Open Access Article
Open Access ArticleDendronic trimaltoside amphiphiles (DTMs) for membrane protein study†
Aiman
Sadaf‡
 a,
Yang
Du‡
a,
Yang
Du‡
 b,
Claudia
Santillan
b,
Claudia
Santillan
 c,
Jonas S.
Mortensen
c,
Jonas S.
Mortensen
 d,
Iago
Molist
d,
Iago
Molist
 e,
Alpay B.
Seven
e,
Alpay B.
Seven
 f,
Parameswaran
Hariharan
f,
Parameswaran
Hariharan
 c,
Georgios
Skiniotis
c,
Georgios
Skiniotis
 f,
Claus J.
Loland
f,
Claus J.
Loland
 d,
Brian K.
Kobilka
d,
Brian K.
Kobilka
 b,
Lan
Guan
b,
Lan
Guan
 c,
Bernadette
Byrne
c,
Bernadette
Byrne
 e and
Pil Seok
Chae
e and
Pil Seok
Chae
 *a
*a
aDepartment of Bionanotechnology, Hanyang University, Ansan, 155-88, Korea. E-mail: pchae@hanyang.ac.kr
bMolecular and Cellular Physiology, Stanford, CA 94305, USA. E-mail: kobilka@stanford.edu
cDepartment of Cell Physiology and Molecular Biophysics, Center for Membrane Protein Research, School of Medicine, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA. E-mail: lan.guan@ttuhsc.edu
dCenter of Neuroscience, University of Copenhagen, DK 2200 Copenhagen, Denmark. E-mail: cllo@sund.ku.dk
eDepartment of Life Sciences, Imperial College London, London, SW7 2AZ, UK. E-mail: b.byrne@imperial.ac.uk
fStructural Biology & Molecular and Cellular Physiology, Stanford, CA 94305, USA. E-mail: yiorgo@stanford.edu
First published on 25th October 2017
Abstract
The critical contribution of membrane proteins in normal cellular function makes their detailed structure and functional analysis essential. Detergents, amphipathic agents with the ability to maintain membrane proteins in a soluble state in aqueous solution, have key roles in membrane protein manipulation. Structural and functional stability is a prerequisite for biophysical characterization. However, many conventional detergents are limited in their ability to stabilize membrane proteins, making development of novel detergents for membrane protein manipulation an important research area. The architecture of a detergent hydrophobic group, that directly interacts with the hydrophobic segment of membrane proteins, is a key factor in dictating their efficacy for both membrane protein solubilization and stabilization. In the current study, we developed two sets of maltoside-based detergents with four alkyl chains by introducing dendronic hydrophobic groups connected to a trimaltoside head group, designated dendronic trimaltosides (DTMs). Representative DTMs conferred enhanced stabilization to multiple membrane proteins compared to the benchmark conventional detergent, DDM. One DTM (i.e., DTM-A6) clearly outperformed DDM in stabilizing human β2 adrenergic receptor (β2AR) and its complex with Gs protein. A further evaluation of this DTM led to a clear visualization of β2AR-Gs complex via electron microscopic analysis. Thus, the current study not only provides novel detergent tools useful for membrane protein study, but also suggests that the dendronic architecture has a role in governing detergent efficacy for membrane protein stabilization.
Introduction
Membrane proteins embedded in cellular membranes account for approximately 30% of total gene transcripts.1 Restricted to lateral motion, these proteins are of pivotal importance in cell physiology. They include cell surface receptors, signal transducers, metabolite transporters and membrane channels. Malfunctions of these bio-macromolecules lead to various health disorders such as cystic fibrosis, Alzheimer's, cancer and neurodegenerative diseases.2 Approximately 60% of current pharmaceutical agents target membrane proteins.3 Despite their biochemical and pharmaceutical significance, the structural and functional study of membrane proteins lags far behind that of soluble proteins. The paucity of membrane protein structures mainly results from the incompatibility of their physicochemical properties with the prerequisite requirements for biophysical analysis.4 As expected from the structure of native membranes, membrane proteins are inserted asymmetrically into the bilayer, with the central transmembrane segment flanked by the two non-identical hydrophilic portions. The amphiphilic nature and preferred orientation contribute to proper functions of these bio-macromolecules in the membrane environment, but hampers their isolation in a soluble and stable state in aqueous media.5Detergent micelles are capable of stabilizing membrane proteins outside the native membranes by encapsulation, leading to formation of protein-detergent complexes (PDCs).6 Thus, detergents are widely used for membrane protein manipulation including solubilization, stabilization and crystallization. Non-ionic amphiphiles are of prime importance for membrane protein study mainly due to their superior ability to stabilize membrane proteins compared to other zwitter-ionic or ionic detergents with charged head groups. This is well illustrated by the fact that ∼75% of structurally characterised α-helical membrane proteins were isolated and manipulated in one of four non-ionic detergents (OG (n-octyl-β-D-glucoside), NG (n-nonyl-β-D-glucoside), DM (n-decyl-β-D-maltoside), or DDM (n-dodecyl-β-D-maltoside)), along with a zwitterionic detergent, LDAO (lauryldimethylamine-N-oxide).7 Despite their widespread use for membrane protein structural studies, these detergents are often suboptimal inducing membrane protein denaturation and/or aggregation.8 These classical detergents are structurally simple, typically having a flexible alkyl tail and a large hydrophilic head group. In contrast, membrane proteins are highly diverse in terms of their three dimensional structures and biophysical properties. The large gap in structure diversity between conventional detergents and membrane proteins indicates that novel detergents distinct from conventional agents in terms of architecture/chemical properties are necessary to advance membrane protein research.9
Over the past two decades, many efforts have been made to develop novel classes of amphiphiles, architecturally different from conventional detergents.10 Peptide amphiphiles with α-helical (e.g., lipopeptide detergents (LPDs))11a or β-sheet structure (β-peptides (BPs))11b have been developed. Polymeric amphiphiles were invented and frequently used alone (e.g., amphipols (Apols) and styrene-maleic acid (SMA) polymer),12a–c or in a combination with a patch of lipid bilayers (e.g., nanodiscs (NDs))12d for membrane protein studies. This disc system, similar to bicelles in architecture,12e provides a mimetic environment closer to the native membrane than detergent micelles.12f However, most of these agents are heterogenous, generally hard to prepare on a large scale, or often inefficient at extracting membrane proteins from the membranes. In this context, small amphipathic agents are advantageous over the peptide- or polymer-based amphiphiles. Representatives of small amphiphiles include rigid hydrophobic group-bearing agents (e.g., glyco-diosgenin (GDN),13a tripod amphiphiles (TPAs),13b–d norbornane-based amphphiles (NBMs),13e resorcinarene-based glucoside amphiphiles (RGAs)13f), neopentyl glycol-based amphiphiles (glucose neopentyl glycols (GNGs), maltose neopentyl glycols (MNGs) and neopentyl glycol-derived triglucosides (NDTs))14a–f and penta-saccharide-bearing amphiphiles (PSEs).15 Among these agents, it is notable that GNG-3 (commercial name: OGNG) and MNG-3 (commercial name: LMNG) have facilitated the elucidation of more than 30 new membrane protein structures including the β2 adrenergic,16a–e acetylcholine16f,g and opioid G-protein coupled receptors16h,i in the last six years, highlighting the importance and potential of novel amphiphile development. In addition, systematic variations in detergent structure have enabled us to investigate multiple detergent design principles essential for novel amphiphile design and to help provide insights into the nature of interactions between detergent molecules and membrane proteins. For instance, the importance of detergent chirality and kinking as well as detergent conformational/configurational flexibility in detergent stabilization efficacy could be explained by the recent studies on butane-tetraol maltosides (BTMs),17 NBMs,13e and RGAs/NDTs.13f,14f In the present study, we have designed and synthesized two sets of novel amphiphiles containing a dendronic hydrophobic group, the first example of this type of group being introduced into a detergent for membrane protein study. Because of the presence of a trimaltoside head and dendronic tail groups, these agents were designated dendronic trimaltoside amphiphiles (DTMs). When these agents were evaluated with several membrane proteins, some of these agents displayed enhanced protein stabilization efficacy compared to a gold standard detergent (DDM). In addition, negative stain electron microscopic (EM) analysis revealed that a representative of the DTMs can be effectively used for clear visualization of membrane protein complexes with multi-domains architecture.
Results and discussion
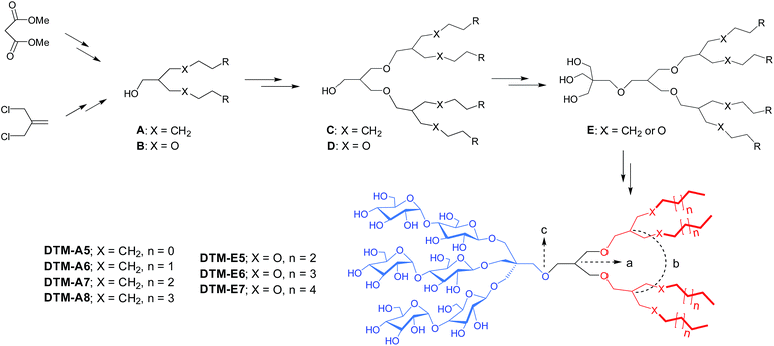
The novel agents examined in this study bear three maltosides and four alkyl chains as the head and tail groups, respectively, connected to each other via a neopentyl glycol linker (Scheme 1). The hydrophobic group of the DTMs has dendronic architecture with two branch points (a and b in Scheme 1). The first branch point (a) was connected to the second point (b in Scheme) by an ether linkage, while the second branch point was connected to the terminal alkyl chains either directly (DTM-As) or through an additional ether linkage (DTM-Es). It is notable that the dendronic hydrophobic group introduced here enabled us to first develop novel amphiphiles containing four alkyl chains diverging from a single focal point (c). Because of the presence of the multiple alkyl chains, these agents have high hydrophobic density and show multi-valency effect.18 These have critical influences on the ability of any detergent to effectively stabilise and solubilize membrane proteins, as supported by results obtained for the TPAs,13b–d mesitylene-cored glucoside amphiphiles (MGAs),19 and adamantane-based amphiphiles (ADAs).20 The DTMs are structurally different from the TPAs, MGAs and ADAs as their lipophilic regions contain neither a tripod unit nor a rigid core/tail group, producing a unique combination of detergent flexibility/hydrophobic density. Detergent alkyl chain length varied from C5 to C8 for the DTM-As and from C5 to C7 for the DTM-Es, numbers used in detergent designation. Variation in the alkyl chain length is essential for optimizing hydrophile-lipophile balance (HLB),21 known to be crucial in detergent efficacy.22 We did not further increase the alkyl chain length for each set of the DTMs due to a high chance of reduced water-solubility. These novel agents were prepared in overall good yields through a synthetic protocol comprising six or seven synthetic steps. The dendronic hydrophobic groups were synthesized by a convergent method (Scheme 1). For preparation of the DTM-As, dialkylated mono-ol derivatives (A) with a branch point were first synthesized from dimethylmalonate via successive dialkylation, Krapcho decarboxylation and reduction reactions. The resulting dialkylated mono-ol derivatives were reacted with methallyl dichloride containing an additional branch point, followed by hydroboration–oxidation to produce tetraalkylated alcohols with dendronic architecture (C). The DTM-Es were prepared by a protocol similar to that used for the preparation of the DTM-As, but in this case methallyl dichloride was used as a starting material to obtain the ether-functionalized dialkylated mono-ol derivatives (B) and tetraalkylated dendronic alcohols (D). The tetraalkylated alcohols (C or D) were coupled with neopentyl glycol to generate tri-ol derivatives (E) which were used as substrates for glycosylation (see ESI Schemes 1 and 2†). Notably, the glycosylation used here would proceed stereo-specifically owing to the use of maltosylbromide with a benzoyl protecting group at 2′-carbon as a glycosyl donor. As the benzoyl protecting group neighbouring anomeric carbon (1′-carbon) is involved in the formation of the cyclic oxonium ion intermediate via an intramolecular reaction, known as anchimeric assistance (Fig. S1a†), the glycosylated products are expected to be β-anomers, which is well supported by 1H NMR spectra of representative DTM-A/Es (Fig. S1†). The coupling constants (J = 8.0 Hz) and chemical shifts (δ = ∼4.35 ppm) estimated from these NMR spectra are typical for β-anomeric protons, which are quite different from the coupling constant (J = 4.0 Hz) and chemical shifts (δ = ∼5.15 ppm) observed for α-anomeric protons.All the DTMs were highly soluble in water (>5%), except DTM-A8 and DTM-E7 that tended to form hydrogels at a low temperature. Micelles formed by the DTMs were characterized in terms of critical micelle concentration (CMC) and hydrodynamic radii (Rh). A fluorescent probe, diphenylhexatriene (DPH), was used to estimate individual CMC values23 as this probe is highly fluorescent upon encapsulation in detergent micelles. Dynamic light scattering experiments were conducted to determine micelle size in terms of hydrodynamic radii (Rh). The summarized data for the DTM-A/Es and DDM is presented in Table 1. All the DTMs gave CMC values ranging from ∼3 to ∼40 μM, much smaller than DDM (170 μM), indicating an increased tendency of these agents to self-assemble. The CMC values of the DTM-As were more or less similar to those of the DTM-Es when compared between a detergent pair with the same alkyl chain length (e.g., DTM-A6 vs. DTM-E6). Detergent CMC values were observed to roughly decrease with increasing alkyl chain length for both sets of DTMs. For instance, DTM-A5 and DTM-A8 with the shortest and longest chain length, respectively, gave the largest and smallest CMC values (∼20 and ∼3 μM) among the DTM-As. The micelle sizes formed by the DTMs with relatively short alkyl chain were comparable to that of DDM (∼3.4 nm) in terms of Rh, as exemplified by DTM-A5/A6 and DTM-E5/E6 (from 3.4 to 3.9 nm). In contrast, the long alkyl chain DTMs (e.g., DTM-A7/A8 and DTM-E7) formed significantly larger micelles with Rh ranging from 16 to 34 nm. This sensitive variation in micelle size depending on detergent alkyl chain length observed here is likely due to substantial changes in the volume of the detergent tail group generated by the small extension of chain length. With the volume of the head group constant (i.e., trimaltoside), the volume of the tail group rapidly enlarges with increasing alkyl chain length as a result of the dendronic scaffold in the lipophilic region; an increase in alkyl chain length by one carbon unit corresponds to the extension of the individual four alkyl chains by that much. A further analysis of the detergent micelles by DLS indicates that most DTMs are homogeneous in terms of micelle size. The exceptions are DTM-A7/A8 whose micelles were observed to be heterogeneous (Fig. S2†).
| Detergent | M.W.a | CMC (μM) | CMC (wt%) | R h (nm) |
|---|---|---|---|---|
| a Molecular weight of detergents. b Detergent hydrodynamic radius measured at 0.5 wt% detergent concentration by dynamic light scattering (DLS). | ||||
| DTM-A5 | 1485.7 | ∼20 | ∼0.0030 | 3.6 ± 0.1 |
| DTM-A6 | 1541.8 | ∼10 | ∼0.0015 | 3.7 ± 0.0 |
| DTM-A7 | 1598.0 | ∼5 | ∼0.0008 | 18.2 ± 0.5 |
| DTM-A8 | 1654.1 | ∼3 | ∼0.0005 | 34.0 ± 0.4 |
| DTM-E5 | 1653.8 | ∼40 | ∼0.0070 | 3.4 ± 0.2 |
| DTM-E6 | 1709.9 | ∼10 | ∼0.0017 | 3.9 ± 0.0 |
| DTM-E7 | 1766.1 | ∼5 | ∼0.0009 | 16.2 ± 0.7 |
| DDM | 510.1 | 170 | 0.0087 | 3.4 ± 0.0 |
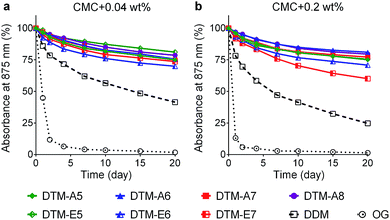
The stabilization efficacy of the DTMs was first evaluated with the photosynthetic superassembly of Rhodobacter (R.) capsulatus, comprising light-sensitive light harvesting complex I (LHI) and reaction centre complex (RC).24 The structurally intact LHI-RC complex strongly absorbs at 875 nm due to the presence of a number of cofactors (e.g., chlorophyll and carotenoids) embedded in the complex interior. Thus, protein stability for this complex could be conveniently assessed by monitoring protein absorbance at 875 nm (A875) over time. The LHI-RC complexes were extracted from the membrane using 1.0% DDM and then purified in 1 × CMC the same detergent (i.e., 0.0087 wt% DDM) via Ni2+-NTA affinity column. The DDM-purified LHI-RC complexes were diluted into buffer solutions containing the individual detergents (OG, DDM, DTM-As, and DTM-Es) so that a final detergent concentration was CMC + 0.04 wt%. Absorption spectra for the complex samples in the individual detergents were collected at regular intervals during a 20 day incubation at room temperature. DDM and OG were used as control agents as these are representatives of maltoside and glucoside detergents, respectively. Overall, absorption data at 875 nm showed clear advantages of all the DTMs over DDM and OG in maintaining the integrity of the complexes (Fig. 1). The DDM-solubilized complexes gradually lost integrity, reaching ∼40% retention after a 20 day incubation, while the OG-solubilized complexes underwent a rapid loss in this regard, giving only ∼10% integrity retention following a 2 day incubation. In contrast, all the DTMs showed 75–85% retention in complex integrity at the end of incubation. The best performance was observed for DTM-E5, but all DTMs were similar. The stability of the complex tended to decrease with increasing detergent concentration, particularly for DDM and OG. These two detergents gave more rapid decrease in A875 when detergent concentration was increased to CMC + 0.2 wt%. A similar trend was observed for DTM-E7, but all the other DTM-solubilized complexes were stable at the increased detergent concentration. Overall, the DTM-A/Es were more effective at maintaining the integrity of the LHI-RC complex compared to the conventional agents.
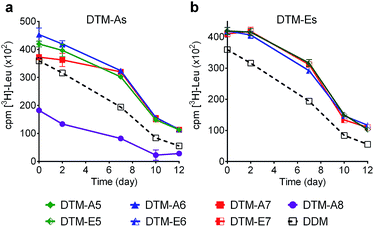
The encouraging result of the DTMs with the LHI-RC complex provoked us to further investigate these agents with the leucine transporter (LeuT) from bacteria Aquifex aeolicus.25,26 For the evaluation, the transporter was first solubilized by 1.0% DDM and purified in 0.05% of the same detergent. The DDM-purified transporter was diluted with buffer solutions supplemented with the individual DTMs to give a final detergent concentration of CMC + 0.04 wt%. Protein stability was assessed by measuring the ability to bind the radio-labelled substrate ([3H]-leucine) via scintillation proximity assay (SPA). Ligand binding activity was monitored at regular intervals over the course of a 12 day incubation at room temperature.27 As shown in Fig. 2, with the exception of the longest alkyl chain DTM-As (i.e., DTM-A8), all the DTM-A/Es were similar to each other and better than DDM at retaining transporter activity. At an increased detergent concentration of CMC + 0.2 wt%, a similar trend was observed. Again, DTM-A8 was worst at maintaining LeuT activity while the other DTM-A/Es are more or less comparable to each other and better than DDM (Fig. S3†).
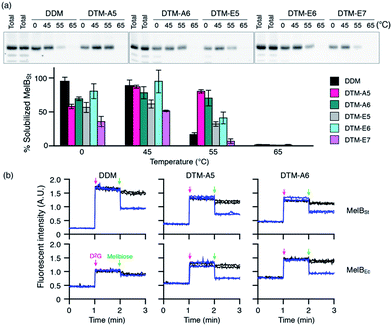
The promising results with the LHI-RC complex and LeuT prompted us to test these agents with melibiose permease of Salmonella typhimurium (MelBSt).28 Five DTMs (DTM-A5, DTM-A6, DTM-E5, DTM-E6 and DTM-E7) with high water-solubility were used to measure MelBSt thermostability. Membranes containing MelBSt were incubated with 1.5% individual detergents for 90 min, and the amounts of MelBSt solubilized by detergent treatment were analysed by SDS-PAGE and Western blotting. As shown in Fig. 3, when carried out at 0 °C, all the DTMs tested here yielded smaller amounts of soluble MelBSt than DDM under the conditions tested. At increasing incubation temperatures of 45, 55, or 65 °C efficiency of membrane protein extraction by detergents tends to be improved. This is mainly due to an increase in membrane dynamics at the elevated temperatures. In contrast, protein stability/water-solubility tends to decrease with increasing temperature as membrane proteins are prone to aggregation/denaturation at a high temperature. Thus, information on detergent efficiency and efficacy for protein extraction and stabilization, respectively, could be obtained by measuring the amount of soluble MelBSt as a function of incubation temperature. At 45 °C, noticeable increases in the amount of soluble MelBSt were observed for all the tested DTMs, indicating that the transporters were efficiently extracted by treatment with the individual detergents and the resulting transporters are stable enough to maintain good water-solubility under the conditions tested. The amounts of soluble MelBSt obtained from DTM-A5/A6 and DTM-E6 were comparable to that of DDM. When incubation temperature was further increased to 55 °C, the amount of soluble MelBSt in DDM was dropped down to ∼15%, indicating that the DDM-solubilized transporter underwent significant protein denaturation/aggregation under the conditions. Similar behaviours were observed for the DTM-Es. For example, DTM-E6, effective at retaining MelBSt solubility at 45 °C, produced only 30–40% soluble transporter at 55 °C. In contrast, two DTM-As (DTM-A5 and DTM-A6) were remarkable at preserving MelBSt solubility even at the high temperature of 55 °C. Thus, the DTM-As appeared to be superior to DTM-Es/DDM. When the incubation temperature was increased further to 65 °C, none of the detergents was able to prevent MelBSt denaturation/aggregation. Because of the outstanding behaviours of DTM-A5 and DTM-A6 for maintaining MelBSt solubility, these two DTMs were further assessed for their ability to maintain MelBSt function. For this purpose, MelBSt was extracted by selected individual detergents (DTM-A5, DTM-A6, and DDM) from the membranes and the functional state of the resulting transporters assessed via melibiose reversal of Förster resonance energy transfer (FRET) from tryptophan (Trp) to the fluorescent ligand, 2′-(N-dansyl)aminoalkyl-1-thio-β-D-galactopyranoside (D2G).28a,d,e In the presence of D2G, functional MelB binds to the fluorescent ligand, producing a strong fluorescent signal produced partly via the efficient energy transfer from Trp to this ligand. This signal is reduced by the addition of an excess amount of competitive non-fluorescent substrate (i.e., melibiose) due to ligand exchange. The fluorescent signal was highly sensitive to both D2G and melibiose added successively into MelBSt solubilized in DDM, indicating that the DDM-solubilized MelBSt is functional. However, protein function was not detected for a less stable MelB homologue from Escherichia coli (MelBEc)28d solubilized in the same detergent. In contrast, DTM-A5 and DTM-A6 provided full functionality of both MelB homologs. Collectively, two DTM representatives, DTM-A5 and DTM-A6, were not only effective at maintaining MelB solubility, but also superior to DDM in preserving MelB function.
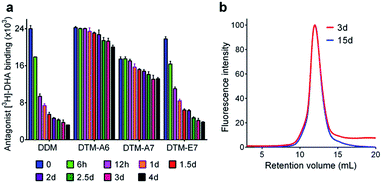
We then evaluated the novel agents with a G-protein coupled receptor (GPCR), the human β2 adrenergic receptor (β2AR).29 The receptor was isolated in 0.1% DDM. The individual DTMs were introduced into the samples containing the DDM-purified β2AR by a dilution method, giving a final detergent concentration of CMC + 0.2 wt%. As a direct assessment of receptor stability in the individual detergents, the ability of the receptor to bind the radioactive antagonist ([3H]-dihydroalprenolol (DHA))30–32 was used. A preliminary result was obtained by measuring initial ligand binding ability of the receptor following 30 min sample dilution. As can be seen in Fig. S4,† detergent efficacy tended to strongly depend on detergent alkyl chain length, with an optimal chain length observed at C6 or C7 for both sets of the DTMs. Next, three well-behaving agents (DTM-A6, DTM-A7 and DTM-E7) were selected to further evaluate these agents with regards to long-term receptor stability. The receptor solubilized in each of these agents was incubated at room temperature and the ligand binding ability was measured at regular intervals over the course of 4 day incubation (Fig. 4a). DTM-E7 behaved similarly to DDM in terms of stabilizing the receptor long-term: initially high ligand binding activity followed by a subsequent rapid decrease in receptor activity. After the 4 day incubation, the receptor solubilized in this DTM retained only 10–20% initial activity. In contrast, significantly improved behaviours were observed for DTM-A6 and DTM-A7. Both novel agents were superior to DDM in retaining receptor stability long term. Particularly, the use of DTM-A6 led to the receptor with high initial activity (t = 0 day) and 80% retention in that activity at the end of incubation (t = 4 day). It is notable that the alkyl versions of the DTMs (i.e., DTM-As) were generally better than the ether counterparts (i.e., DTM-Es). This result indicates the potential utility of these agents, particularly DTM-A6, in GPCR structural study. DTM-A6 was further evaluated with the β2AR-Gs complex. For this purpose, β2AR and Gs protein individually isolated in DDM were combined together to construct β2AR-Gs complex and then DDM was exchanged with DTM-A6. When complex integrity was measured using size exclusion chromatography (SEC), we found that β2AR-Gs complexes purified in DTM-A6 fully maintained their integrity for 15 days (Fig. 4b), which is in contrast to a result obtained from DDM-purified β2AR-Gs complexes in a previous study;33 the complex in DDM showed significant dissociation between the receptor and Gs protein even after a 2 day incubation. The full retention in complex integrity under the detergent-free condition observed here implies that DTM-A6 strongly binds to the surface of the complex, a favourable detergent characteristic for single particle cryo-EM studies.34
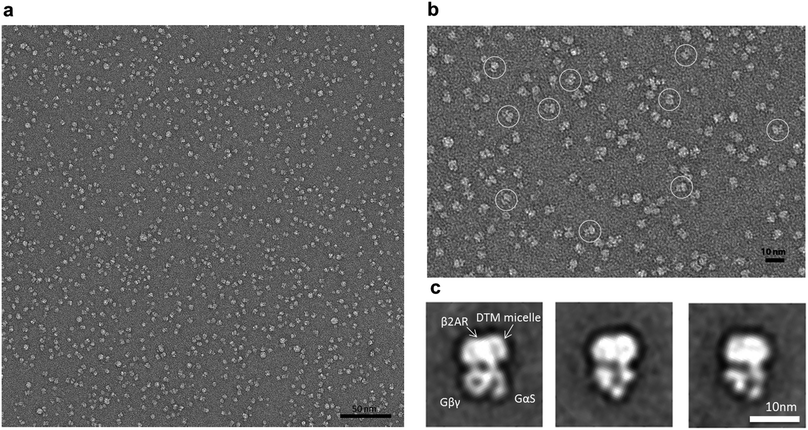
The enhanced complex stability encouraged us to continue to evaluate this agent by visualization of the complex using negative stain electron microscopy (EM).35 The EM images showed that particles generated from DTM-A6-purified β2AR-Gs complexes were highly homogeneous (Fig. 5a), which is quite different from a substantial particle aggregation observed for the DDM-purified complex in a previous study.36 2D classification of particle projections obtained from a single preparation enabled the calculation of class averages where we can readily identify the individual domains of the complex (β2AR, Gαs, and Gβγ), which appear more defined than those displayed by some other recently described novel agents (MNG-3 and PSE-C11) (Fig. 5c & S5†). This result indicates that DTM-A6 is a promising candidate for structure determination of membrane protein complexes and the visualization of their conformational ensemble via cryo-EM. Finally, the DTMs were evaluated with UapA, the uric acid–xanthine/H+ symporter from Aspergillus nidulans.37 The transporter was extracted from the membranes by the individual DTMs or DDM. DTM-A6, DTM-E6 and DTM-E7 gave similar extraction efficiencies to DDM (∼90%). Soluble protein stability was then assessed via fluorescent size exclusion chromatography (FSEC) following a heat treatment at 40 °C for 10 min. When the stability of the extracted transporter was measured, all DTMs were inferior to DDM in preserving the protein in a monodispersed state.
Conclusions
Detergent efficacy toward protein stability tends to be protein-specific and to be highly susceptible to small variation in detergent structure. In this study, the DTM-As and DTM-Es were comparable to each other for LHI-RC and LeuT stability, while the DTM-As were superior to the DTM-Es for β2AR and MelBSt stability. Considering the subtle structural difference between these two sets of DTMs (–CH2– vs. –O–), it seems remarkable that the DTM-As, particularly DTM-A6, were significantly more effective than the DTM-Es at stabilizing some membrane proteins tested here. Interestingly enough, an opposite trend was observed in a previous study15 where PSAs with an alkyl linkage were shown to be less effective than PSEs with an ether linkage at stabilizing several tested membrane proteins including MelBSt and β2AR. This opposite preference for the linker functional group (ether vs. alkyl) between DTM-A/Es and PSA/Es indicates that the optimal functional group could be dependent on detergent architecture. From the current and previous results, the alkyl connection appeared to be preferable for the DTM architecture while the ether linkage was advantageous for the PSA/E architecture.At this stage, it is hard to know the precise reasons for the enhanced efficacy of the DTM-As compared to the DTM-Es, but one notable feature of the DTM-Es is the presence of more ether linkages in the tail region than the DTM-As (7 vs. 3), generating more propyleneglycol units in the hydrophobic group. As this glycol unit is generally considered to behave as a hydrophilic group at room temperature,38 the tail groups of the DTM-Es would be relatively more hydrophilic than their DTM-A counterparts, thereby weakening their interactions with the hydrophobic segment of a membrane protein. Strong hydrophobic interactions of detergent micelles with membrane proteins are essential to prevent protein denaturation/aggregation. Micellar stability, deeply associated with membrane protein stability, may also be reduced due to the presence of the several hydrophilic units in the hydrophobic tail region in the case of DTM-Es as micelle formation is mainly driven by hydrophobic effect. We also believe that the hydrophobic chain length is one of the reasons for the generally favourable behaviours of the DTM-As over DTM-Es. As for the current DTMs with the multiple propyleneglycol units, the hydrophobic chain length could be effectively measured from the ether bond located between the first and second branch points rather than from the focal point. On the basis of this notion, the effective hydrophobic chain length for DTM-A6 is estimated to be C12, more comparable to that of DDM (C12) than that for DTM-E6 (C10). The hydrophobic chain length of detergents is crucial for membrane protein stability as, for effective encapsulation, the chain length should conform to the lipophilic width of a target membrane protein (∼30 Å).39
The most distinct feature of the DTMs compared to previously developed detergents is the presence of the dendronic hydrophobic group. Dendritic architecture is widely used in a variety of applications including drug delivery,40 biochemical sensors,41 fluorescence imaging42 but hasn't yet been incorporated into detergent structure for membrane protein study. One potential reason is associated with the difficulty of synthesis of amphipathic dendrimers with facially segregated head and tail groups. A second reason for the rare use of dendronic architecture in detergent design may be the strict restriction in detergent hydrophobic chain length as discussed above. This restriction in the chain length isn't usually compatible with dendronic architecture because dendrimer dimensions tend to rapidly increase with increasing generation. Third, functional groups utilized for dendrimer preparation are often suboptimal for the current application. Because of synthetic convenience, an amide or amine group has been most popularly used in the preparation of dendrimers, but this functional group is too rigid and/or too polar to favourably interact with target membrane proteins. In contrast, the ether linkage is better in this regard because of its reduced polarity and enhanced flexibility as observed in a previous study,14f but its polarity could still be an issue particularly for a detergent with several ether bonds in the tail region, as exemplified by the DTM-Es. Thus, a dendrimer with an alkyl linkage would be most suited for detergents with enhanced protein stabilization and solubilisation properties, but is generally accompanied by increasing difficulty in preparation. A compromise between synthetic convenience and detergent efficacy is necessary to develop a practical detergent with improved properties, which seems to have been attained by DTM-A6 with three ether bonds in the tail region. Despite the multiple challenges in detergent design with a dendronic tail group, some DTM-As, particularly DTM-A6, showed a clear advantage over DDM in stabilization of the multiple membrane proteins (complexes) tested here, indicative of a significantly optimized architecture of these agents in terms of amphiphilic nature and a functional group choice.
In summary, the current study introduced two sets of the detergents (DTM-As and DTM-Es) as the first novel agents with a dendronic tail group for membrane protein study. These agents were slightly inferior to DDM at extracting MelBSt from the membrane, but representatives were superior to this conventional detergent in stabilizing a few tested membrane proteins except UapA. Among the DTMs, DTM-A6 induced, in general, the greatest stability of the individual membrane proteins and proved effective at allowing visualisation of each component of the β2AR-Gs complex by negative stain EM analysis. This result indicates that this DTM has significant potential for membrane protein structural studies using X-ray crystallography and cryo-EM. In addition, the detailed discussion on the enhanced membrane protein stabilisation efficacy of the DTM-As relative to DTM-Es serves as a design principle essential for future detergent development.
Conflicts of interest
The authors declare the following competing financial interest(s): P. S. C. and A. S. are inventors on a patent application describing the DTMs.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2016R1A2B2011257 to P. S. C. and A. S.).Notes and references
- E. Wallin and G. von Heijne, Protein Sci., 1998, 7, 1029–1038 CrossRef CAS PubMed.
- (a) M. T. Drake, S. K. Shenoy and R. J. Lefkowitz, Circ. Res., 2006, 99, 570–582 CrossRef CAS PubMed; (b) R. Lappano and M. Maggiolini, Nat. Rev. Drug Discovery, 2011, 10, 47–60 CrossRef CAS PubMed.
- (a) C. R. Sanders and J. K. Myers, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 25–51 CrossRef CAS PubMed; (b) J. P. Overington, B. Al-Lazikani and A. L. Hopkins, Nat. Rev. Drug Discovery, 2006, 5, 993–996 CrossRef CAS PubMed.
- J. J. Lacapere, E. Pebay-Peyroula, J. M. Newmann and C. Etchebest, Trends Biochem. Sci., 2007, 32, 259–270 CrossRef CAS PubMed.
- (a) S. H. White and W. C. Wimley, Annu. Rev. Biophys. Biomol. Struct., 1999, 28, 319–365 CrossRef CAS PubMed; (b) J. U. Bowie, Curr. Opin. Struct. Biol., 2001, 11, 397–402 CrossRef CAS PubMed; (c) J. J. Lacapere, E. Pebay-Peyroula, J. M. Neumann and C. Etchebest, Trends Biochem. Sci., 2007, 32, 259–270 CrossRef CAS PubMed; (d) R. Philips, T. Ursell, P. Wiggins and P. Sens, Nature, 2009, 459, 379–385 CrossRef PubMed.
- (a) A. M. Seddon, P. Curnow and P. J. Booth, Biochim. Biophys. Acta, 2004, 1666, 105–117 CrossRef CAS PubMed; (b) Z. Yang, C. Wang, Q. Zhou, J. An, E. Hildebrandt, L. A. Aleksandrov, J. C. Kappes, L. J. DeLucas, J. R. Riordan, I. L. Urbatsch, J. F. Hunt and C. G. Brouillette, Protein Sci., 2014, 23, 769–789 CrossRef CAS PubMed.
- J. L. Parker and S. Newstead, Protein Sci., 2012, 21, 1358–1365 CrossRef CAS PubMed.
- (a) M. J. Serrano-Vega, F. Magnani, Y. Shibata and C. G. Tate, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 877–882 CrossRef CAS PubMed; (b) S. Newstead, S. Ferrandon and S. Iwata, Protein Sci., 2008, 17, 466–472 CrossRef CAS PubMed; (c) Y. He, K. Wang and N. Yan, Protein Cell, 2014, 5, 658–672 CrossRef CAS PubMed.
- (a) G. G. Privé, Methods, 2007, 41, 388–397 CrossRef PubMed; (b) P. S. Chae, P. D. Laible and S. H. Gellman, Mol. BioSyst., 2010, 6, 89–94 RSC; (c) Q. Zhang, H. Tao and W.-X. Hong, Methods, 2011, 55, 318–323 CrossRef CAS PubMed.
- (a) P. S. Chae, M. J. Wander, K. H. Cho, P. D. Laible and S. H. Gellman, Mol. BioSyst., 2013, 9, 626–629 RSC; (b) H. E. Bae, K. Gotfryd, J. Thomas, H. Hussain, M. Ehsan, J. Go, C. J. Loland, B. Byrne and P. S. Chae, ChemBioChem, 2015, 16, 1454–1459 CrossRef CAS PubMed.
- (a) C.-L. McGregor, L. Chen, N. C. Pomroy, P. Hwang, S. Go, A. Chakrabartty and G. G. Privé, Nat. Biotechnol., 2003, 21, 171–176 CrossRef CAS PubMed; (b) H. Tao, S. C. Lee, A. Moeller, R. S. Roy, F. Y. Siu, J. Zimmermann, R. C. Stevens, C. S. Potter, B. Carragher and Q. Zhang, Nat. Methods, 2013, 10, 759–761 CrossRef CAS PubMed.
- (a) C. Tribet, R. Audebert and J.-L. Popot, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 15047–15050 CrossRef CAS PubMed; (b) J. L. Popot, T. Althoff, D. Bagnard, J. L. Banères, P. Bazzacco, E. Billon-Denis, L. J. Catoire, P. Champeil, D. Charvolin, M. J. Cocco, G. Crémel, T. Dahmane, L. M. de la Maza, C. Ebel, F. Gabel, F. Giusti, Y. Gohon, E. Goormaghtigh, E. Guittet, J. H. Kleinschmidt, W. Kühlbrandt, C. Le Bon, K. L. Martinez, M. Picard, B. Pucci, J. N. Sachs, C. Tribet, C. van Heijenoort, F. Wien, F. Zito and M. Zoonens, Annu. Rev. Biophys., 2011, 40, 379–408 CrossRef CAS PubMed; (c) J. M. Dörr, S. Scheidelaar, M. C. Koorengevel, J. J. Dominguez, M. Schäfer, C. A. van Walree and J. A. Killian, Eur. Biophys. J., 2016, 45, 3–21 CrossRef PubMed; (d) A. Nath, W. M. Atkins and S. G. Sligar, Biochemistry, 2007, 46, 2059–2069 CrossRef CAS PubMed; (e) R. Ujwal and J. U. Bowie, Methods, 2011, 55, 337–341 CrossRef CAS PubMed; (f) L. Frey, N.-A. Lakomek, R. Riek and S. Bibow, Angew. Chem., Int. Ed., 2017, 56, 380–383 CrossRef CAS PubMed.
- (a) P. S. Chae, S. G. F. Rasmussen, R. R. Rana, K. Gotfryd, A. C. Kruse, S. Nurva, U. Gether, L. Guan, C. J. Loland, B. Byrne, B. K. Kobilka and S. H. Gellman, Chem.–Eur. J., 2012, 18, 9485–9490 CrossRef CAS PubMed; (b) D. T. McQuade, M. A. Quinn, S. M. Yu, A. S. Polans, M. P. Krebs and S. H. Gellman, Angew. Chem., Int. Ed., 2000, 39, 758–761 CrossRef CAS; (c) P. S. Chae, M. J. Wander, A. P. Bowling, P. D. Laible and S. H. Gellman, ChemBioChem, 2008, 9, 1706–1709 CrossRef CAS PubMed; (d) P. S. Chae, K. H. Cho, M. J. Wander, H. E. Bae, S. H. Gellman and P. D. Labile, Biochim. Biophys. Acta, 2014, 1838, 278–286 CrossRef CAS PubMed; (e) M. Das, Y. Du, O. Ribeiro, P. Hariharan, J. S. Mortensen, D. Patra, G. Skiniotis, C. J. Loland, L. Guan, B. K. Kobilka, B. Byrne and P. S. Chae, J. Am. Chem. Soc., 2017, 139, 3072–3081 CrossRef CAS PubMed; (f) H. Hussain, Y. Du, E. Tikhonova, J. S. Mortensen, O. Ribeiro, C. Santillan, M. Das, M. Ehsan, C. J. Loland, L. Guan, B. K. Kobilka, B. Byrne and P. S. Chae, Chem.–Eur. J., 2017, 23, 6724–6729 CrossRef CAS PubMed.
- (a) P. S. Chae, R. R. Rana, K. Gotfryd, S. G. F. Rasmussen, A. C. Kruse, K. H. Cho, S. Capaldi, E. Carlsso, B. Kobilka, C. J. Loland, U. Gether, S. Banerjee, B. Byrne, J. K. Lee and S. H. Gellman, Chem. Commun., 2013, 49, 2287–2289 RSC; (b) K. H. Cho, H. E. Bae, M. Das, S. H. Gellman and P. S. Chae, Chem.–Asian J., 2014, 9, 632–638 CrossRef CAS PubMed; (c) P. S. Chae, S. G. F. Rasmussen, R. R. Rana, K. Gotfryd, R. Chandra, M. A. Goren, A. C. Kruse, S. Nurva, C. J. Loland, Y. Pierre, D. Drew, J. L. Popot, D. Picot, B. G. Fox, L. Guan, U. Gether, B. Byrne, B. Kobilka and S. H. Gellman, Nat. Methods, 2010, 7, 1003–1008 CrossRef CAS PubMed; (d) K. H. Cho, M. Husri, A. Amin, K. Gotfryd, H. J. Lee, J. Go, C. J. Loland, L. Guan, B. Byrne and P. S. Chae, Analyst, 2015, 140, 3157–3163 RSC; (e) K. H. Cho, B. Byrne and P. S. Chae, ChemBioChem, 2013, 14, 452–454 CrossRef CAS PubMed; (f) A. Sadaf, J. S. Mortensen, S. Capaldi, E. Tikhonova, P. Hariharan, O. Ribeiro, C. J. Loland, L. Guan, B. Byrne and P. S. Chae, Chem. Sci., 2016, 7, 1933–1939 RSC.
- M. Ehsan, Y. Du, N. J. Scull, E. Tikhonova, J. Tarrasch, J. S. Mortensen, C. J. Loland, G. Skiniotis, L. Guan, B. Byrne, B. Kobilka and P. S. Chae, J. Am. Chem. Soc., 2016, 138, 3789–3796 CrossRef CAS PubMed.
- (a) S. G. F. Rasmussen, H. J. Choi, J. J. Fung, E. Pardon, P. Casarosa, P. S. Chae, B. T. DeVree, D. M. Rosenbaum, F. S. Thian, T. S. kobilka, A. Schnapp, I. Konetzki, R. K. Sunahara, S. H. Gellman, A. Pautsch, J. Steyaert, W. I. Weis and B. K. Kobilka, Nature, 2011, 469, 175–180 CrossRef CAS PubMed; (b) D. M. Rosenbaum, C. Zhang, J. Lyons, R. Holl, D. Aragao, D. H. Arlow, S. G. F. Rasmussen, H.-J. Choi, B. T. DeVree, R. K. Sunahara, P. S. Chae, S. H. Gellman, R. O. Dror, D. E. Shaw, W. I. Weis, M. Caffrey, P. Gmeiner and B. K. Kobilka, Nature, 2011, 469, 236–240 CrossRef CAS PubMed; (c) S. G. F. Rasmussen, B. T. DeVree, Y. Zou, A. C. Kruse, K. Y. Chung, T. S. Kobilka, F. S. Thian, P. S. Chae, E. Pardon, D. Calinski, J. M. Mathiesen, S. T. A. Shah, J. A. Lyons, M. Caffrey, S. H. Gellman, J. Steyaert, G. Skiniotis, W. I. Weis, R. K. Sunahara and B. K. Kobilka, Nature, 2011, 477, 549–555 CrossRef CAS PubMed; (d) A. M. Ring, A. Manglik, A. C. Kruse, M. D. Enos, W. I. Weis, K. C. Garcia and B. K. Kobilka, Nature, 2013, 502, 575–579 CrossRef CAS PubMed; (e) A. K. Shukla, G. H. Westfield, K. Xiao, R. I. Reis, L.-Y. Huang, P. T. Shukla, J. Qian, S. Li, A. Blanc, A. N. Oleskie, A. M. Dosey, M. Su, C.-R. Liang, L. L. Gu, J. M. Shan, X. Chen, R. Hanna, M. Choi, X. J. Yao, B. U. Klink, A. W. Kahsai, S. S. Sidhu, S. Koide, P. A. Penczek, A. A. Kossiakoff, V. L. Woods Jr, B. K. Kobilka, G. Skiniotis and R. J. Lefkowitz, Nature, 2014, 512, 218–222 CrossRef CAS PubMed; (f) A. C. Kruse, J. Hu, A. C. Pan, D. H. Arlow, D. M. Rosenbaum, E. Rosemond, H. F. Green, T. Liu, P. S. Chae, R. O. Dror, D. E. Shaw, W. I. Weis, J. Wess and B. K. Kobilka, Nature, 2012, 482, 552–556 CrossRef CAS PubMed; (g) K. Haga, A. C. Kruse, H. Asada, T. Y. Kobayashi, M. Shiroishi, C. Zhang, W. I. Weis, T. Okada, B. K. Kobilka, T. Haga and T. Kobayashi, Nature, 2012, 482, 547–551 CrossRef CAS PubMed; (h) A. Manglik, A. C. Kruse, T. S. Kobilka, F. S. Thian, J. M. Mathiesen, R. K. Sunahara, L. Pardo, W. I. Weis, B. K. Kobilka and S. Granier, Nature, 2012, 485, 321–326 CrossRef CAS PubMed; (i) S. Granier, A. Manglik, A. C. Kruse, T. S. Kobilka, F. S. Thian, W. I. Weis and B. K. Kobilka, Nature, 2012, 485, 400–404 CrossRef CAS PubMed; (j) J. Kellosalo, T. Kajander, K. Kogan, K. Pokharel and A. Goldman, Science, 2012, 337, 473–476 CrossRef CAS PubMed; (k) A. Quigley, Y. Y. Dong, A. C. W. Pike, L. Dong, L. Shrestha, G. Berridge, P. J. Stansfeld, M. S. P. Sansom, A. N. Edwards, C. Bountra, F. Von Delft, A. N. Bullock, N. A. Burgess-Brown and E. P. Carpenter, Science, 2013, 339, 1604–1607 CrossRef CAS PubMed; (l) A. Frick, U. K. Eriksson, F. de Mattia, F. Oberg, K. Hedfalk, R. Neutze, W. J. Grip, P. M. T. Deen and S. T. Horsefield, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6305–6310 CrossRef CAS PubMed.
- M. Das, Y. Du, N. J. S. Mortensen, O. Ribeiro, E. Tikhonova, P. Hariharan, L. Guan, C. J. Loland, B. K. Kobilka, B. Byrne and P. S. Chae, Chem. Sci., 2017, 8, 1169–1177 RSC.
- (a) P. I. Kitov and D. R. Bundle, J. Am. Chem. Soc., 2003, 125, 16271–16284 CrossRef CAS PubMed; (b) S. Basha, P. Rai, V. Poon, A. Saraph, K. Gujraty, M. Y. Go, S. Sadacharan, M. Frost, J. Mogridge and R. S. Kane, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 13509–13513 CrossRef CAS PubMed.
- K. H. Cho, O. Ribeiro, Y. Du, E. Tikhonova, J. S. Mortensen, K. Markham, P. Hariharan, C. J. Loland, L. Guan, B. K. Kobilka, B. Byrne and P. S. Chae, Chem.–Eur. J., 2016, 22, 18833–18839 CrossRef CAS PubMed.
- P. S. Chae, H. E. Bae and M. Das, Chem. Commun., 2014, 50, 12300–12303 RSC.
- J. N. Israelachvili, Intermolecular and surface forces, Academic Press, London, 2nd edn, 1992 Search PubMed.
- P. S. Chae, A. C. Kruse, K. Gotfryd, R. R. Rana, K. H. Cho, S. G. F. Rasmussen, H. E. Bae, U. Gether, L. Guan, C. J. Loland, B. Byrne, B. K. Kobilka and S. H. Gellman, Chem.–Eur. J., 2013, 19, 15645–15651 CrossRef CAS PubMed.
- A. Chattopadhyay and E. London, Anal. Biochem., 1984, 139, 408–412 CrossRef CAS PubMed.
- P. D. Labile, C. Kirmaier, C. S. Udawatte, S. J. Hofman, D. Holten and D. K. Hanson, Biochemistry, 2003, 42, 1718–1730 CrossRef PubMed.
- G. Deckert, P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen and R. V. Swanson, Nature, 1998, 392, 353–358 CrossRef CAS PubMed.
- A. Yamashita, S. K. Singh, T. Kawate, Y. Jin and E. Gouaux, Nature, 2005, 437, 215–223 CrossRef CAS PubMed.
- G. Swaminath, J. Steenhuis, B. Kobilka and T. W. Lee, Mol. Pharmacol., 2002, 61, 65–72 CrossRef CAS PubMed.
- (a) L. Guan, S. Nurva and S. P. Ankeshwarapu, J. Biol. Chem., 2011, 286, 6367–6374 CrossRef CAS PubMed; (b) A. S. Ethayathulla, M. S. Yousef, A. Amin, G. Leblanc, H. R. Kaback and L. Guan, Nat. Commun., 2014, 5, 3009 Search PubMed; (c) A. Amin, A. S. Ethayathulla and L. Guan, J. Bacteriol., 2014, 196, 3134–3139 CrossRef PubMed; (d) A. Amin, P. Hariharan, P. S. Chae and L. Guan, Biochemistry, 2015, 54, 5849–5855 CrossRef CAS PubMed; (e) E. Cordat, I. Mus-Veteau and G. Leblanc, J. Biol. Chem., 1998, 273, 33198–33202 CrossRef CAS PubMed.
- D. M. Rosenbaum, V. Cherezov, M. A. Hanson, S. G. Rasmussen, F. S. Thian, T. S. Kobilka, H. J. Choi, X. J. Yao, W. I. Weis, R. C. Stevens and B. K. Bobilka, Science, 2007, 318, 1266–1273 CrossRef CAS PubMed.
- S. E. Mansoor, H. S. McHaourab and D. L. Farrens, Biochemistry, 2002, 41, 2475–2484 CrossRef CAS PubMed.
- X. Yao, C. Parnot, X. Deupi, V. R. P. Ratnala, G. Swaminath and D. Farrens, Nat. Chem. Biol., 2006, 2, 417–422 CrossRef CAS PubMed.
- G. Swaminath, J. Steenhuis, B. Kobilka and T. W. Lee, Mol. Pharmacol., 2002, 61, 65–72 CrossRef CAS PubMed.
- K. Y. Chung, S. G. F. Rasmussen, T. Liu, S. Li, B. T. DeVree, P. S. Chae, D. Calinski, B. K. Kobilka, V. L. Woods Jr and R. K. Sunnahara, Nature, 2011, 477, 611–615 CrossRef CAS PubMed.
- F. Hauer, C. Gerle, N. Fischer, A. Oshima, K. Shinzawa-Itoh, S. Shimada, K. Yokoyama, Y. Fujiyoshi and H. Stark, Structure, 2015, 23, 1769–1775 CrossRef CAS PubMed.
- A. Peisley and G. Skiniotis, Methods in Molecular Biology, ed. M. Filizola, 2015, vol. 1335, pp. 29–38 Search PubMed.
- H. Hussain, Y. Du, N. J. Scull, J. S. Mortensen, J. Tarrasch, C. J. Loland, B. Byrne, B. K. Kobilka and P. S. Chae, Chem.–Eur. J., 2016, 22, 7068–7073 CrossRef CAS PubMed.
- J. Takano, K. Noguchi, M. Yasumori, M. Kobayashi, Z. Gajdos, K. Miwa, H. Hayashi, T. Yoneyama and T. Fujiwara, Nature, 2002, 420, 337–340 CrossRef CAS PubMed.
- K. Miki, P. Westh and Y. Koga, J. Phys. Chem. B, 2005, 109, 19536–19541 CrossRef CAS PubMed.
- K. H. Cho, P. Hariharan, J. S. Mortensen, Y. Du, A. K. Nielsen, B. Byrne, B. K. Kobilka, C. J. Loland, L. Guan and P. S. Chae, ChemBioChem, 2016, 17, 2334–2339 CrossRef CAS PubMed.
- X. Ma, Z. Zhou, E. Jin, Q. Sun, B. Zhang, J. Tang and Y. Shen, Macromolecules, 2013, 46, 37–42 CrossRef CAS.
- (a) C. M. Lamy, O. Sallin, C. Loussert and J.-Y. Chatton, ACS Nano, 2012, 6, 1176–1187 CrossRef CAS PubMed; (b) N. Zhu, Y. Gu, Z. Chang, P. He and Y. Fang, Electroanalysis, 2006, 21, 2107–2114 CrossRef.
- Y. Kim, S. H. Kim, M. Tanyeri, J. A. Katzenellenbogen and C. M. Shroeder, Biophys. J., 2013, 104, 1566–1575 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc03700g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |