Open Access Article
Open Access ArticleResponsive hetero-organelle partition conferred fluorogenic sensing of mitochondrial depolarization†
Zhongwei
Xue
a,
Hu
Zhao
a,
Jian
Liu
a,
Jiahuai
Han
b and
Shoufa
Han
*a
aState Key Laboratory for Physical Chemistry of Solid Surfaces, Department of Chemical Biology, College of Chemistry and Chemical Engineering, The Key Laboratory for Chemical Biology of Fujian Province, The MOE Key Laboratory of Spectrochemical Analysis & Instrumentation, Innovation Center for Cell Signalling Network, Xiamen University, Xiamen, 361005, China. E-mail: shoufa@xmu.edu.cn; Tel: +86-0592-2181728
bState Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Signalling Network, School of Life Sciences, Xiamen University, Xiamen, 361005, China
First published on 8th November 2016
Abstract
Malfunctioning organelles are often difficult to probe with classical organelle-homing sensors owing to disruption of physiological organelle-probe affinity. We herein report the use of a responsive hetero-organelle partition and signal activable probe (RC-TPP) for detecting mitochondrial depolarization, a pathologically relevant event featuring loss of the electrical potentials across the mitochondrial membrane (ΔΨm). Partitioned in mitochondria to give blue fluorescence, RC-TPP relocates into lysosomes upon mitochondrial depolarization and exhibits red fluorescence triggered by lysosomal acidity, enabling determination of autophagy relevant mitochondrial depolarization and the chronological sequence of mitochondrial depolarization and lysosomal neutralization in distinct cell death signalling pathways. As an alternative to classic homo-organelle specific molecular systems, this hetero-organelle responsive approach provides a new perspective from which to study dysfunctional organelles.
Introduction
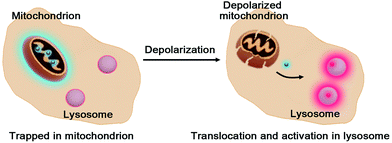
Defining the parameters of dysfunctional organelles is valuable for probing their functionality in diseases. Mitochondria are dynamic organelles hallmarked by electrochemical gradients of protons and negative electrical potentials across the mitochondrial membrane (ΔΨm), which are critical for diverse biological activities such as ATP biosynthesis.1 By contrast, mitochondrial depolarization, owing to loss of ΔΨm, triggers distinct cellular and pathological events, e.g. autophagy and cell death.2Mitochondria are routinely imaged with cationic dyes, e.g. rhodamine 123, which accumulate in mitochondria, driven by ΔΨm.3 Albeit widely used, these ΔΨm-responsive dyes quickly leak from mitochondria upon loss of ΔΨm. Alternatively, cationic dyes with a reactive moiety have been employed to form covalent bioconjugates with intramitochondrial protein sulfhydryls once sequestered inside mitochondria.4 Although the covalent linkage between dyes and proteins prevents loss of intramitochondrial fluorescence on dissipation of ΔΨm, these probes are incapable of discerning depolarized mitochondria. Recently, detection of mitochondrial depolarization was achieved using a bipolar probe featuring a positively charged hydrophilic group and an environment sensitive fluorophore which exhibits an altered fluorescence lifetime related to membrane polarization.5 Herein we report responsive hetero-organelle partition mediated detection of mitochondrial depolarization using RC-TPP, which preferentially accumulates in mitochondria to give blue fluorescence. RC-TPP relocates from depolarized mitochondria into acidic lysosomes to give intense rhodamine fluorescence, allowing a signal-on report of mitochondrial depolarization (Fig. 1).
Results and discussion
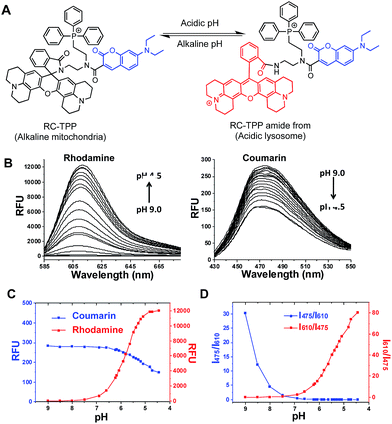
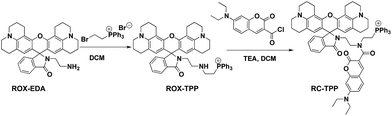
Given the myriad roles of altered ΔΨm in cell homeostasis and death,2 it is imperative to detect mitochondrial depolarization. As escaping of ΔΨm-responsive dyes from depolarized mitochondria is concomitant with loss of mitochondrion-specific fluorescence, we envisioned that conjugation of mitochondria-targetable agents with a profluorophore that is activable by extra-mitochondrial factors could potentially yield a signal-on assay for mitochondrial depolarization. Rhodamine-lactams, a group of nonfluorescent rhodamine derivatives featuring intramolecular spiro-lactam under alkaline conditions, are prone to proton triggered opening of the lactams to give bright rhodamine fluorescence.6 We previously used rhodamine-lactams for imaging of lysosomes.7 As such, RC-TPP was constructed to combine a coumarin fluorophore to indicate the sensor’s intracellular distribution, a ΔΨm responsive TPP moiety widely used to ferry various cargoes into mitochondria,8 and a rhodamine-lactam profluorophore activable by acidic lysosomes, which are the ubiquitous digestive organelles of mammalian cells (Fig. 2A).Acidic pH mediated switched-on fluorescence of RC-TPP
pH titration of RC-TPP shows “always-on” coumarin fluorescence whereas rhodamine fluorescence, negligible at pH 7.0–9.0, occurred in acidic media and intensified as pH decreased (pH 6.5–4.5) (Fig. 2B and C), which is consistent with proton mediated opening of the intramolecular lactam (Fig. 2A). Lysosomes are of acidic pH whereas mitochondria contain alkaline lumen with the pH maintained by the transmembrane proton gradients.9 The ratiometric titration shows distinct blue to red fluorescence patterns of RC-TPP respectively matching pH windows of lysosomes (pH 4.5–6.5) and mitochondria (∼pH 8.0) (Fig. 2D).Accumulation of RC-TPP in polarized mitochondria
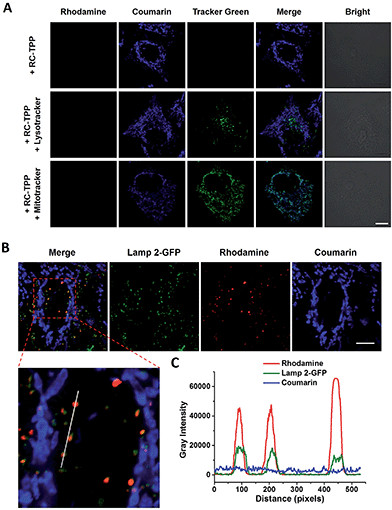
RC-TPP was evaluated for its ability to target mitochondria. HeLa cells stained with RC-TPP (1 μM) display negligible red fluorescence and strong blue fluorescence colocalized with Mitrotracker Green specific for mitochondria (Fig. 3A; ESI, Fig. S1†). Cells that are stained with an elevated concentration of RC-TPP (3 μM) exhibit sparsely punctate rhodamine signals eclipsing lysosomes bearing Lamp2-GFP (GFP-tagged lysosome associated membrane protein 2) (Fig. 3B). These results show that RC-TPP has a high tendency to be partitioned in mitochondria over lysosomes, and is prone to lysosome activation to give red fluorescence. We next probed the structural factor that leads to accumulation of RC-TPP in mitochondria. Rhodamine-coumarin (RC), lacking the TPP moiety, is exclusively located in lysosomes (ESI, Fig. S2†), confirming the critical role of the TPP moiety in mitochondrial partition of RC-TPP.Mitochondrial ΔΨm mediated lysosomal partition of RC-TPP
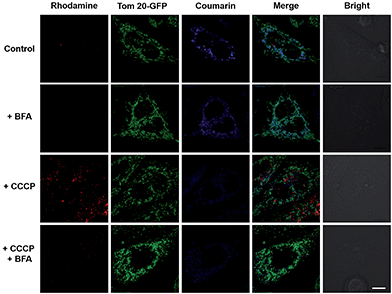
To elucidate the impacts of ΔΨm on the subcellular distribution of RC-TPP, HeLa cells expressing Tom20-GFP were analyzed for lysosomal activation of RC-TPP in response to carbonyl cyanide m-chlorophenylhydrazone (CCCP), a protonophore used to decrease ΔΨm.10 In contrast with Mitotrackers relying on ΔΨm, Tom20 is a constitutive protein of mitochondria, allowing staging of mitochondria by Tom20-GFP irrespective of ΔΨm. As expected, RC-TPP predominantly resides in mitochondria of control cells whereas CCCP-treated cells exhibit attenuated mitochondrial coumarin fluorescence and dramatically elevated rhodamine signals in a dose dependent manner (Fig. 4; ESI, Fig. S3†), showing relocation of RC-TPP from depolarized mitochondria to lysosomes.To ascertain the dependence of rhodamine fluorescence of RC-TPP on lysosomal acidity, HeLa cells were probed with RC-TPP in the presence of bafilomycin A1 (BFA), which inhibits vacuolar ATPase-H1 pump and neutralizes lysosomes.11 The lack of rhodamine fluorescence in BFA-treated cells proves lysosomal acidity triggered rhodamine fluorescence of RC-TPP in live cells (Fig. 4, ESI, Fig. S4†). The demonstrated translocation of RC-TPP into lysosomes is ascribed to loss of affinity of TPP to mitochondria upon depolarization, and protonation of the rhodamine-lactam by lysosomal acidity and possibly the contribution of the tendency of high molecular weight dyes to accumulate in lysosomes.12 Taken together, these data clearly evidence ΔΨm mediated intra-partition of RC-TPP between mitochondria and lysosomes, and lysosomal acidity mediated activation of RC-TPP upon mitochondrial depolarization.
Fluorogenic sensing of autophagy-associated mitochondrial depolarization with RC-TPP
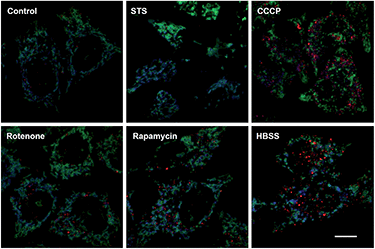
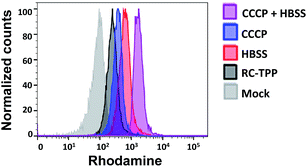
Mitochondrial depolarization correlates with autophagy, a catabolic mechanism mediating cell degradation of unnecessary or dysfunctional cellular components in response to diverse cues such as starvation.13 Cells cultivated in Hanks' balanced salt solution (HBSS) free of amino acids display dramatically enhanced rhodamine fluorescence (Fig. 5), which is consistent with nutrient starvation induced mitochondrial depolarization.13 Compared with red signal-free control cells or cells treated with apoptosis-inducing staurosporine (STS), there is moderate rhodamine fluorescence induced by the autophagy inducers rotenone or rapamycin (Fig. 4),14 demonstrating the utility of RC-TPP in distinguishing the efficacy of autophagy inducers. Flow cytometry analysis confirms enhanced rhodamine fluorescence in CCCP- or HBSS-treated cells, and reveals the synergistic effects of CCCP and HBSS on mitochondrial depolarization (Fig. 6), proving the usefulness of RC-TPP for fluorogenic sensing of mitochondrial depolarization induced by distinct biochemical cues.Historically, distinct methods have been developed to detect autophagy using GFP-tagged LC-3,15 pH-reporting proteins16 or dyes covalently immobilized to mitochondria.17 Complementing these approaches, our method focuses on the analysis of mitochondrial depolarization, a vital event preceding autophagy, using a small molecular probe prone to ΔΨm mediated partition between mitochondria and lysosomes.
Analysis of mitochondrial depolarization during apoptosis and necrosis
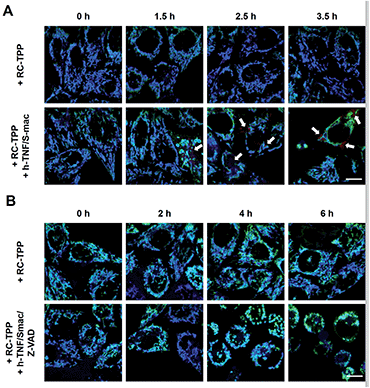
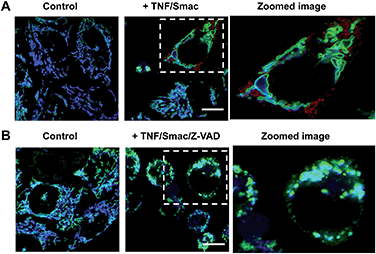
Dying cells undergo mitochondrial depolarization and loss of lysosomal acidity owing to membrane permeabilization.18 As distinct cell death modalities, apoptosis occurs under physiological conditions and is critical for cell number homeostasis whereas necrosis triggered by external factors leads to inflammatory responses.19 To date, the chronological order of mitochondrial depolarization and lysosomal neutralization during cell death has been undefined.Tom20-GFP+ HeLa cells stained with RC-TPP were treated with human Tumor Necrosis Factor-α (TNF) and Smac to trigger apoptosis. HeLa cells lack the endogenous receptor-interacting protein 3 (RIP3), a mediator which is critical for necrosis. Hence RIP3+/Tom20-GFP+ HeLa cells, used to reconstitute necrosis, were loaded with RC-TPP and then treated with TNF, Smac and Z-VAD to trigger necroptosis, which is a programmed form of necrosis.20 Time course imaging reveals subsided localization of RC-TPP in mitochondria in both cell populations, whereas punctate rhodamine signals occurred in apoptotic cells over necroptotic cells (Fig. 7 and 8). Given the dependence of rhodamine fluorescence on lysosomal acidity and translocation of RC-TPP from mitochondria into lysosomes, these results suggest that mitochondrial depolarization precedes lysosome membrane permeabilization in apoptosis, but the succession is swapped during necroptosis.
Conclusions
RC-TPP was developed for signal-on sensing of mitochondrial depolarization via lysosomal acidity activated fluorescence. RC-TPP, accumulated in mitochondria to exhibit blue fluorescence, relocates to acidic lysosomes to give rhodamine fluorescence upon mitochondrial depolarization, enabling detection of mitochondrial depolarization during autophagy and determination of the chronological sequence of mitochondrial depolarization and lysosomal neutralization during distinct cell death pathways. Abnormal organelles are manifested in diverse pathological conditions.21 Defining dysfunctional organelles with classic sensors is challenging owing to frequent loss of physiological organelle-probe affinity. Complementing mono-organelle targeting approaches,22 the hetero-organelle responsive approach offers a new tool to study malfunctioning organelles.Experimental procedure
Materials and methods
Lysotracker green DND-26 and Mitotracker green were purchased from Invitrogen. Bafilomycin A1 was purchased from Selleck. Rotenone and carbonyl cyanide m-chlorophenylhydrazone (CCCP), human tumor necrosis factor-α, Smac mimetic, and benzyloxycarbonyl-Val-Ala-DL-Asp(O-methyl)fluoromethyl-ketone (Z-VAD-FMK) were obtained from Sigma. All other chemicals were obtained from Alfa-Aesar unless specified.7-Diethylamino-4-carboxy-coumarin (2.6 g, 10.0 mmol) was dissolved in anhydrous DCM (40 ml) followed by addition of oxalyl chloride (2.52 g, 20.00 mmol) and a drop of N,N-dimethylformamide. The mixture was stirred at RT for 2 h and then evaporated to remove excess oxalyl chloride. The residue was dissolved in anhydrous DCM, and then added dropwise to the solution of ROX-TPP (0.821 g, 1 mmol) in anhydrous DCM spiked with triethylamine (1 ml). The mixture was stirred at RT for 1 h and then evaporated to remove the solvent. The residue was purified by silica gel column chromatography using DCM/MeOH/TEA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) as the eluent. RC-TPP was obtained as an off-white solid (0.64 g, 60%). 1H-NMR (500 MHz, CD3OD) δ: 7.97–7.63 (m, 18H), 7.51–7.48 (m, 2H), 7.34 (d, J = 8.9 Hz, 1H), 6.92 (d, J = 5.8 Hz, 1H), 6.83 (d, J = 8.8 Hz, 1H), 6.61 (s, 1H), 5.82 (s, 2H), 3.56 (dd, J = 14.0, 6.9 Hz, 8H), 3.37 (t, J = 6.0 Hz, 2H), 3.16–3.02 (m, 10H), 2.78–2.69 (m, 2H), 2.66–2.56 (m, 2H), 2.41 (m, J = 14.3, 7.5 Hz, 2H), 2.38–2.30 (m, 2H), 1.97–1.86 (m, 2H), 1.81 (dt, J = 11.9, 5.8 Hz, 4H), and 1.28 (t, J = 6.9 Hz, 6H); 13C-NMR (126 MHz, MeOD) δ: 170.38, 169.08, 160.98, 158.43, 155.27, 153.49, 149.43, 145.03, 144.88, 136.52, 134.86, 134.78, 134.19, 131.76, 131.66, 131.59, 131.37, 129.43, 125.29, 125.05, 123.53, 119.63, 119.03, 118.94, 115.95, 111.12, 108.72, 108.64, 105.66, 97.97, 67.65, 50.82, 50.30, 45.87, 28.21, 22.97, 22.50, 22.17, and 12.82; MS (C68H66N5O5P2+): calculated (MH+): 1063.4, found: 1066.1.
1, v/v/v) as the eluent. RC-TPP was obtained as an off-white solid (0.64 g, 60%). 1H-NMR (500 MHz, CD3OD) δ: 7.97–7.63 (m, 18H), 7.51–7.48 (m, 2H), 7.34 (d, J = 8.9 Hz, 1H), 6.92 (d, J = 5.8 Hz, 1H), 6.83 (d, J = 8.8 Hz, 1H), 6.61 (s, 1H), 5.82 (s, 2H), 3.56 (dd, J = 14.0, 6.9 Hz, 8H), 3.37 (t, J = 6.0 Hz, 2H), 3.16–3.02 (m, 10H), 2.78–2.69 (m, 2H), 2.66–2.56 (m, 2H), 2.41 (m, J = 14.3, 7.5 Hz, 2H), 2.38–2.30 (m, 2H), 1.97–1.86 (m, 2H), 1.81 (dt, J = 11.9, 5.8 Hz, 4H), and 1.28 (t, J = 6.9 Hz, 6H); 13C-NMR (126 MHz, MeOD) δ: 170.38, 169.08, 160.98, 158.43, 155.27, 153.49, 149.43, 145.03, 144.88, 136.52, 134.86, 134.78, 134.19, 131.76, 131.66, 131.59, 131.37, 129.43, 125.29, 125.05, 123.53, 119.63, 119.03, 118.94, 115.95, 111.12, 108.72, 108.64, 105.66, 97.97, 67.65, 50.82, 50.30, 45.87, 28.21, 22.97, 22.50, 22.17, and 12.82; MS (C68H66N5O5P2+): calculated (MH+): 1063.4, found: 1066.1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
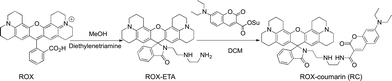
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) as the eluent to give ROX-ETA as an off-white solid (4.88 g, 85%). 1H-NMR (500 MHz, CDCl3) δ: 7.91–7.86 (1H, m), 7.45–7.40 (2H, m), 7.07 (1H, dt, J = 4.6, 3.3 Hz), 5.98 (2H, s), 3.24 (2H, t, J = 6.4 Hz), 3.15–3.11 (4H, t), 3.10–3.06 (4H, t), 2.89 (4H, dt, J = 10.0, 4.8 Hz), 2.60 (2H, t, J = 5.9 Hz), 2.47 (4H, dd, J = 11.1 HZ, 6.1 Hz), 2.41 (4H, t, J = 6.1 Hz), 2.06–1.99 (4H, m), and 1.88–1.82 (4H, m); 13C-NMR (126 MHz, CDCl3) δ: 168.81, 154.03, 148.39, 143.69, 132.47, 131.23, 127.96, 124.73, 124.09, 122.75, 117.41, 107.80, 105.79, 65.90, 52.19, 50.04, 49.60, 47.97, 41.94, 40.45, 27.30, 22.14, 21.62, and 21.35 (Scheme 2).
1, v/v/v) as the eluent to give ROX-ETA as an off-white solid (4.88 g, 85%). 1H-NMR (500 MHz, CDCl3) δ: 7.91–7.86 (1H, m), 7.45–7.40 (2H, m), 7.07 (1H, dt, J = 4.6, 3.3 Hz), 5.98 (2H, s), 3.24 (2H, t, J = 6.4 Hz), 3.15–3.11 (4H, t), 3.10–3.06 (4H, t), 2.89 (4H, dt, J = 10.0, 4.8 Hz), 2.60 (2H, t, J = 5.9 Hz), 2.47 (4H, dd, J = 11.1 HZ, 6.1 Hz), 2.41 (4H, t, J = 6.1 Hz), 2.06–1.99 (4H, m), and 1.88–1.82 (4H, m); 13C-NMR (126 MHz, CDCl3) δ: 168.81, 154.03, 148.39, 143.69, 132.47, 131.23, 127.96, 124.73, 124.09, 122.75, 117.41, 107.80, 105.79, 65.90, 52.19, 50.04, 49.60, 47.97, 41.94, 40.45, 27.30, 22.14, 21.62, and 21.35 (Scheme 2).
To a solution of 7-diethylamino-4-carboxy-coumarin (2.61 g, 10.00 mmol) in anhydrous DCM (40 ml) was added N-hydroxysuccinimide (NHS, 1.73 g, 15.0 mmol) and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (2.9 g, 15.0 mmol). The mixture was maintained at RT for 2 h with stirring. After removal of the solvent, the residue was purified by silica gel column chromatography using DCM as the eluent to yield the NHS ester of 7-diethylamino-coumarin-3-acid as a pale yellow solid (1.88 g, 50%).
To a solution of ROX-ETA (2.4 g, 4.2 mmol) in anhydrous dichloromethane was added the NHS ester of 7-diethylamino-coumarin-carboxylic acid (1.8 g, 5 mmol) with TEA (1 ml). The reaction mixture was stirred at room temperature for 12 h. After rotary evaporation of the solvent, the residue was purified by silica gel flash column chromatography using DCM/PE/TEA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) as the eluent to give RC as a pale gray solid (2.4 g, 70%). 1H-NMR (500 MHz, CDCl3) δ: 8.81 (t, J = 5.3 Hz, 1H), 8.64 (s, 1H), 7.86 (dd, J = 5.8, 2.7 Hz, 1H), 7.39 (dd, J = 8.7, 3.3 Hz, 3H), 7.05 (dd, J = 5.7, 2.6 Hz, 1H), 6.62 (dd, J = 8.9, 2.3 Hz, 1H), 6.47 (d, J = 2.1 Hz, 1H), 5.99 (s, 2H), 3.43 (q, J = 7.1 Hz, 2H), 3.36 (dd, J = 12.2, 6.2 Hz, 4H), 3.23 (t, J = 6.5 Hz, 2H), 3.11 (d, J = 6.4 Hz, 2H), 3.06 (dt, J = 16.3, 5.5 Hz, 4H), 2.93–2.84 (m, 4H), 2.60 (t, J = 6.4 Hz, 2H), 2.55–2.40 (m, 6H), 2.02 (td, J = 12.2, 6.3 Hz, 4H), 1.89–1.80 (td, 4H), and 1.22 (t, J = 7.1 Hz, 6H). 13C-NMR (126 MHz, CDCl3) δ: 168.62, 163.30, 162.63, 157.70, 154.03, 152.52, 148.34, 148.01, 143.66, 132.33, 131.25, 131.15, 127.83, 124.69, 124.01, 122.73, 117.39, 110.59, 109.92, 108.45, 107.76, 105.72, 96.67, 77.36, 65.84, 50.00, 49.55, 48.44, 47.79, 45.15, 40.24, 39.77, 27.25, 22.11, 21.59, 21.30, and 12.52.
1, v/v/v) as the eluent to give RC as a pale gray solid (2.4 g, 70%). 1H-NMR (500 MHz, CDCl3) δ: 8.81 (t, J = 5.3 Hz, 1H), 8.64 (s, 1H), 7.86 (dd, J = 5.8, 2.7 Hz, 1H), 7.39 (dd, J = 8.7, 3.3 Hz, 3H), 7.05 (dd, J = 5.7, 2.6 Hz, 1H), 6.62 (dd, J = 8.9, 2.3 Hz, 1H), 6.47 (d, J = 2.1 Hz, 1H), 5.99 (s, 2H), 3.43 (q, J = 7.1 Hz, 2H), 3.36 (dd, J = 12.2, 6.2 Hz, 4H), 3.23 (t, J = 6.5 Hz, 2H), 3.11 (d, J = 6.4 Hz, 2H), 3.06 (dt, J = 16.3, 5.5 Hz, 4H), 2.93–2.84 (m, 4H), 2.60 (t, J = 6.4 Hz, 2H), 2.55–2.40 (m, 6H), 2.02 (td, J = 12.2, 6.3 Hz, 4H), 1.89–1.80 (td, 4H), and 1.22 (t, J = 7.1 Hz, 6H). 13C-NMR (126 MHz, CDCl3) δ: 168.62, 163.30, 162.63, 157.70, 154.03, 152.52, 148.34, 148.01, 143.66, 132.33, 131.25, 131.15, 127.83, 124.69, 124.01, 122.73, 117.39, 110.59, 109.92, 108.45, 107.76, 105.72, 96.67, 77.36, 65.84, 50.00, 49.55, 48.44, 47.79, 45.15, 40.24, 39.77, 27.25, 22.11, 21.59, 21.30, and 12.52.
RIP3+/Tom 20-GFP+ HeLa cells prestained with RC-TPP (2 μM, 30 min) were cultured for 6 h in DMEM containing no addition or human TNF-α (20 ng m−1)/Smac mimetic (100 nM)/Z-VAD (20 μM) to initiate necroptosis. The cells were analyzed by confocal fluorescence microscopy.
Acknowledgements
This work was supported by grants from NSF China (21572189, 21272196) and the Fundamental Research Funds for the Central Universities (20720160052, 20720150047), Xiamen University; Dr J. Han was supported by grants from NSF China (91429301, 31420103910, 31330047, 31221065), the National Scientific and Technological Major Project (2013ZX10002-002), the Hi-Tech Research and Development Program of China (863 program; 2012AA02A201), the 111 Project (B12001), the Science and Technology Foundation of Xiamen (3502Z20130027) and the National Science Foundation of China for Fostering Talents in Basic Research (J1310027). Boyu Han is thanked for assistance with the English in this paper.Notes and references
- (a) J. R. Friedman and J. Nunnari, Nature, 2014, 505, 335 Search PubMed; (b) D. R. Green, Cell, 1998, 94, 695 Search PubMed.
- (a) M. T. Lin and M. F. Beal, Nature, 2006, 443, 787 Search PubMed; (b) A. Abeliovich, Nature, 2010, 463, 744 Search PubMed.
- B. Chazotte, Cold Spring Harbor Protocols, 2011, 2011, 89 Search PubMed.
- (a) R. P. Haugland, Handbook of Fluorescent Probes and Research Chemicals, Molecular Probes, Inc., Eugene, Oregon, 1999 Search PubMed; (b) S. Wu, Y. Song, Z. Li, J. Han and S. Han, Anal. Methods, 2014, 4, 1699 Search PubMed; (c) M. H. Lee, N. Park, C. Yi, J. H. Han, J. H. Hong, K. P. Kim, D. H. Kang, J. L. Sessler, C. Kang and J. S. Kim, J. Am. Chem. Soc., 2014, 136, 14136 Search PubMed.
- B. Wang, X. Zhang, C. Wang, L. Chen, Y. Xiao and Y. Pang, Analyst, 2015, 140, 5488 Search PubMed.
- (a) W. Zhang, B. Tang, X. Liu, Y. Liu, K. Xu, J. Ma, L. Tong and G. Yang, Analyst, 2009, 134, 367 Search PubMed; (b) T. Hasegawa, Y. Kondo, Y. Koizumi, T. Sugiyama, A. Takeda, S. Ito and F. Hamada, Bioorg. Med. Chem., 2009, 17, 6015 Search PubMed.
- (a) M. Yu, X. Wu, B. Lin, J. Han, L. Yang and S. Han, Anal. Chem., 2015, 87, 6688 Search PubMed; (b) X. Wu, M. Yu, B. Lin, H. Xing, J. Han and S. Han, Chem. Sci., 2015, 6, 798 Search PubMed; (c) Z. Li, S. Wu, J. Han and S. Han, Analyst, 2011, 136, 3698 Search PubMed; (d) X. Wu, B. Lin, M. Yu, L. Yang, J. Han and S. Han, Chem. Sci., 2015, 6, 2002 Search PubMed.
- (a) J. Asin-Cayuela, A. R. Manas, A. M. James, R. A. Smith and M. P. Murphy, FEBS Lett., 2004, 571, 9 Search PubMed; (b) H. Yuan, H. Cho, H. H. Chen, M. Panagia, D. E. Sosnovik and L. Josephson, Chem. Commun., 2013, 49, 10361 Search PubMed; (c) M. F. Ross, G. F. Kelso, F. H. Blaikie, A. M. James, H. M. Cocheme, A. Filipovska, T. Da Ros, T. R. Hurd, R. A. Smith and M. P. Murphy, Biochemistry, 2005, 70, 222 Search PubMed; (d) H. M. Cocheme, G. F. Kelso, A. M. James, M. F. Ross, J. Trnka, T. Mahendiran, J. Asin-Cayuela, F. H. Blaikie, A. R. Manas, C. M. Porteous, V. J. Adlam, R. A. Smith and M. P. Murphy, Mitochondrion, 2007, 7, S94 Search PubMed; (e) S. E. Abu-Gosh, N. Kolvazon, B. Tirosh, I. Ringel and E. Yavin, Mol. Pharm., 2009, 6, 1138 Search PubMed; (f) G. F. Kelso, C. M. Porteous, C. V. Coulter, G. Hughes, W. K. Porteous, E. C. Ledgerwood, R. A. Smith and M. P. Murphy, J. Biol. Chem., 2001, 276, 4588 Search PubMed.
- J. Llopis, J. M. McCaffery, A. Miyawaki, M. G. Farquhar and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6803 Search PubMed.
- P. G. Heytler, Biochemistry, 1963, 2, 357 Search PubMed.
- T. Yoshimori, A. Yamamoto, Y. Moriyama, M. Futai and Y. Tashiro, J. Biol. Chem., 1991, 266, 17707 Search PubMed.
- X. Zhang, C. Wang, Z. Han and Y. Xiao, ACS Appl. Mater. Interfaces, 2014, 6, 21669 Search PubMed.
- M. Frank, S. Duvezin-Caubet, S. Koob, A. Occhipinti, R. Jagasia, A. Petcherski, M. O. Ruonala, M. Priault, B. Salin and A. S. Reichert, Biochim. Biophys. Acta, 2012, 1823, 2297 Search PubMed.
- A. Fleming, T. Noda, T. Yoshimori and D. C. Rubinsztein, Nat. Chem. Biol., 2011, 7, 9 Search PubMed.
- (a) Y. Kabeya, N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi and T. Yoshimori, EMBO J., 2000, 19, 5720 Search PubMed; (b) N. Mizushima, T. Yoshimori and B. Levine, Cell, 2010, 140, 313 Search PubMed.
- H. Katayama, T. Kogure, N. Mizushima, T. Yoshimori and A. Miyawaki, Chem. Biol., 2011, 18, 1042 Search PubMed.
- S. Rodriguez-Enriquez, I. Kim, R. T. Currin and J. J. Lemasters, Autophagy, 2006, 2, 39 Search PubMed.
- (a) P. Boya and G. Kroemer, Oncogene, 2008, 27, 6434 Search PubMed; (b) U. Repnik, M. Hafner Cesen and B. Turk, Mitochondrion, 2014, 19, 49 Search PubMed.
- T. Vanden Berghe, A. Linkermann, S. Jouan-Lanhouet, H. Walczak and P. Vandenabeele, Nat. Rev. Mol. Cell Biol., 2014, 15, 135 Search PubMed.
- S. He, L. Wang, L. Miao, T. Wang, F. Du, L. Zhao and X. Wang, Cell, 2009, 137, 1100 Search PubMed.
- (a) G. Kroemer and M. Jaattela, Nat. Rev. Cancer, 2005, 5, 886 Search PubMed; (b) K. C. Vaughn, L. R. Debonte, K. G. Wilson and G. W. Schaffer, Science, 1980, 208, 196 Search PubMed; (c) P. H. Reddy, P. Mao and M. Manczak, Brain Res. Rev., 2009, 61, 33 CrossRef CAS PubMed; (d) A. Sawa, G. W. Wiegand, J. Cooper, R. L. Margolis, A. H. Sharp, J. F. Lawler Jr, J. T. Greenamyre, S. H. Snyder and C. A. Ross, Nat. Med., 1999, 5, 1194 Search PubMed.
- (a) Y. Chen, C. Zhu, Z. Yang, J. Chen, Y. He, Y. Jiao, W. He, L. Qiu, J. Cen and Z. Guo, Angew. Chem., Int. Ed. Engl., 2013, 52, 1688 Search PubMed; (b) B. C. Dickinson and C. J. Chang, J. Am. Chem. Soc., 2008, 130, 9638 Search PubMed; (c) Y. Koide, Y. Urano, S. Kenmoku, H. Kojima and T. Nagano, J. Am. Chem. Soc., 2007, 129, 10324 Search PubMed; (d) L. Yuan, L. Wang, B. K. Agrawalla, S. J. Park, H. Zhu, B. Sivaraman, J. Peng, Q. H. Xu and Y. T. Chang, J. Am. Chem. Soc., 2015, 137, 5930 Search PubMed; (e) C. Zhang, Q. Hu, G. Feng, R. Zhang, Y. Yuan, X. Lu and B. Liu, Chem. Sci., 2015, 4, 4580 Search PubMed; (f) J. J. Zhou, L. Li, W. Shi, X. Gao, X. Li and H. Ma, Chem. Sci., 2015, 6, 4884 Search PubMed.
- C. Li and R. M. Evans, Nucleic Acids Res., 1997, 25, 4165 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Biodistribution of RC; cytotoxicity of RC-TPP; and analysis spectra of RC-TPP and RC. See DOI: 10.1039/c6sc04158b |
| This journal is © The Royal Society of Chemistry 2017 |