Open Access Article
Open Access ArticleIntegration of G-quadruplex and DNA-templated Ag NCs for nonarithmetic information processing†
Ru-Ru
Gao
,
Tian-Ming
Yao
*,
Xiao-Yan
Lv
,
Yan-Yan
Zhu
,
Yi-Wei
Zhang
and
Shuo
Shi
 *
*
Shanghai Key Laboratory of Chemical Assessment and Sustainability, School of Chemical Science and Engineering, Tongji University, Shanghai, 200092, P. R. China. E-mail: shishuo@tongji.edu.cn; tmyao@tongjji.edu.cn
First published on 6th April 2017
Abstract
To create sophisticated molecular logic circuits from scratch, you may not believe how common the building blocks can be and how diverse and powerful such circuits can be when scaled up. Using the two simple building blocks of G-quadruplex and silver nanoclusters (Ag NCs), we experimentally construct a series of multifunctional, label-free, and multi-output logic circuits to perform nonarithmetic functions: a 1-to-2 decoder, a 4-to-2 encoder, an 8-to-3 encoder, dual transfer gates, a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer, and a 1
1 multiplexer, and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer. Moreover, a parity checker which is capable of identifying odd and even numbers from natural numbers is constructed conceptually. Finally, a multi-valued logic gate (ternary inhibit gate) is readily achieved by taking this DNA/Ag NC system as a universal platform. All of the above logic circuits share the same building blocks, indicating the great prospects of the assembly of nanomaterials and DNA for biochemical logic devices. Considering its biocompatibility, the novel prototypes developed here may have potential applications in the fields of biological computers and medical diagnosis and serve as a promising proof of principle in the not-too-distant future.
2 demultiplexer. Moreover, a parity checker which is capable of identifying odd and even numbers from natural numbers is constructed conceptually. Finally, a multi-valued logic gate (ternary inhibit gate) is readily achieved by taking this DNA/Ag NC system as a universal platform. All of the above logic circuits share the same building blocks, indicating the great prospects of the assembly of nanomaterials and DNA for biochemical logic devices. Considering its biocompatibility, the novel prototypes developed here may have potential applications in the fields of biological computers and medical diagnosis and serve as a promising proof of principle in the not-too-distant future.
Introduction
The beguiling discovery that chemical molecules and their inherent chemical conversions could use logic to transmit and process information,1 just like human brains and electronic computers, has spawned considerable research into intelligent decision-making molecular logic devices. These show great application potential, especially in the field of life sciences, through the processing of binary and multi-valued information for applications such as multi-parameter chemosensing and biosensing,2 prodrug activation,3 medical diagnostics,4 object labelling,5 data storage,6 and improving the cognition of complicated bio-phenomena in humans.7 Acting as an outstanding biochemical material, DNA is endowed with amazing advantages for the construction of biochemical logic circuits since its biological compatibility facilitates practical experiments in vivo, its simple synthesis method reduces operating costs, and the sufficiency of sequence design space and predictability of its molecular behavior provide the potential for construction of various information processing systems.8DNA has been widely employed to construct various basic logic gates such as AND, OR, NOR, NAND, XOR, INH, etc., by identifying and detecting its different secondary structures.9 Nevertheless, it is challenging to individually control each logic component in the circuit. To fulfill the requirements of increasing computational complexity, it is in high demand to physically integrate these basic logic gates in arrays for the construction of complicated logic devices such as adders/subtractors,10 multiplexers/demultiplexers,11 encoders/decoders,12 and flip–flop logics13 which mostly present a multi-output. Although great advancements have been made in the construction of such multi-output multifunctional logic devices, the majority of them are realized with the help of dye-labeled14 or redox-labeled15 DNA strands or the interaction between enzymes and DNA.16 Taking into account the complexity of the modification process and the special conditions required for the enzyme/DNA reaction, a new area based on the assembly of smart functional nanomaterials and DNA has attracted our attention. In particular, DNA has a variety of secondary structures, such as duplex, triplex, hairpin, i-motif, G-quadruplex, three-way and four-way junction, etc., which can be detected by specific fluorescent dyes.17 Additionally, DNA-templated Ag NCs, which exhibit robust and size-dependent fluorescence emission, have been widely employed as a new type of fluorophore in the fields of biosensing18 and construction of logic devices.19
Herein, by employing G-quadruplex, which can be identified by the fluorescent dye N-methylmesoporphyrin IX (NMM),20 and DNA-templated Ag NCs as the two building blocks and using the fluorescence of NMM and Ag NCs as the two outputs, we experimentally construct a series of multifunctional multi-output logic devices to perform nonarithmetic information processing, including a 1-to-2 decoder, a 4-to-2 encoder, an 8-to-3 encoder, dual transfer gates, a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer, and a 1
1 multiplexer, and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer, without the help of enzymes and labeled DNA. Moreover, a parity checker that is capable of identification of odd and even numbers from natural numbers is constructed. Finally, a multi-valued logic gate (ternary inhibit gate) is readily achieved by taking this DNA/Ag NC system as a universal platform.
2 demultiplexer, without the help of enzymes and labeled DNA. Moreover, a parity checker that is capable of identification of odd and even numbers from natural numbers is constructed. Finally, a multi-valued logic gate (ternary inhibit gate) is readily achieved by taking this DNA/Ag NC system as a universal platform.
Results and discussion
Construction of the 1-to-2 decoder
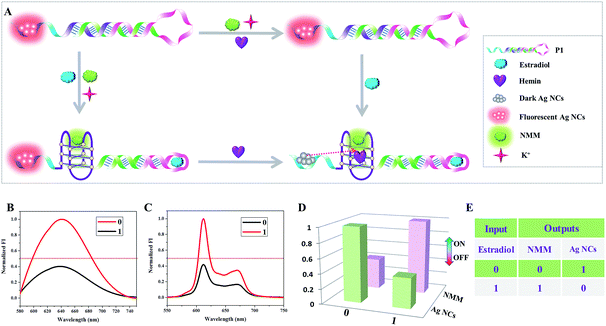
Among the series of multi-output logic circuits, encoders and decoders that are popular in molecular computations because of their ability to convert data to code or otherwise have significant applications in data communication and information storage.21In this paper, a 1-to-2 decoder that converts one input bit into two output bits is first designed based on the interaction between DNA-templated Ag NCs and G-quadruplex/hemin complexes. As depicted schematically in Fig. 1A, the hairpin structure P1 used as the initial state is integrated with the G-quadruplex sequence (purple), the aptamer sequence of estradiol (pink), and C-rich sequence (blue), which is used for the formation of fluorescent Ag NCs. This hairpin design ensures that the aptamer sequence and the G-quadruplex sequence are caged in the duplex domain of the hairpin. In addition, hemin and K+ are also in the solution in the initial state. With respect to the inputs, the presence and absence of estradiol are defined as “1” and “0” respectively. The normalized fluorescence signals of the Ag NCs and NMM are taken as the dual-output, with a threshold value of 0.5. In the absence of estradiol, there is almost no change in the fluorescence value of the Ag NCs and no formation of G-quadruplex, generating the output NMM = 0, Ag NCs = 1. After the introduction of estradiol, formation of the estradiol/aptamer complexes opens the hairpin structure and leads to the self-assembly of the G-quadruplex/hemin complexes which could turn on the fluorescence of NMM, generating the output NMM = 1. Moreover, the fluorescence of the Ag NCs is quenched by the proximity of the G-quadruplex/hemin complexes due to photoinduced electron transfer (PET),22 generating the output Ag NCs = 0. The column bars of normalized fluorescence intensity in Fig. 1D clearly indicate the different responses of NMM and the Ag NCs under various input states of the logic device, conforming to the features of a 1-to-2 decoder.
Construction of the 4-to-2 encoder
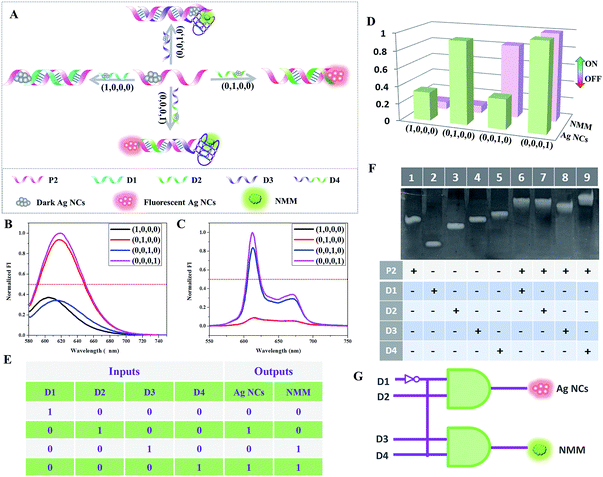
Complementary to a decoder, an encoder is a digital device that can compress information for storage or transportation by converting data signals into codes.21 Herein, a 4-to-2 encoder that compresses four input data signals into two output codes is designed, also based on the two building blocks. Yeh and co-workers demonstrated a useful phenomenon recently in that the fluorescence of darkish DNA/Ag NCs could be enhanced more than 500-fold when placed in proximity to a guanine-rich (G-rich) sequence.23 Inspired by the fluorescence turn-on mode of DNA/Ag NCs by hybridization with a G-rich sequence, Yin and co-workers reported another interesting fluorescence light-up mechanism based on an adjacent DNA/Ag NC probe pair.24 Inspired by these discoveries, we have experimentally proven the fact that the fluorescence would increase 3-fold when placing two darkish DNA/Ag NCs together via the formation of a duplex through their complementary linkers compared to that measured by placing darkish DNA/Ag NCs adjacent to a G-rich sequence. We speculate that this may be due to the surface enhanced fluorescence of silver nanoclusters, which requires further validation. On the basis of this fact, a 4-to-2 encoder is successfully realized based on the integration of G-rich DNA-templated Ag NCs, C-rich DNA-templated Ag NCs, and G-quadruplex.For construction of the 4-to-2 encoder, a P2 containing a Ag NC-nucleation sequence at the 5′-terminal is taken as the template to synthesize the darkish Ag NCs. Fig. 2 presents the schematic illustration of the cascade between the platform and the 4-to-2 encoder. In the presence of D1, which could hybridize with a part of P2 but has neither a G-rich nor C-rich sequence, there is little change in the low fluorescence intensity of Ag NCs and NMM. However, in the presence of D2, which contains the Ag NC-nucleation sequence at the 3′-terminal and a complementary linker sequence for the formation of the duplex, the fluorescence of the Ag NCs increases sharply, which verifies that the fluorescence enhancement can be induced by the adjacent darkish DNA/Ag NC probe pair. When D3 is added, which is integrated with a G-rich sequence at the 3′-terminal, this enhances the fluorescence of the Ag NCs slightly but folds into a G-quadruplex, enhancing the fluorescence intensity of NMM. Finally, upon addition of D4, which is designed with the D2 segment at the 3′-terminal and the D3 segment at the 5′-terminal, the fluorescence intensities of both Ag NCs and NMM achieve high values. The presence of the DNA strands D1, D2, D3, and D4 is defined as “input = 1”; their absence is defined as “input = 0”. Similar to the 1-to-2 decoder, the normalized fluorescence signals of the Ag NCs and NMM are taken as the dual-output, with a threshold value of 0.5. Overall, in the presence of different inputs (D1, D2, D3, and D4), we can obtain the truth table (Fig. 2E) with dual-output “Ag NCs = 0, NMM = 0” for input D1, “Ag NCs = 1, NMM = 0” for input D2, “Ag NCs = 0, NMM = 1” for input D3, and “Ag NCs = 1, NMM = 1” for input D4, fully satisfying the requirements for a 4-to-2 encoder. Fig. 2F shows the polyacrylamide gel analysis of the interaction between the different DNA strands used in the 4-to-2 encoder. Lane 1 to lane 5 show the DNA bands of P2, D1, D2, D3, and D4, respectively. However, new bands appear in lane 6 to lane 9, suggesting the formation of duplex P2/D1, P2/D2, P2/D3, and P2/D4. The PAGE results indicate that the DNA interactions occur as expected.
Construction of the 8-to-3 encoder
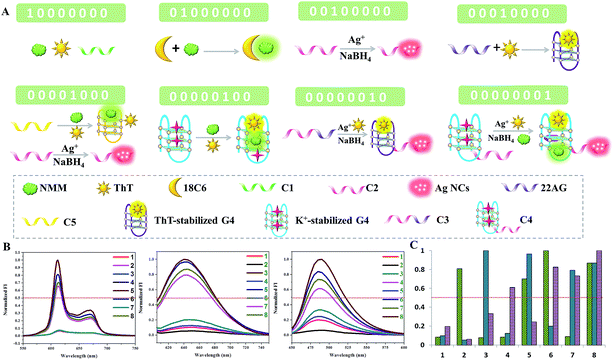
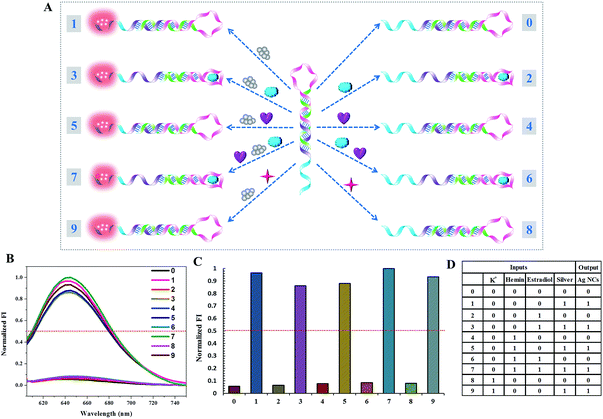
To further demonstrate the feasibility of our molecular logic system, an 8-to-3 encoder is also fabricated. In addition to NMM and the Ag NCs, another fluorescent dye, Thioflavin T (ThT), is employed as the third output signal.25 For the inputs, the presence of C1, 18-crown-6-ether (18C6), C2, 22AG, C5/C2, G4, C3, and C4 is defined as “1”. The eight inputs are compressed into three fluorescent outputs: the optical signals of NMM, Ag NCs, and ThT. C1 is a poly-thymine sequence and has no obvious influence on the three outputs. Upon the addition of 18C6, the fluorescence intensity of NMM is enhanced significantly. Both 18C6 and NMM are macrocyclic compounds, from which we infer that the reaction of NMM and 18C6 may be due to the hydrophilic–hydrophobic interaction, trapping NMM in an anhydrous cavity and blocking fluorescence quenching of NMM by water consequently. C2 is integrated with a Ag NC-nucleation sequence, which is used for the formation of bright Ag NCs but has little influence on the fluorescence of NMM and ThT. When the 22AG sequence is added in the absence of K+, ThT exclusively induces G-quadruplex folding and its fluorescence is turned on. However, 22AG cannot light up the fluorescence of NMM without K+. The fifth input, C5/C2, could light up the fluorescence of NMM instead of ThT. Moreover, it has a template for the synthesis of the fluorescent Ag NCs. When the G-quadruplex induced by K+ is added, both NMM and ThT could insert into its cavity and light up their fluorescence. Similarly, C3 is integrated with the 22AG and C2 sequences and C4 is integrated with the K+-stabilized G-quadruplex and C2 segment. The schematic representation and truth table of the 8-to-3 encoder are presented in Fig. 3, which demonstrates an easy realization of a relatively complex encoder by our multiplexed logic platform.Construction of the dual transfer gates
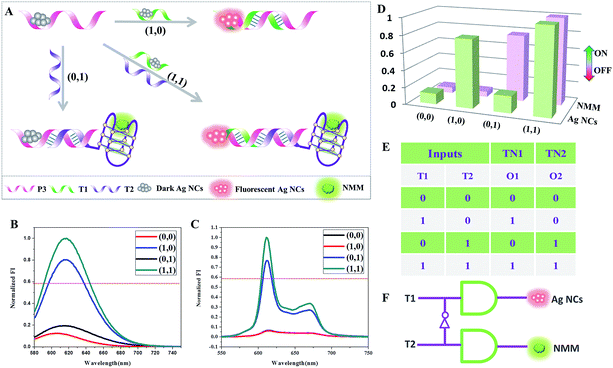
Concatenation of gates is the major challenge in logic systems if they are to be used for performing sophisticated computations.26 Transfer gates merely transfer the state of an input to the state of an output with no logical change (0 is transferred to 0, 1 is transferred to 1).27 They are used in concatenated logic gates through the conversion of the output of the former gate into the input of the next gate.Based on the above 4-to-2 encoder, we design another logic device that can be configured as two transfer gates: TN1 and TN2. As shown in Fig. 4, P3-stabilized darkish Ag NCs are used as the initial state, which contains a Ag NC-nucleation sequence at the 5′-terminal. In the presence of T1, which could hybridize with a part of P3 and also contains a Ag NC-nucleation sequence at the 3′-terminal, high fluorescence intensity of the Ag NCs is obtained. When T2 is added, which is integrated with a G-rich sequence at the 5′-terminal and could hybridize with the other part of P3, it has almost no influence on the fluorescence value of the Ag NCs but folds into the duplex-based G-quadruplex, enhancing the fluorescence intensity of NMM. Finally, in the presence of T1 and T2 together, both fluorescence intensities of the Ag NCs and NMM achieve high values. It is clearly seen in Fig. 4D that two different input combinations (0/0 and 0/1) generate off (0) responses at output O1, whereas both 1/0 and 1/1 generate on (1) responses. For output O2, the input combinations 0/0 and 1/0 generate off (0) responses, whereas both states 0/1 and 1/1 generate on (1) responses. Therefore, TN1 alone is used to generate only O1 and TN2 alone is used to generate only O2. Furthermore, PAGE experiments are conducted to validate all of the above mentioned DNA interactions, see Fig. S1.† From lane 1 to lane 3, the bands indicate the individual ssDNA P3, T1, and T2. New bands appear in lane 4 to lane 5, suggesting the formation of duplex P3/T1 and P3/T2. In the coexistence of P3, T1, and T2, there is still only one band appearing at a different position in lane 6, indicating the formation of duplex – quadruplex junction P3/T1/T2. The PAGE results indicate that the DNA interactions occur as expected.
Construction of the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 1 multiplexer
1 multiplexer
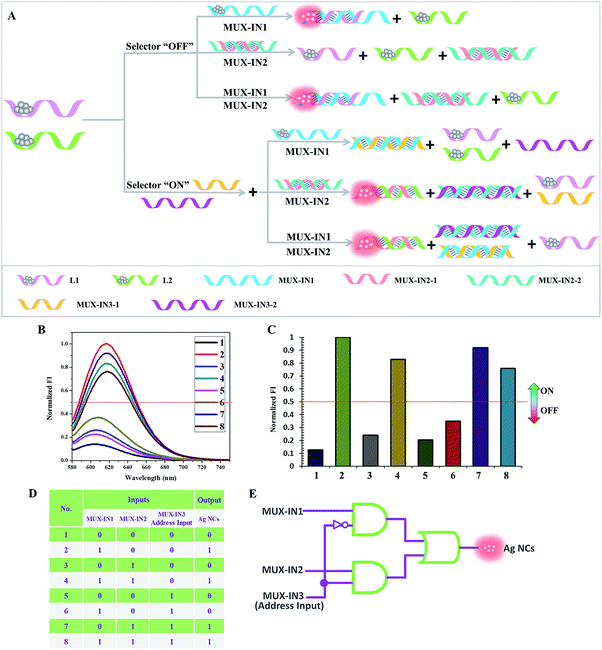
As an important digital device, a multiplexer can act as a mechanical rotary switch to select a binary input and then transmit it into a single output channel.28 A 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer involves two data inputs, one address input, and one output. The selection result is determined by the binary state of the address input. If the address input appears as “0”, the first input is directly transmitted to the output channel regardless of its binary state. However, the directed input to output is taken from another input when the binary state of the address input is “1”.29
1 multiplexer involves two data inputs, one address input, and one output. The selection result is determined by the binary state of the address input. If the address input appears as “0”, the first input is directly transmitted to the output channel regardless of its binary state. However, the directed input to output is taken from another input when the binary state of the address input is “1”.29
Fig. 5 presents the schematic illustration of the cascade between the platform and the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer. L1 (pink) and L2 (green) are taken as templates to synthesize the darkish Ag NCs, which are employed as the initial state. Herein, the fluorescence of the Ag NCs is used as the signal reporter for the single output channel. The designed DNA strands are classified into three inputs: MUX-IN1, MUX-IN2, and MUX-IN3 (address input). In the presence of MUX-IN1, which contains a Ag NCs-nucleation sequence and complementary linker sequence to form the probe pair with L1, the fluorescence of the adjacent darkish DNA/Ag NCs is enhanced greatly. Upon the addition of MUX-IN2, which is a partly complementary duplex, its Ag NCs-nucleation sequence hybridizes with its complementary sequence and has little influence on the L1, L2-stabilized darkish Ag NCs. In the presence of both MUX-IN1 and MUX-IN2, the MUX-IN2 could not obviously affect the formation of a probe pair between MUX-IN1 and L1, producing a high output signal. As shown in Fig. 5, in the case of the address input = 0 (without MUX-IN3), the output reports the state of data input MUX-IN1. If the address input is “1”, the output mirrors the state of data input MUX-IN2. Since MUX-IN3-1 has a higher affinity for MUX-IN1 than L1, the addition of MUX-IN1 could not affect the fluorescence of L1 and L2-stabilized Ag NCs any longer, generating a low fluorescence signal. Once MUX-IN2 is added, MUX-IN3-2 presents a higher affinity for MUX-IN2-2 and MUX-IN2-1 is released, which contains a Ag NC-nucleation sequence and complementary linker sequence, to form the probe pair with L2, resulting in a significant enhancement of fluorescence. In the presence of both MUX-IN1 and MUX-IN2, MUX-IN1 could not obviously affect the formation of the probe pair between MUX-IN2-2 and L2, producing a high fluorescence value. Fig. 5D is obtained according to the defined threshold value (0.5), fully conforming to the features of a 2
1 multiplexer. L1 (pink) and L2 (green) are taken as templates to synthesize the darkish Ag NCs, which are employed as the initial state. Herein, the fluorescence of the Ag NCs is used as the signal reporter for the single output channel. The designed DNA strands are classified into three inputs: MUX-IN1, MUX-IN2, and MUX-IN3 (address input). In the presence of MUX-IN1, which contains a Ag NCs-nucleation sequence and complementary linker sequence to form the probe pair with L1, the fluorescence of the adjacent darkish DNA/Ag NCs is enhanced greatly. Upon the addition of MUX-IN2, which is a partly complementary duplex, its Ag NCs-nucleation sequence hybridizes with its complementary sequence and has little influence on the L1, L2-stabilized darkish Ag NCs. In the presence of both MUX-IN1 and MUX-IN2, the MUX-IN2 could not obviously affect the formation of a probe pair between MUX-IN1 and L1, producing a high output signal. As shown in Fig. 5, in the case of the address input = 0 (without MUX-IN3), the output reports the state of data input MUX-IN1. If the address input is “1”, the output mirrors the state of data input MUX-IN2. Since MUX-IN3-1 has a higher affinity for MUX-IN1 than L1, the addition of MUX-IN1 could not affect the fluorescence of L1 and L2-stabilized Ag NCs any longer, generating a low fluorescence signal. Once MUX-IN2 is added, MUX-IN3-2 presents a higher affinity for MUX-IN2-2 and MUX-IN2-1 is released, which contains a Ag NC-nucleation sequence and complementary linker sequence, to form the probe pair with L2, resulting in a significant enhancement of fluorescence. In the presence of both MUX-IN1 and MUX-IN2, MUX-IN1 could not obviously affect the formation of the probe pair between MUX-IN2-2 and L2, producing a high fluorescence value. Fig. 5D is obtained according to the defined threshold value (0.5), fully conforming to the features of a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer operation.
1 multiplexer operation.
Construction of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 2 demultiplexer
2 demultiplexer
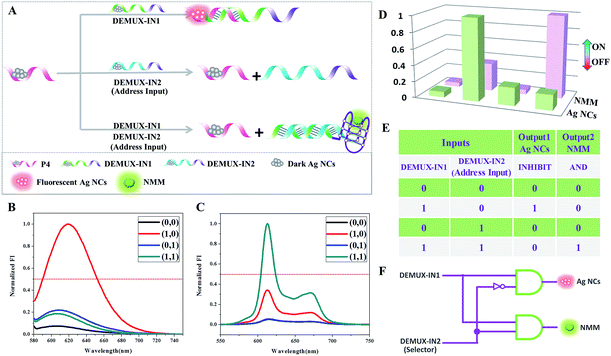
A multiplexer could encode multiple data flows into one single data line for transmission. Contrarily, a demultiplexer converts one single input signal into multiple output channels.30 A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer circuit involves one data input signal and one additional address input which is used to select in which output channel to process the input. The selection of the output channel depends on the binary state of the address input. If the address input appears as “0”, the binary state of the data input is reported to “Output 1”. However, the binary data input is reported to “Output 2” when the binary state of the address input is “1”.31 Precisely one input signal could be transmitted into two possible output channels controlled by an address input that acts as a selector. To implement this function, Ag NCs and NMM are selected as the output reporters. The operational principle is illustrated in Fig. 6A, with P4-stabilized darkish Ag NCs used as the common initial state. DEMUX-IN1 is defined as the single input, and DEMUX-IN2 functions as the address input to select the output channel that the single input is transmitted into. In the presence of DEMUX-IN1, which contains a Ag NCs-nucleation sequence and complementary linker sequence to form the probe pair with P4, the fluorescence of the adjacent darkish DNA/Ag NC probe pair is enhanced greatly. Upon the addition of DEMUX-IN2, which presents a higher affinity for DEMUX-IN1 than that of P4, the P4 is released, resulting in fluorescence quenching. After normalizing the fluorescence intensity of the Ag NCs and NMM, the correspondingly obtained truth table (Fig. 6E) indicates that the logic circuit is composed of “inhibit” and “and” logic gates. PAGE experiments are conducted to validate all of the above mentioned DNA interactions and are shown in Fig. S2.† From lane 1 to lane 3, the bands indicate the individual ssDNA P4, DEMUX-IN1, and DEMUX-IN2. In the presence of P4 and DEMUX-IN1, a new band appears at a different position in lane 4, indicating the formation of a duplex of P4/DEMUX-IN1. In the presence of both P4 and DEMUX-IN2 in lane 5, two bands appear at similar positions to those in lane 1 and lane 3, respectively, suggesting that no hybridization happens between the P4 and DEMUX-IN2. In the presence of P4, DEMUX-IN1, and DEMUX-IN2 two bands are found in lane 6. One band appears at a position that is similar to that of P4, another appears at a position different from any of the three ssDNA, indicating the formation of a duplex of DEMUX-IN1/DEMUX-IN2. The PAGE results indicate that the DNA interactions occur as expected.
2 demultiplexer circuit involves one data input signal and one additional address input which is used to select in which output channel to process the input. The selection of the output channel depends on the binary state of the address input. If the address input appears as “0”, the binary state of the data input is reported to “Output 1”. However, the binary data input is reported to “Output 2” when the binary state of the address input is “1”.31 Precisely one input signal could be transmitted into two possible output channels controlled by an address input that acts as a selector. To implement this function, Ag NCs and NMM are selected as the output reporters. The operational principle is illustrated in Fig. 6A, with P4-stabilized darkish Ag NCs used as the common initial state. DEMUX-IN1 is defined as the single input, and DEMUX-IN2 functions as the address input to select the output channel that the single input is transmitted into. In the presence of DEMUX-IN1, which contains a Ag NCs-nucleation sequence and complementary linker sequence to form the probe pair with P4, the fluorescence of the adjacent darkish DNA/Ag NC probe pair is enhanced greatly. Upon the addition of DEMUX-IN2, which presents a higher affinity for DEMUX-IN1 than that of P4, the P4 is released, resulting in fluorescence quenching. After normalizing the fluorescence intensity of the Ag NCs and NMM, the correspondingly obtained truth table (Fig. 6E) indicates that the logic circuit is composed of “inhibit” and “and” logic gates. PAGE experiments are conducted to validate all of the above mentioned DNA interactions and are shown in Fig. S2.† From lane 1 to lane 3, the bands indicate the individual ssDNA P4, DEMUX-IN1, and DEMUX-IN2. In the presence of P4 and DEMUX-IN1, a new band appears at a different position in lane 4, indicating the formation of a duplex of P4/DEMUX-IN1. In the presence of both P4 and DEMUX-IN2 in lane 5, two bands appear at similar positions to those in lane 1 and lane 3, respectively, suggesting that no hybridization happens between the P4 and DEMUX-IN2. In the presence of P4, DEMUX-IN1, and DEMUX-IN2 two bands are found in lane 6. One band appears at a position that is similar to that of P4, another appears at a position different from any of the three ssDNA, indicating the formation of a duplex of DEMUX-IN1/DEMUX-IN2. The PAGE results indicate that the DNA interactions occur as expected.
Construction of the parity checker
A parity checker, which is a digital device to detect erroneous programs in data storage and transmission, is successfully designed based on the above 1-to-2 decoder. In order to develop this novel application of the two building blocks, we have employed the 1-to-2 decoder with a slight modification to distinguish odd and even numbers from natural numbers in the range of 0 to 9. First of all, these natural numbers are transformed into corresponding binary numbers. Each decimal digit is encoded into a four-bit binary code. Then K+, hemin, estradiol, and silver assigned to the four bits are employed as the inputs with P1 as the initial state. Fig. 7 presents the computational results of the parity checker. As expected, the input states 0/0/0/1, 0/0/1/1, 0/1/0/1, 0/1/1/1, and 1/0/0/1 are transmitted to the true output “1” and the input states 0/0/0/0, 0/0/1/1, 0/1/0/0, 0/1/1/0, and 1/0/0/0 are transmitted to the false output “0”. In other words, the corresponding fluorescence values of all odd numbers produce the true output, whereas these of all even numbers present the false output. As we know, the parity of a natural number depends on the digit in the unit position. Therefore, it can be deduced that the parity checker could distinguish odd and even numbers from all decimal numbers.Construction of the ternary logic gate
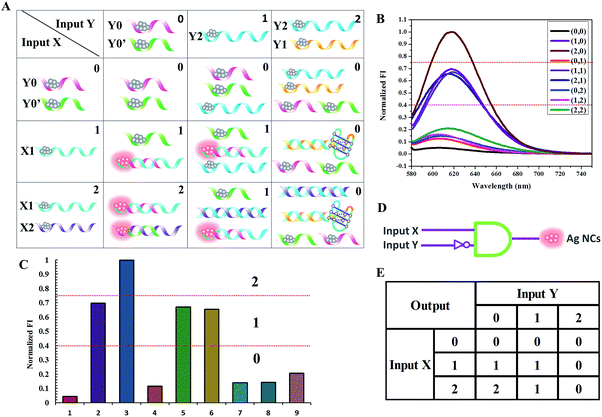
Binary logic in which the output state is “0” or “1”, corresponding to a low or high signal, often struggles to process information because the complexity of information makes it difficult to identify certain and accurate states based on the low/high system. Fortunately, multi-valued logic, which involves the switching between more than two states, shows great promise to make up for the imperfections of binary logic when dealing with uncertain information. It has three different output states: low, medium and high signals, which correspond to logic values of “0”, “1”, and “2”, respectively.32 As shown in Fig. 8, a ternary inhibit logic gate is constructed, utilizing Y0-stabilized darkish Ag NCs and Y0′-stabilized darkish Ag NCs as the initial state. For input X, the value “1” represents the introduction of X1-stabilized darkish Ag NCs and the value “2” represents the mixture of X1-stabilized darkish Ag NCs and X2-stabilized darkish Ag NCs. For input Y, the value “1” represents the introduction of Y2-stabilized darkish Ag NCs and the value “2” represents a mixture of Y1-stabilized darkish Ag NCs and Y2-stabilized darkish Ag NCs. The ssDNA X1 containing 54 bases is designed to form a duplex of 17 base pairs with Y0. The ssDNA X2 containing 47 bases is designed to form a duplex of 17 base pairs with Y0′. The ssDNA Y2, which would form a duplex of 35 base pairs with X2, is designed to initiate a displacement reaction in order to release Y0′. The ssDNA Y1, which would form a duplex of 29 base pairs with X1 accompanying the formation of a G-quadruplex configuration, is designed to initiate a displacement reaction in order to release Y0. Upon the addition of X1 (input state “1/0”), which contains a Ag NC-nucleation sequence and complementary linker sequence to form the probe pair with Y0, the adjacent darkish DNA/Ag NC probe pair is turned on, giving the output value of “1”. When the mixture of X1 and X2 is introduced (input state “2/0”), both of these contain a Ag NC-nucleation sequence and complementary linker sequence to form probe pairs with Y0 and Y0′, respectively, producing the output value of “2”. Upon the addition of X1 and Y2 (input state “1/1”), as Y2 has little effect on the reaction between X1 and Y0, the output state is “1”. However, when a mixture of X1, X2, and Y2 is introduced to the system (input state “2/1”), X2 prefers to hybridize with Y2, leaving the Y0′-stabilized darkish Ag NCs alone, giving the output value of “1”. Similarly, when the mixture of X1, Y1 and Y2 is introduced, (input state “1/2”), X1 prefers to hybridize with Y1, leaving the Y0-stabilized darkish Ag NCs alone, giving the output value of “0”. In the presence of X1, X2, Y1, and Y2, where X1 hybridizes with Y1 and X2 hybridizes with Y2, the fluorescence of the Ag NCs is completely quenched and the output value is “0”. The read-out of the computational results is displayed in Fig. 8C. The output value is defined as “0” when the normalized fluorescence intensity is lower than 0.4, and it is defined as “1” when the normalized fluorescence intensity is between 0.4 and 0.75. The output value “2” is defined as when the normalized fluorescence intensity is higher than 0.75.Perspectives for medical applications
From the viewpoint of practical application, the designed logic gates can not only perform a function in data storage and processing, but also present great potential applications in disease diagnosis. For example, a significant portion of chronic hepatitis B (CHB) patients are in the inactive carrier state, characterized by little viral replication and minimal liver necroinflammatory activity. A confirmed diagnosis of CHB requires both the hepatitis B virus (HBV) DNA and serum alanine aminotransferase (ALT) measurements. Thus, we can use HBV DNA and ALT as the two inputs to construct a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer which is composed of “inhibit” and “and” logic gates. As shown in Fig. 6E, if both HBV DNA and ALT are positive (output 1 = 1, output 2 = 1), a diagnosis of CHB can be made. If only the HBV DNA is positive (output 1 = 1, output 2 = 0), this can diagnose hepatitis B virus carriers (HBVER). The output state output 1 = 0, output 2 = 0 indicates no illness. Even if only ALT is positive, this may be due to the long-term use of medicines. Therefore, we can use the logic system to determine whether the illness is present and further diagnose HBVER or CHB. Moreover, we can use our logic system to indicate the existence of inputs. For example, the 1-to-2 decoder system can be used for the detection of 17β-estradiol, which can be absorbed from external sources, interfere with normal physiological processes, and create many deleterious effects.
2 demultiplexer which is composed of “inhibit” and “and” logic gates. As shown in Fig. 6E, if both HBV DNA and ALT are positive (output 1 = 1, output 2 = 1), a diagnosis of CHB can be made. If only the HBV DNA is positive (output 1 = 1, output 2 = 0), this can diagnose hepatitis B virus carriers (HBVER). The output state output 1 = 0, output 2 = 0 indicates no illness. Even if only ALT is positive, this may be due to the long-term use of medicines. Therefore, we can use the logic system to determine whether the illness is present and further diagnose HBVER or CHB. Moreover, we can use our logic system to indicate the existence of inputs. For example, the 1-to-2 decoder system can be used for the detection of 17β-estradiol, which can be absorbed from external sources, interfere with normal physiological processes, and create many deleterious effects.
Conclusions
In conclusion, combining silver nanoclusters (Ag NCs) and G-quadruplex in different ways with the help of the G-quadruplex-enhanced fluorescence intensity of NMM, we experimentally construct a series of sophisticated logic circuits to perform nonarithmetic functions: a 1-to-2 decoder, a 4-to-2 encoder, an 8-to-3 encoder, a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer, a 1
1 multiplexer, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer and dual transfer gates. Moreover, a parity checker that is capable of identification of odd and even numbers from natural numbers is constructed conceptually. Finally, a multi-valued logic gate (ternary inhibit gate) is readily achieved by taking this DNA/Ag NC system as a universal platform. Though it is improbable that these molecular logic systems will soon be applied in biocomputers, they can be expediently used in data storage and transmission where electronic computers could not be used, such as in object tracking, tumor labelling, procedural drug delivery, molecular self-regulation and self-control, and related functions in biocomputers and biomedicine. Importantly, our label-free and enzyme-free system could save experimental time and costs. Moreover, these logic operations are performed in aqueous media, providing their potential applications in biological and biomedical systems. If molecular computers are implemented one day, the logic systems presented herein may act as a potential building block towards these, because of their unprecedented flexibility and high level of complexity.
2 demultiplexer and dual transfer gates. Moreover, a parity checker that is capable of identification of odd and even numbers from natural numbers is constructed conceptually. Finally, a multi-valued logic gate (ternary inhibit gate) is readily achieved by taking this DNA/Ag NC system as a universal platform. Though it is improbable that these molecular logic systems will soon be applied in biocomputers, they can be expediently used in data storage and transmission where electronic computers could not be used, such as in object tracking, tumor labelling, procedural drug delivery, molecular self-regulation and self-control, and related functions in biocomputers and biomedicine. Importantly, our label-free and enzyme-free system could save experimental time and costs. Moreover, these logic operations are performed in aqueous media, providing their potential applications in biological and biomedical systems. If molecular computers are implemented one day, the logic systems presented herein may act as a potential building block towards these, because of their unprecedented flexibility and high level of complexity.
Experimental details
Chemicals and materials
All oligonucleotides were purchased from Sangon Biotechnology Co., Ltd (Shanghai, China). Silver nitrate (AgNO3), sodium borohydride (NaBH4), and 18-crown-6-ether (18C6) were purchased from Aladdin Ltd. (Shanghai, China). Hemin and N-methylmesoporphyrin IX (NMM) were purchased from Sangon Biotechnology Co., Ltd (Shanghai, China). Both the stock solution of hemin (5.0 μM) and the stock solution of NMM (100.0 μM) were prepared in dimethyl sulfoxide (DMSO) and stored in darkness at −20.0 °C, then diluted to the desired concentration with distilled water. Thioflavin T (ThT) was obtained from Sigma-Aldrich. The water used in the experiments was purified using a Millipore system. Other chemicals were of reagent grade and were used without further purification.Before the operation of logic computation, all of the DNA stock solutions were diluted to a concentration of 100.0 μM with 20.0 mM phosphate buffer (5.0 mM Mg2+, pH = 7.4), and then the solutions were heated at 88.0 °C for 10.0 min and slowly cooled down to room temperature.
For the emission spectra of DNA-stabilized Ag NCs involved in the 1-to-2 decoder, 4-to-2 encoder, dual transfer gates, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer, 1
1 multiplexer, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer, parity checker, and ternary inhibit gate, the excitation wavelength is 560 nm and the spectra are recorded between 580 and 750 nm. For the emission spectra of DNA-stabilized Ag NCs involved in the 8-to-3 encoder, the excitation wavelength is 585 nm and the spectra are recorded between 605 and 750 nm. Both the excitation and emission slit widths are 10 nm and the PMT voltage is 600 V. For the ThT emission spectra, the excitation wavelength is 425 nm and the spectra are recorded between 450 and 600 nm. Both the excitation and emission slit widths are 10 nm and the PMT voltage is 500 V. For the NMM emission spectra, the excitation wavelength is 399 nm and the spectra are recorded between 550 and 750 nm. Both the excitation and emission slit widths are 10 nm and the PMT voltage is 500 V.
2 demultiplexer, parity checker, and ternary inhibit gate, the excitation wavelength is 560 nm and the spectra are recorded between 580 and 750 nm. For the emission spectra of DNA-stabilized Ag NCs involved in the 8-to-3 encoder, the excitation wavelength is 585 nm and the spectra are recorded between 605 and 750 nm. Both the excitation and emission slit widths are 10 nm and the PMT voltage is 600 V. For the ThT emission spectra, the excitation wavelength is 425 nm and the spectra are recorded between 450 and 600 nm. Both the excitation and emission slit widths are 10 nm and the PMT voltage is 500 V. For the NMM emission spectra, the excitation wavelength is 399 nm and the spectra are recorded between 550 and 750 nm. Both the excitation and emission slit widths are 10 nm and the PMT voltage is 500 V.
Synthesis of P1-stabilized fluorescent Ag NCs involved in the 1-to-2 decoder and parity checker
The solution of AgNO3 (6.0 μM) was added into a 1.0 μM P1 template in 20.0 mM phosphate buffer (5.0 mM Mg2+, pH = 7.4), and then the mixture was kept in the dark at room temperature for 45.0 min. Subsequently, NaBH4 (6.0 μM) was added to the above mixture, followed by vigorous shaking for 15.0 s. Finally, the mixture was kept in the dark at room temperature for 4.0 h before use.Synthesis of DNA-stabilized fluorescent Ag NCs involved in the 8-to-3 encoder
The solution of AgNO3 (6.0 μM) was added into a 1.0 μM DNA template in 20.0 mM phosphate buffer (5.0 mM Mg2+, pH = 7.4), and then the mixture was kept in the dark at room temperature for 25.0 min. Subsequently, NaBH4 (6.0 μM) was added to the above mixture, followed by vigorous shaking for 15.0 s. Finally, the mixture was kept in the dark at room temperature for 5.0 h before use.Synthesis of DNA-stabilized fluorescent Ag NCs involved in the 4-to-2 encoder, dual transfer gates, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 1 multiplexer, 1
1 multiplexer, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 2 demultiplexer, and ternary inhibit gate
2 demultiplexer, and ternary inhibit gate
The solution of AgNO3 (6.0 μM) was added into a 1.0 μM DNA template in 20.0 mM phosphate buffer (5.0 mM Mg2+, pH 7.4), and then the mixture was kept in the dark at room temperature for 25.0 min. Subsequently, NaBH4 (6.0 μM) was added to the above mixture, followed by vigorous shaking for 15.0 s. Finally, the mixture was kept in the dark at room temperature for 1.0 h before use.
The purification of the DNA-stabilized Ag NCs
All of the synthesized DNA/Ag NCs were centrifuged using Nanosep Centrifugal Devices (30K, molecular weight cutoffs) at 5000 rpm for 10 min. The obtained solution containing DNA/Ag NCs was prepared for further application.Operation of the 1-to-2 decoder
For the “input = 0” state, hemin (1.0 μM) and K+ (50.0 mM) were added into the prepared P1/Ag NC solution and the mixture was incubated in the dark at room temperature for another 2.0 h. For the “input = 1” state, hemin (1.0 μM), K+ (50.0 mM), and estradiol (1.5 μM) were added into the prepared P1/Ag NC solution and the mixture was incubated in the dark at room temperature for another 2.0 h. Subsequently, the fluorescence intensity of Ag NCs was measured and this was followed by the addition of NMM (2.0 μM) and reacted at room temperature for another 10.0 min. Finally, the fluorescence analysis of NMM was performed.Operation of the 4-to-2 encoder
In the 4-to-2 encoder, each input was added to the platform and kept in the dark at room temperature for 1.0 h. Subsequently, the fluorescence intensity of the Ag NCs was measured and this was followed by the addition of NMM (2.0 μM) and reacted at room temperature for another 10.0 min. Finally, the fluorescence analysis of NMM was performed.Operation of the 8-to-3 encoder
For the 8-to-3 encoder, the 20.0 mM phosphate buffer (5.0 mM Mg2+, pH = 7.4) was used as the initial state and C1 (1.0 μM), 18C6 (50 mM), C2 (1.0 μM), 22AG (1.0 μM), C5 (1.0 μM)/C2 (1.0 μM), G4 (1.0 μM), C3 (1.0 μM), and C4 (1.0 μM) were employed as the eight inputs. Subsequently, the fluorescence intensity of the Ag NCs, NMM (2.0 μM) and ThT (1.0 μM) for each input was measured.Operation of the dual transfer gates
In the dual transfer gates, each input was added to the platform and kept in the dark at room temperature for 1.0 h and this was followed by the addition of NMM (2.0 μM) and reacted at room temperature for another 10.0 min. Subsequently, the fluorescence intensity of the Ag NCs and NMM was measured.Operation of the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 1 multiplexer
1 multiplexer
For each input state, the involved DNA should be kept in the dark at room temperature for 2.0 h before silver deposition in the 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 multiplexer. Subsequently, the DNA-stabilized Ag NCs were added to the platform and kept in the dark at room temperature for another 80.0 min. Finally, the fluorescence analysis of the Ag NCs was performed.
1 multiplexer. Subsequently, the DNA-stabilized Ag NCs were added to the platform and kept in the dark at room temperature for another 80.0 min. Finally, the fluorescence analysis of the Ag NCs was performed.
Operation of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 2 demultiplexer
2 demultiplexer
In the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer, it should be noted that the input state (1/1) is the darkish Ag NCs stabilized by the duplex formed by DEMUX-IN1 and DEMUX-IN2, instead of the DEMUX-IN1-stabilized Ag NCs and DEMUX-IN2-stabilized Ag NCs. The input was added to the platform and kept in the dark at room temperature for 1.0 h and this was followed by the addition of NMM (2 μM) and reacted at room temperature for another 10.0 min. Subsequently, the fluorescence intensity of the Ag NCs and NMM was measured.
2 demultiplexer, it should be noted that the input state (1/1) is the darkish Ag NCs stabilized by the duplex formed by DEMUX-IN1 and DEMUX-IN2, instead of the DEMUX-IN1-stabilized Ag NCs and DEMUX-IN2-stabilized Ag NCs. The input was added to the platform and kept in the dark at room temperature for 1.0 h and this was followed by the addition of NMM (2 μM) and reacted at room temperature for another 10.0 min. Subsequently, the fluorescence intensity of the Ag NCs and NMM was measured.
Operation of the parity checker
The operation of the parity checker is similar to the 1-to-2 decoder.Operation of the ternary inhibit gate
The operation of the ternary inhibit gate is similar to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 demultiplexer.
2 demultiplexer.
Nondenaturing polyacrylamide gel electrophoresis (PAGE)
Nondenaturing polyacrylamide gels (15.0%) were prepared in 1× TBE buffer (89.0 mM Tris, 89.0 mM boric acid, 2.0 mM EDTA, pH = 8.3). 25.0 μL of each sample (5.0 μM) was mixed with 6.0× loading buffer (5.0 μL) and loaded into the gels. Electrophoresis was conducted in 1.0× TBE at a constant voltage of 140.0 V for 50.0 min. The gels were scanned using a UV transilluminator after silver staining.Acknowledgements
This work is financially supported by the National Natural Science Foundation of China (No. 21671150 and 21472139) and the Science and Technology Commission of Shanghai Municipality (14DZ2261100), as well as the Fundamental Research Funds for the Central Universities.Notes and references
- P. A. de Silva, N. H. Q. Gunaratne and C. P. McCoy, Nature, 1993, 364, 42–44 CrossRef.
- D. C. Magri, G. J. Brown, G. D. McClean and A. P. de Silva, J. Am. Chem. Soc., 2006, 128, 4950–4951 CrossRef CAS PubMed; L. Zhi, Z. Wang, J. Liu, W. Liu, H. Zhang, F. Chen and B. Wang, Nanoscale, 2015, 7, 11712–11719 RSC; E. Katz, J. Wang, M. Privman and J. Halámek, Anal. Chem., 2012, 84, 5463–5469 CrossRef PubMed; M. Privman, T. K. Tam, V. Bocharova, J. Halámek, J. Wang and E. Katz, ACS Appl. Mater. Interfaces, 2011, 3, 1620–1623 Search PubMed; M.-C. Chuang, J. R. Windmiller, P. Santhosh, G. V. Ramirez, E. Katz and J. Wang, Chem. Commun., 2011, 47, 3087–3089 RSC.
- R. J. Amir, M. Popkov, R. A. Lerner, C. F. Barbas and D. Shabat, Angew. Chem., 2005, 117, 4452–4455 CrossRef CAS; S. Ozlem and E. U. Akkaya, J. Am. Chem. Soc., 2009, 131, 48–49 CrossRef PubMed; X. Chen, A. H. Soeriyadi, X. Lu, S. M. Sagnella, M. Kavallaris and J. J. Gooding, Adv. Funct. Mater., 2014, 24, 6999–7006 CrossRef; J. Andreasson and U. Pischel, Chem. Soc. Rev., 2015, 44, 1053–1069 RSC; Z. Zhang, D. Balogh, F. Wang and I. Willner, J. Am. Chem. Soc., 2013, 135, 1934–1940 CrossRef PubMed.
- M. Ikeda, T. Tanida, T. Yoshii, K. Kurotani, S. Onogi, K. Urayama and I. Hamachi, Nat. Chem., 2014, 6, 511–518 CrossRef CAS PubMed; M. You, L. Peng, N. Shao, L. Zhang, L. Qiu, C. Cui and W. Tan, J. Am. Chem. Soc., 2014, 136, 1256–1259 CrossRef PubMed; M. Zhou, N. Zhou, F. Kuralay, J. R. Windmiller, S. Parkhomovsky, G. Valdés-Ramírez, E. Katz and J. Wang, Angew. Chem., Int. Ed., 2012, 51, 2686–2689 CrossRef PubMed.
- R. Gui, H. Jin, X. Liu, Z. Wang, F. Zhang, J. Xia, M. Yang and S. Bi, Chem. Commun., 2014, 50, 14847–14850 RSC; J. Ling, G. Naren, J. Kelly, T. S. Moody and A. P. de Silva, J. Am. Chem. Soc., 2015, 137, 3763–3766 CrossRef CAS PubMed.
- J. Kärnbratt, M. Hammarson, S. Li, H. L. Anderson, B. Albinsson and J. Andréasson, Angew. Chem., 2010, 122, 1898–1901 CrossRef CAS; R. Gaber, T. Lebar, A. Majerle, B. Šter, A. Dobnikar, M. Benčina and R. Jerala, Nat. Chem. Biol., 2014, 10, 203–208 CrossRef PubMed.
- K. He, Y. Li, B. Xiang, P. Zhao, Y. Hu, Y. Huang, W. Li, Z. Nie and S. Yao, Chem. Sci., 2015, 6, 3556–3564 RSC; K. M. Manesh, J. Halámek, M. Pita, J. Zhou, T. K. Tam, P. Santhosh, M.-C. Chuang, J. R. Windmiller, D. Abidin, E. Katz and J. Wang, Biosens. Bioelectron., 2009, 24, 3569–3574 CrossRef CAS PubMed.
- Y. Benenson, T. Paz-Elizur, R. Adar, E. Keinan, Z. Livneh and E. Shapiro, Nature, 2001, 414, 430–434 CrossRef CAS PubMed.
- M. N. Stojanovic, T. E. Mitchell and D. Stefanovic, J. Am. Chem. Soc., 2002, 124, 3555–3561 CrossRef CAS PubMed; T. Li, E. Wang and S. Dong, J. Am. Chem. Soc., 2009, 131, 15082–15083 CrossRef PubMed; J. Elbaz, F. Wang, F. Remacle and I. Willner, Nano Lett., 2012, 12, 6049–6054 CrossRef PubMed; R. Orbach, R. Orbach, B. Willner and I. Willner, Chem. Commun., 2015, 51, 4144–4160 RSC; F. Wang, X. Liu and I. Willner, Angew. Chem., Int. Ed., 2015, 54, 1098–1129 CrossRef PubMed; T. Niazov, R. Baron, E. Katz, O. Lioubashevski and I. Willner, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17160–17163 CrossRef PubMed.
- S. Xu, H. Li, Y. Miao, Y. Liu and E. Wang, NPG Asia Mater., 2013, 5, e76 CrossRef CAS; R. Orbach, F. Wang, O. Lioubashevski, R. D. Levine, F. Remacle and I. Willner, Chem. Sci., 2014, 5, 3381–3387 RSC; S. Zhang, K. Wang, C. Huang and T. Sun, Nanoscale, 2015, 7, 20749–20756 RSC; W. Li, F. Zhang, H. Yan and Y. Liu, Nanoscale, 2016, 8, 3775–3784 RSC.
- R. Orbach, F. Remacle, R. D. Levine and I. Willner, Chem. Sci., 2014, 5, 1074–1081 RSC; R. Orbach, B. Willner and I. Willner, Chem. Commun., 2015, 51, 4144–4160 RSC; H. Li, Y. Liu, S. Dong and E. Wang, NPG Asia Mater., 2015, 7, e166 CrossRef CAS.
- J. Andréasson, U. Pischel, S. D. Straight, T. A. Moore, A. L. Moore and D. Gust, J. Am. Chem. Soc., 2011, 133, 11641–11648 CrossRef CAS PubMed; Y. He, Y. Chen, C. Li and H. Cui, Chem. Commun., 2014, 50, 7994–7997 RSC; D. Fan, J. Zhu, Y. Liu, E. Wang and S. Dong, Nanoscale, 2016, 8, 3834–3840 RSC.
- G. de Ruiter, L. Motiei, J. Choudhury, N. Oded and M. E. van der Boom, Angew. Chem., 2010, 122, 4890–4893 CrossRef CAS; P. Remón, M. Bälter, S. Li, J. Andréasson and U. Pischel, J. Am. Chem. Soc., 2011, 133, 20742–20745 CrossRef PubMed; K. MacVittie, J. Halamek and E. Katz, Chem. Commun., 2012, 48, 11742–11744 RSC.
- L. Wang, J. Zhu, L. Han, L. Jin, C. Zhu, E. Wang and S. Dong, ACS Nano, 2012, 6, 6659–6666 CrossRef CAS PubMed; J. Yang, L. Shen, J. Ma, H. I. Schlaberg, S. Liu, J. Xu and C. Zhang, ACS Appl. Mater. Interfaces, 2013, 5, 5392–5396 Search PubMed.
- D. Kang, R. J. White, F. Xia, X. Zuo, A. Vallee-Belisle and K. W. Plaxco, NPG Asia Mater., 2012, 4, e1 CrossRef.
- K. S. Park, C. Jung and H. G. Park, Angew. Chem., Int. Ed., 2010, 49, 9757–9760 CrossRef CAS PubMed; Y. Lin, Y. Tao, F. Pu, J. Ren and X. Qu, Adv. Funct. Mater., 2011, 21, 4565–4572 CrossRef; S. Mailloux, Y. V. Gerasimova, N. Guz, D. M. Kolpashchikov and E. Katz, Angew. Chem., Int. Ed., 2015, 54, 6562–6566 CrossRef PubMed.
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893–5909 RSC; Y. Du and X. Zhou, Chem. Rec., 2013, 13, 371–384 CrossRef CAS PubMed.
- P. Peng, L. Shi, H. Wang and T. Li, Nucleic Acids Res., 2016, 541 Search PubMed; J. Sharma, H.-C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2010, 46, 3280–3282 RSC; N. Enkin, F. Wang, E. Sharon, H. B. Albada and I. Willner, ACS Nano, 2014, 8, 11666–11673 CrossRef CAS PubMed; J. Sharma, H.-C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2011, 47, 2294–2296 RSC.
- F. Pu, J. Ren and X. Qu, Adv. Mater., 2014, 26, 5742–5757 CrossRef CAS PubMed; D.-L. Ma, H.-Z. He, D. S.-H. Chan and C.-H. Leung, Chem. Sci., 2013, 4, 3366–3380 RSC.
- J. M. Nicoludis, S. T. Miller, P. D. Jeffrey, S. P. Barrett, P. R. Rablen, T. J. Lawton and L. A. Yatsunyk, J. Am. Chem. Soc., 2012, 134, 20446–20456 CrossRef CAS PubMed.
- S. Liu, M. Li, X. Yu, C.-Z. Li and H. Liu, Chem. Commun., 2015, 51, 13185–13188 RSC; D. Fan, J. Zhu, Q. Zhai, E. Wang and S. Dong, Chem. Commun., 2016, 52, 3766–3769 RSC.
- L. Zhang, J. Zhu, S. Guo, T. Li, J. Li and E. Wang, J. Am. Chem. Soc., 2013, 135, 2403–2406 CrossRef CAS PubMed.
- H.-C. Yeh, J. Sharma, J. J. Han, J. S. Martinez and J. H. Werner, Nano Lett., 2010, 10, 3106–3110 CrossRef CAS PubMed; J. Zhu, L. Zhang, Y. Teng, B. Lou, X. Jia, X. Gu and E. Wang, Nanoscale, 2015, 7, 13224–13229 RSC; L. Gong, H. Kuai, S. Ren, X.-H. Zhao, S.-Y. Huan, X.-B. Zhang and W. Tan, Chem. Commun., 2015, 51, 12095–12098 RSC.
- B.-C. Yin, J.-L. Ma, H.-N. Le, S. Wang, Z. Xu and B.-C. Ye, Chem. Commun., 2014, 50, 15991–15994 RSC.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed; T. Gupta and M. E. van der Boom, Angew. Chem., 2008, 120, 5402–5406 CrossRef; E. Katz and V. Privman, Chem. Soc. Rev., 2010, 39, 1835–1857 RSC; R.-R. Gao, S. Shi, Y. Zhu, H.-L. Huang and T.-M. Yao, Chem. Sci., 2016, 7, 1853–1861 RSC.
- B. Wang, R. I. Kitney, N. Joly and M. Buck, Nat. Commun., 2011, 2, 508 CrossRef PubMed.
- C. Wu, K. Wang, D. Fan, C. Zhou, Y. Liu and E. Wang, Chem. Commun., 2015, 51, 15940–15943 RSC.
- J. C. Claussen, N. Hildebrandt, K. Susumu, M. G. Ancona and I. L. Medintz, ACS Appl. Mater. Interfaces, 2014, 6, 3771–3778 CAS.
- J. Zhu, L. Zhang, T. Li, S. Dong and E. Wang, Adv. Mater., 2013, 25, 2440–2444 CrossRef CAS PubMed.
- A. J. Genot, J. Bath and A. J. Turberfield, Angew. Chem., Int. Ed., 2013, 52, 1189–1192 CrossRef CAS PubMed.
- X. Ran, F. Pu, J. Ren and X. Qu, Small, 2014, 10, 1500–1503 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7sc00361g |
| This journal is © The Royal Society of Chemistry 2017 |