 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNA-based nanoscale walking devices and their applications
Yikang Xing†
,
Bing Liu†
,
Jie Chao
* and
Lianhui Wang
 *
*
Institute of Advanced Materials (IAM), Jiangsu National Syngerstic Innovation Center for Advanced Materials (SICAM), Nanjing University of Posts & Telecommunications, Nanjing 210023, China. E-mail: iamjchao@njupt.edu.cn; iamlhwang@njupt.edu.cn
First published on 10th October 2017
Abstract
The rapidly growing field of nanodevices is attracting more and more attention on account of their ingenious design and broad application. Taking advantage of the sequence specificity and excellent programmability of DNA molecules, numerous DNA devices with multiple functions have been designed and fabricated such as tweezers, gears, cranes and so on. DNA walking devices are the most sophisticated ones either in design or fabrication. They exhibit a powerful ability to actuate walkers walking on their tracks. Herein we review DNA walking devices based on walking functions including unipedal, bipedal, multipedal, and other novel walking devices, as well as their applications.
1. Introduction
The field of nanodevices is one of the more challenging frontiers at present. It is important for us to understand the law of energy conversion at the nanoscale, as well as the interaction and regulation mechanism between biological molecules.1–3 Many scientists have paid extensive attention to this field in recent years.4–11 As DNA owns the distinguished principle of Watson–Crick base pairing and special physical character, it has been nominated as one of the most versatile and robust building materials, not only to construct controllable nanoarchitectures in nanoscale resolution with excellent sequence programmability and structural rigidity, but also to provide memory components in dynamic structures with high fidelity. DNA origami has been proven as one of the most attractive breakthroughs offering a compelling approach leading toward the precise design of nanostructures with predestinate addressability at sub-10 nm resolution. Since then, numerous DNA origami nanostructures with different shapes have been constructed including rectangles, stars, smiley faces, maps, dolphins and three-dimensional (3D) structures such as honeycomb lattices, nanotubes, boxes and nanoflasks.12–19 Inspired by kinesin movement along a microtubule to perform intracellular transport in biological systems,20–23 researchers have extended the area of DNA nanotechnology from structure to function, that is, converting static DNA nanostructures to dynamic, functional DNA nanodevices such as tweezers, gears, cranes and so on.21,24–26 DNA walking devices are the most sophisticated ones either in design or fabrication.27–33 They have exhibited a powerful ability to actuate walkers walking on their tracks from starting point to destination.34–39Generally, the concept of DNA walking devices refers to a walking body such as a DNA duplex, nanoparticle or nanorod, and a track in multiple dimensions. For synthetic DNA walking devices, the walking process is usually enabled by the combination of strand hybridization,40–43 enzymatic reaction44,45 and environmental stimuli.46–53 Although DNA walking devices remain a mystery with respect to their low fidelity and slow rates, they still have great potential for various applications, such as sensors54–56 and switchable carriers.57–61 Generally, the ample evidence presented enables us to reasonably conclude that the enhancement of complexity regarding scale and speed is an area to be addressed. It can be believed that DNA walking devices will play a significant role in delivering targeted drugs into a patient’s body to cure corresponding diseases in future.
On the basis of summarizing the previous work and combining the latest developmental achievements, herein we review the construction of nanostructures including unipedal, bipedal, multipedal, and other novel walking devices, and the applications of DNA nanomachines are also summarized.
2. Unipedal walking devices
Unipedal walking devices refer to one “foot” travel on a track from a morphological perspective. Single-foot DNA walking devices are probably the simplest formation because a single DNA strand plays the leading role. Wickham et al.62 used a single strand of DNA as a walking device to move autonomously along the full length of a track (Fig. 1a). Single-stranded stators on a DNA origami tile constitute a 100 nm-long DNA track. With the help of a nicking enzyme, the unipedal walking device took up to 16 consecutive steps by branch migration. Later, Endo’s group63 developed a DNA nanodevice with light-fuelled mechanical movements (Fig. 1c). These pyrene-modified walking strands migrated from one cleaved stator to the next on a similar 2D DNA origami tile. It is good to be reminded that these two stepwise walking systems were directly observed by high-speed AFM in real-time, which tended to facilitate the development of systems that could be programmed and routed by instructions encoded in the nucleotide sequences of the track and walking strands.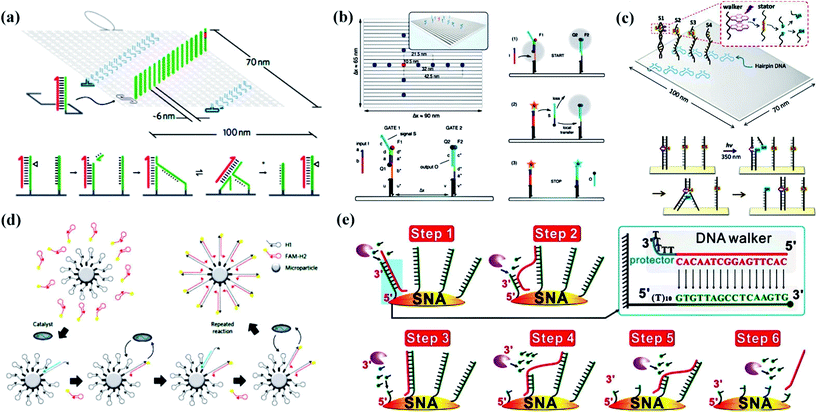 | ||
| Fig. 1 Unipedal DNA walking devices. (a) The layout of the DNA origami tile and walker mechanism.62 A nicking enzyme cuts the motor-bound stator revealing a toehold at the 3′ end of the motor (magenta) that facilitates the transfer of the motor to the adjacent intact stator by branch migration. (b) An illustration of a rectangular single-layered DNA origami platform and the immobilized DNA strand displacement cascade.64 (c) An illustration of a rectangular DNA tile and light-driven walking system.63 (d) A schematic of a catalytic hairpin assembly reaction on a microparticle surface shows how fluorescent FAM-H2 (red hairpins) are catalytically hybridized to surface-immobilized H1 (grey hairpins).65 (e) Schematics for Exo III-powered stochastic DNA walkers that move on SNA surfaces.66 Adapted with permission from ref. 62 and 65, Copyright 2011 and 2016 Nature Publishing Group, ref. 63 and 64, Copyright 2015 and 2014 American Chemical Society, and ref. 66, Copyright 2017 Wiley-VCH Verlag GmbH & Co. KGaA Weinheim. | ||
Generally, these walking nanodevices are mostly fueled by enzymatic reactions or powered by light. The external addition of counterpart strands is also an important class of reaction utilized in dynamic DNA nanotechnology. Simmel and his coworkers64 utilized walking devices based on origami to investigate the effect of colocalization on the performance of DNA strand displacement (DSD) reactions (Fig. 1b). Devoted to immobilizing a two-stage DSD reaction cascade, the system was composed of “sender” and “receiver” gates in a DNA origami platform, in which the additional DNA input strand displaced a signal strand from the sender gate and then transferred to the receiver gate.
In fact, considerable research efforts on single-foot walking devices have been devoted to the development from a 2D track to 3D track. Originally, Jung et al.65 reported a stochastic DNA walking device that traversed a microparticle surface in a creative way (Fig. 1d). The DNA walking device utilized hybridization energy to drive walking on a 3D track, which was the DNA-coated microparticle surface. It showed that the walking traversal could cover more than 30 continuous steps entirely due to autonomous decisions made by the walking device. This system shines a light on the future development of DNA walking devices that travel on the nanoparticle, and has been confirmed by other studies. Recently, Qu and his coworkers66 constructed a single-foot DNA walking device consisting of a 21-base sequence with five consecutive thymines (oligo-T) (Fig. 1e). The 3D track refers to 15 nm-sized gold nanoparticles (AuNPs) decorated with thiol-tagged oligonucleotides. Moreover, the movement avoided randomly hydrolyzing RNA sequences owing to exonuclease III, and the system has great potential for analytical and diagnostic applications.
3. Bipedal walking devices
Compared to unipedal walking devices, it is better to link bipedal walking devices to pincers because two “feet” drive walking sideways. For bipedal walking devices, three substantial problems must be considered. Firstly, we have to take measures to work out the challenge that some random thermal motions always hinder bipedal walking devices from moving in a synchronized way. Omabegho et al.67 reported a bipedal DNA Brownian motor with coordinated legs to cyclically catalyze the hybridization of metastable DNA fuel strands (Fig. 2a). This process led to a chemically ratcheted movement along a track with directional double-crossover (DX). The successive transition and full walking cycle helped to uncover principles behind the design of unidirectional devices that could function without intervention.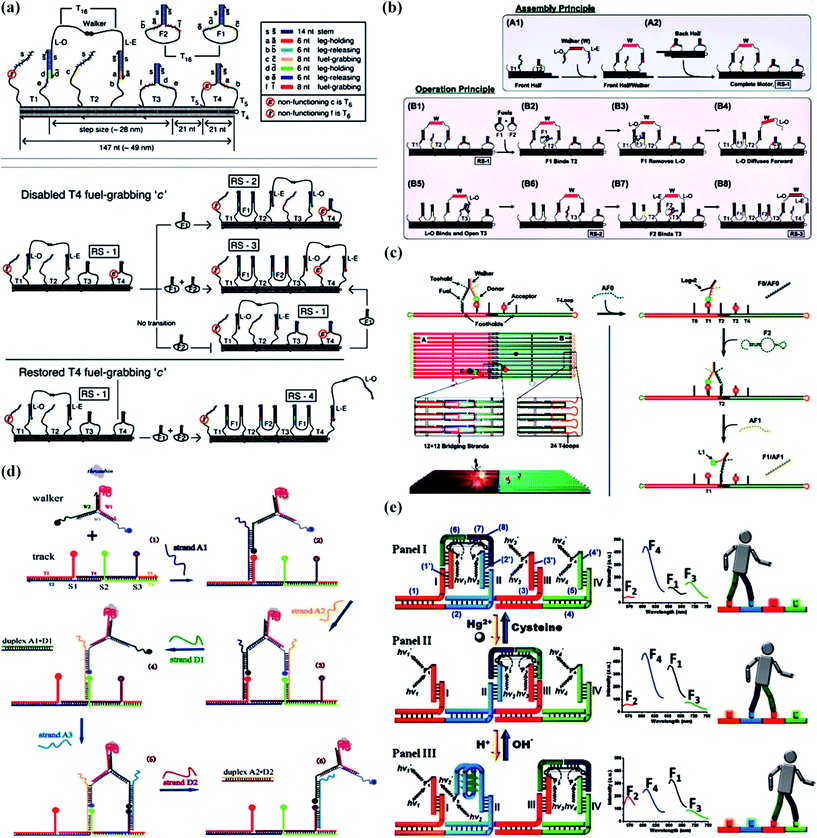 | ||
| Fig. 2 Bipedal DNA walking devices. (a) An illustration of the DX track structure with the walker on it.67 (b) A schematic representation of the motor’s assembly and principle of operation.68 (c) The design of the origami track and the bipedal motor, and the motor operation principle.69 (d) A strategy to speed up bipedal DNA walking devices through the use of DNA catalysts.70 (e) A bipedal DNA walker powered by H+/OH− or Hg2+ ions/cysteine from its environment.71 Adapted with permission from ref. 67, Copyright 2009 American Association for the Advancement of Science, ref. 69, Copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA Weinheim, ref. 68 and 70, Copyright 2012 American Chemical Society, and ref. 71, Copyright 2011 American Chemical Society. | ||
Secondly, it is known that gel electrophoresis, fluorescence microscopy, atomic force microscopy (AFM), and transmission electron microscopy (TEM) techniques are widely applied in studying dynamic movement, but it is difficult to measure time-dependent conformational changes and protect samples from damage. To address this problem, Nir’s group68 presented a detailed single-molecule fluorescence study based on an autonomous bipedal DNA walking device (Fig. 2b). Their work demonstrated that single-molecule fluorescence constituted an excellent tool for chaperoning the development of DNA-based technology, which was used appropriately and efficiently in another paper reported by them three years later.69 The single-molecule fluorescence characterization showed the successful operation of a dynamic DNA walking device, striding back and forth between two origami tiles (Fig. 2c). This bipedal DNA walking device system was programmed by sequential interaction with “fuel” and “antifuel” DNA strands that resulted in movement from one origami tile to another and back again. As the track consisted of two different DNA origami tiles, this study has contributed to disclosing secrets behind the design of DNA walking devices that could stride over micrometer distances.
The third challenge is the speed of the walking devices. In 2012, Wang et al.70 elaborated a strategy to accelerate the DNA walking process on a three foothold molecular track through the use of a DNA catalyst (Fig. 2d). The advantage of using strand-displacement-based catalysts over enzymes is that the former generally have fewer sequence constraints and are less likely to be influenced by environmental conditions, such as pH, temperature, and salt concentrations. The locomotion is about one order of magnitude faster than previous hybridization-based bipedal walking devices.
In addition to DNA hybridization, light powering and enzyme triggering, DNA walking devices can also be activated by H+/OH− or Hg2+ ions/cysteine which was reported by the group of Willner (Fig. 2e).71 Their principle for operating a bipedal walking device was aimed at constructing four nucleic acid footholds on a DNA template and building the signal-triggered motility of a “walker” nucleic acid tethered to two of the footholds in each state of the machine. Importantly, it was a reversible bipedal DNA walking device because one sequence of the complementary nucleic acids responded to Hg2+ ions, which detached from the former foothold and attached to the next, and the other responded to H+ ions, which led to the formation of an i-motif structure. Similarly, the backward movement was activated by OH− ions and cysteine that destroys the i-motif or thymine–Hg2+–thymine complexes.
4. Multipedal walking devices
In dynamic DNA nanotechnology, three-feet DNA walking devices have been constructed infrequently. However, multipedal walking devices comprising an anisotropic nanoparticle as the body and discrete DNA or RNA strands as the feet have been widely studied. Consequently, this suggests that multipedal nanodevices with longer distance and higher fidelity and rates must predominate in the future.In 2010, Seeman’s group72 designed a proximity-based programmable DNA nanoscale assembly line to satisfy a DNA walking device assembling cargos as it travelled (Fig. 3a). The core part of the system was the walker structure, containing three hands and four feet and moving along the track to collect and transfer different cargos (AuNPs). Coincidentally, another three-leg DNA walking device was built by Yan and his colleagues at the same time.73 The molecular spider was made up of one streptavidin molecule as an inert body and three 8–17 bp DNA enzymes as catalytic legs that cleaved oligonucleotide substrates at single ribose sites into shorter strands (Fig. 3c). However, the spider DNA walking device was not fast, efficient, or powerful enough due to the core principle of the catalytic reaction of DNAzymes. In order to speed up the walking process, Salaita’s group74 successfully designed relatively fast DNA walking devices, DNA-modified particles (Fig. 3b). The maximum speed was three orders of magnitude greater than the maximum for conventional DNA motors. RNase H selectively hydrolysed the hybridized RNA modified on a surface to hybridize the DNA-coated spherical particles, which resulted in a rolling movement rather than walking. Also, the nanodevice could travel in a self-avoiding fashion and anisotropic particles could travel linearly without a track or external force.
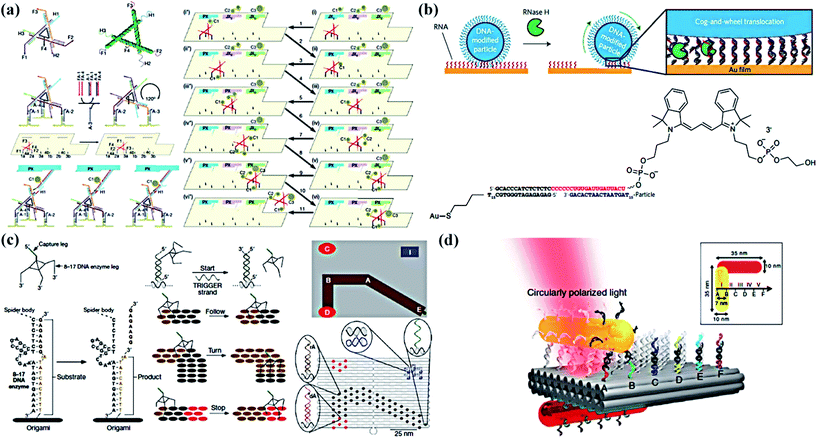 | ||
| Fig. 3 Multipedal DNA walking devices. (a) Details of the three-feet walking device, the molecular assembly line and its operation.72 (b) The approach used to generate RNA-fuelled, enzyme-catalysed autonomous DNA motors.74 (c) A deoxyribozyme-based molecular walker called a molecular spider and the origami prescriptive landscape.73 (d) A schematic of the plasmonic multipeda walking device.75 Adapted with permission from ref. 72 and 73, Copyright 2010 Nature Publishing Group, and ref. 74 and 75, Copyright 2016 and 2015 Nature Publishing Group. | ||
Furthermore, in answer to the formidable challenge of precise transport, Liu et al.75 designed a gold nanorod (AuNR) that can perform stepwise walking directionally and progressively on DNA origami (Fig. 3d). In the active plasmonic system, two AuNRs assembled perpendicularly to one another were placed on two opposite surfaces of a 2D rectangular DNA origami platform. The walking process triggered a series of conformational changes, thus giving rise to immediate spectral response changes that can be read out optically. Therefore, its own walking directions and consecutive steps could be optically reported in situ. Moreover, stepwise walking of the walking device on a 3D origami platform, triangular prism origami, was examined.
5. Other novel walking devices
To date, researchers have paid more attention to individual walking device behavior. However, few attempts have been made to implement multiple artificial walking devices which have the advantage of walking collectively on the same track. In another report from Liu’s group,76 they demonstrated a plasmonic walking device couple system involving two AuNR walking devices which could perform simultaneous walking along the same DNA origami track (Fig. 4a). It was crucial that the number of walking devices was related to the optical response of the system which probably made a contribution to developing advanced artificial nanomachinery with multiple walking devices in concert.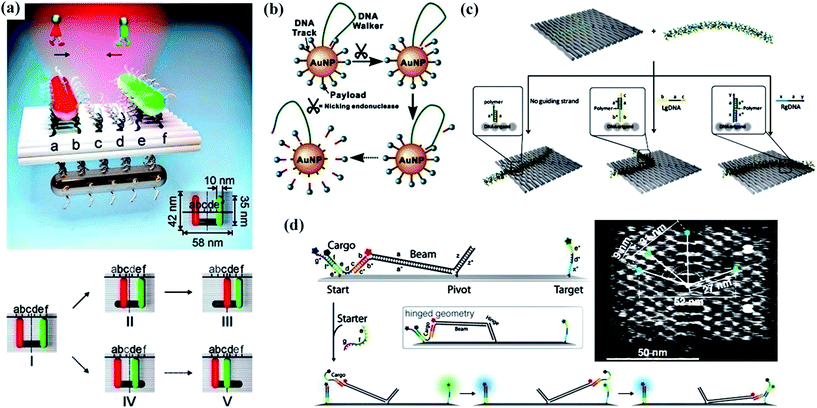 | ||
| Fig. 4 Several novel walking devices. (a) A schematic of the plasmonic walker couple and stepwise walking of the individual walkers.76 (b) A schematic illustrating the principle of the 3D DNA nanomachine constructed by co-conjugating DNA walker and DNA track (substrate) components on a single AuNP.77 (c) An illustration of the control of the conformation of a polymer on predesigned ssDNA tracks of staple strands extending from rectangular DNA origami.79 (d) A schematic side view of a DNA origami platform with a rigid tether fixed in the center and the mechanism of cargo transport and fluorescence labeling.78 Adapted with permission from ref. 76 and 78, Copyright 2015 American Chemical Society, and ref. 77 and 79, Copyright 2016 American Chemical Society. | ||
Most frequently, the autonomous movements of DNA walking devices depend on the cleavage of endonucleases or deoxyribozymes, or photoirradiation, resulting in the relatively slow kinetics and limited steps of movement. Li and co-workers77 creatively built an enzyme-powered 3D DNA nanomachine to execute walking, resolving the task of releasing payloads (Fig. 4b). A DNA-functionalized gold nanoparticle (DNA-AuNP) served as the 3D track, while the long DNA strand which was inserted into the AuNP acted as the walking device. Powered by a nicking endonuclease, the DNA walking device cleaved multiple specific substrates on the track and plentiful payload fell off.
As pointed out in the previous reports, much effort on the movement of DNA walking devices had been emphasized particularly on utilizing a flexible and hinged tether with the help of environmental stimuli in order to achieve fast and efficient transport of molecular cargo along tracks. On the contrary, Simmel and his co-workers78 developed a rigid tether that can rotate to mediate cargo transfer from the start to a target site on the DNA origami structure, which presented very fast, robust and more efficient motion over long distances (Fig. 4d). Gothelf et al.79 redefined DNA walking devices because they described the development of a nanomechanical device that allowed switching of the position of a single-molecule conjugated polymer (Fig. 4c). The DNA polymer conjugate, functionalized with short single-stranded (ss) DNA strands extending from the backbone, could be aligned on DNA origami in three well-defined geometries (straight line, left-turned, and right-turned patterns).
Therefore, it shows that these novel walking devices have been developed on the strength of increasing the number of walking devices on the same track, constructing 3D nanomachine systems, and replacing the walking process with rolling, switching and pivoting. Although the theories of these nanodevices still focus on hybridization, enzymatic reaction and environmental stimuli, these systems always differ from the traditional systems.
6. Applications of DNA walking devices
DNA nanomachines that walk by converting chemical entropy into controlled motion could be of good use in applications such as biosensing,80–82 molecular computing83–85 and drug delivery.86–88 It is nucleic acids that are adept in molecular computation, especially constructing numerous molecular logic gates which have been combined to form various molecular circuits.89–92 In addition, some specific 3D nanostructures, including DNA nanotubes, DNA tetrahedra and DNA origami structures, reveal the promising features to be the use as universal nanocarriers for cargo or drug delivery.Recently, Ding and his coworkers93 designed a target-switched DNA nanotweezer for AND logic gate operation and sensitive enzyme-free biosensing of microRNAs (Fig. 5a). An arched structure as the set strand for the target inputs and two split G-rich DNAs at the termini of the two arms for the signal output play significant roles in the building of the nanotweezer. When a catalytic hairpin assembles, numerous open nanotweezers switch to a closed state owing to these binary target inputs. This logical process directly results in the formation of a proximity-dependent DNAzyme in the presence of hemin to produce a highly sensitive biosensing system.
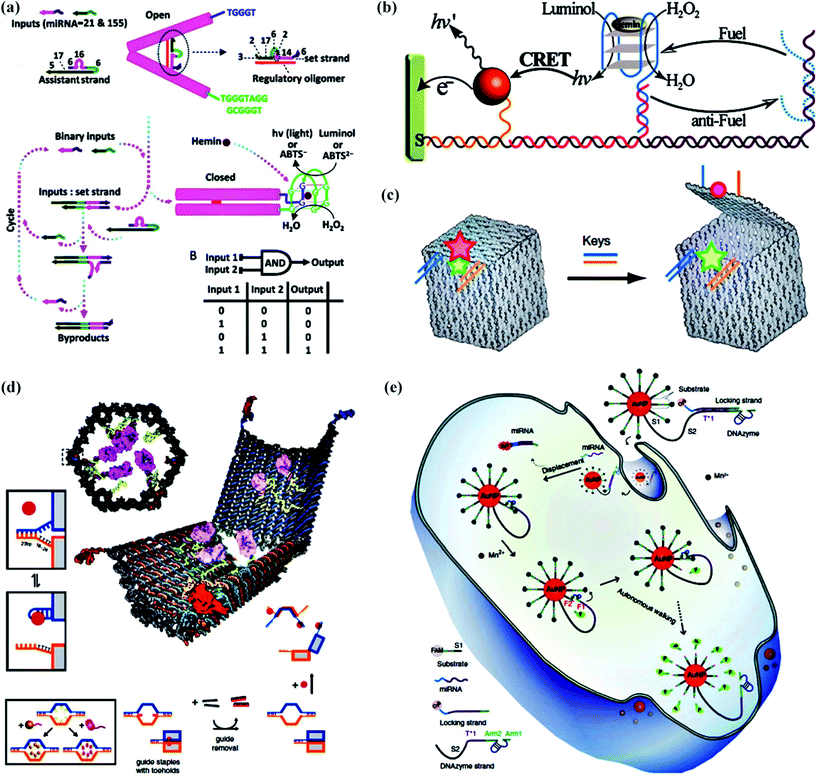 | ||
| Fig. 5 Applications of DNA walking devices. (a) A target-switched DNA nanotweezer for AND logic gate operation and sensitive enzyme-free biosensing of microRNAs.93 (b) The switching photonic and electrochemical functions of a DNAzyme by DNA walking devices.94 (c) A nanoscale DNA box with a controllable lid used to combine a diagnostic sensor of complex signals.95 (d) A logic-gated nanorobot for targeted transport of molecular payloads.96 (e) Intracellular operation of a DNAzyme motor initiated by a specific miRNA in living cells.97 Adapted with permission from ref. 93 and 94, Copyright 2016 and 2013 American Chemical Society, ref. 95 and 97, Copyright 2009 and 2017 Nature Publishing Group, and ref. 96, Copyright 2012 American Association for the Advancement of Science. | ||
In 2013, Willner’s group94 demonstrated the powerful biosensing function of DNA walking devices (Fig. 5b). They described a DNA walking device assembled on nucleic acid scaffolds and triggered by fuel/antifuel strands. The walking device successfully switched formation or dissociation of the hemin/G-quadruplex DNAzyme on the DNA scaffolds, which enabled the chemiluminescence, chemiluminescence resonance energy transfer (CRET), or the electrochemical or photoelectrochemical transduction of the walking processes.
With regards to drug delivery, Gothelf and Kjems’s group95 spared no effort to make a DNA origami box with a controllable lid (Fig. 5c). The lid of this DNA box had a dual lock-key system composed of DNA duplexes with sticky-end extensions as the “toehold” which will be displaced by introducing “key” oligonucleotides and then the lid will be opened. With the help of efficient fluorescence resonance energy transfer (FRET), researchers effectively monitored the operation of closing and opening. This profound work in building a DNA box has the potential to combine a diagnostic sensor of complex signals with the controlled release or access of a payload. Douglas’ group96 took a giant step forward to realize the biological application of a DNA origami nanorobot in the form of a hexagonal barrel with dimensions of 35 nm × 35 nm × 45 nm (Fig. 5d). The barrel consisted of two domains that were covalently attached in the rear by single-stranded scaffold hinges, and could be noncovalently fastened in the front by staples modified with DNA aptamer-based locks. The aptamer-encoded locks function as an AND logic gate responding to molecular inputs (keys) which are expressed by cells. Plenty of practical experiments have been carried out, demonstrating that the robot can inspire new designs with different selectivities and biologically active payloads for cell-targeting tasks.
It should be pointed out that much work has focused on constructing various synthetic DNA motors to mimic the functions of protein motors. All of this effort aims ultimately at the operation of synthetic DNA motors in living cells. To our delight, Peng and his coworkers97 invented a DNAzyme motor that operated in living cells with a response to a specific intracellular target (Fig. 5e). Their DNA walking device referred to a 20 nm AuNP functionalized with hundreds of substrate strands, and dozens of DNAzyme molecules were silenced by a locking strand. After entering a living cell, the target miRNA was hybridized to the locking strand and released the locking strand from the DNAzyme through a strand displacement reaction, which drove the DNAzyme motor to traverse along the AuNP surface. The process of this stepwise walking inside the cell was monitored in real-time through the fluorescence of FAM.
7. Summary and perspectives
From the discussed research above, the complexity of DNA walking devices has been enhanced from one “foot” travel to multipedal movement, and even to complex assemblies involving tweezers,98–103 cranes,104,105 catenanes,106 rotaxanes,107,108 metronomes,109 gears110 and spiders. The track has also been promoted from individual DNA strands to 2D or 3D forms.111–116 Despite the fact that these outstanding DNA nanomachines have been proven to possess exquisite programmability and addressability with the help of FRET,117,118 electrophoretic experiments, or fast AFM measurements on surfaces, there still remain challenges for future development, such as the scale and speed. The good news is that researchers have tried their best to design and manipulate more and faster walking devices in the form of rotating or rolling and larger nanostructures including DNA nanotubes, DNA tetrahedra and DNA origami boxes. Suitable applications for DNA walking devices have been a matter of continuous exploration, including drug delivery, mediating organic synthesis,119 programmed control of RNA expression and computation.It is difficult to predict the practical application of these smart walking devices, but just as Feringa, one of the 2016 Nobel Prize winners for chemistry, said “People were saying, why do we need a flying machine? Now we have a Boeing 747 and an Airbus. That’s a little bit how I feel. The opportunities are great.” It is deserved to imagine that walking devices could be used as tiny robots in people’s bodies to deliver drugs or detect cancerous cells, or as smart materials that could adapt or change depending on external signals. Anyway, it’s time to pay more attention to develop DNA walking devices because they are coming.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Ministry of Science and Technology of China (2017YFA0205302), Sci-tech Support Plan of Jiangsu Province (BE2014719), Program for Changjiang Scholars and Innovative Research Team in University (IRT_15R37), Natural Science Foundation of Jiangsu Province (BK20151504), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, YX03001), Mega-projects of Science and Technology Research (AWS13C007) and NUPTSF (Grant no. 214175).References
- R. D. Astumian, Science, 1997, 276, 917–922 CrossRef CAS PubMed.
- C. Bustamante, D. Keller and G. Oster, Acc. Chem. Res., 2001, 34, 412–420 CrossRef CAS PubMed.
- E. R. Kay, D. A. Leigh and F. Zerbetto, Angew. Chem., Int. Ed., 2007, 46, 72–191 CrossRef CAS PubMed.
- C. Mao, W. Sun, Z. Shen and N. C. Seeman, Nature, 1999, 397, 144–146 CrossRef CAS PubMed.
- S. K. Kufer, E. M. Puchner, H. Gumpp, T. Liedl and H. E. Gaub, Science, 2008, 319, 594–596 CrossRef CAS PubMed.
- W. B. Sherman and N. C. Seeman, Nano Lett., 2004, 4, 1203–1207 CrossRef CAS.
- J. S. Shin and N. A. Pierce, J. Am. Chem. Soc., 2004, 126, 10834 CrossRef CAS PubMed.
- Y. Tian, Y. He, Y. Chen, P. Yin and C. Mao, Angew. Chem., 2005, 117, 4429–4432 CrossRef.
- R. A. Muscat, J. Bath and A. J. Turberfield, Small, 2012, 8, 3593–3597 CrossRef CAS PubMed.
- R. Hou, I. Y. Loh, H. Li and Z. Wang, Phys. Rev. Appl., 2017, 7, 024020 CrossRef.
- J. Pan, T. G. Cha, F. Li, H. Chen, N. A. Bragg and J. H. Choi, Sci. Adv., 2017, 3, e1601600 CrossRef PubMed.
- P. W. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- L. Qian, Y. Wang, Z. Zhang, J. Zhao, D. Pan, Y. Zhang, Q. Liu, C. Fan, J. Hu and L. He, Chin. Sci. Bull., 2006, 51, 2973–2976 CrossRef CAS.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, A. Lindthomsen, W. Mamdouh, K. V. Gothelf, F. Besenbacher and J. Kjems, ACS Nano, 2008, 2, 1213 CrossRef CAS PubMed.
- S. M. Douglas, H. Dietz, T. Liedl, B. Hogberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 CrossRef CAS PubMed.
- S. M. Douglas, J. J. Chou and W. M. Shih, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 6644–6648 CrossRef CAS PubMed.
- D. Han, S. Pal, J. Nangreave, Z. Deng, Y. Liu and H. Yan, Science, 2011, 332, 342–346 CrossRef CAS PubMed.
- K. E. Dunn, F. Dannenberg, T. E. Ouldridge, M. Kwiatkowska, A. J. Turberfield and J. Bath, Nature, 2015, 525, 82–86 CrossRef CAS PubMed.
- Y. He, T. Ye, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Nature, 2008, 452, 198–201 CrossRef CAS PubMed.
- M. Schliwa and G. Woehlke, Nature, 2003, 422, 759–765 CrossRef CAS PubMed.
- H. Liu and D. Liu, Chem. Commun., 2009, 2625–2636, 10.1039/b822719e.
- R. D. Vale and R. A. Milligan, Science, 2000, 288, 88 CrossRef CAS PubMed.
- R. D. Vale, Cell, 2003, 112, 467–480 CrossRef CAS PubMed.
- M. J. Campolongo, S. J. Tan, J. Xu and D. Luo, Adv. Drug Delivery Rev., 2010, 62, 606–616 CrossRef CAS PubMed.
- Y. Lu and J. Liu, Curr. Opin. Biotechnol., 2006, 17, 580–588 CrossRef CAS PubMed.
- O. I. Wilner and I. Willner, Chem. Rev., 2012, 112, 2528–2556 CrossRef CAS PubMed.
- J. Cheng, S. Sreelatha, I. Y. Loh, M. Liu and Z. Wang, Methods, 2014, 67, 227–233 CrossRef CAS PubMed.
- J. Cheng, S. Sreelatha, R. Hou, A. Efremov, R. Liu, J. R. van der Maarel and Z. Wang, Phys. Rev. Lett., 2012, 109, 238104 CrossRef PubMed.
- M. You, F. Huang, Z. Chen, R. W. Wang and W. Tan, ACS Nano, 2011, 6, 7935–7941 CrossRef PubMed.
- S. J. Green, J. Bath and A. J. Turberfield, Phys. Rev. Lett., 2008, 101, 238101 CrossRef CAS PubMed.
- P. Yin, A. J. Turberfield and J. H. Reif, Lect Notes Comput. Sci., 2004, 41, 410–425 Search PubMed.
- J. Bath, S. J. Green, K. E. Allen and A. J. Turberfield, Small, 2009, 5, 1513–1516 CrossRef CAS PubMed.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- S. F. Wickham, J. Bath, Y. Katsuda, M. Endo, K. Hidaka, H. Sugiyama and A. J. Turberfield, Nat. Nanotechnol., 2012, 7, 169–173 CrossRef CAS PubMed.
- R. A. Muscat, J. Bath and A. J. Turberfield, Nano Lett., 2011, 11, 982–987 CrossRef CAS PubMed.
- R. Pei, S. K. Taylor, D. Stefanovic, S. Rudchenko, T. E. Mitchell and M. N. Stojanovic, J. Am. Chem. Soc., 2006, 128, 12693 CrossRef CAS PubMed.
- F. C. Simmel, ChemPhysChem, 2009, 10, 2593–2597 CrossRef CAS PubMed.
- Z. Wang, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17921–17926 CrossRef CAS PubMed.
- P. Yin, H. Yan, X. G. Daniell, A. J. Turberfield and J. H. Reif, Angew. Chem., Int. Ed., 2004, 43, 4906–4911 CrossRef CAS PubMed.
- S. Venkataraman, R. M. Dirks, P. W. Rothemund, E. Winfree and N. A. Pierce, Nat. Nanotechnol., 2007, 2, 490–494 CrossRef PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- X. Olson, S. Kotani, B. Yurke, E. Graugnard and W. L. Hughes, J. Phys. Chem. B, 2017, 121, 2594–2602 CrossRef CAS PubMed.
- P. Gong, K. Wang, Y. Liu, K. Shepard and R. Levicky, J. Am. Chem. Soc., 2010, 132, 9663–9671 CrossRef CAS PubMed.
- J. Bath, S. J. Green and A. J. Turberfield, Angew. Chem., 2005, 117, 4432–4435 CrossRef.
- J. H. Reif and S. Sahu, Theor. Comput. Sci., 2009, 410, 1428–1439 CrossRef.
- C. Wang, J. Ren and X. Qu, Chem. Commun., 2011, 47, 1428–1430 RSC.
- O. Mendoza, J. L. Mergny, J. P. Aime and J. Elezgaray, Nano Lett., 2016, 16, 624–628 CrossRef CAS PubMed.
- L. K. Bruetzel, T. Gerling, S. M. Sedlak, P. U. Walker, W. Zheng, H. Dietz and J. Lipfert, Nano Lett., 2016, 16, 4871–4879 CrossRef CAS PubMed.
- D. Liu and S. Balasubramanian, Angew. Chem., Int. Ed., 2003, 42, 5734–5736 CrossRef CAS PubMed.
- M. You, Y. Chen, X. Zhang, H. Liu, R. Wang, K. Wang, K. R. Williams and W. Tan, Angew. Chem., Int. Ed., 2012, 51, 2457–2460 CrossRef CAS PubMed.
- I. Y. Loh, J. Cheng, S. R. Tee, A. Efremov and Z. Wang, ACS Nano, 2014, 8, 10293 CrossRef CAS PubMed.
- J. Elbaz, B. Shlyahovsky and I. Willner, Chem. Commun., 2008, 13, 1569–1571 RSC.
- M. Liu, R. Hou, J. Cheng, I. Y. Loh, S. Sreelatha, J. N. Tey, J. Wei and Z. Wang, ACS Nano, 2014, 8, 1792–1803 CrossRef CAS PubMed.
- C. Zhu, Y. Wen, D. Li, L. Wang, S. Song, C. Fan and I. Willner, Chemistry, 2009, 15, 11898–11903 CrossRef CAS PubMed.
- E. Johann, M. Michal, S. Bella and W. Itamar, Chemistry, 2009, 15, 3411–3418 CrossRef PubMed.
- C. Zhu, Y. Wen, H. Peng, Y. Long, Y. He, Q. Huang, D. Li and C. Fan, Anal. Bioanal. Chem., 2011, 399, 3459–3464 CrossRef CAS PubMed.
- S. Ko, H. Liu, Y. Chen and C. Mao, Biomacromolecules, 2008, 9, 3039–3043 CrossRef CAS PubMed.
- A. S. Walsh, H. Yin, C. M. Erben, M. J. Wood and A. J. Turberfield, ACS Nano, 2011, 5, 5427–5432 CrossRef CAS PubMed.
- J. Li, H. Pei, B. Zhu, L. Liang, M. Wei, Y. He, N. Chen, D. Li, Q. Huang and C. Fan, ACS Nano, 2011, 5, 8783 CrossRef CAS PubMed.
- H. Lee, A. K. Lytton-Jean, Y. Chen, K. T. Love, A. I. Park, E. D. Karagiannis, A. Sehgal, W. Querbes, C. S. Zurenko, M. Jayaraman, C. G. Peng, K. Charisse, A. Borodovsky, M. Manoharan, J. S. Donahoe, J. Truelove, M. Nahrendorf, R. Langer and D. G. Anderson, Nat. Nanotechnol., 2012, 7, 389–393 CrossRef CAS PubMed.
- J. Fu and H. Yan, Nat. Biotechnol., 2012, 30, 407 CrossRef CAS PubMed.
- S. F. Wickham, M. Endo, Y. Katsuda, K. Hidaka, J. Bath, H. Sugiyama and A. J. Turberfield, Nat. Nanotechnol., 2011, 6, 166–169 CrossRef CAS PubMed.
- Y. Yang, M. A. Goetzfried, K. Hidaka, M. You, W. Tan, H. Sugiyama and M. Endo, Nano Lett., 2015, 15, 6672–6676 CrossRef CAS PubMed.
- M. Teichmann, E. Kopperger and F. C. Simmel, ACS Nano, 2014, 8, 8487 CrossRef CAS PubMed.
- C. Jung, P. B. Allen and A. D. Ellington, Nat. Nanotechnol., 2016, 11, 157–163 CrossRef CAS PubMed.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 CrossRef CAS PubMed.
- T. Omabegho, R. Sha and N. C. Seeman, Science, 2009, 324, 67–71 CrossRef CAS PubMed.
- R. Masoud, R. Tsukanov, T. E. Tomov, N. Plavner, M. Liber and E. Nir, ACS Nano, 2012, 6, 6272 CrossRef CAS PubMed.
- M. Liber, T. E. Tomov, R. Tsukanov, Y. Berger and E. Nir, Small, 2015, 11, 568–575 CrossRef CAS PubMed.
- C. Wang, Y. Tao, G. Song, J. Ren and X. Qu, Langmuir, 2012, 28, 14829–14837 CrossRef CAS PubMed.
- Z. G. Wang, J. Elbaz and I. Willner, Nano Lett., 2011, 11, 304–309 CrossRef CAS PubMed.
- H. Gu, J. Chao, S. J. Xiao and N. C. Seeman, Nature, 2010, 465, 202–205 CrossRef CAS PubMed.
- K. Lund, A. J. Manzo, N. Dabby, N. Michelotti, A. Johnson-Buck, J. Nangreave, S. Taylor, R. Pei, M. N. Stojanovic, N. G. Walter, E. Winfree and H. Yan, Nature, 2010, 465, 206–210 CrossRef CAS PubMed.
- K. Yehl, A. Mugler, S. Vivek, Y. Liu, Y. Zhang, M. Fan, E. R. Weeks and K. Salaita, Nat. Nanotechnol., 2016, 11, 184–190 CrossRef CAS PubMed.
- C. Zhou, X. Duan and N. Liu, Nat. Commun., 2015, 6, 8102 CrossRef CAS PubMed.
- M. J. Urban, C. Zhou, X. Duan and N. Liu, Nano Lett., 2015, 15, 8392–8396 CrossRef CAS PubMed.
- X. Yang, Y. Tang, S. D. Mason, J. Chen and F. Li, ACS Nano, 2016, 10, 2324–2330 CrossRef CAS PubMed.
- E. Kopperger, T. Pirzer and F. C. Simmel, Nano Lett., 2015, 15, 2693–2699 CrossRef CAS PubMed.
- A. Krissanaprasit, M. Madsen, J. B. Knudsen, D. Gudnason, W. Surareungchai, V. Birkedal and K. V. Gothelf, ACS Nano, 2016, 10, 2243–2250 CrossRef CAS PubMed.
- Y. Zhang, L. Wang, F. Luo, B. Qiu, L. Guo, Z. Weng, Z. Lin and G. Chen, Chem. Commun., 2017, 53, 2910–2913 RSC.
- L. Peng, P. Zhang, Y. Chai and R. Yuan, Anal. Chem., 2017, 89, 5036–5042 CrossRef CAS PubMed.
- H. Cho, D. Alcantara, H. Yuan, R. A. Sheth, H. H. Chen, P. Huang, S. B. Andersson, D. E. Sosnovik, U. Mahmood and L. Josephson, ACS Nano, 2013, 7, 2032–2041 CrossRef CAS PubMed.
- Y. Peng, R. J. Turberfield, S. Sahu and J. H. Reif, Design of an autonomous DNA nanomechanical device capable of universal computation and universal translational motion, Springer, Berlin, Heidelberg, 2004 Search PubMed.
- F. Dannenberg, M. Kwiatkowska, C. Thachuk and A. J. Turberfield, Nat. Comput., 2014, 14, 195–211 CrossRef.
- F. Pu, Z. Liu, X. Yang, J. Ren and X. Qu, Chem. Commun., 2011, 47, 6024–6026 RSC.
- J. W. Keum and H. Bermudez, Chem. Commun., 2009, 45, 7036–7038 RSC.
- Z. Xia, P. Wang, X. Liu, T. Liu, Y. Yan, J. Yan, J. Zhong, G. Sun and D. He, Biochemistry, 2016, 55, 1326–1331 CrossRef CAS PubMed.
- V. J. Schüller, S. Heidegger, N. Sandholzer, P. C. Nickels, N. A. Suhartha, S. Endres, C. Bourquin and T. Liedl, ACS Nano, 2011, 5, 9696 CrossRef PubMed.
- M. N. Stojanovic, P. de Prada and D. W. Landry, Nucleic Acids Res., 2000, 28, 2915–2918 CrossRef CAS PubMed.
- M. N. Stojanovic, P. de Prada and D. W. Landry, ChemBioChem, 2001, 2, 411–415 CrossRef CAS PubMed.
- M. N. Stojanovic, T. E. Mitchell and D. Stefanovic, J. Am. Chem. Soc., 2002, 124, 3555–3561 CrossRef CAS PubMed.
- R. Yashin, S. Rudchenko and M. N. Stojanovic, J. Am. Chem. Soc., 2007, 129, 15581 CrossRef CAS PubMed.
- D. Li, W. Cheng, Y. Li, Y. Xu, X. Li, Y. Yin, H. Ju and S. Ding, Anal. Chem., 2016, 88, 7500–7506 CrossRef CAS PubMed.
- X. Liu, A. Niazov-Elkan, F. Wang and I. Willner, Nano Lett., 2013, 13, 219–225 CrossRef CAS PubMed.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Nature, 2009, 459, 73–76 CrossRef CAS PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- Y. Chen, M. Wang and C. Mao, Angew. Chem., Int. Ed., 2004, 43, 3554–3557 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills, F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- M. Liu, J. Fu, C. Hejesen, Y. Yang, N. W. Woodbury, K. Gothelf, Y. Liu and H. Yan, Nat. Commun., 2013, 4, 2127 Search PubMed.
- J. Elbaz, M. Moshe and I. Willner, Angew. Chem., Int. Ed., 2009, 48, 3834–3837 CrossRef CAS PubMed.
- X. Liu, C. H. Lu and I. Willner, Acc. Chem. Res., 2014, 47, 1673–1680 CrossRef CAS PubMed.
- G. Song, M. Chen, C. Chen, C. Wang, D. Hu, J. Ren and X. Qu, Biochimie, 2010, 92, 121–127 CrossRef CAS PubMed.
- F. Wang, B. Willner and I. Willner, Top. Curr. Chem., 2014, 354, 279–338 CrossRef CAS PubMed.
- M. K. Beissenhirtz and I. Willner, Org. Biomol. Chem., 2006, 4, 3392–3401 CAS.
- J. Elbaz, Angew. Chem., Int. Ed., 2012, 51, 2349–2353 CrossRef CAS PubMed.
- D. Ackermann, T. L. Schmidt, J. S. Hannam, C. S. Purohit, A. Heckel and M. Famulok, Nat. Nanotechnol., 2010, 5, 436 CrossRef CAS PubMed.
- J. Weigandt, C. L. Chung, S. S. Jester and M. Famulok, Angew. Chem., Int. Ed., 2016, 55, 5512–5516 CrossRef CAS PubMed.
- C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef CAS PubMed.
- Y. Tian and C. Mao, J. Am. Chem. Soc., 2004, 126, 11410–11411 CrossRef CAS PubMed.
- Z. G. Wang, J. Elbaz and I. Willner, Angew. Chem., 2012, 51, 4322 CrossRef CAS PubMed.
- R. Chhabra, J. Sharma, Y. Liu and H. Yan, Nano Lett., 2006, 6, 978–983 CrossRef CAS PubMed.
- T. G. Cha, J. Pan, H. Chen, J. Salgado, X. Li, C. Mao and J. H. Choi, Nat. Nanotechnol., 2014, 9, 39–43 CrossRef CAS PubMed.
- M. von Delius, E. M. Geertsema and D. A. Leigh, Nat. Chem., 2010, 2, 96–101 CrossRef CAS PubMed.
- T. G. Cha, J. Pan, H. Chen, H. N. Robinson, X. Li, C. Mao and J. H. Choi, J. Am. Chem. Soc., 2015, 137, 9429–9437 CrossRef CAS PubMed.
- T. E. Tomov, R. Tsukanov, Y. Glick, Y. Berger, M. Liber, D. Avrahami, D. Gerber and E. Nir, ACS Nano, 2017, 11, 4002–4008 CrossRef CAS PubMed.
- T. E. Tomov, R. Tsukanov, M. Liber, R. Masoud, N. Plavner and E. Nir, J. Am. Chem. Soc., 2013, 135, 11935–11941 CrossRef CAS PubMed.
- R. Tsukanov, T. E. Tomov, M. Liber, Y. Berger and E. Nir, Acc. Chem. Res., 2014, 47, 1789–1798 CrossRef CAS PubMed.
- Y. He and D. R. Liu, Nat. Nanotechnol., 2010, 5, 778–782 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2017 |




