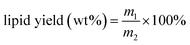Open Access Article
Open Access ArticleInfluence of a facile pretreatment process on lipid extraction from Nannochloropsis sp. through an enzymatic hydrolysis reaction†
Qingtai Chen‡
 a,
Dong Liu‡*a,
Chongchong Wu‡b,
Airong Xu‡c,
Wei Xia‡a,
Zhaowen Wang‡d,
Fushan Wen‡e and
Daoyong Yu‡a
a,
Dong Liu‡*a,
Chongchong Wu‡b,
Airong Xu‡c,
Wei Xia‡a,
Zhaowen Wang‡d,
Fushan Wen‡e and
Daoyong Yu‡a
aState Key Laboratory of Heavy Oil Processing, College of Chemical Engineering, China University of Petroleum, Qingdao, Shandong 266580, China. E-mail: Liudong@upc.edu.cn; Tel: +86 532-86984629
bDepartment of Chemical and Petroleum Engineering, University of Calgary, T2N 1N4, Calgary, Alberta, Canada
cSchool of Chemical Engineering and Pharmaceutics, Henan University of Science and Technology, Luoyang, Henan 471003, China
dDongying Environmental Protection Bureau, Dongying, Shandong 257091, China
eCollege of Science, China University of Petroleum, Qingdao, Shandong 266580, China
First published on 20th November 2017
Abstract
Microalgal lipid is being considered as an alternative source of ω-3 polyunsaturated fatty acids (ω-3 PUFAs); however, the extraction of ω-3 PUFA-rich algal lipid still needs further research. In this study, single enzyme and their combinations were screened based on their cell wall disrupting capability and lipid recovery yield, and economical and feasible extraction parameters were determined. Lipid recovery could reach 22.18 ± 0.26 wt% when algae was treated with an 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) algae/cocktail enzyme ratio and a 2
1 (w/w) algae/cocktail enzyme ratio and a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 cellulase/lysozyme mass ratio at 50 °C under stirring for 5 h at a pH of 4.0, and the content of PUFAs was as high as 23.30%. In addition, alkaline pretreatment efficiently facilitates the degradation of algal cell wall in the enzymatic hydrolysis, and it has been verified that cell wall ruptures via two steps: swelling by a weak alkali pretreatment and decomposition by enzymatic hydrolysis. Furthermore, all the extraction operations can take place under facile conditions. These results imply that algal lipid extracted by a weak alkali pretreatment aiding enzymatic hydrolysis, is the most appropriate raw material for ω-3 PUFAs.
1 cellulase/lysozyme mass ratio at 50 °C under stirring for 5 h at a pH of 4.0, and the content of PUFAs was as high as 23.30%. In addition, alkaline pretreatment efficiently facilitates the degradation of algal cell wall in the enzymatic hydrolysis, and it has been verified that cell wall ruptures via two steps: swelling by a weak alkali pretreatment and decomposition by enzymatic hydrolysis. Furthermore, all the extraction operations can take place under facile conditions. These results imply that algal lipid extracted by a weak alkali pretreatment aiding enzymatic hydrolysis, is the most appropriate raw material for ω-3 PUFAs.
1. Introduction
Polyunsaturated fatty acids (ω-3 PUFAs) are health-beneficial nutrients that are widely used in the food and pharmaceutics fields.1 Besides lowering cholesterol, they can also promote the immune system, prevent cancer and cardiovascular diseases, and contribute to the development of brain and cognition health.2,3 Although representative compounds, including γ-linolenic acid (ALA), eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA), of ω-3 PUFAs mainly exist in marine fish and microalgae, their commercial source is mainly deep-sea fish.2,4 However, ω-3 PUFAs derived from fish oils have many limitations such as fishy smell, heavy metal contamination, and inadequate supplies.5 Therefore, the lipid extracted from microalgae has become a promising alternative source for ω-3 PUFAs owing to the high content of ω-3 PUFAs in it; thus, these microalgae have attracted significant attention and interest in academia and industry.6 However, ω-3 PUFAs extracted from algal lipid via practical processes are greatly restricted because of their poor stability and easy oxidation in the traditional extraction methods.7,8 Furthermore, the extraction of algal lipid is particularly difficult since the lipid is stored in lipid vesicles,3 which are protected by robust cell walls. Cell walls of microalgae are composed of microfibril networks of polysaccharides, including cellulose and pectin, and algaenans, with complicated interactions between them, and the entangled network is implanted in a matrix of pectin along with cross-linked algaenans;9,10 thus, they severely hamper conventional solvent access to the lipid vesicles. Therefore, it is essential to develop more efficient and effective approaches to break the algal cell wall for lipid extraction.Compared to the conventional disruption technologies,11 enzymatic hydrolysis is being considered as a promising wall-breaking method for algal lipid extraction because of its inherent advantages12 such as mild conditions, low-energy, and ability to preserve the most labile compounds. However, it is difficult to obtain a desirable extraction efficiency for an individual enzyme. For example, the yield of the lipid extracted with free and immobilized cellulase from Chlorella vulgaris was 73% and 56%, respectively.13,14 To degrade the algae cell wall more completely and obtain more ω-3 PUFAs, cocktail enzymes have been employed in the hydrolysis reaction. Moreover, enzymatic hydrolysis in combination with a pretreatment, which includes hydrothermal reaction, ultra-sonication, and acidic or alkaline reaction, could cause an obvious increase in the lipid recovery.15 It was concluded that acidic environment could obviously enhance the disruption efficiency of algal cells in the enzymatic hydrolysis and improve the recovery of sugar,16 and the alkaline hydrothermal reaction could greatly increase the dissolution of cellulose present in cell wall in the enzymatic hydrolysis because of hydrogen bond rupture.17,18 Wu et al. developed a process via an alkaline pretreatment (at 110 °C and pH 10.5), which assisted enzymatic hydrolysis to extract algal lipid, and the yield exceeded 90%.19 However, oxidation easily occurs above 100 °C of the pretreatment temperature for algal lipids, which are released in the early stage owing to the presence of a great amount of unsaturated double bonds.20 Moreover, significant research has been focused on the extraction of algal lipid, which is used as a feedstock for biodiesel. Furthermore, to the best of our knowledge, the extraction technology of algal lipid aimed at separating ω-3 PUFAs has not been investigated to date. In this study, the marine microalgae Nannochloropsis sp. was selected as the raw material for algal lipid extraction; the main reason for using this microalgae was its ability to accumulate large amounts of ω-3 PUFAs and EPA.21 The disruption process of algal cell wall via weak alkali pretreatment-aided enzymatic hydrolysis (WAEH) for the extraction of the lipid aimed at separating ω-3 PUFAs was developed, and the high efficiency cell wall-breaking of enzyme based on the yield of lipid recovery was investigated. The suitable reaction conditions, including reaction temperature and time, pH value, the ratio (w/w) of cocktail enzymes, and mass ratio between enzyme and algae, were determined, and the highest extraction yield of the algal lipid was obtained. Microalgal cells treated with different methods and lipids extracted by several methods were characterized using Fourier transform infrared spectroscopy (FT-IR), ultraviolet-visible spectroscopy (UV-Vis), scanning electron microscopy (SEM), and fluorescence microscopy. The composition and content of fatty acids obtained from algal lipid were analyzed using gas chromatography (GC). The effect of the weak alkali pretreatment (WAP) on the algal cell wall rupture and lipid extraction was investigated.
2. Materials and methods
2.1 Chemicals and reagents
Nannochloropsis sp. was purchased from Yantai Hairong Biology Technology Co. Ltd. (Yantai, China). Cellulase, pectinase, lysozyme, and neutral protease were obtained from Nanjing Oddfoni Biology Technology Co. Ltd. (Nanjing, China). Hexane, ethanol, methanol, chloroform, acetic acid, and sodium acetate were purchased from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China), and the purity of these solvents was analytical grade.2.2 The extraction of algal lipid
Soxhlet extraction was conducted using 1.0 g of microalgae and a mixed solvent of chloroform and methanol (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), and the temperature was set at 90 °C. The reaction continues until the solvent stored in the extractor is almost colorless and transparent. After the solvent is removed, the remnant is dissolved and filtered with n-hexane, and algal lipid, which is defined as the n-hexane soluble fraction, is acquired after the filtrate is obtained and evaporated.
1, v/v), and the temperature was set at 90 °C. The reaction continues until the solvent stored in the extractor is almost colorless and transparent. After the solvent is removed, the remnant is dissolved and filtered with n-hexane, and algal lipid, which is defined as the n-hexane soluble fraction, is acquired after the filtrate is obtained and evaporated.
On the other hand, in the Bligh and Dyer extraction, 100 mg of microalgae, which was ground using a high-pressure homogenizer, was added to a mixture of chloroform, methanol, and water (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.9, v/v/v) and stirred for 12 h. Then, the mixture was centrifuged, and the supernatant was transferred to a flask. The process was repeated two more times using a stirring time of 2 h. After removing the solvent from the above collected supernatant, the residual material was dissolved using n-hexane, and algal lipid was obtained from the filtrate by vacuum distillation. The weight of the algal lipid was measured gravimetrically, and the data were determined through the mean value obtained from the abovementioned two extraction methods.
0.9, v/v/v) and stirred for 12 h. Then, the mixture was centrifuged, and the supernatant was transferred to a flask. The process was repeated two more times using a stirring time of 2 h. After removing the solvent from the above collected supernatant, the residual material was dissolved using n-hexane, and algal lipid was obtained from the filtrate by vacuum distillation. The weight of the algal lipid was measured gravimetrically, and the data were determined through the mean value obtained from the abovementioned two extraction methods.
(1) Weak alkali pretreatment. Herein, 3.0 g of microalgae was added to a flask that contained 10 mL of deionized water. The pH value of the mixture was adjusted to 9.0 with sodium hydroxide. After being sealed with parafilm, the flask was incubated in an oil bath (DF-101S, Gongyi Yingyu Instrument Factory) with a magnetic stirrer for 5 h at 80 °C. After the reaction, the flask was removed from the oil bath and cooled to ambient temperature.
(2) Enzymatic hydrolysis. After WAP, the pH value of the reaction solution was adjusted to the acid range with acetic acid. Different dosages of single or cocktail enzymes were added to the mixture. Then, the algae suspension was stirred for various lengths of time at different temperatures. Then, the mixtures were heated in an oil bath at 95 °C for 10 min to deactivate the enzyme. After this, 15 mL of ethanol was added to the mixture prior to lipid extraction with n-hexane (30 mL) by centrifuging at 5000 rpm for 25 min. The obtained organic phases were evaporated by a rotary evaporator (Shanghai Yarong Biochemical Instrument Factory) under vacuum, and the residues were dried for 6 h under vacuum at 40 °C. Finally, the lipid was stored in a vacuum dryer.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w) ratio of the binary mixed enzymes was adopted, and the proportion of 1
1 (w/w) ratio of the binary mixed enzymes was adopted, and the proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was adopted when three enzymes were involved in the reaction; moreover, the mass ratio of four enzymes participating in the reaction was 1
1 was adopted when three enzymes were involved in the reaction; moreover, the mass ratio of four enzymes participating in the reaction was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.The extraction conditions for algal lipid were optimized in enzymatic hydrolysis, and cocktail enzymes, including cellulase and lysozyme, were used as candidates. The influence of the reaction time on lipid recovery was investigated, and 1 h, 2 h, 3 h, 4 h, 5 h, 6 h, 7 h, 8 h… were set as the reaction times. Moreover, the temperature, pH value, cellulase/lysozyme mass ratio, and algae/enzyme mass ratio of the reaction was 50 °C, 4.0, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 8
1, and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. In addition, 30 °C, 35 °C, 40 °C, 45 °C, 50 °C, 55 °C, 60 °C, 65 °C, and 70 °C reaction temperatures were used for this research, and the other reaction conditions i.e. reaction time, pH, cellulase/lysozyme ratio, and algae/enzyme mass ratio were 7 h, 4.0, 2
1, respectively. In addition, 30 °C, 35 °C, 40 °C, 45 °C, 50 °C, 55 °C, 60 °C, 65 °C, and 70 °C reaction temperatures were used for this research, and the other reaction conditions i.e. reaction time, pH, cellulase/lysozyme ratio, and algae/enzyme mass ratio were 7 h, 4.0, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w), and 8
1 (w/w), and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. The effect of pH on lipid recovery was investigated when the reaction pH value was set as 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, and 7.0, and other conditions of the reaction were 50, 7 h, 2
1, respectively. The effect of pH on lipid recovery was investigated when the reaction pH value was set as 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, and 7.0, and other conditions of the reaction were 50, 7 h, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w), and 8
1 (w/w), and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w). The relationship of mixed ratio with the binary enzymes and lipid recovery was investigated, and the mass ratio of cellulase and lysozyme was designed as 6
1 (w/w). The relationship of mixed ratio with the binary enzymes and lipid recovery was investigated, and the mass ratio of cellulase and lysozyme was designed as 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5
1, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and 1
3, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, and other reaction conditions including reaction temperature, pH value, reaction time, and algae/enzyme mass ratio were 50 °C, 4.0, 7 h, and 8
6, and other reaction conditions including reaction temperature, pH value, reaction time, and algae/enzyme mass ratio were 50 °C, 4.0, 7 h, and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. The influence of enzyme dosage on the yield of lipid recovery was determined, and the quantity of mixed enzymes (cellulase and lysozyme) increases in the following order: 160
1, respectively. The influence of enzyme dosage on the yield of lipid recovery was determined, and the quantity of mixed enzymes (cellulase and lysozyme) increases in the following order: 160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 80
1, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 40
1, 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 20
1, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 8
1, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2.6
1, 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 2
1, and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Temperature, pH, time, and the cellulase/lysozyme mass ratio of the reaction were 50 °C, 4.0, 7 h, and 2
1. Temperature, pH, time, and the cellulase/lysozyme mass ratio of the reaction were 50 °C, 4.0, 7 h, and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. Moreover, the control test was performed without the pretreatment of WAP under the same conditions.
1, respectively. Moreover, the control test was performed without the pretreatment of WAP under the same conditions.
2.3 The yield of algal lipid recovery
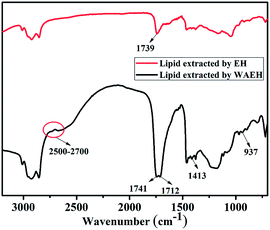
The weight of algal lipid was then determined gravimetrically, and the yield of lipid recovery was calculated using the equation as follows:where m1 is the dry weight of lipid recovery, mg g−1 DCW (dry cell weight), and m2 is the dry weight of microalgae, mg g−1 DCW.
2.4 Analytical methods
An acidic catalyst of the methyl esterification reaction was prepared by mixing acetyl chloride and methanol at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v) at ambient temperature, and the reaction mixture was stirred for a period of time until the reactant was cooled to atmospheric temperature. Then, 50 mg of lipid sample, 5 mL of n-hexane, and 4 mL of the abovementioned solution were added to a flask in which the mixture was refluxed for 2 h in an oil bath at 75 °C. The reaction was quenched by adding 4 mL of water. The hexane layer was transferred into a pre-weighed glass tube and evaporated to dryness under a nitrogen stream at 37 °C. Finally, FAMEs were stored under a nitrogen atmosphere.
10 (v/v) at ambient temperature, and the reaction mixture was stirred for a period of time until the reactant was cooled to atmospheric temperature. Then, 50 mg of lipid sample, 5 mL of n-hexane, and 4 mL of the abovementioned solution were added to a flask in which the mixture was refluxed for 2 h in an oil bath at 75 °C. The reaction was quenched by adding 4 mL of water. The hexane layer was transferred into a pre-weighed glass tube and evaporated to dryness under a nitrogen stream at 37 °C. Finally, FAMEs were stored under a nitrogen atmosphere.
The composition of FAMEs was determined using GC (6890A, HP-Agilent Technologies, USA) equipped with a DB-23 capillary column (30 m × 0.25 mm × 0.25 μm). All the ingredients of FAMEs were identified by comparing their retention times with a reference standard comprising 37 components. The injection and flame ionization detector (FID) temperature were maintained at 240 °C, and the time–temperature program was as follows: 100–180 °C at 10°C min−1 and 180–240 °C at 2°C min−1. Helium was used as the carrier gas at a flow rate of 1 mL min−1. The fatty acids content was determined by adding an internal standard of undecanoic acid (known quantity) to the oil sample before extraction, and the peak areas were quantified using Chromcard for Windows software. The areas of the peaks were summed and compared with the area of the internal standard (C11:0).
2.5 Statistical analysis
All experiments were conducted in triplicate. The data were evaluated using one-way analysis of variance (ANONA) followed by SPSS. P value ≤ 0.05 was regarded as significant, and data were presented as mean ± standard deviation (SD).3. Results and discussion
3.1 Screening enzymes
Considering two factors that cell walls rigidity of Nannochloropsis sp. is three times stronger than that of plant and the activity of enzyme is specificity, enzymes with stronger wall-breaking capability, which include cellulase, lysozyme, pectinase, and neutral protease, were selected as candidates for the screening enzyme.25,26 Moreover, a series of reactions of individual or cocktail enzymes was designed to screen the suitable enzymes that have a better cell wall breaching efficiency. The most suitable enzymes were determined based on the lipid recovery after enzymatic hydrolysis, and the relevant results are shown in Table 1.| Enzymes | Lipid recovery yield (wt%) | |||
|---|---|---|---|---|
| Cellulase | Lysozyme | Pectinase | Neutral protease | |
| a Note: +, the enzyme was added; −, the enzyme was not added. | ||||
| + | − | − | − | 15.55 ± 0.23 |
| − | + | − | − | 18.16 ± 0.03 |
| − | − | + | − | 13.82 ± 0.15 |
| − | + | − | + | 10.18 ± 0.07 |
| + | + | − | − | 22.18 ± 0.26 |
| + | − | + | − | 20.49 ± 0.07 |
| + | − | − | + | 15.14 ± 0.19 |
| − | + | + | − | 18.55 ± 0.10 |
| − | + | − | + | 20.97 ± 0.27 |
| − | − | + | + | 13.40 ± 0.09 |
| − | + | + | + | 17.05 ± 0.13 |
| + | + | + | − | 22.46 ± 1.34 |
| + | − | + | + | 18.04 ± 0.22 |
| + | + | − | + | 21.17 ± 0.08 |
| + | + | + | + | 20.89 ± 0.06 |
As seen in Table 1, for an individual enzyme, the lipid recovery followed the order lysozyme > cellulase > pectinase > neutral protease. The yields of lipid extracted separately with the binary, ternary, and quaternary mixed enzymes were higher than those of lipid extracted with a single enzyme, and higher lipid yields were obtained when lysozyme was among the cocktail enzymes, which was consistent with the reported results.19 Furthermore, 22.46 ± 1.34 wt% of the lipid recovery for cocktail enzymes, containing lysozyme, cellulase, and pectinase, was the highest among that of all the enzyme combinations. There are two possible reasons: one is that the cocktail enzymes containing lysozyme, cellulase, and pectinase can lead to a stronger degradation capability of algal cell wall in the hydrolysis reaction; the other reason is that the cocktail enzymes enable the fast liberation of the lipid that exists in the subcellular structure to the slurry when it is extracted by complex solvents.27 However, it is noteworthy that the lipid recovery declined slightly in the presence of neutral protease. The reason for this is that the activity of neutral protease is inhibited to some extent, and the concentration of effective cocktail enzymes is reduced equivalently since the hydrolysis medium pH is 4.0, whereas the optimized pH value of neutral-protease hydrolysis is 7.0.28 Although the lipid recovery yield obtained after utilizing the cocktail enzymes cellulase and lysozyme was 0.28 wt% lower than that obtained after using cellulase, lysozyme, and pectinase, the most suitable cocktail enzymes were chosen to be lysozyme and cellulase for lipids extracted from Nannochloropsis sp. taking into consideration the lipid recovery and economic feasibility.
3.2 The effect of enzymatic hydrolysis conditions on lipid recovery
Enzymatic hydrolysis conditions including enzyme dosage, temperature, time, pH, and mass ratio of cocktail enzymes were investigated (Fig. 1). As shown in Fig. 1, the yield of algal lipid recovery increased with an increase in enzymatic dosage and remained almost at the same level when the mass ratio between the microalgae and cocktail enzymes was larger than 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The highest yield of lipid could reach 22.18 ± 0.26 wt% when the hydrolysis reaction took place at 50 °C for 5 h at a pH value of 4.0 with a 2
1. The highest yield of lipid could reach 22.18 ± 0.26 wt% when the hydrolysis reaction took place at 50 °C for 5 h at a pH value of 4.0 with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio of cellulase and lysozyme, and this result was more than the yield of the algal lipid extracted by the Bligh–Dyer method and Soxhlet extraction; the data was 22.09 ± 0.18 wt%. The reason is that algal lipid extracted by WAEH contains more free fatty acids as compared to that extracted by the Bligh and Dye and Soxhlet extraction, and the relevant evidence has been provided in Section 3.3. The abovementioned results indicate that sufficient quantity of cocktail enzymes is necessary for the complete degradation of algal cell walls. However, the curve of the lipid yield was almost flat when the mass ratio between the algae and cocktail enzymes was increased to 8
1 mass ratio of cellulase and lysozyme, and this result was more than the yield of the algal lipid extracted by the Bligh–Dyer method and Soxhlet extraction; the data was 22.09 ± 0.18 wt%. The reason is that algal lipid extracted by WAEH contains more free fatty acids as compared to that extracted by the Bligh and Dye and Soxhlet extraction, and the relevant evidence has been provided in Section 3.3. The abovementioned results indicate that sufficient quantity of cocktail enzymes is necessary for the complete degradation of algal cell walls. However, the curve of the lipid yield was almost flat when the mass ratio between the algae and cocktail enzymes was increased to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Therefore, 8
1. Therefore, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mass ratio between microalgae and cocktail enzymes was appropriate for the abovementioned enzymatic hydrolysis considering the extraction costs. Furthermore, lipid recovery increased with an increase in the mass ratio between cellulase and lysozyme when the ratio was higher than 2
1 mass ratio between microalgae and cocktail enzymes was appropriate for the abovementioned enzymatic hydrolysis considering the extraction costs. Furthermore, lipid recovery increased with an increase in the mass ratio between cellulase and lysozyme when the ratio was higher than 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, whereas the yield declined with an increase in the ratio when this ratio was below 2
1, whereas the yield declined with an increase in the ratio when this ratio was below 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Based on the abovementioned discussion, the highest yield of lipid recovery could be obtained under the following conditions: temperature: 50 °C, the reaction time: 5 h, pH: 4.0, the ratio of microalgae to enzyme: 8
1. Based on the abovementioned discussion, the highest yield of lipid recovery could be obtained under the following conditions: temperature: 50 °C, the reaction time: 5 h, pH: 4.0, the ratio of microalgae to enzyme: 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w), and mass ratio between cellulase and lysozyme: 2
1 (w/w), and mass ratio between cellulase and lysozyme: 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
3.3 Analysis of the lipid
The compositions and content of fatty acids in algal lipid were measured by a GC with FID detector, and test samples were extracted separately by EH and WAEH. A detailed analysis of fatty acids is displayed in Table 2 and ESI Fig. S1.† It was observed that the content of total fatty acids obtained from algal lipid extracted by WAEH was much more than that extracted by EH, whereas the amount of unsaturated fatty acids was more than that of saturated fatty acids in the abovementioned lipids.29 In addition, it is worth noting that EPA and PUFA account for 13.21 ± 0.06% and 23.3 ± 0.31% total fatty acids in algal lipid, respectively, and these results are higher than the data (10.30% and 17.10%) reported in previous literature.19 Furthermore, EPA, which accounts for 56.69% of PUFAs, is the most abundant ingredient in ω-3 PUFAs.30 This suggests that the lipid extracted by WAEH is recognized as a promising feedstock for separating PUFAs. Moreover, the extraction yields of palmitic acid and palmitoleic acid are 33.23 ± 0.02% and 23.90 ± 0.04%, respectively, similar to the result reported in the literature.31 The abovementioned results demonstrate that the weak alkaline hydrothermal treatment-aided enzymatic hydrolysis reaction is more suitable for the extraction of algal lipid, and the extracted lipid can be used as the feedstock for PUFA separation.| Classifications of fatty acids | Content of fatty acids in algal lipid (mg g−1 DCW) | Item | Fraction (%) | ||
|---|---|---|---|---|---|
| WAEH | EH | WAEH | EH | ||
| a Note: each sample was analyzed in duplicate, in addition to duplicate injections into the gas chromatograph, and mean values are shown.b Not detected or relative content less than 0.4%. | |||||
| Saturate | 220.1 ± 0.97 | 71.1 ± 0.99 | C6:0 caproic acid | 0.05 ± 0.00 | —b |
| C8:0 octanoic acid | 0.25 ± 0.00 | — | |||
| C10:0 decanoic acid | 0.17 ± 0.00 | 0.12 ± 0.00 | |||
| C12:0 dodecylic acid | 0.88 ± 0.09 | 0.51 ± 0.00 | |||
| C13:0 tridecanoic acid | 0.31 ± 0.01 | — | |||
| C14:0 tetradecanoic acid | 7.22 ± 0.02 | 6.54 ± 0.00 | |||
| C15:0 pentadecanoic acid | 0.55 ± 0.02 | 0.56 ± 0.00 | |||
| C16:0 palmitic acid | 33.23 ± 0.02 | 30.21 ± 0.03 | |||
| C17:0 heptadecanoic acid | 0.38 ± 0.00 | 0.35 ± 0.00 | |||
| C18:0 stearic acid | 0.88 ± 0.01 | 0.75 ± 0.00 | |||
| C20:0 arachidic acid | 0.14 ± 0.00 | — | |||
| C22:0 docosanoic acid | 0.31 ± 0.03 | — | |||
| Mono-unsaturated fatty acids (MUFAs) | 160.3 ± 0.85 | 56.1 ± 0.70 | C15:1 pentadecylenic acid | 0.11 ± 0.00 | — |
| C16:1 palmitoleic acid | 23.91 ± 0.04 | 22.61 ± 0.00 | |||
| C17:1 heptadecenoic acid | 0.58 ± 0.01 | 0.65 ± 0.00 | |||
| C18:1 trans-octadecenoic acid | 6.78 ± 0.01 | 0.75 ± 0.00 | |||
| C18:1 cis-octadecenoic acid | 0.91 ± 0.00 | 6.45 ± 0.03 | |||
| C20:1 eicosenoic acid | 0.14 ± 0.00 | — | |||
| C22:1 erucic acid | — | 0.21 ± 0.00 | |||
| Poly-unsaturated fatty acids (PUFAs) | 115.9 ± 1.27 | 55.2 ± 1.45 | C18:2 trans-octadecadienoic acid | 3.65 ± 0.02 | 3.63 ± 0.03 |
| C18:2 cis-octadecadienoic acid | 0.63 ± 0.08 | — | |||
| C18:3 octadecatrienoic acid | 1.73 ± 0.00 | 1.87 ± 0.01 | |||
| C20:3 epoxyeicosatrienoic acid | 0.22 ± 0.00 | — | |||
| C20:4 arachidonic acid (AA) | 3.76 ± 0.15 | 4.47 ± 0.00 | |||
| C20:5 eicosapentaenoic acid (EPA) | 13.21 ± 0.06 | 20.32 ± 0.03 | |||
| Total fatty acids (TFAs) | 496.3 ± 3.09 | 182.4 ± 3.13 | |||
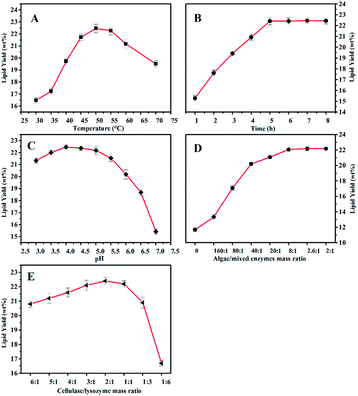
3.4 Influence of weak alkali pretreatment on cell wall disruption
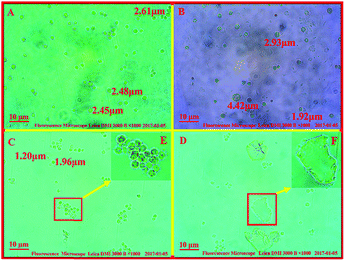
It was reported that the cell wall disruption of microalgae that was processed by freeze-drying was difficult because the cell surface of the algae powder was covered with wrinkles and the algal cell was more unsusceptible to enzyme and water.32 However, an algal cell is more easily degraded by enzymatic hydrolysis after being treated by an acidic or alkaline hydrothermal reaction.33 To investigate the influence of the weak alkali pretreatment on algal cell wall disruption and lipid extraction, morphologies of the algal cell treated with different methods were investigated (Fig. 2). Upon comparing Fig. 2A with Fig. 2B, it was deduced that the particle size of an algal cell shown in Fig. 2B was larger than that shown in Fig. 2A; this might be caused by the swelling of algal cells during the pretreatment process.34 In addition, small amounts of cracks on the cell surface were found, as shown in Fig. 2B, since the swollen cell might be destroyed during the agitation process. Compared to that of raw algal cell, there was no significant change in the morphology of the algal cell treated by EH, whereas a handful of cell debris was found, as shown in Fig. 2C and E, and the surface of freeze-dried algal powders after EH presented small amounts of collapse (ESI Fig. S2†). Herein, one explanation of this could be that algae cells were damaged by enzymes in the process of hydrolysis; then, the algal lipid was released from the broken cell with the permeation of organic solvent, and the rupture of algal cells with the internal cavity was caused by external forces during the agitation process.35 Cell fragments increased significantly when algal cells were treated with weak alkali before the enzymatic hydrolysis as compared to those of the original algal cell and those after treatment by EH; the detailed information can be seen in Fig. 2C and F. Furthermore, the damage to tiny amounts of algal cells is severe such that the intracellular components are completely released, and only a layer of membrane remained in the slurry. In addition, it was observed that the particle size of the algal cell treated with WAEH decreased significantly as compared to that of the original algae. The reason for this is that the swelling of algal cells is more easily degraded by enzyme in the hydrolysis reaction, and the degree of algal cell rupture is enhanced remarkably after the liberation of the lipid. The abovementioned results suggested that the more complete degradation of the algal cell after WAEH was caused by the high degradation capabilities of the cocktail enzymes toward the algal cell, which were treated by weal alkali in the hydrolysis reaction.Interestingly, as depicted in the ESI Fig. S3,† although greater than 10 wt% of the lipid could be extracted by EH, a larger amount of pigments, such as chlorophyll, were obtained as compared to the case of WAEH.36 In addition, the content of total fatty acids obtained from algal lipid extracted by EH was much smaller than that obtained from algal lipid extracted by WAEH (Table 2); this further confirmed that EH could extract more pigments than WAEH. The possible reason is that a great quantity of algal cells are not disrupted after EH; this hinders the permeation of the extracting agent into the cell and hampers the extraction of the lipid. Furthermore, the components of fatty acids in algal lipids extracted by WAEH were more than those extracted by EH, and the result was verified by FTIR spectra of the algal lipid extracted by EH and WAEH (see Fig. 3). It is observed that both the absorption peaks at about 1739 cm−1 and 1741 cm−1 can be assigned to the stretching vibration of the carbonyl group from the glyceryl ester, whereas the absorption at 1712 cm−1 may be associated with the vibration peak of the carbonyl group of free fatty acids.37 This may be ascribed to the movement of the absorption peak of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O towards lower wavenumbers because of the formation of fatty acid dimers in which hydrogen bonds are generated.38 The absorption peaks at 1413 cm−1 and 937 cm−1 are separately attributed to the in-plane and out-of-plane bending vibration of O–H from the carboxylic acid dimer, and several weak peaks at 2500 cm−1–2700 cm−1 are assigned to the coupled vibration of frequency doubling and combination of frequency resulting from stretching and bending of the hydroxyl group in carboxylic acid.37,39 These results indicate that algal lipids extracted by WAEH contain a certain amount of free fatty acids,40 whereas no or lower content is found in that extracted by EH. However, the compositions of algal lipid extracted separately by EH and WAEH are on the whole similar, and the relevant evidence is shown in the Table 2. Therefore, it is confirmed that algal cell swelling occurs in the process of WAP, following which the cell wall is decomposed via enzymatic hydrolysis. The constitutions of fatty acids obtained from algal lipid extracted separately by EH and WAEH are roughly same, even if there is a greater difference between their composition and content.
O towards lower wavenumbers because of the formation of fatty acid dimers in which hydrogen bonds are generated.38 The absorption peaks at 1413 cm−1 and 937 cm−1 are separately attributed to the in-plane and out-of-plane bending vibration of O–H from the carboxylic acid dimer, and several weak peaks at 2500 cm−1–2700 cm−1 are assigned to the coupled vibration of frequency doubling and combination of frequency resulting from stretching and bending of the hydroxyl group in carboxylic acid.37,39 These results indicate that algal lipids extracted by WAEH contain a certain amount of free fatty acids,40 whereas no or lower content is found in that extracted by EH. However, the compositions of algal lipid extracted separately by EH and WAEH are on the whole similar, and the relevant evidence is shown in the Table 2. Therefore, it is confirmed that algal cell swelling occurs in the process of WAP, following which the cell wall is decomposed via enzymatic hydrolysis. The constitutions of fatty acids obtained from algal lipid extracted separately by EH and WAEH are roughly same, even if there is a greater difference between their composition and content.
3.5 Investigation of the extraction technology for algal lipid
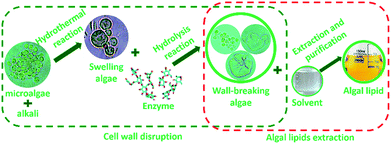
To further investigate the extraction process of algal lipid, more comprehensive understanding of the wall-breaking process by weak alkaline treatment-aided enzymatic hydrolysis is quite essential. Based on the abovementioned results, an extraction protocol of algal lipid by WAEH is schematically illustrated in Fig. 4. It can be seen that the disruption of cell wall is an imperative procedure to extract lipid from microalgae, which needs two steps of cell swelling and cell wall rupture. As far as the former is concerned, the surface of the original algal cell is not very smooth because of the loss of moisture during the drying process. However, the wrinkled cell walls gradually become smooth since microalgae absorb some amount of moisture during the reaction of alkaline hydrothermal, and subsequent algae cell begins to swell because hydrogen bond networks of native polysaccharide existing in the cell wall are destroyed by alkali.41 Furthermore, as is well known, swollen algal cells are more easily subjected to the targeted attack derived from enzyme.42 For this reason, as the fundamental structural unit building, the cell wall of microalgae, micro-fibrous network within a gel-like protein matrix, can be destroyed with cellulose and lysozyme breaking β (1–4) glucosidic linkages in the polysaccharide skeleton during the hydrolysis, which reduces polysaccharides to soluble oligosaccharides and monosaccharides.43,44 Furthermore, cell membrane consisting of phospholipid and glycolipid can be degraded in the hydrolysis reaction since cellulase is a composite enzyme containing multiple hydrolase.45 After this, by adding the solvent to the reaction, damaged algal cells are disrupted or deconstructed under the interaction of the intracellular osmotic pressure,46 and algal lipids are released into the slurry from the lipid vesicle when the complex solvent permeates into the damaged algal cell. Then, the released lipids are achieved via centrifuging the slurry and distilling under vacuum. These results suggest that weak alkali treatment can only make cell wall swell, whereas enzymatic hydrolysis is the crucial procedure for the disruption of algal cell wall. Furthermore, all experimental manipulations of algal lipid extracted by WAEH were carried out at lower temperatures, using a weaker base, and at atmospheric pressure. Therefore, the technology of WAEH makes algal lipid extraction become more complete and economically viable, and the extracted lipid is more suitable as the raw material of the separation of ω-PUFAs.4. Conclusions
A conclusion that the disruption process of cell wall was composed of swelling by weak alkali and decomposition by enzyme was verified through exploring the effect of WAP on cell wall-breaking. Single and mixed enzymes were screened for superior disruption capacity, and the highest yield (22.18 ± 0.26 wt%) of algal lipid recovery was obtained when the temperature, time, pH value, algae/enzyme ratio, and mass ratio of cellulase and lysozyme during the enzymatic hydrolysis were 50 °C, 5 h, 4.0, 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (w/w), and 2
1 (w/w), and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. Compositional analysis of fatty acids demonstrates that algal lipid extracted by WAEH is applicable as a raw material for the separation of ω-PUFAs.
1, respectively. Compositional analysis of fatty acids demonstrates that algal lipid extracted by WAEH is applicable as a raw material for the separation of ω-PUFAs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities, China (No. 15CX05009A), Shandong Provincial Key Project of Research and Development, China (No. 2016GGX102017), Natural Science Foundation of Shandong Province, China (No. ZR2015BM003), Natural Science Foundation of Shandong Province, China (No. ZR2014BQ012), National Natural Science Foundation of China (No. 21176259), and National Natural Science Foundation of China (No. 21406269).Notes and references
- A. T. Elmira, J. Muriel, G. Claire, I. Muhammad, D. Stephane and L. Michel, Trends Food Sci. Technol., 2012, 25, 24–33 CrossRef.
- Y. J. He, J. B. Li, K. Sitharam, B. Chen and Z. Guo, Food Chem., 2017, 219, 230–239 CrossRef CAS PubMed.
- J. T. Dillon, J. C. Aponte, R. Tarozo and Y. Huang, J. Chromatogr. A, 2013, 1312, 18–25 CrossRef CAS PubMed.
- M. Li, P. J. Pham, T. Wang, C. U. Pittman and T. Li, Sep. Purif. Technol., 2009, 66, 1–8 CrossRef CAS.
- R. N. Rubio, S. M. De-Diego, S. Beltrán, I. Jaime, M. T. Sanz and J. Rovira, J. Food Eng., 2012, 109, 238–248 CrossRef.
- M. M. Rahman, A. M. Pourkhesalian, M. I. Jahirul, S. Stevanovic, P. X. Pham, H. Wang, A. R. Masri, R. J. Brown and Z. D. Ristovski, Fuel, 2014, 134, 201–208 CrossRef CAS.
- B. P. Nobre, F. Villalobos, B. E. Barragán, A. C. Oliveira, A. P. Batista, P. A. S. S. Marques, R. L. Mendes, H. Sovová, A. F. Palavra and L. Gouveia, Bioresour. Technol., 2013, 135, 128–136 CrossRef CAS PubMed.
- R. Halim, M. K. Danquah and P. A. Webley, Biotechnol. Adv., 2012, 30, 709–732 CrossRef CAS PubMed.
- L. R. Adetunji, A. Adekunle, V. Orsat and V. Raghavan, Food Hydrocolloids, 2017, 62, 239–250 CrossRef CAS.
- P. Blokker, S. Schouten, H. Van-den-Ende, J. W. De-Leeuw, P. G. Hatcher and J. S. Sinninghe-Damsté, Org. Geochem., 1998, 29, 1453–1468 CrossRef CAS.
- E. T. Rodrigo, Green Chem., 2012, 14, 419–427 RSC.
- Y. Zheng, R. Xiao and M. Roberts, Algal Res., 2016, 14, 100–108 CrossRef.
- C. C. Fu, T. C. Hung, J. Y. Chen, C. H. Su and W. T. Wu, Bioresour. Technol., 2010, 101, 8750–8754 CrossRef CAS PubMed.
- H. S. Cho, Y. K. Oh, S. C. Park, J. W. Lee and J. Y Park, Renewable Energy, 2013, 54, 156–160 CrossRef CAS.
- Z. H. Liu, L. Qin, B. Z. Li and Y. J. Yuan, ACS Sustainable Chem. Eng., 2015, 3, 140–146 CrossRef CAS.
- T. Yuan, X. Li, S. Xiao, Y. Guo, W. Zhou, J. Xu and Z. Yuan, Bioresour. Technol., 2016, 220, 530–536 CrossRef CAS PubMed.
- Z. Yang, R. Guo, X. Xu, X. Fan and S. Luo, Appl. Energy, 2011, 88, 3468–3472 CrossRef CAS.
- Z. M. Xue, M. G. Ma, Z. H. Li and T. C. Mu, RSC Adv., 2016, 6, 98874–98892 RSC.
- C. Wu, Y. Xiao, W. Lin, J. Li, S. Zhang, J. Zhu and J. Rong, Bioresour. Technol., 2017, 223, 312–316 CrossRef CAS PubMed.
- J. S. Elmore, M. M. Campo, M. Enser and D. S. Mottram, J. Agric. Food Chem., 2002, 50, 1126–1132 CrossRef CAS PubMed.
- O. Kilian, C. S. E. Benemann, K. K. Niyogi and B. Vick, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 21265–21269 CrossRef CAS PubMed.
- S. F. Sing, A. Isdepsky, M. A. Borowitzka and D. M. Lewis, Bioresour. Technol., 2014, 161, 47–54 CrossRef PubMed.
- S. Jensen, L. Haggberg, H. Jorundsdottir and G. Odham, J. Agric. Food Chem., 2003, 51, 5607–5611 CrossRef CAS PubMed.
- M. R. Zakaria, S. Hirata and M. A. Hassan, Bioresour. Technol., 2014, 169, 236–243 CrossRef CAS PubMed.
- A. Zuorro, S. Miglietta, G. Familiari and R. Lavecchia, Bioresour. Technol., 2016, 212, 35–41 CrossRef CAS PubMed.
- H. P. Guo, H. M. Chen, L. Fan, A. Linklater, B. S. Zheng, D. Jiang and W. S. Qin, Renewable Energy, 2017, 109, 195–201 CrossRef CAS.
- W. C. Huang and J. D. Kim, Bioresour. Technol., 2017, 232, 142–145 CrossRef CAS PubMed.
- H. Taher, S. Al-Zuhair, A. H. Al-Marzouqi, Y. Haik and M. Farid, Biomass Bioenergy, 2014, 66, 159–167 CrossRef CAS.
- I. Suppavorasatit, E. G. D. Mejia and K. R. Cadwallader, J. Agric. Food Chem., 2011, 59, 11621–11628 CrossRef CAS PubMed.
- R. D. Chakrborty, K. Chakraboty and E. V. Radhakrishnan, J. Agric. Food Chem., 2007, 55, 4043–4051 CrossRef PubMed.
- E. D. Frankel, T. S. Gracia, A. S. Meyer and J. B. German, J. Agric. Food Chem., 2002, 50, 2094–2099 CrossRef CAS PubMed.
- D. Swanson, R. Block and S. A. Mousa, Adv. Nutr., 2012, 3, 1–7 CrossRef CAS PubMed.
- E. Günerken, E. D'Hondt, M. H. M. Eppink, L. Garcia-Gonzalez, K. Elst and R. H. Wijffels, Biotechnol. Adv., 2015, 303, 243–260 CrossRef PubMed.
- J. S. Graves and J. Gutknecht, J. Gen. Physiol., 1976, 67, 579–597 CrossRef CAS PubMed.
- D. R. Schmitt and M. D. Zoback, New Phytol., 1986, 102, 521–529 Search PubMed.
- E. Santillan-Jimenez, R. Pace, S. Marques, T. Morgan, C. McKelphin, J. Mobley and M. Crocker, Fuel, 2016, 180, 668–678 CrossRef CAS.
- M. J. Lerma-García, E. F. Simó-Alfonso, A. Bendini and L. Cerretani, J. Agric. Food Chem., 2009, 57, 9365–9369 CrossRef PubMed.
- Z. H. Liu, X. F. Sun, M. Y. Hao, C. Y. Huang, Z. M. Xue and T. C. Mu, Carbohydr. Polym., 2015, 117, 99–105 CrossRef CAS PubMed.
- Y. B. Che-Man and G. Setiowaty, Food Chem., 1999, 66, 109–114 CrossRef.
- M. Demuez, A. Mahdy, E. Tomas-Pejo, C. Gonzalez-Fernandez and M. Ballesteros, Biotechnol. Bioeng., 2015, 112, 1955–1966 CrossRef CAS PubMed.
- X. F. Sun, Z. H. Liu, Z. M. Xue, Y. W. Zhang and T. C. Mu, Green Chem., 2015, 17, 2719–2722 RSC.
- X. F. Sun, C. Y. Huang, Z. M. Xue, C. Y. Yan and T. C. Mu, Energy Fuels, 2015, 29, 2564–2570 CrossRef CAS.
- S. Y. Lee, J. M. Cho, Y. K. Chang and Y. K. Oh, Bioresour. Technol., 2017, 244, 1317–1328 CrossRef CAS PubMed.
- Z. M. Xue, X. H. Zhao, R. C. Sun and T. C. Mu, ACS Sustainable Chem. Eng., 2016, 4, 3864–3870 CrossRef CAS.
- C. A. Wang, S. H. Chou and G. L. Guo, U. S. Pat., 9,297,001 B2, 2016, vol. 3, p. 29.
- A. Pellegrini, U. Thomas, F. R. Von Fellenberg and P. Wild, J. Appl. Bacteriol., 1992, 72, 180–187 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The compositional analysis of fatty acids in algal lipid extracted separately by EH and WAEH, morphology of algal cell, and the content of chlorophyll in algal lipid extracted separately by WAEH and EH can be found in the ESI, and detailed data can be found in Fig. S1–S3. See DOI: 10.1039/c7ra11483d |
| ‡ Contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2017 |