 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Arabidopsis thaliana SKP1 interacting protein 11 (At2g02870) negatively regulates the release of green leaf volatiles†
Muhammad Naeem-ul-Hassan *ac,
Zamri Zainalab,
Chew Jin Kiatab,
Hossein Hosseini Monfareda and
Ismanizan Ismail*ab
*ac,
Zamri Zainalab,
Chew Jin Kiatab,
Hossein Hosseini Monfareda and
Ismanizan Ismail*ab
aSchool of Bioscience and Biotechnology, Faculty of Science and Technology, University Kebangsaan Malaysia, Malaysia
bInstitute of Systems Biology (INBIOSIS), University Kebangsaan Malaysia, Bangi, 43600, Selangor, Malaysia. E-mail: maniz@ukm.edu.my; ismanizan@gmail.com; Tel: +60 389214546
cDepartment of Chemistry, University of Sargodha, Sargodha, Pakistan. E-mail: sobheel@yahoo.com; Tel: +92 3338967518
First published on 8th December 2017
Abstract
Green leaf volatiles (GLV) are important factors in plant defense, and plant–plant and plant–insect interactions. Kelch-repeat containing F-box proteins (KFB) are plant-specific proteins, which recruit substrates for protein degradation. The present study was conducted to evaluate the role of AtSKIP11, an Arabidopsis KFB, in the expression of hydroperoxide lyase (HPL) pathway genes and the formation of products by creating transgenic Arabidopsis thaliana plants expressing the AtSKIP11 open reading frame and its antisense sequence. We found that production of C5 volatiles was greatly reduced in these transgenic plants. The production of C6 GLV was reduced by 49% in AtSKIP11 overexpressing plants, whereas the production of these compounds was increased by 12% in the AtSKIP11-antisense suppression plants. Real time quantitative polymerase chain reaction (RT-qPCR) results showed that expression of LOX2, HPL and ADH1 genes was reduced by 1.8, 1.3 and 2.5 fold, respectively, in the AtSKIP11 overexpressing plants, compared with wild-type plants. In the AtSKIP11-antisense plants, expression of LOX2 was reduced 1.2 fold, but HPL and ADH1 genes were up-regulated by 1.2 fold and 1.7 fold, respectively, over the wild-type samples. Our results suggest the involvement of AtSKIP11 in negatively regulating the HPL pathway, especially the expression of ADH1, possibly through degradation of the transcription factors.
Introduction
The sessile nature of the plant kingdom does not allow these creatures to move towards a safe or a favourable place. Moreover, these organisms lack the specialized immune system animals use to combat diseases. Alternatively, plants have developed various other mechanisms to combat a variety of threats. One way of doing this is through inducing the production of defense-related proteins and the release of toxic chemicals. These chemical defenses are the products of the secondary metabolism of plants and they obstruct the growth of and potential damage by different pathogens, including viruses, fungi, bacteria and herbivores. Thousands of different kinds of secondary metabolites are synthesized in plants by a large number of inducible pathways. The oxylipin pathway is among the major inducible pathways for the production of defense responses through activation of its genes in response to different biotic and abiotic stresses.1,2 These stresses can activate the oxylipin pathway for the release of higher amounts of jasmonates and green leaf volatiles (GLV), which are otherwise released in very low concentrations under normal conditions.3Plant oxylipins constitute a diverse class of bioactive lipids that are produced by the oxidation of α-linolenic acid (ALA, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3Δ (ref. 9, 12 and 15)) and linoleic acid (LA, 18
3Δ (ref. 9, 12 and 15)) and linoleic acid (LA, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2Δ (ref. 9 and 12)). Products of the oxylipin pathway include 12-oxo-phytodienoic acid, jasmonic acid (JA), methyl jasmonate (MeJA), and GLV.4–6 Although jasmonates are well known for their role in the regulation of many physiological processes in plant development, they are particularly regarded as specialists for host immunity and plant defense.7–9 The production of GLV is induced under conditions of stress in order to protect the plants against herbivores and microbial pathogens.10 Under normal physiological conditions, only traces of GLV are released. However, the production of these compounds abruptly increases in response to tissue disruption resulting from biotic or abiotic stresses.3
2Δ (ref. 9 and 12)). Products of the oxylipin pathway include 12-oxo-phytodienoic acid, jasmonic acid (JA), methyl jasmonate (MeJA), and GLV.4–6 Although jasmonates are well known for their role in the regulation of many physiological processes in plant development, they are particularly regarded as specialists for host immunity and plant defense.7–9 The production of GLV is induced under conditions of stress in order to protect the plants against herbivores and microbial pathogens.10 Under normal physiological conditions, only traces of GLV are released. However, the production of these compounds abruptly increases in response to tissue disruption resulting from biotic or abiotic stresses.3
The hydroperoxide lyase (HPL) branch of the oxylipin pathway takes its share from the polyunsaturated fatty acids (PUFA) hydroperoxide substrates, generated by lipoxygenase (LOX) enzymes due to activation of the pathway genes under the stressed conditions.11 The products of LOX are divided between the two competing branches of the oxylipin pathway in a coordinated manner.12,13 Allene oxide synthase converts these intermediates to jasmonates and the HPL enzyme releases volatile C9 and C6 aldehydes. These aldehydes are further converted to other aldehydes, alcohols and their acetylated derivatives.14 Among the HPL downstream enzymes are included (3Z):(2E)-hexenal isomerase, responsible for the conversion of Z-3-hexenal to its E-2 isomer, and NAD-dependent alcohol dehydrogenase (ADH), that catalyzes the reduction of aldehydes to their respective alcohols in order to confer higher stability.15–17 All these saturated and unsaturated aldehydes and their downstream products synthesized in the plant's vegetative tissue are volatile in nature and are collectively named as GLV. The role of GLV in the plant's response to various stresses, like microbial pathogens and herbivory attack, has been studied since the early 1990s. But they have been getting the special attention of researchers around the world for the last decade and a half because of their emerging role as aerial messengers for plant–plant communication.18,19
The regulation of cellular processes through the degradation of regulatory proteins by the ubiquitin 26S proteasome system (UPS) followed by recognition via specific F-box proteins is a common signaling mechanism. F-box is an emerging family of signaling proteins in eukaryotes and is thought to be one of the largest superfamilies of plant proteins.20 The majority of F-box proteins serve as a substrate-recruiting component of SCF (SKP1, Cullin 1, RING-BOX 1 and an F-box protein ) complexes. Nearly two decades since the recognition of and giving the name ‘F-box’ to a domain in cyclin-F protein, our understanding of the role of F-box proteins, SCF biology and the molecular mechanisms employed by UPS is steadily developing.21 F-box proteins are comprised of a conserved 40–50 amino acids F-box domain on the N-terminal, while the C-terminals contain one or more secondary motifs. These motifs, which include WD40-repeats, leucine rich-repeats (LRR), kelch-repeats and some other motifs, may be the same or variable in a particular protein and are involved in protein–protein interactions.22,23 Researchers around the world have identified many F-box proteins since the discovery of cyclin-F, especially in plants. Since hundreds of SCF complexes have been characterized, the number of F-box proteins serving these SCF complexes is also quite large. However, most of these F-box proteins either remain ‘orphans’ or, until now, could not be completely characterized for their function.24
F-box proteins containing the kelch-repeat motifs (KFB) are among the major plant F-box proteins and are considered to be plant specific. The fact that only a couple of KFB have been characterized in non-plant organisms suggests that the KFB subfamily of F-box proteins is exclusively a plant-specific family of genes.25,26 This indicates that some very crucial roles have been assigned to these proteins in the plant cell. In recent years, the pace of research involving plant KFB in molecular signaling has advanced at a reasonable rate, but there is still a lot more to be explored. Several KFB have been reported for their signaling roles in plant defense, plant physiology, circadian mechanisms, secondary metabolism, growth and development, but still the majority of these proteins remain ‘orphans’ or under investigation.24
Although several recent studies have highlighted the role of different KFB in plant physiology, not even a single KFB has been reported for any role in the HPL branch of the oxylipin pathway.27–29 A couple of recent studies identified and characterized a novel KFB gene in Persicaria minor, named PmF-box1. This gene was found to respond to jasmonic acid elicitation and co-up-regulated with PmADH1 and PmLOX1 upon exogenous treatment with jasmonic acid.30,31 BLAST searches were run to find a PmF-box1 homolog in Arabidopsis thaliana. The results returned by the BLAST search show that SKP1 interacting protein 11 (SKIP11) possesses the highest (69%) sequence similarity with PmF-box1. AtSKIP11 (At2g02870) is a KFB consisting of 467 amino acid residues with five kelch repeats. Although no specific function has been identified for AtSKIP11 until now, the protein has been found to interact with the Arabidopsis thaliana ASK1 and ASK2 proteins.29,32 The present study highlights the role of the AtSKIP11 gene in the HPL branch of the oxylipin pathway for the production of GLV. A full length AtSKIP11 gene sequence was cloned to the pB2GW7 gateway destination vector to form overexpression and antisense suppression constructs. Both the constructs were transferred to Arabidopsis thaliana wild-type (At-WT) plants by Agrobacterium tumefaciens mediated transformation using the floral dip method. Functional analysis was performed on the T2 generation of the transgenic plants, along with the At-WT plants (control plants). Our results have shown that the AtSKIP11 overexpressing plants produced a lower amount of GLV as compared to the wild-type Arabidopsis plants. Whereas, the production of GLV was found to be enhanced in the transgenic Arabidopsis plants in which the AtSKIP11 gene transcript was suppressed by the antisense strategy. These results suggest that AtSKIP11 is involved in the biosynthesis of GLV, possibly affecting the transcription factors, and negatively regulates the HPL pathway.
Experimental
Plant material and bacterial cultures
A. thaliana ecotype Columbia-0 seeds were obtained from Dr Noor Azmi Shaharudin (Universiti Putra Malaysia). Chemically competent E. coli strain TOP10 cells and the GV3101 strain of A. tumefaciens were obtained from Plant Biotechnology Lab, Universiti Kebangsaan Malaysia.Chemicals and reagents
All the chemicals and reagents used in this study were of analytical grade or higher. The chemicals used for the molecular work and as the reference standards were obtained from Sigma-Aldrich (USA). For the tissue culture work, chemicals were obtained from Duchefa Biochemie B.V. (Netherlands). The QIAquick Gel Extraction Kit and the DNeasy Plant Mini Kit were from QIAGEN (UK). The Gateway Cloning kit, PureLink Quick Plasmid Miniprep Kit, Plant RNA Reagent, Platinum Taq DNA Polymerase, Ambion TURBO DNase and DNA Ladders were obtained from Life Technologies (Carlsbad, CA, USA). KAPA SYBR FAST qPCR Kit, KAPA Taq and KAPA Taq HotStart DNA Polymerase were from Kapa Biosystems (Wilmington, Massachusetts). Restriction enzymes and the ProtoScript First Strand cDNA Synthesis Kit were from New England Biolabs (MA, USA).Construction of AtSKIP11 overexpression and antisense recombinant plasmid
For the cloning of the AtSKIP11 gene and its antisense sequence, the full length open reading frame was amplified by a standard PCR reaction, using A. thaliana cDNA as the template. The primers for the isolation of the AtSKIP11 gene with flanking attB sites were designed as guided by the Gateway Cloning system and verified by the OligoAnalyzer 3.1 tool from Integrated DNA Technologies (IDT). Using these primer pairs, fragments of 1555 bps were amplified by PCR for each AtSKIP11 open reading frame and its antisense sequence plus Gateway attB sites (Table 1).
| Primer | Sequence (5′–3′) | Amplicon size |
|---|---|---|
| At-SKIP11-attB1 (F) | GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGTTTGGGATTGGGATTGATCTGA | |
| At-SKIP11-attB2 (R) | GGGGACCACTTTGTACAAGAAAGCTGGGTGGAGGGTCTTCAAATGGATAGTCGT | 1555 bps |
| At-SKIP11-attB1 antisense (F) | GGGGACAAGTTTGTACAAAAAAGCAGGCTGGAGGGTCTTCAAATGGATAGTCGT | |
| At-SKIP11-attB2 antisense (R) | GGGGACCACTTTGTACAAGAAAGCTGGGTGGGTTTGGGATTGGGATTGATCTGA | 1555 bps |
The PCR reaction was run with Platinum Taq DNA Polymerase from Life Technologies using Arabidopsis cDNA as the template. Two separate 50 μL PCR reactions were performed on an Eppendorf Mastercycler machine with the following program: initial denaturation of 1 cycle at 94 °C for 2 min; and 30 cycles of: denaturation at 94 °C for 15 s, primers annealing at 55 °C for 30 s, extension at 68 °C for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 min and the final hold at 10 °C. The PCR products were analyzed by 1% (w/v) agarose-EtBr gel electrophoresis and purified by a Purelink PCR purification kit (Life Technologies) according to the manufacturer's protocol. The quality of the purified PCR product was verified by NanoDrop and then stored at −20 °C until the next step.
30 min and the final hold at 10 °C. The PCR products were analyzed by 1% (w/v) agarose-EtBr gel electrophoresis and purified by a Purelink PCR purification kit (Life Technologies) according to the manufacturer's protocol. The quality of the purified PCR product was verified by NanoDrop and then stored at −20 °C until the next step.
Generation of transgenic Arabidopsis thaliana plants
Agrobacterium tumefaciens strain GV3101 harbouring pB2GW7::AtSKIP11 and pB2GW7::AtSKIP11-antisense recombinant plasmids was used to transform the A. thaliana (ecotype Columbia) plants by the floral dip method.33 The plants were grown under standard conditions and selected by the herbicide ‘glufosinate’ (basta) due to the presence of the bar gene in the recombinant plasmids. The putative transgenic plants, which survived the herbicide treatment, were confirmed for the integration of the transgenes by PCR, using the genomic DNA as the templates. The PCR was run with Kapa Polymerase Master Mix and the primers listed in Table 2.| Primer | Sequence (5′–3′) | Amplicon size |
|---|---|---|
| b2sk-F1 (pB2GW7 specific) | CCGTGAAGACTGGCGAACA | |
| b2skox-R1 (AtSKIP11 specific) | GCCAGCTCTTTCAGGCAATCTT | ≈1830 bps |
| b2skanti-R1 (AtSKIP11-antisense specific) | CCGATATGGAAGTGAGGAAGTACGA | ≈960 bps |
The PCR Master Mix was prepared as given in the manufacturer's manual. The following PCR program was employed: 1 cycle of initial denaturation at 95 °C for 3 min; and 30 cycles of: denaturation at 98 °C for 30 s, primers annealing at 62 °C for 30 s, primers extension at 72 °C for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 min and 1 cycle of final extension at 72 °C for 3 min. The products of PCR were analyzed by running 1% agarose-EtBr gel electrophoresis and comparing the bands with a 1 kb DNA ladder to confirm the integration of the DNA insert.
30 min and 1 cycle of final extension at 72 °C for 3 min. The products of PCR were analyzed by running 1% agarose-EtBr gel electrophoresis and comparing the bands with a 1 kb DNA ladder to confirm the integration of the DNA insert.
Molecular analysis of transgenic A. thaliana
Molecular analysis of transgenic A. thaliana plants was initiated by growing the third generation. Three lines were selected from each type of transgenic plant for the analysis of metabolic products and the gene expression of the HPL pathway. To ensure the growth of only the desired transgenic plants and to avoid the growth of any non-transgenic contaminants, the young seedlings were again subjected to the Basta selection. Confirmed transgenic plants were grown in the growth chamber until six weeks of age under standard conditions before sampling. Two hours prior to sampling, the plants were treated with 250 μM methyl jasmonate aqueous solution to activate the plant defence related gene expression and the production of plant defence compounds. After two hours the samples were collected for real-time quantitative PCR (RT-qPCR) and solid phase micro-extraction GC-MS (SPME-GC-MS) analysis.RT-qPCR analysis
Traces of any methyl jasmonate, sprayed two hours before, were adsorbed from the leaf surface using a clean tissue paper. The upper parts of 4–5 healthy leaves (≈300 μg) were cut with a clean pair of scissors and the plant material was quickly transferred to labelled 1.5 mL RNase free microfuge tubes and immediately frozen in liquid nitrogen. Total RNA was then extracted by using the Purelink Plant RNA Reagent (Life Technologies, USA) according to the manufacturer's instructions. After removing any contaminating DNA, first strand cDNA synthesis was accomplished by a ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs) using 500 ng of purified total RNA as the template.Analysis for the expression of Arabidopsis Actin2 (Act2), LOX2, HPL and ADH1 genes was performed by RT-qPCR, using a KAPA SYBR FAST qPCR Kit (Kapa Biosystems, USA). For each gene, a pair of gene-specific primers was designed, following the instructions from the manufacturer of the kit. These primers were verified using OligoAnalyzer 3.1 software (IDT) to amplify a short fragment from each gene (Table 3). Act2, a reference gene, was used as the calibrator for the normalization.34
| Target gene/primers | Sequence (5′–3′) | Amplicon size |
|---|---|---|
| Act2-forward | CTTGCACCAAGCAGCATGAA | 68 |
| Act2-reverse | CCGATCCAGACACTGTACTTCCTT | |
| Lox2-forward | CGTGCACGCCAAAGTCTTGTCAA | 130 |
| Lox2-reverse | CGGGTAAGCCTTCCTGGTCAA | |
| Hpl-forward | CGCACGTGCGAGGAAAGA | 88 |
| Hpl-reverse | CGGCTGATAACCACAAAGAAGCTCA | |
| Adh-forward | GGAGTGGAGTACAGCTTCGAGTGT | 130 |
| Adh-reverse | GGATAGGGAGAAGCTGTGGGCTT |
In order to optimize various parameters, such as the template concentration and the primer concentration to achieve a reliable comparison among the samples and the controls, calibration curves were constructed for all the genes under study. Using the control sample cDNAs as templates, five or four dilution points, each point representing a 10-fold dilution were plotted against the respective CT values to calculate the efficiency, R2 and slope. For the quantification, three lines of the transgenic plants (biological replicates) were used and each line was analysed in triplicate (technical replicates). In addition, non-template controls (NTC) were also prepared for each primer pair. The 15 μL PCR reaction components include; 30 ng of first strand cDNA template (final conc. 2 ng μL−1), 7.5 μL Kapa SYBR Fast qPCR Master Mix (2×), 0.125 μM of each primer and nuclease-free water to a total of 15 μL. The PCR program adjusted for the amplification of target fragments, using a Bio-Rad IQ5 optical detection system, consists of: one cycle at 95 °C for 3 min; and 40 cycles of: 95 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s. The amplification curve program was followed by a melting curve program that consisted of 61 cycles, increasing the temperature in each cycle by 0.5 °C and dwelling for 6 s from 65 °C to 95 °C. The PCR amplification of the reference and the target genes from transgenic A. thaliana cDNA templates, as well as cDNA templates from the wild-type plants, was performed in separate reactions. Template cDNA from the wild-type plants was used as a control, to calculate the fold change in gene expression levels by determining the threshold cycle (Ct) values of the target and the calibrator genes from the control and the test samples using the Livak method.35 The PCR products were also analysed on 1.2% (w/v) agarose gel electrophoresis to identify the amplification of any non-specific sequences.
Statistical analysis
Statistical significance was assessed by using GraphPad Prism 5. Data is presented as mean ± SD and the comparison among the three groups was assessed by using one-way ANOVA followed by Dunnett's ultiple comparison test. Differences under P < 0.05 were considered statistically significant.Extraction of GLV for SPME-GC-MS analysis
Pre-labelled 20 mL glass vials from Agilent Technologies (USA) were used to collect the samples for this purpose. In each vial, 4 glass beads were added to disrupt the plant tissue for the release of GLV. About 1–2 g of healthy leaves were cut from each pot of plants with a razor blade and quickly transferred to the respective vials. The leaves were further cut into small pieces by the same blade during shifting of plant material to the vial, so as to facilitate the leaves' disruption process. The vials were sealed with parafilm and the crimp-top caps were tightened before weighing the vials.The GLV emitted from the Arabidopsis leaf samples were analysed by extracting these volatiles through the headspace solid-phase micro extraction (HS-SPME) technique and GC-MS, following a recently published protocol from Matsui et al. (2012) with some modifications.36 The tissues were completely disrupted by vigorously vortexing the vials for 5–10 min. Then the vials were incubated at 25 °C for 10 min followed by the addition of 1 mL of saturated solution of calcium chloride to deactivate any enzyme activity.
SPME-GC-MS analysis
GLV formed by completely disrupted plant material were analyzed by exposing an SPME fiber (50/30 mm DVB/Carboxen/PDMS, Sigma-Aldrich, USA) in the headspace of the vials at 40 °C for 30 min. Subsequently, the fiber was inserted into the insertion port of a GC (Agilent 7890A) containing a 0.25 mm by 30 m BP20 (WAX) column (SGE Analytical Science, Australia) for thermal desorption of the volatiles. The GC was directly coupled to the MS of an Agilent 5975C inert MSD with a triple-axis detector. To completely remove any compound absorbed on the fiber matrix, the fiber was held in the injection port for 10 min. The temperature of the injector was 200 °C and the interface was at 230 °C. The column temperature program followed was: 40 °C for 1 min, rising at a rate of 15 °C per min to 230 °C and then kept there for 1 min. Helium was used as the carrier gas and delivered at a flow rate of 1 mL per min. The analysis was performed by splitless injection with a sampling time of 1 min. The MSD Chemstation was used to find all the peaks in the raw GC chromatogram. A library search was carried out for all the peaks using NIST/EPA/NIH version 2.0, and the results were combined in a single peak table. Among all the GLV, Z-3-hexen-1-ol can serve as the best compound for the analysis of the HPL pathway because it is the first C6 alcohol formed in the pathway. A standard curve was constructed for Z-3-hexen-1-ol consisting of four concentration points at 5, 50, 500 and 1500 nanomolar (nM) concentrations.Results and discussion
The physiological role of AtSKIP11 in the Arabidopsis HPL pathway was determined by generating transgenic A. thaliana plants, integrating the overexpression and the antisense suppression constructs of the AtSKIP11 gene. Three of the transgenic lines from each of the AtSKIP11 overexpressing plants and the AtSKIP11-antisense plants were selected for further analysis by RT-qPCR and SPME-GC-MS. Wild-type plants were also grown along with the transgenic plants. Seeds from each of the above-mentioned lines were planted on soil and the plants grown for six weeks under standard conditions. The plants were treated with methyl-jasmonate, to activate the Arabidopsis defence-related pathways, including the oxylipin pathway genes, two hours prior to collecting the samples for analysis.37 The leaf samples were analyzed to measure any changes in the levels of oxylipin gene expression and the GLV formed in transgenic plants against the control plants.RT-qPCR analysis
Total RNA was extracted from 100 mg of the leaf powder by PureLink Plant RNA Reagent (Life Technologies), according to the manufacturer's protocol. cDNA was synthesized with a ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs), according to the manufacturer's protocol, using 500 ng of purified total RNA as the template. The cDNA product was diluted with nuclease-free water to a final concentration of 2 ng μL−1, based on the concentration of the RNA.The cDNAs of the AtSKIP11 overexpressing and the AtSKIP11-antisense sequence expressing transgenic lines were used as templates to determine any change in the gene expression levels by RT-qPCR, using a pair of gene-specific primers for the target genes and the calibrator gene (reference gene). The relative expression of the genes of the HPL pathway was calculated by averaging the threshold cycle (CT) values of each line and the fold changes in the expression levels of the target genes were calculated by applying the 2−ΔΔCt method (Appendix A, ESI†). Changes in the gene expression levels for the transgenic plant cDNAs were plotted on a graph to highlight the differences (Fig. 1).
Reduction in the expression of LOX2, HPL and ADH1 was shown by the AtSKIP11 overexpressing plant cDNA templates. The highest change was observed in the expression of ADH1, in which more than 2-fold down-regulation was recorded as compared with the control samples. On the other hand, the plants expressing the AtSKIP11-antisense sequence gave a different pattern of expression. In these cDNA samples, LOX2 was down-regulated by about 1.2-fold, while the HPL expression was increased by the same value and the ADH1 was up-regulated by more than 1.7-fold as compared to control plants. The data assessed by using one-way ANOVA followed by Dunnett's multiple comparison Test shows that the difference in expression levels of ADH1 is statistically significant with a p-value of 0.0013.
SPME-GC-MS analysis of GLV
Products of the HPL pathway were analyzed by GC-MS after extracting the volatiles, produced from the transgenic A. thaliana plants integrating the AtSKIP11 gene and the AtSKIP11-antisense sequence by HS-SPME. The GC-MS chromatograms yielded well-resolved peaks for different volatiles with similar patterns (Appendix B, ESI†).Amounts of different GLV produced by each line of transgenic A. thaliana plants and the wild-type A. thaliana plants were calculated by averaging the mass abundance of each compound per gram of fresh leaf material from the respective chromatograms and the RSD was determined (Appendix B, ESI†). Average mass abundance values of each volatile compound for the transgenic and control plants are computed in Table 4 along with their retention times.
| GLV | RT (min) | At-WT plants | AtSKIP11 overexpress | AtSKIP11 antisense |
|---|---|---|---|---|
| Hexanal | 4.80 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 282 282![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 321 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 185 185![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 543 543 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 974 974![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550 550 |
| (E)-2-pentenal | 6.27 | 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 611 611![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 380 380 |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 430 430![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 781 781 |
62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 141 141![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 721 721 |
| 1-Penten-3-ol | 7.60 | 313![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 233 233![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 340 |
47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 704 704![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 923 923 |
102![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 645 645![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 089 089 |
| 1-Pentanol | 10.36 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968 968![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 059 059 |
7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 383 383![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 709 709 |
9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 437 437![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 418 418 |
| 1-Hexanol | 13.42 | 64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 605 605![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 639 639 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 187 187![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 494 494 |
61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 574 574![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 701 701 |
| (Z)-3-Hexen-1-ol | 14.27 | 274![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 028 028![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 143 143 |
124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 994 994![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 804 804 |
337![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579 579![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 587 587 |
The amounts of different GLV per gram of fresh leaf material and their RSD values were plotted on a graph, to compare the levels of volatiles produced in the transgenic plants and the wild-type plants (Fig. 2 and 3).
 | ||
| Fig. 2 Relative mass abundance of different C5 volatiles per gram of fresh leaf material in transgenic plants and wild-type plants. | ||
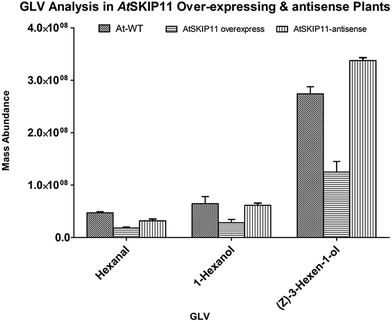 | ||
| Fig. 3 Relative mass abundance of different GLV per gram of fresh leaf material in transgenic plants and wild-type plants. | ||
The synthesis of all target GLV compounds was found to be greatly reduced in the AtSKIP11 overexpressing plants as compared to the control plants, with the highest reduction observed in the formation of two C5 GLV, (E)-2-pentenal and 1-penten-3-ol (Fig. 2). The AtSKIP11-antisense sequence expressing plants also produced lower amounts of some of the volatile compounds, except (Z)-3-hexen-1-ol and 1-hexanol that were either produced in higher concentrations in the transgenic plants or remained almost the same in both types of plants, respectively (Fig. 3). The amounts of (Z)-3-hexen-1-ol released by the AtSKIP11 overexpressing plants was about 55% less, but that released by the AtSKIP11-antisense sequence expressing plants was about 20% higher than that produced by the control plants. Concentrations of (Z)-3-hexen-1-ol, the key GLV in this study, released by the AtSKIP11 overexpressing and the AtSKIP11-antisense expressing transgenic plants were calculated to be 89 nM and 313 nM per g of fresh leaf, respectively. Shiojiri et al. (2012) reported that the maximum concentration of (Z)-3-hexenal, (E)-2-hexenal, and (Z)-3-hexenol released by completely damaged tissue was calculated to be 496 nM GLV per g of the fresh weight.38
The reduction in the levels of C5 volatiles to a very significant degree show that the gene may be affecting LOX1 more than LOX2 in the Arabidopsis. The genes involved in the biosynthesis of C5 volatiles have not been identified until now and, moreover, synthesis of C5 volatiles is found to be independent of HPL.39 Shen et al. (2014) have elucidated the role of TomloxC, a tomato 13-LOX in the biosynthesis of C5 volatiles.40 Arabidopsis LOX1 catalyses the oxygenation at carbon number 9 of the hydrocarbon backbone in LA and ALA to yield (9S,10E,12Z)-9-hydroperoxyoctadeca-10,12-dienoic acid (9-HPOD) and (9S,10E,12Z,15Z)-9-hydroperoxy-10,12,15-octadecatrienoic acid (9-HPOT), respectively.41 The AtSKIP11 protein has five kelch-repeats and interacts with the substrate proteins possibly through the blades of its β-propeller, thus forming a SCFAt2g02870 (SCFAtSKIP11) complex.29
The overall picture obtained from the results of these transgenic plants has highlighted the involvement of AtSKIP11 in the HPL branch of the oxylipin pathway. The protein is suggested to up-regulate a certain portion of the pathway through degradation of some transcription factors or other regulatory proteins. Overall, these results suggest that AtSKIP11 is involved at some stage, most probably downstream of the HPL enzyme, of the HPL biosynthetic pathway in a ‘negatively regulating’ manner. This means that higher the levels of AtSKIP11, the lower will be the production of GLV and vice versa. Recently, a few KFBs have been reported to participate in plant physiological phenomena by degrading the inhibitors of the pathway. In a recent report, a novel KFB encoding gene from rice, an inhibitor for brown furrows1 (IBF1) was cloned to show that IBF1 is involved, as a negative regulator in the biosynthesis of flavonoids and to reduce the accumulation of brown pigments in rice hulls.42 Similarly, a KFB encoding gene in rice, LARGER PANICLE (LP) was found to be involved in the regulation of panicle architecture. This study reported that LP is an endoplasmic reticulum localized protein and possibly modulates the cytokinin levels by interacting with rice SKP1-like protein. Moreover, the mutant plants were found to have increased panicle size, better plant architecture and improved grain yield per plant.43 The role of the KMD family of proteins was recently reported in the regulation of Arabidopsis cytokinin signalling. In the SCFKMD complex, KMD proteins were suggested to physically interact with some key transcription factors in the cytokinin signalling. KMD loss of function mutants were more sensitive to cytokinin, while transgenic lines overexpressing KMD were insensitive to cytokinin, showing that KMD proteins negatively regulate the cytokinin signalling in Arabidopsis.44 Recently, a couple of similar studies have highlighted the role of a family of four similar KFBs in Arabidopsis in the regulation of the phenylpropanoid biosynthetic pathway. Zhang et al. (2013 and 2014) have shown that these proteins regulate the biosynthesis of phenylpropanoids by interaction with and proteolytic turn-over of four phenylalanine ammonia-lyase (PAL) isozymes that catalyse the first and the committed step in phenylpropanoid biosynthesis.45,46
Using the results of present study in the context of these published results, a mechanism for the function of AtSKIP11 is proposed here. AtSKIP11 interacts with SKP1 through its N-terminal F-box domain and interacts with the target proteins, like LOX2, HPL, ADH1 or the regulatory elements of these genes, through its C-terminal domain. The C-terminal of AtSKIP11 consists of a five kelch-repeats containing domain, which is thought to form a bladed β-propeller. Each blade of the β-propeller consists of a single kelch-repeat that is folded into four sheets and may be capable of independently interacting with other different proteins.25 Therefore, it is suggested that multiple interacting partners may exist for the SCFAtSKIP11 complex. The results of the present study show that some proteins, including the enzymes of the oxylipin pathway do interact with the SCFAtSKIP11 complex. Degradation of these proteins, that may be enzymes themselves or regulatory proteins, such as transcription factors, reduce the formation and release of certain GLV, as shown by the transgenic plants overexpressing the AtSKIP11 gene. The levels of various GLV were found to be reduced in these plants and the expression of the HPL pathway genes was also found to be decreased as compared with the control plants (Fig. 1–3). On the other hand, in the transgenic plants expressing the antisense suppression construct, levels of some GLV were found to be enhanced and also the expression of the LOX2 and HPL genes was slightly up-regulated, while the expression of ADH1 was significantly up-regulated (Fig. 1–3). That means that the suppression of the translation of the AtSKIP11 gene transcript leads to formation of a reduced amount of SCFAtSKIP11 complex. This is in accordance with the results of some recent studies, in which certain KFB overexpressing plants resulted in the down-regulation of the metabolic pathways under study while the mutant plants came up with up-regulation of the pathways.24 As a result, the degradation of the protein targets for the SCFAtSKIP11 complex will be decreased. The reduced or the retarded protein degradation, of either the enzymes or the transcription factors, will allow the production of more GLV and the up-regulation of certain enzymes of the pathway. According to our proposed mechanism, the impaired or lower expression of AtSKIP11 leads to decreased production of SCFAtSKIP11 complex and thus less degradation of GLV-producing proteins.
It is obvious from the literature review of the GLV biosynthetic pathway that the pathway is quite complex. Release of PUFAs by the degradation of membrane lipids is still not completely understood. LA and ALA, released by phospholipases, serve as substrates for the oxylipin pathway, but there may exist some other PUFA candidates as well. Although LOX2 contributes more towards the generation of oxygenated intermediates, the role of the regulatory components switching between the two types of LOX may also contribute significantly. The most important step in the pathway is the distribution of oxygenated substrates between the HPL and the AOS branches of the pathway. The importance of jasmonic acid is not only to rescue the plant during any type of stress, but it is involved in almost every aspect of plant growth and development.9 While only trace amounts of GLV are released by plants under normal physiological conditions, they are formed very rapidly under stressful conditions. Numerous studies have suggested that the two competing branches of the oxylipin pathway crosstalk during metabolism and the stress response.14 The role of the regulatory proteins at this stage of the pathway is of great importance, in which SCF complexes may be very crucial. The degradation of certain proteins will allow the pathway to switch between the two branches of the pathway. Among the downstream genes, ADH1 appears to be more responsive towards the KFB of the present study. Either ADH1 itself or the transcription factors that enhance its expression seem to be degraded by over-expressed AtSKIP11 (Appendix C, ESI†).
Conclusions and future prospects
The results of the analysis of transgenic plants manipulating the AtSKIP11 gene, which is endogenous to Arabidopsis and its antisense suppression sequence, suggest a role for the KFB AtSKIP11 in the Arabidopsis HPL branch of the oxylipin pathway. The release of a higher concentration of GLV under stressed conditions led us to conclude that gene expression for the GLV biosynthetic pathway is induced by signals from infection or stress. Therefore, the nature of stress signals and the mechanisms by which these signals give rise to the generation of the defence response remain elusive. Although many studies have shown encouraging results, most are limited to laboratory experiments that were performed using synthetic GLV.14 It is the need of the hour to devise new strategies and to design novel substrates for identifying the ‘missing link’ between the perception of a stress signal and the generation of a defence response. The proposed mechanism from our results suggests that the reduced expression of AtSKIP11 leads to decreased production of the SCFAtSKIP11 complex and thus less degradation of GLV-producing proteins and vice versa. So, AtSKIP11 may be one of the ‘missing links’ in the Arabidopsis thaliana HPL pathway and may serve as a molecular switch for controlling the production of GLV.Conflicts of interest
The authors declare no conflicts of interest.Abbreviations
| ALA | α-Linolenic acid. |
| DES | Divinyl ether synthase. |
| DGL | DONGLE. |
| GLV | Green leaf volatiles. |
| HIPVs | Herbivore-induced plant volatiles. |
| 13-HOT | 13-Hydroxytridecanoic acid. |
| 13-HPOD | (9Z,11E,13S)-13-Hydroperoxyoctadeca-9,11-dienoic acid |
| 13-HPOT | (9Z,11E,15Z)-13-Hydroperoxyoctadeca-9,11,15-trienoic acid. |
| JA | Jasmonic acid. |
| LA | Linoleic acid. |
| PUFAs | Polyunsaturated fatty acids |
| KFBs | Kelch-repeat containing F-box proteins |
| SPME | Solid phase micro-extraction. |
| MeJA | Methyl jasmonate. |
Acknowledgements
The project was partially supported by the UKM grant No. FRGS/1/2015/SG03/UKM/01/1.References
- E. Blée, Impact of phyto-oxylipins in plant defense, Trends Plant Sci., 2002, 7, 315–322 CrossRef.
- S.-C. Lee, M.-H. Lim, J. A. Kim, S.-I. Lee, J. S. Kim, M. Jin, S.-J. Kwon, J.-H. Mun, Y.-K. Kim and H. U. Kim, Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K oligo microarray, Mol. Cells, 2008, 26, 595–605 CAS.
- S. Allmann and I. T. Baldwin, Insects betray themselves in nature to predators by rapid isomerization of green leaf volatiles, Science, 2010, 329, 1075–1078 CrossRef CAS PubMed.
- I. T. Baldwin, R. Halitschke, A. Paschold, C. C. von Dahl and C. A. Preston, Volatile Signaling in Plant-Plant Interactions: “Talking Trees” in the Genomics Era, Science, 2006, 311, 812–815 CrossRef CAS PubMed.
- R. Liechti and E. E. Farmer, Jasmonate Biochemical Pathway, 2006 Search PubMed.
- K. Matsui, Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism, Curr. Opin. Plant Biol., 2006, 9, 274–280 CrossRef CAS PubMed.
- V. Balbi and A. Devoto, Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios, New Phytol., 2008, 177, 301–318 CrossRef CAS PubMed.
- A. Kessler, R. Halitschke and I. T. Baldwin, Silencing the Jasmonate Cascade: Induced Plant Defenses and Insect Populations, Science, 2004, 305, 665–668 CrossRef CAS PubMed.
- C. Wasternack and B. Hause, Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany, Ann. Bot., 2013, 111, 1021–1058 CrossRef CAS PubMed.
- K. Shiojiri, R. Ozawa, K. Matsui, K. Kishimoto, S. Kugimiya and J. Takabayashi, Role of the lipoxygenase/lyase pathway of host-food plants in the host searching behavior of two parasitoid species, Cotesia glomerata and Cotesia plutellae, J. Chem. Ecol., 2006, 32, 969–979 CrossRef CAS PubMed.
- C. Wasternack, Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development, Ann. Bot., 2007, 100, 681–697 CrossRef CAS PubMed.
- S. A. Christensen, A. Nemchenko, E. Borrego, I. Murray, I. S. Sobhy, L. Bosak, S. DeBlasio, M. Erb, C. A. Robert and K. A. Vaughn, The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack, Plant J., 2013, 74, 59–73 CrossRef CAS PubMed.
- X. Liu, F. Li, J. Tang, W. Wang, F. Zhang, G. Wang, J. Chu, C. Yan, T. Wang, C. Chu and C. Li, Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice, PLoS One, 2012, 7, e50089 CAS.
- M. N. Hassan, Z. Zainal and I. Ismail, Green leaf volatiles: biosynthesis, biological functions and their applications in biotechnology, Plant Biotechnol. J., 2015, 13, 727–739 CrossRef PubMed.
- M. L. Fauconnier, A. Mpambara, J. Delcarte, P. Jacques, P. Thonart and M. Marlier, Conversion of green note aldehydes into alcohols by yeast alcohol dehydrogenase, Biotechnol. Lett., 1999, 21, 629–633 CrossRef CAS.
- A. Hatanaka, The biogeneration of green odour by green leaves,, Phytochemistry, 1993, 34, 1201–1218 CrossRef CAS.
- E. A. Spyropoulou, H. L. Dekker, L. Steemers, J. H. van Maarseveen, C. G. de Koster, M. A. Haring, R. C. Schuurink and S. Allmann, Identification and Characterization of (3Z):(2E)-Hexenal Isomerases from Cucumber, Front Plant Sci., 2017, 8, 1342 CrossRef PubMed.
- J. Engelberth, H. T. Alborn, E. A. Schmelz and J. H. Tumlinson, Airborne signals prime plants against insect herbivore attack, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1781–1785 CrossRef CAS PubMed.
- J. Gershenzon, Plant volatiles carry both public and private messages, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 5257–5258 CrossRef CAS PubMed.
- R. M. Clark, G. Schweikert, C. Toomajian, S. Ossowski, G. Zeller, P. Shinn, N. Warthmann, T. T. Hu, G. Fu, D. A. Hinds, H. Chen, K. A. Frazer, D. H. Huson, B. Scholkopf, M. Nordborg, G. Ratsch, J. R. Ecker and D. Weigel, Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana, Science, 2007, 317, 338–342 CrossRef CAS PubMed.
- C. Bai, P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper and S. J. Elledge, SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box, Cell, 1996, 86, 263–274 CrossRef CAS PubMed.
- J. M. Gagne, B. P. Downes, S. H. Shiu, A. M. Durski and R. D. Vierstra, The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11519–11524 CrossRef CAS PubMed.
- Y. Sun, X. Zhou and H. Ma, Genome-wide Analysis of Kelch Repeat-containing F-box Family, J. Integr. Plant Biol., 2007, 49, 940–952 CrossRef CAS.
- M. N. U. Hassan, Z. Zainal and I. Ismail, Plant kelch containing F-box proteins, RSC Adv., 2015, 5, 42808–42814 RSC.
- C. H. Gray, L. C. McGarry, H. J. Spence, A. Riboldi-Tunnicliffe and B. W. Ozanne, Novel β-propeller of the BTB-Kelch protein Krp1 provides a binding site for Lasp-1 that is necessary for pseudopodial extension, J. Biol. Chem., 2009, 284, 30498–30507 CrossRef CAS PubMed.
- N. Schumann, A. Navarro-Quezada, K. Ullrich, C. Kuhl and M. Quint, Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats, Plant Physiol., 2011, 155, 835–850 CrossRef CAS PubMed.
- E. Lechner, P. Achard, A. Vansiri, T. Potuschak and P. Genschik, F-box proteins everywhere, Curr. Opin. Plant Biol., 2006, 9, 631–638 CrossRef CAS PubMed.
- D. Marino, N. Peeters and S. Rivas, Ubiquitination during plant immune signaling, Plant Physiol., 2012, 160, 15–27 CrossRef CAS PubMed.
- E. P. Risseeuw, T. E. Daskalchuk, T. W. Banks, E. Liu, J. Cotelesage, H. Hellmann, M. Estelle, D. E. Somers and W. L. Crosby, Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis, Plant J., 2003, 34, 753–767 CrossRef CAS PubMed.
- M. H. C. Othman, N. A. Hadi, Z. Zainal, C. J. Kiat, M. Naeem-ul-Hassan, C. R. C. M. Zain and I. Ismail, Expression profile of gene encoding Kelch repeat containing F-box protein (PmF-box1) in relation to the production of green leaf volatiles, Aust. J. Crop Sci., 2017, 11, 406–418 CrossRef.
- M. C. Gor, I. Ismail, W. A. W. Mustapha, Z. Zainal, N. M. Noor, R. Othman and Z. A. M. Hussein, Identification of cDNAs for jasmonic acid-responsive genes in Polygonum minus roots by suppression subtractive hybridization, Acta Physiol. Plant., 2011, 33, 283–294 CrossRef CAS.
- H. Kuroda, Y. Yanagawa, N. Takahashi, Y. Horii and M. Matsui, A comprehensive analysis of interaction and localization of Arabidopsis SKP1-like (ASK) and F-box (FBX) proteins,, PLoS One, 2012, 7, e50009 CAS.
- S. J. Clough and A. F. Bent, Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana, Plant J., 1998, 16, 735–743 CrossRef CAS PubMed.
- T. Czechowski, M. Stitt, T. Altmann, M. K. Udvardi and W.-R. Scheible, Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis, Plant Physiol., 2005, 139, 5–17 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- K. Matsui, K. Sugimoto, J. i. Mano, R. Ozawa and J. Takabayashi, Differential metabolisms of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements, PLoS One, 2012, 7, e36433 CAS.
- M. Heil and J. Ton, Long-distance signalling in plant defence, Trends Plant Sci., 2008, 13, 264–272 CrossRef CAS PubMed.
- K. Shiojiri, R. Ozawa, K. Matsui, M. W. Sabelis and J. Takabayashi, Intermittent exposure to traces of green leaf volatiles triggers a plant response, Sci. Rep., 2012, 2, 378 CrossRef PubMed.
- J. J. Salas, D. L. García-González and R. Aparicio, Volatile compound biosynthesis by green leaves from an Arabidopsis thaliana hydroperoxide lyase knockout mutant, J. Agric. Food Chem., 2006, 54, 8199–8205 CrossRef CAS PubMed.
- J. Shen, D. Tieman, J. B. Jones, M. G. Taylor, E. Schmelz, A. Huffaker, D. Bies, K. Chen and H. J. Klee, A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato, J. Exp. Bot., 2014, 65, 419–428 CrossRef CAS PubMed.
- A. Liavonchanka and I. Feussner, Lipoxygenases: occurrence, functions and catalysis, J. Plant Physiol., 2006, 163, 348–357 CrossRef CAS PubMed.
- T. Shao, Q. Qian, D. Tang, J. Chen, M. Li, Z. Cheng and Q. Luo, A novel gene IBF1 is required for the inhibition of brown pigment deposition in rice hull furrows, Theor. Appl. Genet., 2012, 125, 381–390 CrossRef CAS PubMed.
- M. Li, D. Tang, K. Wang, X. Wu, L. Lu, H. Yu, M. Gu, C. Yan and Z. Cheng, Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice, Plant Biotechnol. J., 2011, 9, 1002–1013 CrossRef CAS PubMed.
- H. J. Kim, Y.-H. Chiang, J. J. Kieber and G. E. Schaller, SCFKMD controls cytokinin signaling by regulating the degradation of type-B response regulators, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 10028–10033 CrossRef CAS PubMed.
- X. Zhang, M. Gou and C.-J. Liu, Arabidopsis Kelch Repeat F-Box Proteins Regulate Phenylpropanoid Biosynthesis via Controlling the Turnover of Phenylalanine Ammonia-Lyase, Plant Cell, 2013, 25, 4994–5010 CrossRef CAS PubMed.
- X. Zhang, M. Guo, C. Guo, H. Yang and C.-J. Liu, Down-regulation of the kelch domain-containing F-box protein in Arabidopsis enhances the production of (poly) phenols and tolerance to ultraviolet-radiation, Plant Physiol., 2014, 167, 337–350 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09895b |
| This journal is © The Royal Society of Chemistry 2017 |

