Topographical control of cell behaviour: II. multiple grooved substrata, Development, 1990, 108, 635–644 CAS.
- J. Mai, C. S. Sun, S. Li and X. Zhang, A microfabricated platform probing cytoskeleton dynamics using multidirectional topographical cues, Biomed. Microdevices, 2007, 9, 523–531 CrossRef PubMed.
- K. M. Beussman, M. L. Rodriguez, A. Leonard, N. Taparia, C. R. Thompson and N. J. Sniadecki, Micropost arrays for measuring stem cell-derived cardiomyocyte contractility, Methods, 2016, 94, 43–50 CrossRef CAS PubMed.
- D. S. Sutherland, M. Broberg, H. Nygren and B. Kasemo, Influence of nanoscale surface topography and chemistry on the functional behaviour of an adsorbed model macromolecule, Macromol. Biosci., 2001, 1, 270–273 CrossRef CAS.
- W. B. Tsai, Y. C. Ting, J. Y. Yang, J. Y. Lai and H. L. Liu, Fibronectin modulates the morphology of osteoblast-like cells (MG-63) on nano-grooved substrates, J. Mater. Sci.: Mater. Med., 2009, 20, 1367–1378 CrossRef CAS PubMed.
- E. A. Bremus-Koebberling and S. Beckemper, Nano structures via laser interference patterning for guided cell growth of neuronal cells, J. Laser Appl., 2012, 24, 042013 CrossRef.
- M. Ochesner, M. R. Dusseiller, H. M. Grandin, S. Luna-Morris, M. Textor, V. Vogel and M. L. Smith, Micro-well arrays for 3D shape control and high resolution analysis of single cells, Lab Chip, 2007, 7, 1074–1077 RSC.
- D. Falconnet, G. Csucs, H. M. Grandin and M. Textor, Surface engineering approaches to micropattern surfaces for cell-based assays, Biomaterials, 2006, 27, 3044–3063 CrossRef CAS PubMed.
- V. N. Truskett and M. P. C. Watts, Trends in imprint lithography for biological applications, Trends Biotechnol., 2006, 24, 312–317 CrossRef CAS PubMed.
- A. M. Ross and J. Lahann, Surface engineering the cellular microenvironment via patterning and gradients, J. Polym. Sci., Part B: Polym. Phys., 2013, 51, 775–794 CrossRef CAS.
- X. Liu and P. X. Ma, Polymeric scaffolds for bone tissue engineering, Ann. Biomed. Eng., 2004, 32, 477–486 CrossRef PubMed.
- V. A. Schulte, M. Díez, M. Möller and M. C. Lensen, Surface topography induces fibroblast adhesion on intrinsically nonadhesive poly(ethylene glycol) substrates, Biomacromolecules, 2009, 10, 2795–2801 CrossRef CAS PubMed.
- A. I. Teixeira, P. F. Nealey and C. J. Murphy, Responses of human keratocytes to micro- and nanostructured substrates, J. Biomed. Mater. Res., Part A, 2004, 71, 369–376 CrossRef PubMed.
- X. F. Walboomers, H. J. E. Croes, L. A. Ginsel and J. A. Jansen, Growth behavior of fibroblasts on microgrooved polystyrene, Biomaterials, 1998, 19, 1861–1868 CrossRef CAS PubMed.
- J. L. Charest, M. T. Eliason, A. J. García and W. P. King, Combined microscale mechanical topography and chemical patterns on polymer cell culture substrates, Biomaterials, 2006, 27, 2487–2494 CrossRef CAS PubMed.
- W. A. Loesberg, J. te Riet, F. C. M. J. M. van Delft, P. Schön, C. G. Figdor, S. Speller, J. J. W. A. van Loon, X. F. Walboomers and J. A. Jansen, The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion, Biomaterials, 2007, 28, 3944–3951 CrossRef CAS PubMed.
- E. Lamers, X. F. Walboomers, M. Domanski, J. te Riet, F. C. M. J. M. van Delft, R. Luttge, L. A. J. A. Winnubst, H. J. G. E. Gardeniers and J. A. Jansen, The influence of nanoscale grooved substrates on osteoblast behavior and extracellular matrix deposition, Biomaterials, 2010, 31, 3307–3316 CrossRef CAS PubMed.
- J. Chen, Q. Wang, S. Chen, S. A. Wickline and S. K. Song, In vivo diffusion tensor MRI of the mouse retina: a noninvasive visualization of tissue organization, NMR Biomed., 2011, 24, 447–451 CrossRef PubMed.
- T. Banerjee, S. Mukherjee, S. Ghosh, M. Biswas, S. Dutta, S. Pattari, S. Chatterjee and A. Bandyopadhyay, Clinical significance of markers of collagen metabolism in rheumatic mitral valve disease, PLoS One, 2014, 9, e90527 CrossRef PubMed.
- M. Ghibaudo, L. Trichet, J. Le Digabel, A. Richert, P. Hersen and B. Ladoux, Substrate topography induces a crossover from 2D to 3D behavior in fibroblast migration, Biophys. J., 2009, 97, 357–368 CrossRef CAS PubMed.
- B. Li, L. Xie, Z. C. Starr, Z. Yang, J. S. Lin and J. H. Wang, Development of micropost force sensor array with culture experiments for determination of cell traction forces, Cell Motil. Cytoskeleton, 2007, 64, 509–518 CrossRef PubMed.
- M. L. Rodriguez, B. T. Graham, L. M. Pabon, S. J. Han, C. E. Murry and N. J. Sniadecki, Measuring the contractile forces of human induced pluripotent stem cell-derived cardiomyocytes with arrays of microposts, J. Biomech. Eng., 2014, 136, 051005 CrossRef PubMed.
- X. F. Walboomers, W. Monaghan, A. S. G. Curtis and J. A. Jansen, Attachment of fibroblasts on smooth and microgrooved polystyrene, J. Biomed. Mater. Res., 1999, 46, 212–220 CrossRef CAS PubMed.
- M. T. Frey, I. Y. Tsai, T. P. Russell, S. K. Hanks and Y. L. Wang, Cellular responses to substrate topography: Role of myosin II and focal adhesion kinase, Biophys. J., 2006, 90, 3774–3782 CrossRef CAS PubMed.
- Z. Li, J. Song, G. Mantini, M. Y. Lu, H. Fang, C. Falconi, L. J. Chen and Z. L. Wang, Quantifying the traction force of a single cell by aligned silicon nanowire array, Nano Lett., 2009, 9, 3575–3580 CrossRef CAS PubMed.
- J. Fu, Y. K. Wang, M. T. Yang, R. A. Desai, X. Yu, Z. Liu and C. S. Chen, Mechanical regulation of cell function with geometrically modulated elastomeric substrates, Nat. Methods, 2010, 7, 733–736 CrossRef CAS PubMed.
- S. Huang, C. S. Chen and D. E. Ingber, Control of cyclin D1, p27Kip1, and cell cycle progression in human capillary endothelial cells by cell shape and cytoskeletal tension, Mol. Biol. Cell, 1998, 9, 3179–3193 CrossRef CAS PubMed.
- A. K. Harris, P. Wild and D. Stopak, Silicone rubber substrata: a new wrinkle in the study of cell locomotion, Science, 1980, 208, 177–179 CrossRef CAS PubMed.
- K. Burton, J. H. Park and D. L. Taylor, Keratocytes generate traction forces in two phases, Mol. Biol. Cell, 1999, 10, 3745–3769 CrossRef CAS PubMed.
- A. D. Bershadsky, N. Q. Balaban and B. Geiger, Adhesion-dependent cell mechanosensitivity, Annu. Rev. Cell Dev. Biol., 2003, 19, 677–695 CrossRef CAS PubMed.
- V. Karantalis, W. Balkan, I. H. Schulman, K. E. Hatzistergos and J. M. Hare, Cell-based therapy for prevention and reversal of myocardial remodeling, Am. J. Physiol. Heart Circ. Physiol., 2012, 303, H256–H270 CrossRef CAS PubMed.
- C. Dambrot, R. Passier, D. Atsma and C. L. Mummery, Cardiomyocyte differentiation of pluripotent stem cells and their use as cardiac disease models, Biochem. J., 2011, 434, 25–35 CrossRef CAS PubMed.
- M. Grskovic, A. Javaherian, B. Strulovici and G. Q. Daley, Induced pluripotent stem cells-opportunities for disease modelling and drug discovery, Nat. Rev. Drug Discovery, 2011, 10, 915–929 CAS.
- C. S. Chen, J. L. Alonso, E. Ostuni, G. M. Whitesides and D. E. Ingber, Cell shape provides global control of focal adhesion assembly, Biochem. Biophys. Res. Commun., 2003, 307, 355–361 CrossRef CAS PubMed.
- J. L. Tan, J. Tien, D. M. Pirone, D. S. Gray, K. Bhadriraju and C. S. Chen, Cells lying on a bed of microneedles: An approach to isolate mechanical force, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1484–1489 CrossRef CAS PubMed.
- C. H. Choi, S. H. Hagvall, B. M. Wu, J. C. Y. Dunn, R. E. Beygui and C. J. Kim, Cell interaction with three-dimensional sharp-tip nanotopography, Biomaterials, 2007, 28, 1672–1679 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS PubMed.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. C. Oreffo and M. J. Dalby, Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency, Nat. Mater., 2011, 10, 637–644 CrossRef CAS PubMed.
- A. S. G. Curtis, N. Gadegaard, M. J. Dalby, M. O. Riehle, C. D. W. Wilkinson and G. Aitchison, Cells react to nanoscale order and symmetry in their surroundings, IEEE Trans Nanobioscience, 2004, 3, 61–65 CrossRef CAS PubMed.
- A. S. G. Curtis, B. Casey, J. O. Gallagher, D. Pasqui, M. A. Wood and C. D. W. Wilkinson, Substratum nanotopography and the adhesion of biological cells. Are symmetry or regularity of nanotopography important?, Biophys. Chem., 2001, 94, 275–283 CrossRef CAS PubMed.
- J. O. Gallagher, K. F. McGhee, C. D. W. Wilkinson and M. O. Riehle, Interaction of animal cells with ordered nanotopography, IEEE Trans Nanobioscience, 2002, 1, 24–28 CrossRef CAS PubMed.
- C. C. Berry, G. Campbell, A. Spadiccino, M. Robertson and A. S. G. Curtis, The influence of microscale topography on fibroblast attachment and motility, Biomaterials, 2004, 25, 5781–5788 CrossRef CAS PubMed.
- J. Tan, H. Shen and W. M. Saltzman, Micron-scale positioning of features influences the rate of polymorphonuclear leukocyte migration, Biophys. J., 2001, 81, 2569–2579 CrossRef CAS PubMed.
- M. Werner, S. B. G. Blanquer, S. P. Haimi, G. Korus, J. W. C. Dunlop, G. N. Duda, D. W. Grijpma and A. Petersen, Surface curvature differentially regulates stem cell migration and differentiation via altered attachment morphology and nuclear deformation, Adv. Sci., 2017, 4, 1600347 CrossRef PubMed.
- M. Singh, D. A. Close, S. Mukundan, P. A. Johnston and S. Sant, Production of uniform 3D microtumors in hydrogel microwell arrays for measurement of viability, morphology, and signaling pathway activation, Assay Drug Dev. Technol., 2015, 13, 570–583 CrossRef CAS PubMed.
- M. Singh, S. Mukundan, M. Jaramillo, S. Oesterreich and S. Sant, Three-dimensional breast cancer models mimic hallmarks of size-induced tumor progression, Cancer Res., 2016, 76, 1–12 Search PubMed.
- J. Casey, X. Yue, T. D. Nguyen, A. Acun, V. R. Zellmer, S. Zhang and P. Zorlutuna, 3D hydrogel-based microwell arrays as a tumor microenvironment model to study breast cancer growth, Biomed. Mater., 2017, 12, 025009 CrossRef PubMed.
- S. Sant and P. A. a. Johnston, The production of 3D tumor spheroids for cancer drug discovery, Drug Discovery Today: Technol., 2017, 23, 27–36 CrossRef PubMed.
- N. I. Martins, M. P. Sousa, C. A. Custódio, V. C. Pinto, P. J. Sousa, G. Minas, F. Cleymand and J. F. Mano, Multilayered membranes with tuned well arrays to be used as regenerative patches, Acta Biomater., 2017, 57, 313–323 CrossRef CAS PubMed.
- T. Tzvetkova-Chevolleau, A. Stéphanou, D. Fuard, J. Ohayon, P. Schiavone and P. Tracqui, The motility of normal and cancer cells in response to the combined influence of the substrate rigidity and anisotropic microstructure, Biomaterials, 2008, 29, 1541–1551 CrossRef CAS PubMed.
- B. J. Papenburg, E. D. Rodrigues, M. Wessling and D. Stamatialis, Insights into the role of material surface topography and wettability on cell–material interactions, Soft Matter, 2010, 6, 4377–4388 RSC.
- E. Kim; J. Lee; S. Ahn; H. Jeon and K. Lee Cell culture over nanopatterned surface fabricated by holographic lithography and nanoimprint lithography, 2008 3rd IEEE International Conference on Nano/Micro Engineered and Molecular Systems, 2008, pp. 725–728.
- J. Sun, Y. Ding, N. J. Lin, J. Zhou, H. Ro, C. L. Soles, M. T. Cicerone and S. Lin-Gibson, Exploring cellular contact guidance using gradient nanogratings, Biomacromolecules, 2010, 11, 3067–3072 CrossRef CAS PubMed.
- B. A. Dalton, M. D. M. Evans, G. A. McFarland and J. G. Steele, Modulation of corneal epithelial stratification by polymer surface topography, J. Biomed. Mater. Res., 1999, 45, 384–394 CrossRef CAS PubMed.
- D. D. Deligianni, N. D. Katsala, P. G. Koutsoukos and Y. F. Missirlis, Effect of surface roughness of hydroxyapatite on human bone marrow cell adhesion, proliferation, differentiation and detachment strength, Biomaterials, 2000, 22, 87–96 CrossRef.
- D. D. Deligianni, N. Katsala, S. Ladas, D. Sotiropoulou, J. Amedee and Y. F. Missirlis, Effect of surface roughness of the titanium alloy Ti-6Al-4V on human bone marrow cell response and on protein adsorption, Biomaterials, 2001, 22, 1241–1251 CrossRef CAS PubMed.
- A. Ranella, M. Barberoglou, S. Bakogianni, C. Fotakis and E. Stratakis, Tuning cell adhesion by controlling the roughness and wettability of 3D micro/nano silicon structures, Acta Biomater., 2010, 6, 2711–2720 CrossRef CAS PubMed.
- R. A. Gittens, T. McLachlan, R. Olivares-Navarrete, Y. Cai, S. Berner, R. Tannenbaum, Z. Schwartz, K. H. Sandhage and B. D. Boyan, The effects of combined micron-/submicron-scale surface roughness and nanoscale features on cell proliferation and differentiation, Biomaterials, 2011, 32, 3395–3403 CrossRef CAS PubMed.
- L. G. Villa-Diaz, A. M. Ross, J. Lahann and P. H. Krebsbach, Concise review: the evolution of human pluripotent stem cell culture: from feeder cells to synthetic coatings, Stem Cells, 2013, 31, 1–7 CrossRef CAS PubMed.
- H. Shadpour and N. L. Allbritton, In situ roughening of polymeric microstructures, ACS Appl. Mater. Interfaces, 2010, 2, 1086–1093 CrossRef CAS PubMed.
- H. V. Unadkat, M. Hulsman, K. Cornelissen, B. J. Papenburg, R. K. Truckenmüller, A. E. Carpenter, M. Wessling, G. F. Post, M. Uetz, M. J. T. Reinders, D. Stamatialis, C. A. van Blitterswijk and J. de Boer, An algorithm-based topographical biomaterials library to instruct cell fate, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16565–16570 CrossRef CAS PubMed.
- A. I. Teixeira, G. A. Abrams, P. J. Bertics, C. J. Murphy and P. F. Nealey, Epithelial contact guidance on well-defined micro- and nanostructured substrates, J. Cell Sci., 2003, 116, 1881–1892 CrossRef CAS PubMed.
- F. Klein, T. Striebel, J. Fischer, Z. Jiang, C. M. Franz, G. von Freymann, M. Wegener and M. Bastmeyer, Elastic fully three-dimensional microstructure scaffolds for cell force measurements, Adv. Mater., 2010, 22, 868–871 CrossRef CAS PubMed.
- M. R. Dusseiller, M. L. Smith, V. Vogel and M. Textor, Microfabricated three-dimensional environments for single cell studies, Biointerphases, 2006, 1, P1–P4 CrossRef CAS PubMed.
- J. N. Hanson Shepherd, S. T. Parker, R. F. Shepherd, M. U. Gillette, J. A. Lewis and R. G. Nuzzo, 3D microperiodic hydrogel scaffolds for robust neuronal cultures, Adv. Funct. Mater., 2011, 21, 47–54 CrossRef CAS PubMed.
- F. Klein, B. Richter, T. Stribel, C. M. Franz, G. von Freymann, M. Wegener and M. Bastmeyer, Two-component polymer scaffolds for controlled three-dimensional cell culture, Adv. Mater., 2011, 23, 1341–1345 CrossRef CAS PubMed.
- A. M. Greiner, F. Klein, T. Gudzenko, B. Richter, T. Striebel, B. G. Wundari, T. J. Autenrieth, M. Wegener, C. M. Franz and M. Bastmeyer, Cell type-specific adaptation of cellular and nuclear volume in micro-engineered 3D environments, Biomaterials, 2015, 69, 121–132 CrossRef CAS PubMed.
- V. Nandakumar, L. Kelbauskas, K. F. Hernandez, K. M. Lintecum, P. Senechal, K. J. Bussey, P. C. W. Davies, R. H. Johnson and D. R. Meldrum, Isotropic 3D nuclear morphometry of normal, fibrocystic and malignant breast epithelial cells reveals new structural alterations, PLoS One, 2012, 7, e29230 CrossRef CAS PubMed.
- B. Richter, V. Hahn, S. Bertels, T. K. Claus, M. Wegener, G. Delaittre, C. Barner-Kowollik and M. Bastmeyer, Guiding cell attachment in 3D microscaffolds selectively functionalized with two distinct adhesion proteins, Adv. Mater., 2017, 29, 1604342 CrossRef PubMed.
- M. Li, N. Hakimi, R. Perez, S. Waldman, J. A. Kozinski and D. K. Hwang, Microarchitecture for a three-dimensional wrinkled surface platform, Adv. Mater., 2015, 27, 1880–1886 CrossRef CAS PubMed.
- P. Kim, M. Abkarian and H. A. Stone, Hierarchical folding of elastic membranes under biaxial compressive stress, Nat. Mater., 2011, 10, 952–957 CrossRef CAS PubMed.
- D. M. Dean, A. P. Napolitano, J. Youssef and J. R. Morgan, Rods, tori, and honeycombs: The directed self-assembly of microtissues with prescribed microscale geometries, FASEB J., 2007, 21, 4005–4012 CrossRef CAS PubMed.
- A. P. Napolitano, P. Chai, D. M. Dean and J. R. Morgan, Dynamics of the self-assembly of complex cellular aggregates on micromolded nonadhesive hydrogels, Tissue Eng., 2007, 13, 2087–2094 CrossRef CAS PubMed.
- H. Karlsson, M. Fryknäs, R. Larsson and P. Nygren, Loss of cancer drug activity in colon cancer HCT-116 cells during spheroid formation in a new 3D spheroid cell culture system, Exp. Cell Res., 2012, 318, 1577–1585 CrossRef CAS PubMed.
- K. C. Murphy, S. Y. Fang and J. K. Leach, Human mesenchymal stem cell spheroids in fibrin hydrogels exhibit improved cell survival and potential for bone healing, Cell Tissue Res., 2014, 357, 91–99 CrossRef CAS PubMed.
- S. S. Ho, K. C. Murphy, B. Y. K. Binder, C. B. Vissers and J. K. Leach, Increased survival and function of mesenchymal stem cell spheroids entrapped in instructive alginate hydrogels, Stem Cells Transl. Med., 2016, 5, 773–781 CrossRef CAS PubMed.
- Y. B. Lee, E. M. Kim, H. Byun, H.-k. Chang, K. Jeong, Z. M. Aman, Y. S. Choi, J. Park and H. Shin, Engineering spheroids potentiating cell–cell and cell–ECM interactions by self-assembly of stem cell microlayer, Biomaterials, 2018, 165, 105–120 CrossRef CAS PubMed.
- S. C. Ramaiahgari, M. W. den Braver, B. Herpers, V. Terpstra, J. N. M. Commandeur, B. van de Water and L. S. Price, A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies, Arch. Toxicol., 2014, 88, 1083–1095 CAS.
- G. Mehta, A. Y. Hsiao, M. Ingram, G. D. Luker and S. Takayama, Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy, J. Controlled Release, 2012, 164, 192–204 CrossRef CAS PubMed.
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. Yang, Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors, Assay Drug Dev. Technol., 2014, 12, 207–218 CrossRef CAS PubMed.
- K. Moshksayan, N. Kashaninejad, M. Ebrahimi Warkiani, J. G. Lock, H. Moghadas, B. Firoozabadi, M. S. Saidi and N. T. Nguyen, Spheroids-on-a-chip: Recent advances and design considerations in microfluidic platforms for spheroid formation and culture, Sens. Actuators, B, 2018, 263, 151–176 CrossRef CAS.
- H. Huang, Y. Ding, X. S. Sun and T. A. Nguyen, Peptide hydrogelation and cell encapsulation for 3D culture of MCF-7 breast cancer cells, PLoS One, 2013, 8, e59482 CrossRef CAS PubMed.
- B. M. Baker and C. S. Chen, Deconstructing the third dimension – how 3D culture microenvironments alter cellular cues, J. Cell Sci., 2012, 125, 3015–3024 CrossRef CAS PubMed.
- T. Weiß, R. Schade, T. Laube, A. Berg, G. Hildebrand, R. Wyrwa, M. Schnabelrauch and K. Liefeith, Two-photon polymerization of biocompatible photopolymers for microstructured 3D biointerfaces, Adv. Mater., 2011, 13, B264–B273 Search PubMed.
- M. Wolun-Cholewa, K. Langer, K. Szymanowski, A. Glodek, A. Jankowska, W. Warchol and J. Langer, An efficient 3D cell culture method on biomimetic nanostructured grids, PLoS One, 2013, 8, e72936 CrossRef CAS PubMed.
- T. Xu, P. Molnar, C. Gregory, M. Das, T. Boland and J. J. Hickman, Electrophysiological characterization of embryonic hippocampal neurons cultured in a 3D collagen hydrogel, Biomaterials, 2009, 30, 4377–4383 CrossRef CAS PubMed.
- S. Li, J. Lao, B. P. C. Chen, Y. S. Li, Y. Zhao, J. Chu, K. D. Chen, T. C. Tsou, K. Peck and S. Chien, Genomic analysis of smooth muscle cells in 3-dimensional collagen matrix, FASEB J., 2003, 17, 97–99 CrossRef CAS PubMed.
- E. M. Chandler, C. M. Berglund, J. S. Lee, W. J. Polacheck, J. P. Gleghorn, B. J. Kirby and C. Fischbach, Stiffness of photocrosslinked RGD-alginate gels regulates adipose progenitor cell behavior, Biotechnol. Bioeng., 2011, 108, 1683–1692 CrossRef CAS PubMed.
- P. Kasten, I. Beyen, P. Niemeyer, R. Luginbühl, M. Bohner and W. Richter, Porosity and pore size of β-tricalcium phosphate scaffold can influence protein production and osteogenic differentiation of human mesenchymal stem cells: An in vitro and in vivo study, Acta Biomater., 2008, 4, 1904–1915 CrossRef CAS PubMed.
- P. R. Baraniak and T. C. McDevitt, Scaffold-free culture of mesenchymal stem cell spheroids in suspension preserves multilineage potential, Cell Tissue Res., 2012, 347, 701–711 CrossRef CAS PubMed.
- Y. Yamaguchi, J. Ohno, A. Sato, H. Kido and T. Fukushima, Mesenchymal stem cell spheroids exhibit enhanced in vitro and in vivo osteoregenerative potential, BMC Biotechnol., 2014, 14, 105 CrossRef PubMed.
- T. S. Ramasamy, J. S. L. Yu, C. Selden, H. Hodgson and W. Cui, Application of three-dimensional culture conditions to human embryonic stem cell-derived definitive endoderm cells enhances hepatocyte differentiation and functionality, Tissue Eng., Part A, 2013, 19, 360–367 CrossRef CAS PubMed.
- H. Tanaka, S. Tanaka, K. Sekine, S. Kita, A. Okamura, T. Takebe, Y. W. Zheng, Y. Ueno, J. Tanaka and H. Taniguchi, The generation of pancreatic β-cell spheroids in a simulated microgravity culture system, Biomaterials, 2013, 34, 5785–5791 CrossRef CAS PubMed.
- P. Prang, R. Muller, A. Eljaouhari, K. Heckmann, W. Kunz, T. Weber, C. Faber, M. Vroemen, U. Bogdahn and N. Weidner, The promotion of oriented axonal regrowth in the injured spinal cord by alginate-based anisotropic capillary hydrogels, Biomaterials, 2006, 27, 3560–3569 CAS.
- S. Pedron, E. Becka and B. A. Harley, Spatially gradated hydrogel platform as a 3D engineered tumor microenvironment, Adv. Mater., 2015, 27, 1567–1572 CrossRef CAS PubMed.
- Y. C. Tung, A. Y. Hsiao, S. G. Allen, Y.-s. Torisawa, M. Ho and S. Takayama, High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array, Analyst, 2011, 136, 473–478 RSC.
- A. Abbott, Biology's new dimension, Nature, 2003, 424, 870–872 CrossRef CAS PubMed.
- L. A. Gurski, N. J. Petrelli, X. Jia and M. C. Farach-Carson, 3D matrices for anti-cancer drug testing and development, Oncol. Issues, 2010, 25, 20–25 CrossRef.
- S. J. Fey and K. Wrzesinski, Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line, Toxicol. Sci, 2012, 127, 403–411 CrossRef CAS PubMed.
- Q. Meng, Three-dimensional culture of hepatocytes for prediction of drug-induced hepatotoxicity, Expert Opin. Drug Metab. Toxicol., 2010, 6, 733–746 CrossRef CAS PubMed.
- J. Yin, Q. Meng, G. Zhang and Y. Sun, Differential methotrexate hepatotoxicity on rat hepatocytes in 2D monolayer culture and 3D gel entrapment culture, Chem. – Biol. Interact., 2009, 180, 368–375 CrossRef CAS PubMed.
- S.-h. Hsu and P. S. Hsieh, Self-assembled adult adipose-derived stem cell spheroids combined with biomaterials promote wound healing in a rat skin repair model, Wound Repair Regen., 2015, 23, 57–64 CrossRef PubMed.
- T. P. Amadeu and B. Coulomb, Cutaneous wound healing: Myofibroblastic differentiation and in vitro models, Int. J. Lower Extremity Wounds, 2003, 2, 60–68 CrossRef PubMed.
- Y. Duan, Z. Liu, J. O'Neill, L. Q. Wan, D. O. Freytes and G. Vunjak-Novakovic, Hybrid gel composed of native heart matrix and collagen induces cardiac differentiation of human embryonic stem cells without supplemental growth factors, J. Cardiovasc. Transl. Res., 2011, 4, 605–615 CrossRef PubMed.
- S. K. Goh, S. Bertera, P. Olsen, J. E. Candiello, W. Halfter, G. Uechi, M. Balasubramani, S. A. Jahnson, B. M. Sicari, E. Kollar, S. F. Badylak and I. Banerjee, Perfusion-decellularized pancreas as a natural 3D scaffold for pancreatic tissue and whole organ engineering, Biomaterials, 2013, 34, 6760–6772 CrossRef CAS PubMed.
- S. L. Spear, P. M. Parikh, E. Reisin and N. G. Menon, Acellular dermis-assisted breast reconstruction, Aesthetic Plast. Surg., 2008, 32, 418–425 CrossRef CAS PubMed.
- R. Langer and D. A. Tirrell, Designing materials for biology and medicine, Nature, 2004, 428, 487–492 CrossRef CAS PubMed.
- N. Huebsch and D. J. Mooney, Inspiration and application in the evolution of biomaterials, Nature, 2009, 462, 426–432 CrossRef CAS PubMed.
- M. W. Tibbitt and K. S. Anseth, Hydrogels as extracellular matrix mimics for 3D cell culture, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS PubMed.
- M. Matsusaki, C. P. Case and M. Akashi, Three-dimensional cell culture technique and pathophysiology, Adv. Drug Delivery Rev., 2014, 74, 95–103 CrossRef CAS PubMed.
- H. Wang and S. C. Heilshorn, Adaptable hydrogel networks with reversible linkages for tissue engineering, Adv. Mater., 2015, 27, 3717–3736 CrossRef CAS PubMed.
- S. R. Caliari and J. A. Burdick, A practical guide to hydrogels for cell culture, Nat. Methods, 2016, 13, 405–414 CrossRef CAS PubMed.
- A. M. Rosales and K. S. Anseth, The design of reversible hydrogels to capture extracellular matrix dynamics, Nat. Rev. Mater., 2016, 1, 15012 CrossRef CAS PubMed.
- G. A. Silva, C. Czeisler, K. L. Niece, E. Beniash, D. A. Harrington, J. A. Kessler and S. L. Stupp, Selective differentiation of neural progenitor cells by high-epitope density nanofibers, Science, 2004, 303, 1352–1355 CrossRef CAS PubMed.
- V. Jayawarna, M. Ali, T. A. Jowitt, A. F. Miller, A. Saiani, J. E. Gough and R. V. Ulijn, Nanostructured hydrogels for three-dimensional cell culture through self-assembly of fluorenylmethoxycarbonyl-dipeptides, Adv. Mater., 2006, 18, 611–614 CrossRef CAS.
- N. Li, Q. Zhang, S. Gao, Q. Song, R. Huang, L. Wang, L. Liu, J. Dai, M. Tang and G. Cheng, Three-dimensional graphene foam as a biocompatible and conductive scaffold for neural stem cells, Sci. Rep., 2013, 3, 1604 CrossRef PubMed.
- L. Jin, Z. Feng, T. Wang, Z. Ren, S. Ma, J. Wu and D. Sun, A novel fluffy hydroxylapatite fiber scaffold with deep interconnected pores designed for three dimensional cell culture, J. Mater. Chem. B, 2014, 2, 129–136 RSC.
- R. Jacob, D. Ghosh, P. K. Singh, S. K. Basu, N. Nath Jha, S. Das, P. K. Sukul, S. Patil, S. Sathaye, A. Kumar, A. Chowdhury, S. Malik, S. Sen and S. K. Maji, Self healing hydrogels composed of amyloid nano fibrils for cell culture and stem cell differentiation, Biomaterials, 2015, 54, 97–105 CrossRef CAS PubMed.
- B. Zhang, M. Mongomery, L. Davenport-Huyer, A. Korolj and M. Radisic, Platform technology for scalable assembly of instantaneously functional mosaic tissues, Sci. Adv., 2015, 1, e1500423 CrossRef PubMed.
- Q. Feng, K. Wei, S. Lin, Z. Xu, Y. Sun, P. Shi, G. Li and L. Bian, Mechanically resilient, injectable, and bioadhesive supramolecular gelatin hydrogels crosslinked by weak host-guest interactions assist cell infiltration and in situ tissue regeneration, Biomaterials, 2016, 101, 217–228 CrossRef CAS PubMed.
- X. Dou and C. Feng, Amino acids and peptide-based supramolecular hydrogels for three-dimensional cell culture, Adv. Mater., 2017, 29, 1604062 CrossRef PubMed.
- S. Knowlton, S. Anand, T. Shah and S. Tasoglu, Bioprinting for neural tissue engineering, Trends Neurosci., 2018, 41, 31–46 CrossRef CAS PubMed.
- T. Billiet, E. Gevaert, T. De Schryver, M. Cornelissen and P. Dubruel, The 3D printing of gelatin methacrylamide cell–laden tissue-engineered constructs with high cell viability, Biomaterials, 2014, 35, 49–62 CrossRef CAS PubMed.
- W. Aljohani, M. W. Ullah, X. Zhang and G. Yang, Bioprinting and its applications in tissue engineering and regenerative medicine, Int. J. Biol. Macromol., 2018, 107, 261–275 CrossRef CAS PubMed.
- T. Q. Huang, X. Qu, J. Liu and S. Chen, 3D printing of biomimetic microstructures for cancer cell migration, Biomed. Microdevices, 2014, 16, 127–132 CrossRef PubMed.
- J. U. Lind, T. A. Busbee, A. D. Valentine, F. S. Pasqualini, H. Yuan, M. Yadid, S. J. Park, A. Kotikian, A. P. Nesmith, P. H. Campbell, J. J. Vlassak, J. A. Lewis and K. K. Parker, Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing, Nat. Mater., 2017, 16, 303–308 CrossRef CAS PubMed.
- S. Knowlton, A. Joshi, B. Yenilmez, I. T. Ozbolat, C. K. Chua, A. Khademhosseini and S. Tasoglu, Advancing cancer research using bioprinting for tumor-on-a-chip platforms, Int. J. Bioprint., 2016, 2, 3–8 CAS.
- V. E. Santo, S. P. Rebelo, M. F. Estrada, P. M. Alves, E. Boghaert and C. Brito, Drug screening in 3D in vitro tumor models: Overcoming current pitfalls of efficacy read-outs, Biotechnol. J., 2017, 12, 1600505 CrossRef PubMed.
- J. Norman, R. D. Madurawe, C. M. V. Moore, M. A. Khan and A. Khairuzzaman, A new chapter in pharmaceutical manufacturing: 3D-printed drug products, Adv. Drug Delivery Rev., 2017, 108, 39–50 CrossRef CAS PubMed.
- V. Mironov, T. Boland, T. Trusk, G. Forgacs and R. R. Markwald, Organ printing: computer-aided jet-based 3D tissue engineering, Trends Biotechnol., 2003, 21, 157–161 CrossRef CAS PubMed.
- T. Boland, V. Mironov, A. Gutowska, E. A. Roth and R. R. Markwald, Cell and organ printing 2: Fusion of cell aggregates in three-dimensional gels, Anat. Rec., Part A, 2003, 272, 497–502 CrossRef PubMed.
- N. E. Fedorovich, J. Alblas, W. E. Hennink, F. C. Öner and W. J. A. Dhert, Organ printing: the future of bone regeneration?, Trends Biotechnol., 2011, 29, 601–606 CrossRef CAS PubMed.
- S. V. Murphy and A. Atala, 3D bioprinting of tissues and organs, Nat. Biotechnol., 2016, 32, 773–785 CrossRef PubMed.
- K. Markstedt, A. Mantas, I. Tournier, H. M. Ávila, D. Hägg and P. Gatenholm, 3D bioprinting human chondrocytes with nanocellulose-alginate bioink for cartilage tissue engineering applications, Biomacromolecules, 2015, 16, 1489–1496 CrossRef CAS PubMed.
- M. S. Mannoor, Z. Jiang, T. James, Y. L. Kong, K. A. Malatesta, W. O. Soboyejo, N. Verma, D. H. Gracias and M. C. McAlpine, 3D printed bionic ears, Nano Lett., 2013, 13, 2634–2639 CrossRef CAS PubMed.
- B. Duan, E. Kapetanovic, L. A. Hockaday and J. T. Butcher, Three-dimensional printed trileaflet valve conduits using biological hydrogels and human valve interstitial cells, Acta Biomater., 2014, 10, 1836–1846 CrossRef CAS PubMed.
- X. Cui and T. Boland, Human microvasculature fabrication using thermal inkjet printing technology, Biomaterials, 2009, 30, 6221–6227 CrossRef CAS PubMed.
- R. Lozano, L. Stevens, B. C. Thompson, K. J. Gilmore, R. Gorkin, E. M. Stewart, M. in het Panhuis, M. Romero-Ortega and G. G. Wallace, 3D printing of layered brain-like structures using peptide modified gellan gum substrates, Biomaterials, 2015, 67, 264–273 CrossRef CAS PubMed.
- C. X. F. Lam, X. M. Mo, S. H. Teoh and D. W. Hutmacher, Scaffold development using 3D printing with a starch-based polymer, Mater. Sci. Eng., C, 2002, 20, 49–56 CrossRef.
- B. Leukers, H. Gülkan, S. H. Irsen, S. Milz, C. Tille, M. Schieker and H. Seitz, Hydroxyapatite scaffolds for bone tissue engineering made by 3D printing, J. Mater. Sci.: Mater. Med., 2005, 16, 1121–1124 CrossRef CAS PubMed.
- S. Yang, K. F. Leong, Z. Du and C. K. Chua, The design of scaffolds for use in tissue engineering. Part I. Traditional factors, Tissue Eng., 2001, 7, 679–689 CrossRef CAS PubMed.
- S. Yang, K. F. Leong, Z. Du and C. K. Chua, The design of scaffolds for use in tissue engineering. Part II. Rapid prototyping techniques., Tissue Eng., 2002, 8, 1–11 CrossRef CAS PubMed.
- H. N. Chia and B. M. Wu, Recent advances in 3D printing of biomaterials, J. Biol. Eng., 2015, 9, 4 CrossRef PubMed.
- C. Mandrycky, Z. Wang, K. Kim and D. H. Kim, 3D bioprinting for engineering complex tissues, Biotechnol. Adv., 2016, 34, 422–434 CrossRef CAS PubMed.
- M. C. Echave, P. Sánchez, J. L. Pedraz and G. Orive, Progress of gelatin-based 3D approaches for bone regeneration, J. Drug Delivery Sci. Technol., 2017, 42, 63–74 CrossRef CAS.
- D. Baksh, R. Yao and R. S. Tuan, Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow, Stem Cells, 2007, 25, 1384–1392 CrossRef CAS PubMed.
- B. Chuenjitkuntaworn, T. Osathanon, N. Nowwarote, P. Supaphol and P. Pavasant, The efficacy of polycaprolactone/hydroxyapatite scaffold in combination with mesenchymal stem cells for bone tissue engineering, J. Biomed. Mater. Res., Part A, 2016, 104, 264–271 CrossRef PubMed.
- Y. Reinwald and A. J. El Haj, Hydrostatic pressure in combination with topographical cues affects the fate of bone marrow-derived human mesenchymal stem cells for bone tissue regeneration, J. Biomed. Mater. Res., Part A, 2018, 106, 629–640 CrossRef CAS PubMed.
- S.-W. Kuo, H.-I. Lin, J. H.-C. Ho, Y.-R. V. Shih, H.-F. Chen, T.-J. Yen and O. K. Lee, Regulation of the fate of human mesenchymal stem cells by mechanical and stereo-topographical cues provided by silicon nanowires, Biomaterials, 2012, 33, 5013–5022 CrossRef CAS PubMed.
- J. E. Frith, B. Thomson and P. G. Genever, Dynamic three-dimensional culture methods enhance mesenchymal stem cell properties and increase therapeutic potential, Tissue Eng., Part C, 2010, 16, 735–749 CrossRef CAS PubMed.
- W. Wang, K. Itaka, S. Ohba, N. Nishiyama, U.-i. Chung, Y. Yamasaki and K. Kataoka, 3D spheroid culture system on micropatterned substrates for improved differentiation efficiency of multipotent mesenchymal stem cells, Biomaterials, 2009, 30, 2705–2715 CrossRef CAS PubMed.
- W.-J. Li, R. Tuli, C. Okafor, A. Derfoul, K. G. Danielson, D. J. Hall and R. S. Tuan, A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells, Biomaterials, 2005, 26, 599–609 CrossRef CAS PubMed.
- S. U. Kim and J. de Vellis, Stem cell-based cell therapy in neurological diseases: A review, J. Neurosci. Res., 2009, 87, 2183–2200 CrossRef CAS PubMed.
- V. Jeevanantham, M. Butler, A. Saad, A. Abdel-Latif, E. K. Zuba-Surma and B. Dawn, Adult bone marrow cell therapy improves survival and induces long-term improvement in cardiac parameters: A systematic review and meta-analysis, Circulation, 2012, 126, 551–568 CrossRef PubMed.
- L. Li, H. Hui, X. Jia, J. Zhang, Y. Liu, Q. Xu and D. Zhu, Infusion with human bone marrow-derived mesenchymal stem cells improves β-cell function in patients and non-obese mice with severe diabetes, Sci. Rep., 2016, 6, 37894 CrossRef CAS PubMed.
- E. M. Horwitz, P. L. Gordon, W. K. K. Koo, J. C. Marx, M. D. Neel, R. Y. McNall, L. Muul and T. Hofmann, Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 8932–8937 CrossRef CAS PubMed.
- O. N. Koç, J. Day, M. Nieder, S. L. Gerson, H. M. Lazarus and W. Krivit, Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH), Bone Marrow Transplant., 2002, 30, 215–222 CrossRef PubMed.
- J. Reiser, X. Y. Zhang, C. S. Hemenway, D. Mondal, L. Pradhan and V. F. La Russa, Potential of mesenchymal stem cells in gene therapy approaches for inherited and acquired diseases, Expert Opin. Biol. Ther., 2005, 5, 1571–1584 CrossRef CAS PubMed.
- J. F. Enders, T. H. Weller and F. C. Robbins, Cultivation of the lansing strain of poliomyelitis virus in cultures of various human embryonic tissues, Science, 1949, 109, 85–87 CrossRef CAS PubMed.
- S. L. Plotkin and S. A. Plotkin in Vaccines, ed. S. A. Plotkin, W. A. Orenstein and P. A. Offit, Elsevier, Saunders, 5th edn, 2008, ch. A short history of vaccination Search PubMed.
- P. N. Barrett, W. Mundt, O. Kistner and M. K. Howard, Vero cell platform in vaccine production: moving towards cell culture-based viral vaccines, Expert Rev. Vaccines, 2009, 8, 607–618 CrossRef CAS PubMed.
- J. A. Tree, C. Richardson, A. R. Fook, J. C. Clegg and D. Looby, Comparison of large-scale mammalian cell culture systems with egg culture for the production of influenza virus A vaccine strains, Vaccine, 2001, 19, 3444–3450 CrossRef CAS PubMed.
- G. Andrei, Three-dimensional culture models for human viral diseases and antiviral drug development, Antiviral Res., 2006, 71, 96–107 CrossRef CAS PubMed.
- G. Lee, E. P. Papapetrou, H. Kim, S. M. Chambers, M. J. Tomishima, C. A. Fasano, Y. M. Ganat, J. Menon, F. Shimizu, A. Viale, V. Tabar, M. Sadelain and L. Studer, Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs, Nature, 2009, 461, 402–406 CrossRef CAS PubMed.
- L. Yang, M. H. Soonpaa, E. D. Adler, T. K. Roepke, S. J. Kattman, M. Kennedy, E. Henckaerts, K. Bonham, G. W. Abbott, R. M. Linden, L. J. Field and G. M. Keller, Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population, Nature, 2008, 453, 524–528 CrossRef CAS PubMed.
- J. I. Luna, J. Ciriza, M. E. Garcia-Ojeda, M. Kong, A. Herren, D. K. Lieu, R. A. Li, C. C. Fowlkes, M. Khine and K. E. McCloskey, Multiscale biomimetic topography for the alignment of neonatal and embryonic stem cell-derived heart cells, Tissue Eng., Part C, 2011, 17, 579–588 CrossRef PubMed.
- J. Wang, A. Chen, D. K. Lieu, I. Karakikes, G. Chen, W. Keung, C. W. Chan, R. J. Hajjar, K. D. Costa, M. Khine and R. A. Li, Effect of engineered anisotropy on the susceptibility of human pluripotent stem cell-derived ventricular cardiomyocytes to arrhythmias, Biomaterials, 2013, 34, 8878–8886 CrossRef CAS PubMed.
- M. T. Lam, W. C. Clem and S. Takayama, Reversible on-demand cell alignment using reconfigurable microtopography, Biomaterials, 2008, 29, 1705–1712 CrossRef CAS PubMed.
- M. Guvendiren and J. A. Burdick, Stem cell response to spatially and temporally displayed and reversible surface topography, Adv. Healthcare Mater., 2013, 2, 155–164 CrossRef CAS PubMed.
- J. Webster, P. Bollen, H. Grimm and M. Jennings, Ethical implications of using the minipig in regulatory toxicology studies, J. Pharmacol. Toxicol. Methods, 2010, 62, 160–166 CrossRef CAS PubMed.
- D. R. Jung, R. Kapur, T. Adams, K. A. Giuliano, M. Mrksich, H. G. Craighead and D. L. Taylor, Topographical and physicochemical modification of material surface to enable patterning of living cells, Crit. Rev. Biotechnol., 2001, 21, 111–154 CrossRef CAS PubMed.
- A. Astashkina, B. Mann and D. W. Grainger, A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity, Pharmacol. Ther., 2012, 134, 82–106 CrossRef CAS PubMed.
- T. H. Qazi, D. J. Mooney, G. N. Duda and S. Geissler, Biomaterials that promote cell–cell interactions enhance the paracrine function of MSCs, Biomaterials, 2017, 140, 103–114 CrossRef CAS PubMed.
- F. Xu, J. Wu, S. Wang, N. G. Durmus, U. A. Gurkan and U. Demirci, Microengineering methods for cell-based microarrays and high-throughput drug-screening applications, Biofabrication, 2011, 3, 034101 CrossRef PubMed.
- dit M. A. Faute, L. Laurent, D. Ploton, M.-F. Poupon, J.-C. Jardillier and H. Bobichon, Distinctive alterations of invasiveness, drug resistance and cell–cell organization in 3D-cultures of MCF-7, a human breast cancer cell line, and its multidrug resistant variant, Clin. Exp. Metastasis, 2002, 19, 161–167 CrossRef.
- K. Bhadriraju and C. S. Chen, Engineering cellular microenvironments to improve cell-based drug testing, Drug Discovery Today, 2002, 7, 612–620 CrossRef CAS PubMed.
- A. Albanese, A. K. Lam, E. A. Sykes, J. V. Rocheleau and W. C. W. Chan, Tumour-on-a-chip provides an optical window into nanoparticle tissue transport, Nat. Commun., 2013, 4, 2718 CrossRef PubMed.
- L. Y. Wu, D. Di Carlo and L. P. Lee, Microfluidic self-assembly of tumor spheroids for anticancer drug discovery, Biomed. Microdevices, 2008, 10, 197–202 CrossRef CAS PubMed.
- B. R. Hughes, M. Mirbagheri, S. D. Waldman and D. K. Hwang, Direct cell–cell communication with three-dimensional cell morphology on wrinkled microposts, Acta Biomater., 2018, 78, 89–97 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |





