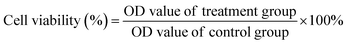Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
MoS2 quantum dots featured fluorescent biosensor for multiple detection of cancer†
Yuhong Liu,
Jinzha Zhang,
Yang Shen,
Jinduo Yan,
Zaiying Hou,
Chun Mao and
Wenbo Zhao
and
Wenbo Zhao *
*
National and Local Joint Engineering Research Center of Biomedical Functional Materials, Jiangsu Key Laboratory of Biofunctional Materials, School of Chemistry and Materials Science, Nanjing Normal University, Nanjing, 210023, China. E-mail: zhaowenbo@njnu.edu.cn
First published on 29th November 2017
Abstract
Transition metal ions, such as those generated through MoS2 material, possess an intrinsic fluorescence quenching property towards organic dye molecules; thus, they can be used to construct biosensors as quenchers. However, we found that the conventional bulk MoS2 blocks the view of fluorescence imaging, and is incapable of tracing and visualizing mucin 1-overexpression cancer cells. Herein, a FAM fluorophore-labeled ssDNA fluorescent probe (P0-FAM) stacked on the surface of MoS2 quantum dots (QDs) was used to construct a MoS2 QDs–P0-FAM biosensor. MoS2 QDs exhibit a high fluorescence quenching ability towards fluorescent dyes, possess large specific surface area and a large number of active sites to adsorb and quench more fluorescent probes, promoting sensitivity between quenching and the recovery signal. In addition, the lighter color of unstack-MoS2 QDs is beneficial to define the location of cancer cells compared to MoS2 nanosheets. The novel MoS2 QDs-based biosensor demonstrates high sensitivity to MUC1 with a detection limit of 0.5 nM, and may become an important tool toward the detection of cancer cells.
Introduction
Breast cancer is, by far, the most frequent cause of cancer deaths in women.1 For this reason, early detection and early diagnosis are critical factors to guarantee treatability and curability.2 As studies have reported, MUC1 is a transmembrane glycoprotein, which is abnormally expressed in all stages of development of human adenocarcinomas.3–5 Overexpression of MUC1 in mammary glandular cells, compared with normal cells, is likely to alter its function and affect the behavior of cancer cells. Therefore, MUC1, as a biomarker indicator, is widely used for early detection of breast cancer.6,7General techniques for detecting MUC1 and cancer cells include flow cytometry, DNA chip technology and PCR techniques; however, these methods usually involve complex instruments and lack efficiency.8 In addition, the fluorescence method has characteristics of high sensitivity, strong anti-interference and rapid response, as well as tracking and visualizing analytes via fluorescence imaging.9–11 To overcome the drawbacks of time-consumption, high cost and complexity of fluorescent biosensors,12–14 we require to construct a simple fluorescent biosensor with sensitive signal and strong specificity recognition abilities to detect MUC1 and MCF-7. Recently, transient metal quantum dots were employed as nanoprobes for biological applications due to their fluorescent, paramagnetic properties, radio-opacity, and quenching ability.15–17 As far as we know, transition metal ions possess an intrinsic fluorescence quenching property towards organic dye molecules.18 Typically, MoS2, as an ultrathin direct bandgap semiconductor, has found wide spread applications in optoelectronics, nanoelectronics, and energy harvesting,19–21 and can act as a fluorescence quencher due to its capacity of quenching dye-labeled single-stranded DNA (ssDNA) via van der Waals force or coordination,22–24 which opens new analytical opportunities.25 However, fluorescent imaging applications of frequently-used quencher MoS2 nanosheets remain significantly challenging. The multi-layer stack of MoS2 nanosheets blocks the view of fluorescence imaging making it hard to clearly observe the location of cancer cells. In this study, we chose MoS2 QDs to solve this problem.
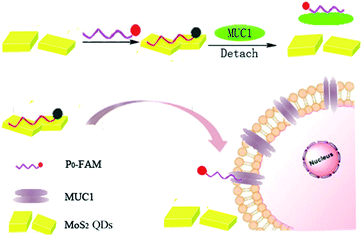
Herein, we constructed a novel, simple and sensitive MoS2 QDs-based sensing platform for the assay of MUC1. The biosensor is composed of a fluorescent probe (FAM fluorophore-labeled ssDNA, defined as P0-FAM) and a quencher (MoS2 QDs). MoS2 QDs can recognize complementary oligonucleotides or aptamers as recognition units.26,27 In addition, they can spontaneously adsorb P0-FAM via van der Waals force between the nucleobases of ssDNA and the surface of MoS2 QDs. The intrinsic fluorescent quenching property of MoS2 to organic dye molecules causes fluorescence quenching of P0-FAM when P0-FAM is adsorbed on MoS2 QDs, while the fluorescence recovery of P0-FAM occurs under the attack of MUC1, which is attributed to the exposure of P0-FAM due to the detachment of P0-FAM from MoS2 QDs with a stronger affinity between P0-FAM and MUC1.28–31 The employment of MoS2 QDs is beneficial in fluorescence imaging to detect the location, the size of the tumor and the treatment region due to larger specific surface area, more active sites to adsorb more fluorescent probes, and the lighter color of unstack-MoS2 QDs compared to MoS2 nanosheets. The MoS2 QDs–P0-FAM-based fluorescent biosensor with sensitive signal and strong specificity recognition abilities to MUC1 is expected to provide a new perspective for the detection and diagnosis of breast cancer.
Materials and reagents
Molybdenum(IV) sulfide (99%) was supplied by Energy Chemical. Ethanol absolute, sodium chloride, sodium dihydrogen phosphate, dehydrate, disodium phosphate dibasic dodecahydrate and potassium chloride were all supplied by Sinopharm Chemical Reagent Co. Ltd. Thiazolyl blue tetrazolium bromide (MTT) was obtained from Aladdin Industrial Co. Ltd. Dulbecco's modified Eagle's medium (DMEM)/high glucose, trypsin–EDTA solution, fetal bovine serum, phosphate buffer solution (NaCl 136.89 mM, KCl 2.67 mM, Na2HPO4 10.15 mM, KH2PO4 1.76 mM, pH 7.2–7.4), penicillin–streptomycin solution (100×) and P0-FAM (5′ to 3′: GCAGTTGATCCTTTGGAT-ACCCTGG, 5′ decorated with FAM fluorophore) were purchased from Sangon Biotech Co. Ltd. (Shanghai, China). Porcine bone marrow mesenchymal stem cells (PBMSC) were provided by Nanjing Gulou Hospital. Human mammary carcinoma (MCF-7) cells and mouse fibroblast cells (L929) were purchased from Shanghai Cell Bank. All the chemicals were of analytical grade and used without further purification.MUC1 (N → C: PDTRPAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHGVTSA) was obtained from China Peptides Co. Ltd. (Shanghai, China).
Instrumentation
A draught drying cabinet, numerical control ultrasonic cleaners and a medical centrifuge were used to prepare the MoS2 QDs. Transmission electron micrograph (TEM) was obtained using an H-7650 TEM instrument (Hitachi, Japan). The X-ray diffraction (XRD) pattern was recorded on a D/max 2005VL/PC X-ray diffractometer (Rigaku, Germany). X-ray photoelectron spectroscopy (XPS) was conducted with PHI Quantera II. Zeta potential analysis was performed on a dynamic light scatter (DLS, NANO-ZS920, Malvern, UK). Fluorescence spectra were recorded on an F-4600 spectrofluorometer (HITACHI, Japan) equipped with a xenon lamp, λex = 490 nm, λem = 520 nm. The PMT voltage was 620 V and the slits for both the excitation and the emission were set at 10 nm. The MTT assay was obtained using a Varioskan flash microplate reader (Thermo Scientific) at 490 nm. The confocal microscopy experiments were conducted using a MRC-1024 (Bio-Rad, Ltd., USA).Synthesis of water-soluble MoS2 QDs
MoS2 QDs were prepared by a modified mixed solvent strategy for liquid exfoliation.32 Initially, 60 mg MoS2 powder was mixed with 20 mL of ethanol/water with an ethanol volume fraction of 45% in a 50 mL flask. The sealed flask with the mixture was ultrasonicated for 24 h and a dark green suspension was obtained. The dispersion was centrifuged at 2000 rpm for 10 min three times to remove the aggregates. Following this, the supernatant was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and collected at 60 °C in a drying cabinet to remove the ethanol and water absolutely. Next, the product was dissolved in deionized water and centrifuged at 2000 rpm for 10 min to remove the larger MoS2 nanoparticles. Finally, the supernatant was filtered through a 0.22 μm Millipore membrane filter and collected at 60 °C in a drying cabinet.
000 rpm for 10 min and collected at 60 °C in a drying cabinet to remove the ethanol and water absolutely. Next, the product was dissolved in deionized water and centrifuged at 2000 rpm for 10 min to remove the larger MoS2 nanoparticles. Finally, the supernatant was filtered through a 0.22 μm Millipore membrane filter and collected at 60 °C in a drying cabinet.
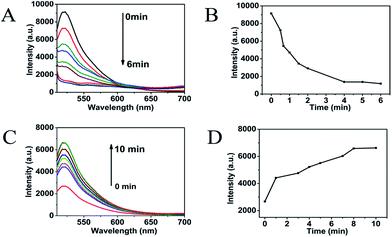
Selection of MoS2 QDs concentration and kinetic assay
The MoS2 QDs solution was diluted by PBS buffer (pH 7.4) to a final concentration of 500 μg mL−1. Different volumes (0–200 μL) of MoS2 QDs solution (500 μg mL−1) were mixed with 100 μL of 100 nM P0-FAM in a 2.0 mL centrifugal tube; different volumes of PBS buffer were added to make 500 μL of each solution. The final P0-FAM concentration was 20 nM, and the concentrations of MoS2 QDs were 0, 5, 10, 15, 20, 30, 50, 100, and 200 μg mL−1. Then, these mixtures were allowed to react for 30 min at 37 °C. Finally, fluorescence measurements were performed at room temperature.Kinetic assay was performed on the fluorescence quenching and fluorescence recovery. For fluorescence quenching, 100 μL of MoS2 QDs solution (500 μg mL−1) was mixed with 100 μL of 100 nM P0-FAM in a cuvette. The fluorescence measurements were performed at different times (0–6 min) at room temperature. For fluorescence recovery, 100 μL of MoS2 QDs solution (500 μg mL−1) was mixed with 100 μL of 100 nM P0-FAM in the cuvette; after 15 min at room temperature, 250 μL MUC1 and 50 μL PBS were added into the reaction mixture. The fluorescence measurements were performed at different times (0–10 min) at room temperature.
Assay for MUC1 in aqueous buffer
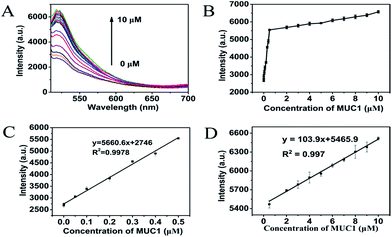
100 μL of MoS2 QDs solution (500 μg mL−1) was mixed with 100 μL of 100 nM P0-FAM in a test tube; then, the mixed solution was allowed to react for 6 min at room temperature. Following this, different volumes of MUC1 (20 μM) in PBS buffer (0–250 μL) were added. Finally, different volumes of PBS buffer (pH 7.4) were introduced to prepare 500 μL of each reaction solution and the final MUC1 concentration (0, 0.001, 0.005, 0.010, 0.050, 0.100, 0.500, 2, 3, 4, 5, 6, 7, 8, 9, and 10 μM). The final mixed solution was allowed to react for 10 min at 37 °C. The fluorescence spectra were measured at room temperature.Cell culture and MTT experiments
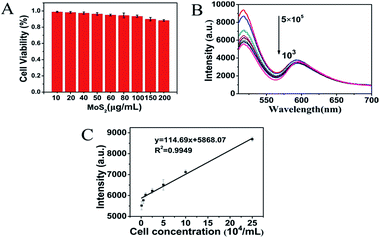
MCF-7 cells, PBMSC and L929 cells were cultured in a cell culture flask in Dulbecco's modified Eagle's medium (DMEM)/high glucose containing 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin solution (100×) at 37 °C under an incubator containing 5% CO2.MCF-7 was incubated with different concentrations of MoS2 QDs (10–200 μg mL−1) at 37 °C and in 5% CO2 for 24 h. Further, the cell viability experiments were conducted using a Varioskan flash microplate reader (Thermo Scientific) at 490 nm. The cell viability was then assessed using the equation below:
Assay for MCF-7 cells in DMEM
Initially, 1 mL of MoS2 QDs solution (500 μg mL−1) was mixed with 1 mL of 100 nM P0-FAM in a test tube; the mixed solution was allowed to react for 6 min at room temperature. Then, 200 μL of the mixed solution was added to different concentrations of MCF-7 (0 to 5 × 105 cells per mL). After incubation at 37 °C for 30 min, the fluorescence spectra were measured at room temperature.Selectivity assays
The selectivity assays were tested by comparing the fluorescence signal changes of samples containing glucose oxidase (GOD), cytochrome c (CyC), myoglobin (Mb), Lys and bovine serum albumin (BSA). Initially, 1 mL of MoS2 QDs solution (500 μg mL−1) was mixed with 1 mL of P0-FAM (100 nM) in a test tube; the mixed solution was allowed to react for 10 min at room temperature. Then, 200 μL of the mixed solution was added to the sensing systems containing 10 μM MUC1, 100 μM glucose oxidase (GOD), 100 μM cytochrome c (CyC), 100 μM myoglobin (Mb), 100 μM Lys and 100 μM bovine serum albumin (BSA). After incubation at 37 °C for 15 min, the fluorescence spectra were measured at room temperature.Results and discussion
Characterization of MoS2 QDs
The morphology of MoS2 was studied using TEM (Fig. 1). The synthetic MoS2 QDs (Fig. 1B and C) are dispersed evenly in aqueous solution compared to MoS2 nanosheets in aqueous solution (Fig. 1A). TEM images revealed that the average size of smaller MoS2 QDs, which were used for further experiments, were about 3 nm (Fig. 1C and D). The XRD patterns (Fig. 2A) of the samples matched well with that of 2H-MoS2 (JCPDS: 24-513). As can be observed, the primary diffraction peaks at 14.4°, 33.2° and 58.4° were attributed to the (002), (100) and (110) planes of the hexagonal MoS2, respectively, indicating the high purity of the obtained smaller MoS2 QDs.33 Raman spectrum was used to further confirm that smaller MoS2 QDs were obtained (Fig. 2B). The Raman spectrum of bulk MoS2 was well-known with two main modes, the A1g and E2g, corresponding to the out-plane vibrations and in-plane vibrations as located at 408 and 382 cm−1, respectively.34 This was represented by the black line shown in Fig. 2B. It could be observed that the A1g mode of MoS2 QDs slightly blue shifted to 405.5 cm−1, which proved that we had successfully prepared MoS2 QDs.35 As shown in Fig. 2C, Mo 3d3/2 and Mo 3d5/2 peaks could be observed at 232.2 eV and 231.4 eV and 229.1 eV and 228.5 eV, respectively. The S 2p peaks at the binding energies of 162.0 and 163.3 eV arise from S 2p3/2 and S 2p1/2, respectively (Fig. 2D). The XPS spectra were consistent with those in previously reported literatures, indicating the dominant 2H MoS2 phase in the MoS2 QDs.36,37 The zeta potential of the MoS2 QDs were determined to be −27.8 mV (Fig. S1†), suggesting the great colloidal stability of the MoS2 QDs in aqueous media. | ||
| Fig. 1 TEM pictures of MoS2 (A) nanosheets, (B) large size (QDs), (C) small size (QDs), and (D) the enlarged view of (C). | ||
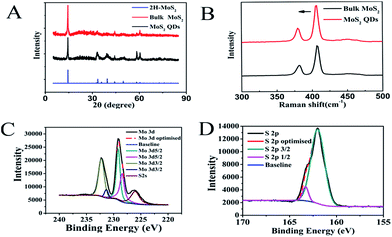 | ||
| Fig. 2 (A) XRD pattern of MoS2 QDs. (B) Raman spectra of MoS2 QDs. XPS survey of (C) Mo atom and (D) S atom in as-prepared MoS2 QDs. | ||
Mechanism of the fluorescent biosensor
Scheme 1 displays the principle diagram of the DNA biosensor composed of the quencher (MoS2 QDs) and the fluorescent probe (P0-FAM). The fluorescence quenching of P0-FAM occurred when MoS2 QDs adsorbed P0-FAM via the van der Waals force between the nucleobases of ssDNA and the surface of MoS2 QDs owing to the possible transfer of electrons or energy between the closely connected dye molecules and the MoS2 QDs (Fig. S2†). Interestingly, in the presence of MUC1, the fluorescence recovery of P0-FAM could be observed because MoS2–P0-FAM adopted a rigid and definite tertiary structure owing to the specific binding between ssDNA and MUC1. The affinity of ssDNA with MUC1 was stronger than that of MoS2 QDs, resulting in the release of the P0-FAM from the QDs surface and recovery of the fluorescence signal. In contrast, without MoS2 QDs, P0-FAM was primarily in the unfolded and flexible state in the presence of MUC1. The FL signal did not drastically change, indicating that MoS2 QDs as a quencher played a crucial role in turn-on FL biosensor for the sensitive detection of MUC1 in cancer cells.Optimization of detection conditions
To evaluate the fluorescence-quenching ability of MoS2 QDs toward P0-FAM, the fluorescence signal changes were recorded upon mixing P0-FAM and the prepared MoS2 QDs. As shown in Fig. 3A, the quenching of FAM fluorescence by MoS2 depended on the concentration of the quenchers. In the presence of 100 μg mL−1 MoS2 QDs, the emission of the FAM was almost quenched with 90% quenching efficiency (Fig. 3B), revealing a high quenching efficiency of MoS2 QDs toward the aptamer biosensor. The observed background fluorescence, as shown in Fig. 3B, corresponding to the fluorescence of 200 μg mL−1 MoS2 QDs, could be attributed to the existence of the secondary structure of P0-FAM at the detection conditions. Fig. 4A and B show the adsorption kinetics of the dye-labeled aptamer biosensor on the MoS2 QDs. The quenching was rapid and achieved equilibrium in about 4 min. This suggested that the interaction of P0-FAM with MoS2 QDs was quite strong and the MoS2 QDs possessed a high fluorescence-quenching ability. The MoS2 QDs exhibited robust quenching efficiency possibly because of the better water dispersivity. Furthermore, the fluorescence recovery kinetics was performed and the best fluorescence recovery efficiency was obtained within 8 min when MUC1 was added into the mixture solution (Fig. 4C and D). This suggested that the designed MUC1 biosensor system works successfully and can deliver high performance. In order to achieve more effective detection, 15 min was chosen as the optimal reaction time.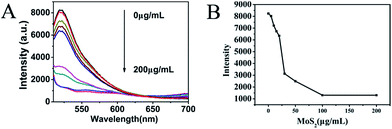 | ||
| Fig. 3 Fluorescence intensity (A) and trend chart (B) of P0-FAM (20 nM) after addition of MoS2 QDs with different concentrations in PBS buffer. | ||
Quantitative analysis of MUC1 in aqueous buffer
As shown in Fig. 5, a new simple and sensitive assay for MUC1 was successfully designed. The fluorescence intensity depended on the concentration of MUC1 over a range of 0–10 μM when the concentration of MoS2 QDs was 100 μg mL−1 (Fig. 5A and B). As shown in Fig. 5C, the fluorescence intensity increases rapidly as the concentration of MUC1 increases from 0 μM to 0.5 μM (R2 = 0.9978). However, it exhibited another linear relationship as shown in Fig. 5D (R2 = 0.997) when the concentration of MUC1 changed from 0.5 μM to 10 μM, where the fluorescence intensity increased more slowly with the increase in MUC1 concentration. The reason for this phenomenon was probably that P0-FAM in the state of random coil single strand was correspondingly abundant when MUC1 was less than 0.5 μM in the system, and MoS2 QDs could strongly adsorb these random coil single-strands FAM-ssDNA onto its surface owing to the weak adsorption competition among the free FAM-ssDNA. When MUC1 exceeded 0.5 μM in the system, free FAM-ssDNA in the state of random coil single strand became scarce. The adsorption competition among them became more intense, so the fluorescence intensity changed more slowly as the MUC1 concentration increased. This method could be applied to detect MUC1 concentrations as low as 0.5 nM (3 times the standard deviation rule) in aqueous buffer. Moreover, the detection range was wide, ranging from 0 μM to 10 μM. Combined with the data listed in Table S1,† the results demonstrated that the fluorescent detection of biosensor for MUC1 was feasible for a relatively broad detection range and low detection limit. From this perspective, the proposed method towards MUC1 detection had its own uniqueness, that is, the present method was much simpler and more effective to detect MUC1.Performance of MCF-7 detection and selectivity assays of FL biosensor
The MTT assays of cell viability studies suggested that MoS2 QDs did not impose a considerable toxicity towards MCF-7 cells as compared to the control (Fig. 6A). The above results indicated that the as-prepared MoS2 QDs could be promising biosensors in cell detection and imaging. As shown in Fig. 6B, the fluorescence intensity was dependent on the concentration of MCF-7 cells over a range of 0 to 5 × 105 cells per mL, when the concentration of MoS2 QDs was 100 μg mL−1. As illustrated in Fig. 6C, a linear relationship between peak intensity at 520 nm and MCF-7 cells concentrations was obtained in the concentration range from 103 to 5 × 105 cells per mL (R2 = 0.9949) with a detection limit of 50 cells per mL (according to the rule of three times the standard deviation corresponding to the blank sample detection). Due to the specific binding between P0-FAM and MUC1, the MoS2 QDs-based biosensor was insensitive to the interfering proteins such as GOD, CyC, Mb, Lys and BSA as shown in Fig. S3.† The good selectivity, biocompatibility and the intrinsic optical properties of the biosensor can be used to construct an excellent bioimaging system and recognition system.Intracellular imaging analysis
Fluorescence microscope images of MCF-7 cells loaded with the MoS2–P0-FAM (MoS2 QDs) biosensor for 1 h at 37 °C showed green fluorescence on cytomembrane (Fig. 7). However, the control experiment on cells without the MoS2–P0-FAM biosensor gave no green fluorescence in the same exposure condition. These results demonstrated the specific recognition of MoS2–P0-FAM biosensor to MUC1 in MCF-7 cells. In contrast, the fluorescence for PBMSC and L929 cells exhibited no green fluorescence due to non-overexpression of MUC1 in PBMSC and L929 cells, proving that the MoS2 QDs-based biosensor can be applied to bioimaging and recognition of MCF-7. By comparison, MCF-7 cells incubated with the MoS2 sheets–P0-FAM biosensor showed extremely weak green fluorescence (Fig. S4†), indicating that the multi-layer stacking of MoS2 nanosheets would block the view of fluorescence imaging and affect the detection of cancer cells. Hence, the MoS2 QDs-based biosensor was superior to that of MoS2 sheets.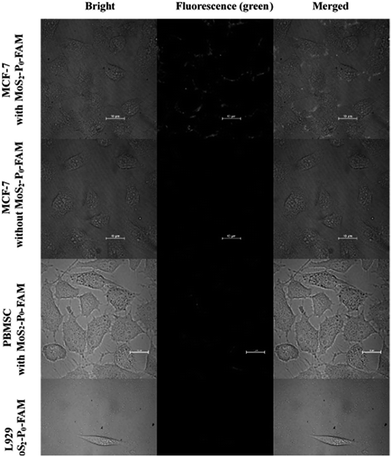 | ||
| Fig. 7 Confocal fluorescence microphotograph of different cells incubated with MoS2 QDs featured fluorescent biosensor for 1 h. Scale: 10 μm. | ||
Conclusion
In summary, this study presented a sensitive MoS2 QDs based fluorescent sensing platform for MUC1. In particular, we applied MoS2 QDs to MCF-7 detection and cellular imaging, which is extremely rare in the application of MoS2. MoS2 QDs exhibited a high fluorescence quenching ability towards fluorescent dyes; therefore, they were exploited as carrier and quencher for a fluorescent dye-labeled DNA aptamer (P0-FAM) to construct a biosensor. The obtained results showed that the detection range of MUC1 in the solution was in the range of 1 nM to 10 μM and the detection limit was 0.5 nM. The detection range of MCF-7 was in the range of 103 to 5 × 105 cells per mL and the detection limit was 50 cells per mL. The biosensor also had the advantages of high sensitivity and specificity. Furthermore, we expect that this strategy based on MoS2 QDs as a fluorescent quencher may offer a new approach in the sensitive and selective detection of a wide spectrum of analytes and cancer cells.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21571104), Major projects of Natural Sciences of University in Jiangsu Province of China (14KJA150006), Projects of Jiangsu Collaborative Innovation Center of Biomedical Functional Materials, and the Priority Academic Program Development of Jiangsu Higher Education Institution.Notes and references
- Y. Li, Y. Zhang, M. Zhao, Q. Zhou, L. Wang, H. Wang, X. Wang and L. Zhan, Chem. Commun., 2016, 52, 3959–3961 RSC.
- R. Beatson, V. Tajadura-Ortega, D. Achkova, G. Picco, T. D. Tsourouktsoglou, S. Klausing, M. Hillier, J. Maher, T. Noll, P. R. Crocker, J. Taylor-Papadimitriou and J. M. Burchell, Nat. Immunol., 2016, 17, 1273–1281 CrossRef CAS PubMed.
- Y. He, Y. Lin, H. W. Tang and D. W. Pang, Nanoscale, 2012, 4, 2054–2059 RSC.
- M. Hiraki, T. Maeda, A. Bouillez, M. Alam, A. Tagde, K. Hinohara, Y. Suzuki, T. Markert, M. Miyo, K. Komura, R. Ahmad, H. Rajabi and D. Kufe, Oncogene, 2017, 36, 2791–2801 CrossRef CAS PubMed.
- A. Lui and J. Lewis-Wambi, Cancer Res., 2016, 76, 2918 Search PubMed.
- X. Y. Jiang, H. J. Wang, H. J. Wang, R. Yuan and Y. Q. Chai, Anal. Chem., 2016, 88, 9243–9250 CrossRef CAS PubMed.
- A. K. H. Cheng, H. Su, A. Wang and H. Z. Yu, Anal. Chem., 2009, 81, 6130–6139 CrossRef CAS PubMed.
- W. Ma, W. Zheng, D. Cui, Y. Song and Q. Wu, Acta Biochim. Biophys. Sin., 2000, 32, 285–289 CAS.
- E. Climent, M. D. Marcos and K. Rurack, Angew. Chem., Int. Ed., 2013, 52, 8938–8942 CrossRef CAS PubMed.
- C. L. Zhu, C. H. Lu, X. Y. Song, H. H. Yang and X. R. Wang, J. Am. Chem. Soc., 2011, 133, 1278–1281 CrossRef CAS PubMed.
- K. Cherukula, K. M. Lekshmi, S. Uthaman, K. Cho, C. S. Cho and I. K. Park, Nanomaterials, 2016, 6, 76 CrossRef PubMed.
- P. Wu, X. D. Hou, J. J. Xu and H. Y. Chen, Nanoscale, 2016, 8, 8427–8442 RSC.
- Q. Wu, M. Q. Chu, Y. X. Shao, F. J. Wo and D. L. Shi, Carbon, 2016, 108, 21–37 CrossRef CAS.
- A. Rammohan, G. Mishra, B. Mahaling, L. Tayal, A. Mukhopadhyay, S. Gambhir, A. Sharma and S. Sivakumar, ACS Appl. Mater. Interfaces, 2016, 8, 350–362 CAS.
- S. Santra, H. Yang, P. H. Holloway, J. T. Stanley and R. A. Mericle, J. Am. Chem. Soc., 2005, 127, 1656–1657 CrossRef CAS PubMed.
- H. M. Fan, M. Olivo, B. Shuter, J. B. Yi and R. Bhuvaneswari, J. Am. Chem. Soc., 2010, 132, 14803–14811 CrossRef CAS PubMed.
- G. W. Walker, V. C. Sundar, C. M. Rudzinski, A. W. Wun and M. G. Bawendi, Appl. Phys. Lett., 2003, 83, 3555–3557 CrossRef CAS.
- Y. Huang, Y. Shi, H. Y. Yang and Y. Ai, Nanoscale, 2015, 7, 2245–2249 RSC.
- B. Radisavljevic, A. Radenovic, J. Brivio, V. Giacometti and A. Kis, Nat. Nanotechnol., 2011, 6, 147 CrossRef CAS PubMed.
- K. F. Mak, C. Lee, J. Hone, J. Shan and T. F. Heinz, Phys. Rev. Lett., 2010, 105, 136805 CrossRef PubMed.
- H. Li, Z. Y. Yin, Q. Y. He, X. Huang, G. Lu, D. W. H. Fam, A. I. Y. Tok, Q. Zhang and H. Zhang, Small, 2012, 8, 63 CrossRef CAS PubMed.
- D. Jariwala, V. K. Sangwan, L. J. Lauhon, T. J. Marks and M. C. Hersam, ACS Nano, 2014, 8, 1102–1120 CrossRef CAS PubMed.
- Y. Shi, W. Zhou, A. Y. Lu, W. Fang, Y. H. Lee, A. L. Hsu, S. M. Kim, K. K. Kim, H. Y. Yang, L. J. Li, J. C. Idrobo and J. Kong, Nano Lett., 2012, 12, 2784–2791 CrossRef CAS PubMed.
- N. Goswami, A. Giri and S. K. Pal, Langmuir, 2013, 29, 11471–11478 CrossRef CAS PubMed.
- H. Deng, X. Yang and Z. Gao, Analyst, 2015, 140, 3210–3215 RSC.
- L. Mo, J. Li, Q. Liu, L. Qiu and W. Tan, Biosens. Bioelectron., 2017, 89, 201–211 CrossRef CAS PubMed.
- B. L. Li, J. Wang, H. L. Zou, S. Garaj, C. T. Lim, J. Xie, N. B. Li and D. T. Leong, Adv. Funct. Mater., 2016, 26, 7034–7056 CrossRef CAS.
- X. Q. Liu, R. Aizen, R. Freeman, O. Yehezkeli and I. Willner, ACS Nano, 2012, 6, 3553–3563 CrossRef CAS PubMed.
- J. Yang, B. Dou, R. Yuan and Y. Xiang, Anal. Chem., 2016, 88, 8218–8223 CrossRef CAS PubMed.
- X. Q. Liu, F. Wang, R. Aizen, O. Yehezkeli and I. Willner, J. Am. Chem. Soc., 2013, 135, 11832–11839 CrossRef CAS PubMed.
- X. Zhang, Z. C. Lai, C. L. Tan and H. Zhang, Angew. Chem., Int. Ed., 2016, 55, 8816–8838 CrossRef CAS PubMed.
- K. G. Zhou, N. N. Mao, H. X. Wang, Y. Peng and H. L. Zhang, Angew. Chem., Int. Ed., 2011, 50, 10839–10842 CrossRef CAS PubMed.
- F. E. Wickman and D. K. Smith, Am. Mineral., 1970, 55, 1843–1856 CAS.
- D. Gopalakrishnan, D. Damien and M. M. Shaijumon, ACS Nano, 2014, 8, 5297–5303 CrossRef CAS PubMed.
- S. Xu, D. Li and P. Wu, Adv. Funct. Mater., 2015, 25, 1127–1136 CrossRef CAS.
- W. Dai, H. Dong, B. Fugetsu, Y. Cao, H. Lu, X. Ma and X. Zhang, Small, 2015, 11, 4158–4164 CrossRef CAS PubMed.
- W. Gu, Y. Yan, C. Zhang, C. Ding and Y. Xian, ACS Appl. Mater. Interfaces, 2016, 8, 11272 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c7ra09300d |
| This journal is © The Royal Society of Chemistry 2017 |